Abstract
Inactivating mutations of the von Hippel-Lindau (VHL) tumor suppressor gene are associated with inherited VHL syndrome, which is characterized by susceptibility to a variety of neoplasms, including central nervous system hemangioblastoma and clear cell renal cell carcinoma (CCRCC). Mutations in the VHL gene are also found in the majority of sporadic clear cell renal carcinoma, the most common malignant neoplasm of the human kidney. Inactivation of VHL ubiquitin ligase is associated with normoxic stabilization of hypoxia-inducible factor-1α and 2-α (HIF-1α and HIF-2α), transcriptional regulators of tumor angiogenesis, invasion, survival, and glucose utilization. HIF-2α has been particularly implicated in the development of CCRCC. Although several inhibitors of HIF-1α have been described, these drugs typically have a minimal affect on HIF-2α. 786-O is a VHL-deficient CCRCC cell line that constitutively expresses only HIF-2α and is therefore suitable for the screening of novel HIF-2α inhibitors. Using this cell line, we have identified emetine as a specific inhibitor of HIF-2α protein stability and transcriptional activity. Without altering HIF-2α mRNA level, emetine rapidly and dramatically down-regulated HIF-2α protein expression in 786-O cells. HIF-2α down-regulation was accompanied by HIF-2α ubiquitination and was reversed by protea-some inhibition. Emetine-induced HIF-2α down-regulation was confirmed in three additional VHL-renal cancer cell lines, was insensitive to the prolyl hydroxylase inhibitor dimethyloxaloyl glycine, and did not require neural precursor cell expressed developmentally down-regulated-8, suggesting that emetine accesses a previously undescribed cullin-independent protea-some degradation pathway for HIF-2α. These data support the use of emetine or structurally related compounds as useful leads for the identification of novel HIF-2α inhibitors.
Introduction
Inactivating mutations of the von Hippel-Lindau (VHL) tumor-suppressor gene are associated with inherited VHL syndrome, which is characterized by a variety of neoplasms, including central nervous system hemangioblastoma and clear cell renal cell carcinomas (CCRCCs) (Krieg et al., 2000; Rankin et al., 2006; Kaelin, 2009). Mutations in the VHL gene are also found in sporadic CCRCCs, the most common malignant neoplasm of the human kidney (Kaelin 2009). VHL inactivation results in the deregulated expression of HIF-1α and HIF-2α transcription factors (Krieg et al., 2000; Wiesener et al., 2001; Rankin et al., 2005, 2006; Block et al., 2007; Kaelin, 2009).
Hypoxia-inducible factor (HIF) is a central regulator in tumor angiogenesis, invasion, cell survival, and glucose metabolism (Kaur et al., 2005; Semenza 2010). Hypoxia-inducible HIF-1α and HIF-2α dimerize with constitutively expressed HIF-1β, also termed aryl hydrocarbon receptor nuclear translocator (ARNT), to activate the transcription of hypoxia-responsive genes. In normoxia, HIF-α proteins are rapidly degraded by proteasomes after their oxygen-dependent ubiquitination that is dependent on VHL recognition (Isaacs et al., 2002; Brahimi-Horn et al., 2005; Semenza, 2009). VHL recognition requires hydroxylation of proline residues in HIF-α, and HIF proline hydroxylases use molecular oxygen and 2-oxoglutarate as cosubstrates for this purpose (Sudarshan et al., 2007; Kaelin, 2009). In hypoxia, or in the case of VHL inactivation, HIF-α proteins are stabilized leading to transcriptional activation of genes whose promoter regions contain hypoxia response elements (Jung et al., 2003; Isaacs et al., 2004; Smaldone and Maranchie, 2009). These target genes include vascular endothelial growth factor (VEGF), glucose transporter-1 (GLUT-1), transforming growth factor-α (TGF-α), platelet-derived growth factor, and erythropoietin (Smaldone and Maranchie, 2009).
Even though HIF-2α, also termed endothelial PAS domain protein 1, is 48% identical in sequence to HIF-1α, and like HIF-1α is induced by hypoxia (Kaur et al., 2005) and stabilized by loss of VHL (Maxwell et al., 1999; Kaelin, 2009), HIF-1α and HIF-2α are believed to have distinct functions, both in normal cellular physiology and in tumorigenesis (Hu et al., 2003; Wang et al., 2005; Löfstedt et al., 2007; Rankin et al., 2008; Imamura et al., 2009). HIF-2α mRNA is abundantly expressed in lung, heart, liver, and other organs under normoxia, whereas HIF-1α mRNA is normally expressed at a much lower level (Krieg et al., 2000; Hu et al., 2003). HIF-1α and HIF-2α have been reported to have contrasting properties in the VHL-deficient CCRCC cell lines 786-O and RCC4, with HIF-1α retarding and HIF-2α enhancing the growth of tumor xenografts in mice (Raval et al., 2005). Likewise, two earlier studies showed that 786-O CCRCC cells stably expressing HIF-1α were not tumorigenic in mice, whereas forced expression of HIF-2α in the same background was tumorigenic (Kondo et al., 2002; Maranchie et al., 2002). In contrast, however, Imamura et al. reported that HIF-1α promoted the growth of colon cancer cells, whereas HIF-2α restrained their growth (Imamura et al., 2009). Taken together, these data suggest that the cellular function of both transcription factors is likely to be context-dependent.
