SUMMARY
The miR-16 family, which targets genes important for the G1-S transition, is a known modulator of the cell cycle, and members of this family are often deleted or down-regulated in many types of cancers. Here we report the reciprocal relationship—that of the cell cycle controlling the miR-16 family. Levels of this family increase rapidly as cells are arrested in G0. Conversely, as cells are released from G0 arrest, levels of the miR-16 family rapidly decrease. Such rapid changes are made possible by the unusual instabilities of several family members. The repression mediated by the miR-16 family is sensitive to these cell cycle changes, which suggests that the rapid up-regulation of the miR-16 family reinforces cell cycle arrest in G0. Upon cell cycle re-entry, the rapid decay of several members allows levels of the family to decrease, alleviating repression of target genes and allowing proper resumption of the cell cycle.
INTRODUCTION
MicroRNAs (miRNAs), ~22-nucleotide RNAs found in higher eukaryotes, are involved in numerous biological processes. Watson-Crick base-pairing between the 5' nucleotide region of the miRNA—the so-called ‘seed region’—and sites within a 3' untranslated region (UTR) direct the miRNA-silencing complex to this target messenger RNA (mRNA) and elicit its repression (Bartel, 2009), primarily by mRNA degradation (Baek et al., 2008; Guo et al.; Hendrickson et al., 2009). As the seed region is the main determinant of target specificity, those miRNAs with the same seed repress the same mRNAs and are thus said to constitute a miRNA family.
Several miRNA families have attracted attention for their involvement in regulating the cell cycle and for their misregulation in cancer cells. One particularly well-studied example is the highly conserved miR-16 family, comprised of six mature miRNAs (miR15a/b, miR-16, miR-195, miR-424 and miR-497) that are transcribed from four genomic loci. The deletion or down-regulation of both miR-15a and miR-16 in cases of B cell chronic lymphocytic leukemias (B-CLL) first suggested an association of this family with cancer (Calin et al., 2002). Further supporting a broad role as potential tumor suppressors are reports that members of this family are often down-regulated in other types of cancer, such as colorectal cancer and lung adenomacarcinomas (Bandi et al., 2009; Calin et al., 2005; Klein et al., 2010; Liu et al., 2010). Indeed, although miR-195 is down-regulated in colorectal cancer, restoration of its expression in cell lines is sufficient to repress tumorigenicity (Liu et al., 2010). Inversely, deletion of the genomic 13q14 region in mouse models, which encodes the mir-15a~16-1 locus, recapitulates B-CLL phenotypes seen in humans (Klein et al., 2010). This role for the miR-16 family can be explained by several of its mRNA targets: the anti-apoptotic gene BCL2; genes involved in the MAP kinase pathway, such as MAPK3, MAP2K1 and MAP3K4; and numerous genes involved in the G1-S transition, such as CYCLIN D1/2/3, CYCLIN E1, CDC25A and CDK6 (Bonci et al., 2008; Cimmino et al., 2005; Linsley et al., 2007; Liu et al., 2008; Marasa et al., 2009). Consistent with these targets and a corresponding ability for this family to repress the G1-S transition, over-expression of any family member is sufficient to induce an accumulation of cells in G1 (Linsley et al., 2007; Liu et al., 2008).
Perturbations of the cell cycle are also known to regulate miRNAs. For example, upon DNA damage or oncogenic stress, the miR-34 family is highly up-regulated (He et al., 2007). This up-regulation is mediated by p53 and is thought to reinforce cell cycle arrest, perhaps by targeting the 3' UTRs of CYCLIN E2 and CDK6. Underscoring the importance of this response, recent work has shown that a p53-independent pathway, mediated by p38 signaling, also activates miR-34c expression in response to DNA damage (Cannell et al., 2010).
