Abstract
It is likely that histone H1 is involved in the condensation of chromatin in eukaryotes. However, both the presence of histone H1 in yeast and the extent of yeast chromatin condensation are controversial. A 20 kD protein copurifies with yeast chromatin and was shown by other investigators to have characteristics of histone H1 protein. In an attempt to obtain a positive identification of the 20 kD protein, we purified the protein to homogeneity and raised antibodies against it. We show here by immunofluorescence that the 20 kD protein does not localize to the nucleus but to cytoplasmic particles resembling mitochondria. Furthermore, we show by Western-blot analysis that anti-20 kD protein antibodies react to protein isolated from purified mitochondria. Finally, we present evidence based on size, charge, amino acid composition and immunological cross reactivity to suggest that the yeast 20 kD protein is likely to be the mitochondrial DNA-binding HM protein. This leaves no candidate for histone H1 in yeast.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Electron microscopic observations on the meiotic karyotype of diploid and tetraploid Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1975 Dec;72(12):5056–5060. doi: 10.1073/pnas.72.12.5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron F., Jacq C., Rouvière-Yaniv J. Characterization of a histone-like protein extracted from yeast mitochondria. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4265–4269. doi: 10.1073/pnas.76.9.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certa U., von Ehrenstein G. Reversed-phase high-performance liquid chromatography of histones. Anal Biochem. 1981 Nov 15;118(1):147–154. doi: 10.1016/0003-2697(81)90171-8. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereford L. M., Rosbash M. Number and distribution of polyadenylated RNA sequences in yeast. Cell. 1977 Mar;10(3):453–462. doi: 10.1016/0092-8674(77)90032-0. [DOI] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lohr D., Hereford L. Yeast chromatin is uniformly digested by DNase-I. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4285–4288. doi: 10.1073/pnas.76.9.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
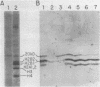
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Pastink A., Berkhout T. A., Mager W. H., Planta R. J. Analysis of histones from the yeast Saccharomyces carlsbergensis. Biochem J. 1979 Mar 1;177(3):917–923. doi: 10.1042/bj1770917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner J. B., Saunders C., Davie J. R., Hamkalo B. A. Ultrastructural organization of yeast chromatin. J Cell Biol. 1982 Apr;93(1):217–222. doi: 10.1083/jcb.93.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Sommer Yeast chromatin: search for histone H1. Mol Gen Genet. 1978 May 31;161(3):323–331. doi: 10.1007/BF00331008. [DOI] [PubMed] [Google Scholar]
- Spiker S., Isenberg I. Cross-complexing pattern of plant histones. Biochemistry. 1977 May 3;16(9):1819–1826. doi: 10.1021/bi00628a009. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis J. W., Rykowski M., Grunstein M. Yeast histone H2B containing large amino terminus deletions can function in vivo. Cell. 1983 Dec;35(3 Pt 2):711–719. doi: 10.1016/0092-8674(83)90104-6. [DOI] [PubMed] [Google Scholar]
- Weber S., Isenberg I. High mobility group proteins of Saccharomyces cerevisiae. Biochemistry. 1980 May 13;19(10):2236–2240. doi: 10.1021/bi00551a037. [DOI] [PubMed] [Google Scholar]