Although several small-molecule inhibitors of HIF-1α have been described previously (Lin et al., 2004; Chau et al., 2005; Zhou et al., 2005; Choi et al., 2006; Faivre et al., 2006; Thomas et al., 2006), reports of HIF-2α inhibitors are less frequent. Because 786-O CCRCC cells express HIF-2α but not HIF-1α (Wiesener et al., 2001; Hu et al., 2003; Semenza 2003; Isaacs et al., 2004; Löfstedt et al., 2007), this cell line is useful for the identification of novel HIF-2α inhibitors.
Emetine is a protein synthesis inhibitor that blocks the translocation of peptidyl-tRNA from the acceptor site to the donor site on the ribosome (Grollman and Huang 1973). Emetine, extracted from Cephaelis ipecacuanha, has been used as a drug for the treatment of amoebiasis, as an antibacterial or antiviral agent, and as an emetic (Cushny 1918; Zhou et al., 2005). Emetine also has been evaluated in Phase II clinical studies for the treatment of solid tumors (Zhou et al., 2005). Recently, emetine was reported to specifically inhibit hypoxia-induced HIF-1α in breast tumor cells; however, its mechanism of action and possible impact on HIF-2α were not described previously (Zhou et al., 2005). In this study, we have examined the ability of emetine to inhibit HIF-2α in several VHL-CCRCC cell lines, and we have investigated its mechanism of action.
Materials and Methods
Chemicals and Antibodies. Emetine dihydrochloride was purchased from Calbiochem (San Diego, CA). Calpain inhibitor I [N-acetyl-Leu-Leu-norleucinal (ALLN)], calpain inhibitor II [N-acetyl-l-leucyl-l-leucyl-l-methioninal (ALLM)], calpastatin peptide, and the proteasome inhibitor N-benzoyloxycarbonyl (Z)-Leu-Leu-leucinal (MG132) were purchased from Sigma-Aldrich (St. Louis, MO). The proteasome inhibitor bortezomib (PS-341) was obtained from Millennium Pharmaceuticals (Cambridge, MA). The protein synthesis inhibitor cycloheximide was purchased from Sigma-Aldrich. The prolyl hydroxylase inhibitor dimethyloxaloyl glycine (DMOG) was purchased from AG Scientific (San Diego, CA). Mouse monoclonal anti-HIF-1α antibody (1:300 final dilution) was from BD Biosciences Transduction Laboratories (San Jose, CA). Rabbit polyclonal anti-HIF-2α antibody and mouse monoclonal anti-ARNT antibody (1:1000 final dilution) were from Novus Biologicals, Inc. (Littleton, CO). Rabbit polyclonal anti-topoisomerase II antibody (1:1000 final dilution) and rabbit polyclonal anti-CREB antibody (1:2000 final dilution) were from Cell Signaling Technology (Danvers, MA). Rabbit polyclonal anti-ubiquitin antibody (1:500 final dilution) was from Sigma-Aldrich.s
Cell Culture and Transient Transfection. The CCRCC cell lines 786-O, UOK220, UOK122, and UOK130 all lack VHL expression and constitutively express HIF-2α. These cell lines were provided by Dr. Donald P. Bottaro (National Cancer Institute, Bethesda, MD) and cultured in Dulbecco’s modified Eagle’s medium (Mediatech, Herndon, VA). Medium was supplemented with 10% fetal bovine serum, glutamine, HEPES, sodium pyruvate, and penicillin/streptomycin. For NEDD8 siRNA transfection, 786-O cells were transfected with 50 nM NEDD8 siRNA or control siRNA (Dharmacon RNA Technologies, Lafayette, CO) in DharmaFECT4 transfection reagent (Dharmacon RNA Technologies) according to the manufacturer’s instructions. After 72 h, cells were treated with emetine, and quantitative RT-PCR and Western blotting analysis were performed.
Western Blotting and Immunoprecipitation. Cells were lysed, and nuclear extracts were prepared by using the method of Isaacs et al. (2002). To prepare nuclear extracts, adherent cells were washed with chilled phosphate-buffered saline and incubated with cold low-salt lysis buffer (10 mM HEPES, 10 mM KCl, 0.1 mM EDTA, and protease inhibitors) on ice for 10 min. The cells were scraped off the dish and transferred to microtubes. Tergitol-type NP-40 (10%) was added to achieve 0.5% final concentration. The microtubes were vortexed vigorously for 10 s and spun down at 3000 rpm at 4°C for 1 min. Supernatants were removed, and nuclear pellets were washed with low-salt lysis buffer. Nuclear pellets were lysed with high-salt lysis buffer (20 mM HEPES, 400 mM NaCl, 1 mM EDTA, and protease inhibitors) at 4°C for 30 min and clarified by centrifugation at 13,000 rpm at 4°C for 15 min. To prepare whole-cell lysates, cells were washed with cold PBS and lysed in TNES buffer (50 mM Tris-HCl, pH 7.5, 1% NP-40, 1 mM EDTA, 150 mM NaCl, and protease inhibitors) after scraping. Protein concentration was determined using the BCA protein assay (Pierce Chemical, Rockford, IL). Equal amounts of protein were loaded onto 7.5% SDS-polyacrylamide gel electrophoresis gels (Bio-Rad Laboratories, Hercules, CA), transferred to nitrocellulose membrane, blocked in 5% milk, and immunoblotted with primary antibodies. After blotting with horseradish peroxidase-linked secondary antibodies, all blots were developed with SuperSignal chemiluminescence substrate (Pierce Chemical).