However, a key issue for understanding the relationship between the cell cycle and miRNAs remains: upon resumption of the cell cycle, what is the fate of these up-regulated miRNAs and their targets? It has long been hypothesized that most miRNAs are highly stable and are down-regulated primarily through dilution (van Rooij et al., 2007). Such a process, though, would be insufficient to rapidly down-regulate miRNAs upon cell cycle re-entry. Although recent work in plants, Caenorhabditis elegans and mammalian systems indicates that some miRNAs are subject to specific degradation (Ameres et al. 2010; Bail et al., 2010; Burns et al., 2011; Hwang et al., 2007; Krol et al.; Ramachandran and Chen, 2008), the involvement and importance of such miRNA decay processes during the cell cycle have thus far been unstudied.
In order to investigate this issue, we profiled miRNAs from cells arrested in G0 by serum starvation and from cells released back into the cell cycle. Among those miRNAs displaying the most rapid down-regulation was miR-503, a member of the extended miR-16 family. Other canonical members of the family were also down-regulated. The dramatic decrease in levels of miR-503 was mediated by active turnover of the miRNA, which we show to be constitutive and dependent on nucleotides in the seed region, combined with presumed transcriptional repression. Arrest in G0, but not in other points of the cell cycle, dramatically increased levels of this miRNA family. Taken together, these data indicate that the miR-16 extended family, which has been known to modulate the cell cycle, is itself dynamically regulated by the cell cycle with coordinated transcriptional regulation and constitutive miRNA decay. Moreover, the observed changes in miRNA levels are consequential for the repression mediated by this miRNA family. We propose that by differentially repressing the activity of several genes known to promote the G1-S transition, the changes in levels of the miR-16 family reinforce other cell cycle pathways.
RESULTS
miR-503, a member of the extended miR-16 family, decreases 10-fold during cell cycle re-entry
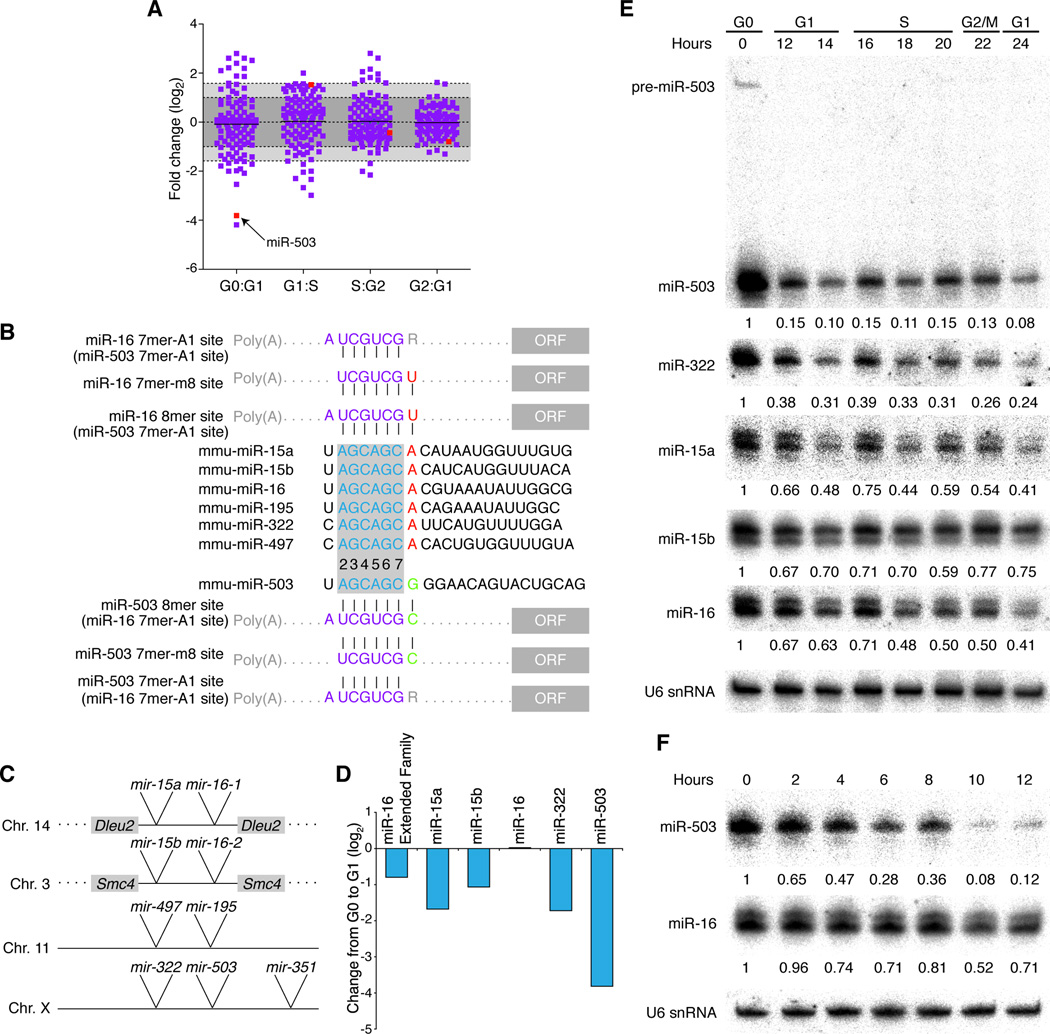
In order to profile the expression of miRNAs during cell cycle re-entry, we synchronously released NIH3T3 cells, which had been arrested in G0 by serum starvation, back into the cell cycle with the addition of serum. Cells were harvested at various time points, and their cell cycle distribution was confirmed by propidium iodide staining and FACS analysis (Figure S1A). With high-throughput sequencing we profiled small RNAs in cell samples arrested in G0, as well as those progressing through G1, S, G2/M and the subsequent G1. Several miRNAs displayed dynamic expression patterns, in some cases changing more than 3-fold between two successive phases of the cell cycle (Figure 1A). miR-10a and miR-503 were the most down-regulated upon cell cycle re-entry, decreasing to less than a tenth of their G0 levels. As this transition from G0 to G1 occurred in 12 hours, such a decrease suggested that these miRNAs were unusually unstable and down-regulated by active degradation. We followed up on miR-503 because miR-10a was lowly expressed, and its sister miRNA, miR-10b, was substantially more abundant and not down-regulated during the G0–G1 transition.
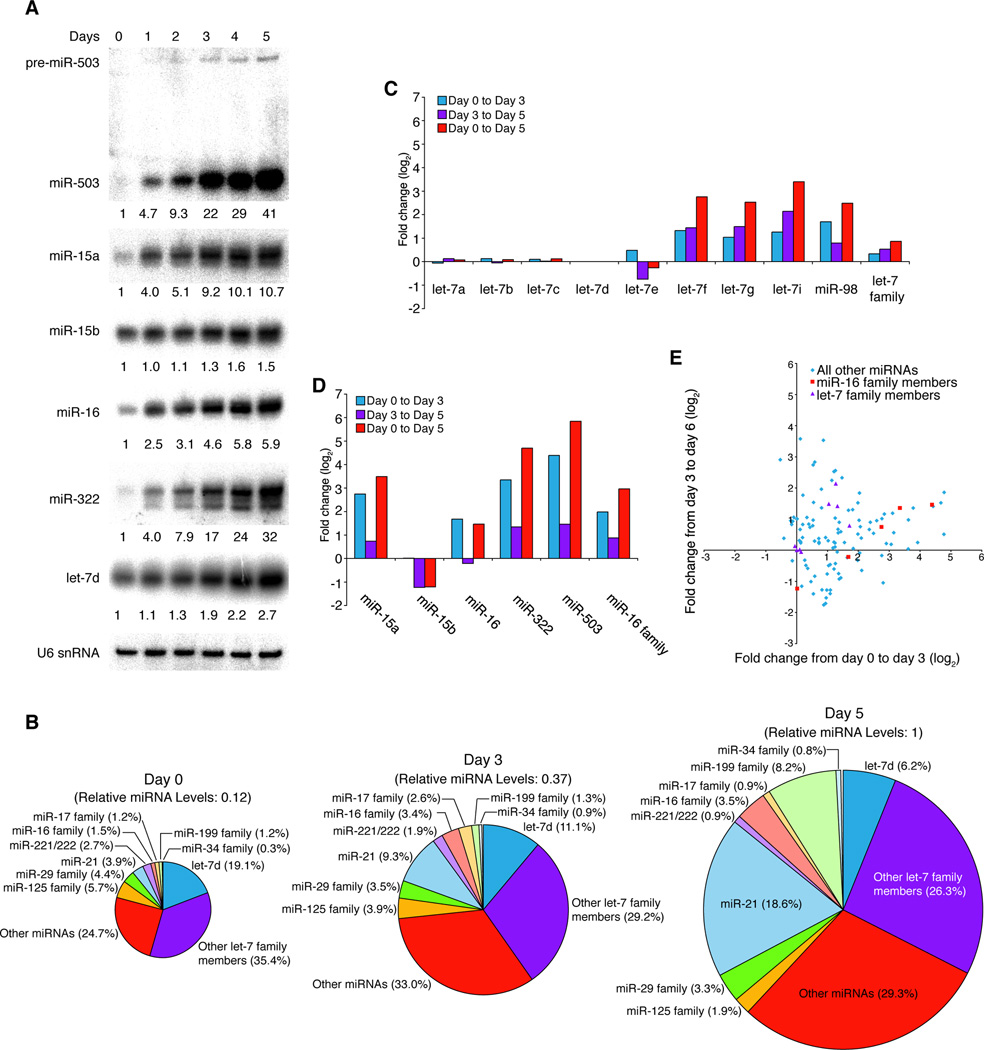
Figure 1. The miR-16 family is dynamically regulated in response to cell cycle changes.