To detect HIF-2α association with ARNT, lysates (1 mg of protein) were incubated with anti-HIF-2α antibody at 4°C for 16 h. Protein G-Sepharose beads (Invitrogen, Carlsbad, CA) were added and incubated at 4°C for 2 h. After low-speed centrifugation, the supernatants were discarded, and remaining beads were washed four times with lysis buffer containing TNES buffer plus 20 mM Na2VO4 and subjected to Western blot using anti-ARNT antibody. For ubiquitinated HIF-2α determination, lysates (1 mg of protein) were incubated with anti-HIF-2α antibody at 4°C for 16 h. Protein Gamma-Bind Plus Sepharose beads (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) were added, incubated at 4°C for 2 h, and samples were processed for Western blot using anti-ubiquitin antibody as described.
Real-Time RT-PCR Analysis. Total mRNA was extracted using RNeasy Mini Kit (QIAGEN, Valencia, CA). Real-time RT-PCR was performed as described previously (Isaacs et al., 2005). In brief, cDNA was prepared using total RNA and the TaqMan Reverse Transcription Kit (Applied Biosystems, Foster City, CA). For reverse transcription, 200 ng of total RNA was used in 20 μl of reaction mixture containing 1× TaqMan RT buffer (TaqMan RT-PCR kit; Applied Biosystems), 5.5 mM magnesium chloride, 500 μM concentration of each dNTP, 2.5 μM random hexamer, 0.4 U/μl RNase inhibitor, and 1.25 U/μl Multiscribe Reverse Trasnscriptase. Reverse transcription was performed for 10 min at 25°C, 30 min at 48°C, and 5 min at 95°C using the PE9700 thermal cycler (Applied Biosystems). The reaction mixture was diluted to 100 μl of with Tris-EDTA. Five microliters of diluted cDNA was used for real-time PCR. Real-time PCR primers were designed using Primer Express software (Applied Biosystems). Analysis of mRNA expression was carried out using the ABI Prism 7700 Sequence Detection System (Applied Bio-systems). Each 25 μl of amplification reaction contained 5 μl of sample, 5.5 μl of distilled H2O, 1 μl of each of 5 μM forward primer and reverse primer, and 12.5 μl of SYBR Green PCR master mix (Applied Biosystems). PCR cycling conditions were as follows: 10 min at 95°C, followed by 40 two-step cycles of 95°C for 15 s and 60°C for 60 s. Detection of the PCR product was monitored by measuring the increase in fluorescence caused by the binding of SYBR Green dye to double-stranded DNA. Emission spectra of all tubes were collected every cycle in the last 60 s of the primer annealing/elongation step in the ABI Prism7700 Sequence Detection System (Applied Biosystems). All samples were normalized to the level of 18S ribosomal RNA (rRNA). The number of PCR cycles to reach the fluorescence threshold value is the cycle threshold (Ct). Ct values for the control (18S rRNA) were determined, and relative RNA levels were calculated by the comparative Ct method as described by the manufacturer. Experiments were performed in duplicate.
VEGF and TGF-α ELISA. 786-O cells were seeded in 10-cm dishes 1 day before experiments. Serum-free medium and the indicated drug treatments were supplied to the cells for 24 h. Secreted VEGF level in conditioned media was determined using a Human VEGF Immunoassay kit (R&D Systems, Minneapolis, MN), and TGF-α level in total cell lysates was determined using Human TGF-α Immunoassay kit (R&D Systems) following the manufacturer’s instructions.