(A) miR-503 levels decrease after cell-cycle re-entry. After NIH3T3 cells were arrested in G0 by serum starvation, cells were released synchronously into the cell cycle by the addition of serum, and miRNAs were profiled at various phases in the cell cycle. Plotted are log2-fold changes in miRNA levels between consecutive cell cycle phases, as determined by Illumina high-throughput sequencing, where each square represents one miRNA and the black line is the median fold change. Dark gray denotes a fold change of two or less; light gray, a fold change between two and three. The red squares represent the fold change of miR-503 between successive phases. (B) Sequences of the canonical members of miR-16 family and miR-503. The miRNA seed, nucleotides 2–7, is outlined in grey. The eighth nucleotide, which differs between canonical miR-16 family members and miR-503, is shown in red (for canonical members) and in green (for miR-503). Various classes of targets are shown with sites highlighted in purple; those nucleotides that interact specifically with canonical members or miR-503 are highlighted in red and green, respectively. (C) Schematics of the four loci encoding the extended miR-16 family. mir-351 is likely part of the same transcriptional unit as mir-322 and mir-503. (D) Overall levels of the miR-16 family decrease upon cell-cycle re-entry. Log2-fold changes observed upon cell cycle re-entry for each expressed member of the extended miR-16 family as well as the entire family. (E, F) Members of the miR-16 family respond to the cell-cycle re-entry as determined by northern blotting. As in (A), NIH3T3 cells were arrested by serum starvation and synchronously released into the cell cycle. Samples were harvested at various points after the addition of serum. Levels of indicated miRNAs were normalized to U6 snRNA (bottom panel).