Results
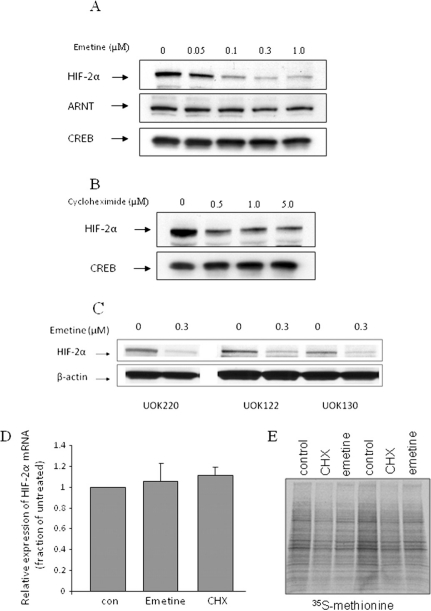
Emetine Promotes VHL-Independent HIF-2α Down-Regulation. The terpenoid tetrahydroisoquinoline alkaloid emetine has been reported to inhibit hypoxic activation of HIF-1α in the breast tumor cell line T47D, although the mechanism responsible for this effect was not identified (Zhou et al., 2005). Emetine is a protein synthesis inhibitor and it has been explored previously as a chemotherapeutic agent (Cushny, 1918; Zhou et al., 2005). We sought to investigate whether emetine could also inhibit HIF-2α. We exposed 786-O CCRCC cells, which express HIF-2α under normoxia and are deficient in both HIF-1α and VHL (Maxwell et al., 1999; Kaelin 2009), to emetine and we monitored HIF-2a mRNA and protein expression. As shown in Fig. 1A (top), HIF-2α protein was highly expressed in untreated normoxic cells. Emetine dose-dependently down-regulated HIF-2α protein after 4 h of treatment, at concentrations as low as 0.05 μM. These results were confirmed in three additional CCRCC cell lines that also lack VHL expression (Fig. 1C). Because HIF-2α dimerization with ARNT can affect HIF-2α protein stability (Isaacs et al., 2004), we examined the possible impact of emetine on ARNT protein expression (Fig. 1A, middle). ARNT levels were not affected by emetine treatment. We next determined whether emetine might affect HIF-2α protein expression by inhibiting its synthesis. We treated 786-O cells with 0.5 to 5.0 μM cycloheximide (CHX) for 4 h to address this question. The impact of CHX on HIF-2α protein expression was much less dramatic than that of emetine (Fig. 1B). The data suggest that emetine-induced down-regulation of HIF-2α protein is not likely to involve protein synthesis inhibition. This hypothesis is supported by data obtained from pulsing cells briefly (30 min) with [35S]methionine to detect total protein synthesis. When 786-O cells were so treated during the final 30 min of a 4-h exposure to either emetine (0.1 μM) or CHX (1 μM), both drugs had a small but equivalent impact on methionine incorporation, although neither drug caused toxicity at the concentrations and incubation times used (Fig. 1E). We also investigated whether emetine-stimulated down-regulation of HIF-2α might be mediated at the level of HIF-2α transcription. We measured HIF-2α mRNA by quantitative real-time RT-PCR, and we found that neither emetine nor CHX had any effect (Fig. 1D).
Fig. 1.
Emetine down-regulates HIF-2α protein expression in 786-O cells. A, 786-O cells were treated with indicated concentrations of emetine for 4 h. HIF-2α and ARNT protein levels in nuclear lysates were analyzed by Western blotting. CREB expression is shown as a loading control. B, 786-O cells were treated with the indicated concentrations of cycloheximide for 4 h. HIF-2α protein level in nuclear lysates was analyzed by Western blotting. CREB expression is shown as a loading control. C, HIF-2α sensitivity to emetine was confirmed in three additional VHL-deficient CCRCC cell lines. Cells were treated and analyzed as in A. D, 786-O cells were treated with 0.3 μM emetine or 40 μg/ml CHX for 4 h, and HIF-2α mRNA was analyzed by quantitative RT-PCR. Each bar represents mean ± S.D. (n = 3). E, 786-O cells were pulsed with [35S]methionine for the final 30 min of a 4-h incubation with either 0.1 μM emetine or 1 μM CHX. Total protein synthesis was monitored by SDS-polyacrylamide gel electrophoresis of labeled cell lysates. Each condition was run in duplicate.
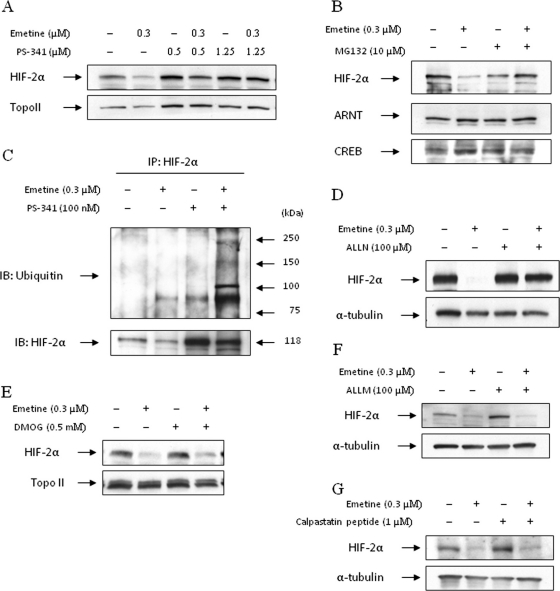
Emetine-Induced HIF-2α Degradation Is Protea-some-Dependent. To examine whether emetine-induced HIF-2α degradation required proteasome activity, we pre-treated 786-O cells with the proteasome inhibitors PS-341, MG132, or ALLN before emetine addition. As shown in Fig. 2, A, B, and D, when we pretreated cells with three different proteasome inhibitors for 1 h and then treated with emetine for 4 h, HIF-2α protein in nuclear lysates of emetine-treated cells was protected from emetine. These data show that emetine, in the absence of VHL, nevertheless promotes degradation of HIF-2α in a proteasome-dependent manner. To further demonstrate that emetine-mediated HIF-2α degradation proceeds via the proteasome, we determined whether emetine influenced the intensity of HIF-2α ubiquitination in proteasome-inhibited 786-O cells. As shown in Fig. 2C, HIF-2α protein immunoprecipitated from cells treated for 4 h with both emetine and a proteasome inhibitor was highly ubiquitinated compared with the other treatment groups. Although it is proteasome-mediated, emetine-induced HIF-2α degradation does not require HIF proline hydroxylation. Treatment of cells with the prolyl hydroxylase inhibitor DMOG did not block emetine-induced HIF-2α degradation (Fig. 2E). Thus, unlike VHL, the E3 ligase responsible for targeting HIF-2α to the proteasome in emetine-treated cells does not require HIF hydroxylation as a recognition motif. Finally, emetine-induced degradation of HIF-2α was not blocked by either calpain inhibitor II (ALLM) or the calpain inhibitory peptide calpastatin (Fig. 2, F and G).
Fig. 2.
Emetine-induced HIF-2α degradation is proteasome-dependent. A, 786-O cells were treated with indicated concentrations of the proteasome inhibitor, PS-341 (1-h pretreatment), 0.3 μM emetine, or with a combination of these agents, for 4 h. HIF-2α protein level in nuclear lysates was measured by Western blotting. Topoisomerase II (Topo II) expression is shown as a loading control. B, 786-O cells were treated with the proteasome inhibitor MG132 (10 μM, 1-h pretreatment), 0.3 μM emetine, or with a combination of these agents, for 4 h. HIF-2α and ARNT protein levels in nuclear lysates were measured by Western blotting. CREB expression is shown as a loading control. C, 786-O cells were treated with 100 nM PS-341, 0.3 μM emetine, or with a combination of these agents for 4 h. Protein lysates (1 mg) were immunoprecipitated with anti-HIF-2α antibody, and resultant blots were probed with an anti-ubiquitin antibody. After stripping, the membrane was reprobed with HIF-2α antibody. D, 786-O cells were pretreated with 100 μM ALLN for 1 h and then treated with 0.3 μM emetine for 4 h or with each agent separately. HIF-2α protein level in whole-cell lysates was measured by Western blotting; α-tubulin level is shown as a loading control. E, 786-O cells were pretreated with 0.5 mM DMOG for 1 h and then exposed to 0.3 μM emetine for 4 h. HIF-2α protein was measured by Western blotting; α-tubulin is shown as loading control. F and G, cells were pretreated for 1 h with 100 μM ALLM (F) or 1 μM calpastatin peptide (G) and then were treated and analyzed as in E; α-tubulin is shown as a loading control.
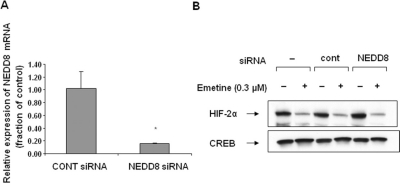
Emetine-Induced HIF-2α Degradation Does Not Require Cullin Neddylation. Oxygen-dependent degradation of HIF-1α and HIF-2α requires HIF-α recognition by VHL, leading to ubiquitination by the VHL, Cullin, and Elongin B/C E3 ubiquitin ligase complex (Sufan et al., 2004; Tan et al., 2008). Activation of cullin-containing E3 ubiquitin ligases requires covalent modification of a conserved lysine residue in the cullin protein by the ubiquitin-like protein NEDD8 (Chew and Hagen, 2007). In the VHL, Cullin, and Elongin B/C complex, Elongin C bridges VHL to Cul2 and Cul2 associates with elongin C, NEDD8, and Rbx1 (Sufan et al., 2004). Although it does not require VHL for its activity, we next asked whether emetine-induced HIF-2α degradation requires cullin neddylation. We transfected 786-O cells with NEDD8 (or control) siRNA. After 72 h (to allow for silencing of NEDD8), we treated cells with emetine for an additional 4 h. The level of NEDD8 mRNA was decreased by approximately 90% in NEDD8-silenced cells (Fig. 3A). However, NEDD8 knockdown had no impact on the ability of emetine to efficiently down-regulate HIF-2α protein (Fig. 3B). These data suggest that emetine-induced HIF-2α ubiquitination, unlike HIF-α ubiquitination mediated by VHL, does not use a cullin-containing E3 ligase.
Fig. 3.
Cullin neddylation is not required for emetine-induced HIF-2α degradation. 786-O cells were transfected with either control siRNA or NEDD8 siRNA. A, to confirm NEDD8 silencing, NEDD8 mRNA was analyzed by quantitative RT-PCR 72 h after transfection. Statistical significance was calculated using SigmaPlot 9.0 (*, p < 0.05) (Systat Software Inc., San Jose, CA). Each bar represents the mean ± S.D. (n = 3). B, 786-O cells were transfected with control or NEDD8 siRNA as in A, and after 72 h, the cells were treated with 0.3 μM emetine for an additional 4 h. HIF-2α protein level was assessed in nuclear lysates by Western blotting. CREB expression is shown as a loading control.