Although no other annotated miRNA has the same seed region as miR-503, the closest seed sequence is that of the miR-16 family, which differs only at nucleotide eight: this nucleotide is a guanosine in miR-503 and an adenosine in the miR-16 family (Figure 1B). Such similarities lead to an overlap in target mRNAs—8mer sites for miR-503 are recognized as 7mer-A1 sites by canonical miR-16 family members and vice versa (Figure 1B, Figure S1H, I). We note that because positions 7 and 8 pair to a CG dinucleotide (which is strongly depleted in mammalian mRNAs), miR-503 has few predicted targets containing 7mer-m8 sites and thus has few predicted targets not also recognized by the miR-16 family.
Members of the miR-16 family are expressed from four loci as pairs of hairpins, closely spaced with linker regions of 150–200 nucleotides (Figure 1C). Taken with similar tissue expression (Chiang et al., 2010), the genomic organization of mir-322 (which is known as hsa-mir-424 in humans) and mir-503 suggested that they are part of the same transcriptional unit (Figure 1C), which also likely includes mir-351, encoding a miRNA of a different seed family (data not shown). The seed-region similarities, as well as the genomic organization, led us to classify miR-503 as part of the extended miR-16 family.
The extended miR-16 family coordinately decreases during cell cycle re-entry
In considering the ramifications of miR-503 degradation upon cell cycle re-entry, we examined the response of other family members during the transition to G1 (Figure 1D). Overall, during this twelve-hour transition, the number of sequenced RNAs from the extended family decreased by 43%. In addition to the reduced levels of miR-503, this overall decrease was attributed to decreases in miR-15a/b and miR-322: the levels of miR-15a and miR-322 in G1 were one-third of those in G0, suggesting that these two miRNAs might also be unstable. miR-16 reads were unchanged, and miR-195 and miR-497 were not expressed. In contrast, the let-7 family did not show coordinate regulation, and levels of this family were unchanged in comparing G0 and G1 (Figure S1D, E).
In order to confirm these results from small-RNA sequencing, we repeated the synchronization experiment and characterized the expression of the miR-16 family by northern blotting (Figure 1E, Figure S1F). Consistent with the high-throughput sequencing data, miR-16 family members decreased after the addition of serum, and, in the case of miR-503, levels decreased 10-fold upon cell cycle re-entry. The changes in miR-503 were rapid. When looking at the first 12 hours after serum addition, we observed that the levels decreased nearly 4-fold even within 6 hours, indicating an apparent half-life of 3.6 hours for miR-503 (Figure 1F). miR-322 also displayed rapid decreases and had an apparent half-life of 5.8 hours (Figure S1G). These half lives, which are unusually short for miRNAs, represent upper limits on the stability of these miRNAs, since residual synthesis of the miRNAs would imply even shorter half-lives.
miR-503 is constitutively unstable
After the addition of serum, pre-miR-503 was no longer visible by northern blotting (Figure 1E), suggesting that production of the miRNA—either at the level of transcription or generation of the pre-miRNA—might be cell cycle regulated and inhibited upon serum addition. Our initial observations of the behavior of miR-503 also indicated that miRNA was unstable and subject to active degradation. We conceived of two alternative models to unify these observations. In one model, miR-503 is constitutively unstable, and, due to this instability, its levels are highly dynamic, rapidly changing after changes in transcription and/or processing. In the other model, the instability of miR-503 is regulated coordinately with the transcription/processing control such that miR-503, while stable in G0, becomes unstable upon addition of serum.