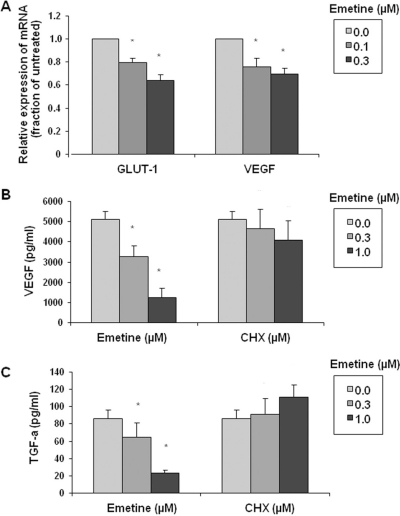
HIF-2α-Mediated Transcription Is Inhibited by Emetine. GLUT-1, VEGF, and TGF-α are HIF target genes in CCRCC (Smaldone and Maranchie 2009). We monitored GLUT-1 and VEGF mRNA levels in 786-O cells after exposure to emetine and found the transcription of both genes to be significantly and dose-dependently reduced by emetine (Fig. 4A). To investigate this phenomenon further at the protein level, we treated 786-O cells with the indicated concentrations of either emetine or CHX for 24 h, and then we determined the amount of secreted VEGF protein in the conditioned media. Emetine, but not CHX, caused a significant dose-dependent decrease in secreted VEGF protein (Fig. 4B). Treatment with 1.0 μM emetine resulted in a 75% decrease of secreted VEGF protein compared with untreated cells. TGF-α protein expression in cell lysate was similarly inhibited by emetine but not by cycloheximide (Fig. 4C). These data show that emetine-induced HIF-2α protein degradation is accompanied by the inhibition of HIF-2α-dependent transcription.
Fig. 4.
Emetine-mediated inhibition of HIF-dependent transcription. A, 786-O cells were untreated or exposed to the indicated concentrations of emetine for 4 h, and mRNA expression for GLUT-1 and VEGF were analyzed by quantitative RT-PCR. Results shown are representative of three separate experiments. B and C, 786-O cells were untreated or exposed to the indicated concentrations of emetine or CHX for 24 h in serum-free medium. The conditioned media were subjected to a VEGF ELISA (B), and TGF-α protein was measured in whole-cell lysates with a TGF-α ELISA (C). Statistical significance was calculated using SigmaPlot 9.0 (*, p < 0.05). Each bar represents the mean ± S.D. (n = 3).
Discussion
786-O CCRCC cells are VHL-deficient and constitutively express HIF-2α but not HIF-1α. Therefore, these cells provide an excellent platform to screen for VHL- and oxygen-independent inhibitors of HIF-2α (Wiesener et al., 2001; Hu et al., 2003; Semenza, 2003; Isaacs et al., 2004). In this study, we have demonstrated that emetine is such an inhibitor, affecting both HIF-2α protein stability and its transcriptional regulation of endogenous target genes. We confirmed these findings using three additional HIF-2α-expressing, VHL-deficient CCRCC cell lines. Emetine has been reported previously to inhibit hypoxia-induced HIF-1α activation in breast tumor cells (Zhou et al., 2005). We confirmed that emetine reduced HIF-1α expression in several cancer cell lines, including MCF7 (VHL wild-type breast cancer), PC3 (VHL wild-type prostate cancer), and UMRC2 cells (VHL-deficient CCRCC that expresses HIF-1α) (data not shown). Thus, emetine possesses the relatively unique property of inhibiting both HIF-1α and HIF-2α expression independently of VHL.
Although HIF-2α inhibition by emetine is oxygen- and VHL-independent, it is accomplished by proteasome-mediated degradation that is preceded by enhanced HIF-2α ubiquitination. Although we have not identified the E3 ubiquitin ligase responsible for emetine-induced HIF-2α ubiquitination, it is unlikely to be a cullin-containing complex (as is the VHL complex) because these E3 ligases require the covalent modification of a conserved lysine residue in the cullin protein with the ubiquitin-like protein NEDD8. We found that NEDD8 molecular silencing had no effect on emetine-induced down-regulation of HIF-2α protein in 786-O cells.
HIF prolyl hydroxylation is necessary to create the recognition motif for VHL binding (Sudarshan et al., 2007; Kaelin, 2009; Semenza, 2009). The calcium-dependent protease cal-pain has also been reported to regulate HIF-1α stability under certain circumstances (Zhou et al., 2006). However, we found that emetine-mediated degradation of HIF-2α was cal-pain-independent because two different calpain inhibitors failed to block emetine-induced HIF-2α depletion. Furthermore, the emetine effect on HIF-2α was also insensitive to the prolyl hydroxylase inhibitor DMOG, offering additional support to the hypothesis that emetine accesses a previously undescribed calcium-dependent proteasome degradation pathway that is distinct from the one mediated by VHL.
By down-regulating HIF-2α protein, emetine affected the transcription of several endogenous HIF-dependent genes in 786-O cells. Both GLUT-1 and VEGF mRNA expression were reduced after emetine treatment, as was the synthesis of VEGF and TGF-α proteins. These gene products are essential HIF-regulated proangiogenic and pro-survival proteins that mediate CCRCC tumorigenicity. Thus, our data support further medicinal chemistry investigation of emetine as a useful platform for the identification of novel HIF-2α inhibitors.
Acknowledgments
We thank Young-Mi Kim, Yun-Jin Jung, and members of their laboratory, as well as Wanping Xu and Eun Joo Chung for helpful discussions and technical assistance.