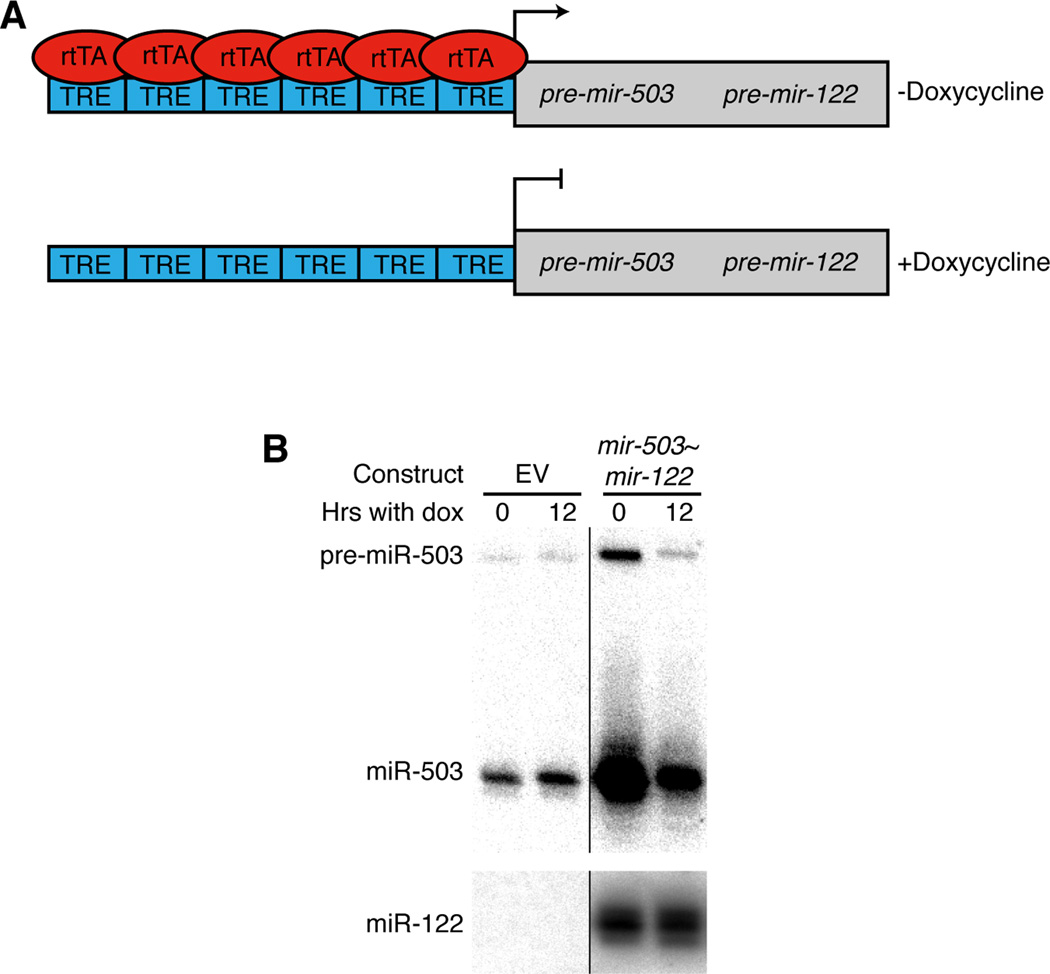
In order to distinguish between these possibilities, a Tet-Off system was used to investigate the instability of miR-503 during arrest in G0. We generated retroviral constructs with a bicistronic cassette of pre-mir-503 and pre-hsa-mir-122 (a miRNA stable in NIH3T3 cells (data not shown)) under a Tet-responsive promoter (Figure 2A). These were then used to create stable lines derived from parental Tet-Off MEF/3T3 cells, which expressed the doxycycline-responsive rtTA transactivator, responded similarly to serum starvation/re-addition and showed similar miR-503 dynamics (Figure S2). In the absence of doxycycline the derived cells expressed both exogenous miRNAs, but, upon the addition of doxycycline, transcription of the bicistronic construct was shut off.
Figure 2. miR-503 is constitutively unstable.
(A) Schematic of the bicistronic miRNA construct. In the presence of doxycycline, transcription of the construct is repressed. TRE, Tet-responsive element. rtTA, Tet-controlled transactivator. (B) miR-503 levels decrease dramatically after transcriptional shut-off. Stable cell lines, derived from Tet-Off MEF/3T3 cells, either expressed the bicistronic miRNA construct or contained an empty vector control. These were arrested by serum starvation and then, while serum starved, treated with doxycycline for 12 hours. Levels of miR-503 (upper panel) and miR-122 (lower panel) were determined by northern blotting.
Stable lines, expressing either the bicistronic cassette or an empty vector control, were first arrested in G0 by serum withdrawal in the absence of doxycycline, and transcription of the exogenous miRNAs was then repressed for 12 hours by addition of doxycycline. Using northern blotting to determine levels of miR-503 and miR-122, we observed, as expected, that in the absence of doxycycline, both the pre-miRNA (detectable in the case of miR-503) and the mature miRNAs were over-expressed relative to the empty vector control line (Figure 2B). As judged by the reduced signal for pre-miR-503, the 12 hour doxycycline treatment repressed transcription of the cassette. Importantly, while levels of miR-122 were unchanged after the 12-hour time course, miR-503 levels were dramatically reduced such that, after normalization to miR-122, only approximately 10% of the exogenously-derived miR-503 remained (Figure 2B). Additional experiments ruled out the possibility that the miRNA was secreted rather than degraded (Figure S1H). Thus, miR-503 is unstable in G0. Moreover, the dynamics observed during these transcriptional shut-off experiments were similar to those upon cell cycle re-entry, further suggesting that the majority of the differential regulation upon cell cycle re-entry occurred at transcriptional and/or processing steps.
The instability of miR-503 is dependent on the seed region and nucleotides in the 3' end
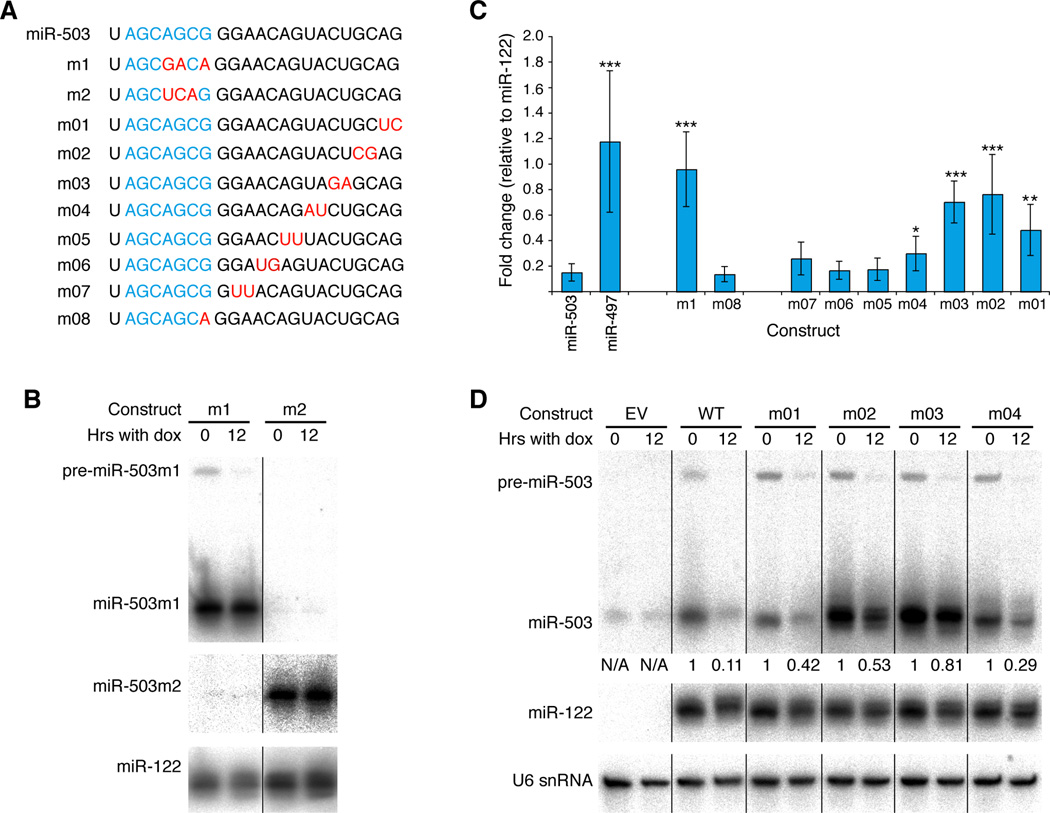
The majority of miRNAs are thought to be stable (van Rooij et al., 2007). Hypothesizing that specific nucleotides within miR-503 mediated its instability, we first generated a miR-503 derivative (m1), in which three nucleotides within the seed region were mutated as well as the corresponding nucleotides in the star strand so as to maintain hairpin structure (Figure 3A, Figure S3). This mutated hairpin was placed within the bicistronic construct described above and used to generate stable cell lines derived from parental Tet-Off MEF/3T3 cells. After arresting these stable cell lines in G0 by serum starvation, we repeated the transcription shut-off experiment. Unlike wild-type miR-503 levels, levels of the mutant were unchanged after the transcriptional shut-off (Figure 3B, C). Indeed, miR-503m1 was now as stable as the control miRNA, miR-122, and significantly more stable than wild-type miR-503 (p = 1E-6, Students t-test). When a second derivative, containing different mutations within the seed region, was tested, similar stabilization was observed (Figure 3B). These data showed that the seed region of miR-503 is required for its instability.
Figure 3. The seed and 3' end of miR-503 are necessary for its instability.