ABBREVIATIONS:
- VHL
von Hippel-Lindau
- CCRCC
clear cell renal cell carcinoma
- HIF
hypoxia-inducible factor
- ARNT
aryl hydrocarbon receptor nuclear translocator
- CREB
cAMP-response element-binding protein
- VEGF
vascular endothelial growth factor
- GLUT-1
glucose transporter-1
- TGF-α
transforming growth factor-α
- DMOG
dimethyloxaloyl glycine
- NEDD8
neural precursor cell expressed developmentally down-regulated-8
- ALLN
N-acetyl-Leu-Leu-norleucinal
- ALLM
N-acetyl-l-leucyl-l-leucyl-l-methioninal
- PCR
polymerase chain reaction
- siRNA
small interfering RNA
- RT-PCR
reverse transcriptase-polymerase chain reaction
- ELISA
enzyme-linked immunosorbent assay
- CHX
cycloheximide
- PS-341
bortezomib
- MG132
N-benzoyloxycarbonyl (Z)-Leu-Leu-leucinal.
Footnotes
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
References
- Block K, Gorin Y, Hoover P, Williams P, Chelmicki T, Clark RA, Yoneda T, Abboud HE. NAD(P)H oxidases regulate HIF-2alpha protein expression. J Biol Chem. 2007;282:8019–8026. doi: 10.1074/jbc.M611569200. [DOI] [PubMed] [Google Scholar]
- Brahimi-Horn C, Mazure N, Pouysségur J. Signalling via the hypoxiainducible factor-1alpha requires multiple posttranslational modifications. Cell Signal. 2005;17:1–9. doi: 10.1016/j.cellsig.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Chau NM, Rogers P, Aherne W, Carroll V, Collins I, McDonald E, Workman P, Ashcroft M. Identification of novel small molecule inhibitors of hypoxiainducible factor-1 that differentially block hypoxia-inducible factor-1 activity and hypoxia-inducible factor-1alpha induction in response to hypoxic stress and growth factors. Cancer Res. 2005;65:4918–4928. doi: 10.1158/0008-5472.CAN-04-4453. [DOI] [PubMed] [Google Scholar]
- Chew EH, Hagen T. Substrate-mediated regulation of cullin neddylation. J Biol Chem. 2007;282:17032–17040. doi: 10.1074/jbc.M701153200. [DOI] [PubMed] [Google Scholar]
- Choi H, Chun YS, Kim SW, Kim MS, Park JW. Curcumin inhibits hypoxia-inducible factor-1 by degrading aryl hydrocarbon receptor nuclear trans-locator: a mechanism of tumor growth inhibition. Mol Pharmacol. 2006;70:1664–1671. doi: 10.1124/mol.106.025817. [DOI] [PubMed] [Google Scholar]
- Cushny AR. A Text-Book of Pharmacology and Therapeutics; or, The Action of Drugs in Health and Disease. Lea & Febiger; Philadelphia: 1918. [Google Scholar]
- Faivre S, Kroemer G, Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov. 2006;5:671–688. doi: 10.1038/nrd2062. [DOI] [PubMed] [Google Scholar]
- Grollman AP, Huang MT. Inhibitors of protein synthesis in eukaryotes: tools in cell research. Fed Proc. 1973;32:1673–1678. [PubMed] [Google Scholar]
- Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T, Kikuchi H, Herraiz MT, Park DY, Mizukami Y, Mino-Kenduson M, Lynch MP, Rueda BR, Benita Y, Xavier RJ, et al. HIF-1alpha and HIF-2alpha have divergent roles in colon cancer. Int J Cancer. 2009;124:763–771. doi: 10.1002/ijc.24032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs JS, Jung YJ, Mimnaugh EG, Martinez A, Cuttitta F, Neckers LM. Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 alpha-degradative pathway. J Biol Chem. 2002;277:29936–29944. doi: 10.1074/jbc.M204733200. [DOI] [PubMed] [Google Scholar]
- Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, Merino M, Trepel J, Zbar B, Toro J, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–153. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Isaacs JS, Jung YJ, Neckers L. Aryl hydrocarbon nuclear translocator (ARNT) promotes oxygen-independent stabilization of hypoxia-inducible factor-1alpha by modulating an Hsp90-dependent regulatory pathway. J Biol Chem. 2004;279:16128–16135. doi: 10.1074/jbc.M313342200. [DOI] [PubMed] [Google Scholar]
- Jung Y, Isaacs JS, Lee S, Trepel J, Liu ZG, Neckers L. Hypoxia-inducible factor induction by tumour necrosis factor in normoxic cells requires receptor-interacting protein-dependent nuclear factor kappa B activation. Biochem J. 2003;370:1011–1017. doi: 10.1042/BJ20021279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG., Jr Treatment of kidney cancer: insights provided by the VHL tumor-suppressor protein. Cancer. 2009;115(10 Suppl):2262–2272. doi: 10.1002/cncr.24232. [DOI] [PubMed] [Google Scholar]
- Kaur B, Khwaja FW, Severson EA, Matheny SL, Brat DJ, Van Meir EG. Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angio-genesis. Neuro Oncol. 2005;7:134–153. doi: 10.1215/S1152851704001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- Krieg M, Haas R, Brauch H, Acker T, Flamme I, Plate KH. Up-regulation of hypoxia-inducible factors HIF-1alpha and HIF-2alpha under normoxic conditions in renal carcinoma cells by von Hippel-Lindau tumor suppressor gene loss of function. Oncogene. 2000;19:5435–5443. doi: 10.1038/sj.onc.1203938. [DOI] [PubMed] [Google Scholar]
- Lin S, Tsai SC, Lee CC, Wang BW, Liou JY, Shyu KG. Berberine inhibits HIF-1alpha expression via enhanced proteolysis. Mol Pharmacol. 2004;66:612–619. [PubMed] [Google Scholar]
- Löfstedt T, Fredlund E, Holmquist-Mengelbier L, Pietras A, Ovenberger M, Poel-linger L, Påhlman S. Hypoxia inducible factor-2alpha in cancer. Cell Cycle. 2007;6:919–926. doi: 10.4161/cc.6.8.4133. [DOI] [PubMed] [Google Scholar]
- Maranchie JK, Vasselli JR, Riss J, Bonifacino JS, Linehan WM, Klausner RD. The contribution of VHL substrate binding and HIF1-alpha to the pheno-type of VHL loss in renal cell carcinoma. Cancer Cell. 2002;1:247–255. doi: 10.1016/s1535-6108(02)00044-2. [DOI] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Rankin EB, Higgins DF, Walisser JA, Johnson RS, Bradfield CA, Haase VH. Inactivation of the arylhydrocarbon receptor nuclear translocator (Arnt) suppresses von Hippel-Lindau disease-associated vascular tumors in mice. Mol Cell Biol. 2005;25:3163–3172. doi: 10.1128/MCB.25.8.3163-3172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin EB, Rha J, Unger TL, Wu CH, Shutt HP, Johnson RS, Simon MC, Keith B, Haase VH. Hypoxia-inducible factor-2 regulates vascular tumorigenesis in mice. Oncogene. 2008;27:5354–5358. doi: 10.1038/onc.2008.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin EB, Tomaszewski JE, Haase VH. Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer Res. 2006;66:2576–2583. doi: 10.1158/0008-5472.CAN-05-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval RR, Lau KW, Tran MG, Sowter HM, Mandriota SJ, Li JL, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin Cancer Biol. 2009;19:12–16. doi: 10.1016/j.semcancer.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaldone MC, Maranchie JK. Clinical implications of hypoxia inducible factor in renal cell carcinoma. Urol Oncol. 2009;27:238–245. doi: 10.1016/j.urolonc.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Sudarshan S, Linehan WM, Neckers L. HIF and fumarate hydratase in renal cancer. Br J Cancer. 2007;96:403–407. doi: 10.1038/sj.bjc.6603547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sufan RI, Jewett MA, Ohh M. The role of von Hippel-Lindau tumor suppressor protein and hypoxia in renal clear cell carcinoma. Am J Physiol Renal Physiol. 2004;287:F1–F6. doi: 10.1152/ajprenal.00424.2003. [DOI] [PubMed] [Google Scholar]
- Tan M, Gu Q, He H, Pamarthy D, Semenza GL, Sun Y. SAG/ROC2/RBX2 is a HIF-1 target gene that promotes HIF-1 alpha ubiquitination and degradation. Oncogene. 2008;27:1404–1411. doi: 10.1038/sj.onc.1210780. [DOI] [PubMed] [Google Scholar]
- Thomas GV, Tran C, Mellinghoff IK, Welsbie DS, Chan E, Fueger B, Czernin J, Sawyers CL. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nat Med. 2006;12:122–127. doi: 10.1038/nm1337. [DOI] [PubMed] [Google Scholar]
- Wang V, Davis DA, Haque M, Huang LE, Yarchoan R. Differential gene up-regulation by hypoxia-inducible factor-1alpha and hypoxia-inducible factor-2alpha in HEK293T cells. Cancer Res. 2005;65:3299–3306. doi: 10.1158/0008-5472.CAN-04-4130. [DOI] [PubMed] [Google Scholar]
- Wiesener MS, Münchenhagen PM, Berger I, Morgan NV, Roigas J, Schwiertz A, Jürgensen JS, Gruber G, Maxwell PH, Löning SA, et al. Constitutive activation of hypoxia-inducible genes related to overexpression of hypoxiainducible factor-1alpha in clear cell renal carcinomas. Cancer Res. 2001;61:5215–5222. [PubMed] [Google Scholar]
- Zhou J, Köhl R, Herr B, Frank R, Brüne B. Calpain mediates a von Hippel-Lindau protein-independent destruction of hypoxia-inducible factor-1alpha. Mol Biol Cell. 2006;17:1549–1558. doi: 10.1091/mbc.E05-08-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YD, Kim YP, Mohammed KA, Jones DK, Muhammad I, Dunbar DC, Nagle DG. Terpenoid tetrahydroisoquinoline alkaloids emetine, klugine, and isocephaeline inhibit the activation of hypoxia-inducible factor-1 in breast tumor cells. J Nat Prod. 2005;68:947–950. doi: 10.1021/np050029m. [DOI] [PMC free article] [PubMed] [Google Scholar]