(A) Sequences of mutant derivatives of miR-503. The seed is in blue, and mutated residues, red. (B, C, D) The effect of various mutations on the stability of miR-503. As in Figure 2, stable derivatives of Tet-Off MEF/3T3 cells, each expressing a bicistronic cassette of a mutant miR-503 derivative and miR-122, were arrested in G0 by serum starvation in the absence of doxycycline. Transcription was shut-off for 12 hours by the addition of doxycycline. Levels of miR-503, miR-122 and U6 snRNA were determined by northern blotting. In order to calculate fold change of exogenously expressed miR-503, levels were normalized to those of miR-122. For those mutant derivatives (m01, m02, m03 and m04) that could not be distinguished from endogenous, wild-type miR-503 by northern blotting, the contribution of endogenous miR-503 was estimated using U6 snRNA. (C) Plotted are the fold changes for various mutant derivatives of miR-503 upon doxycycline addition with error bars representing standard deviation. *, p< 0.05; **, p<0.005; ***, p<0.0005; n≥3.
Given the importance of the seed for the instability of miR-503, we next asked whether other members of the miR-16 family also exhibited similar instability. As miR-497 was not appreciably expressed in NIH3T3 cells, the stability of this miRNA was determined using the same approach as that detailed above. Interestingly, miR-497 was significantly more stable than miR-503 (p = 0.001, Student’s t-test), again with a stability similar to that of miR-122 (Figure 3C). Although this result suggested that the canonical miR-16 family seed region was not sufficient to confer instability, it remained formally possible that nucleotide 8, which differs between miR-503 and the canonical members, was important for the instability of miR-503. However, when the half-life of the corresponding mutant miR-503 (m08) was assayed (Figure 3C), there was no significant difference from that observed with the wild-type miRNA (p = 0.39, Student’s t-test). Together these data indicate that, although the seed region is required for the instability of miR-503, it is not sufficient and other regions of the miRNA are also important for its instability.
To find the instability determinants outside the seed, we analyzed additional miR-503 mutants, generated by swapping dinucleotide pairs between the mature and star strand of the miRNA hairpin (Figure 3A, Figure S3A). While mutations in the central region had no effect on the instability of miR-503, mutations at the 3' end (m01–m04) significantly stabilized the miRNA (Figure 3C, D; p = 0.002, p = 0.0002, p = 2E-5 and p = 0.02, respectively, Student’s t-test), suggesting that nucleotides in this region were partially required for the degradation of miR-503. Of course, because the seed mutants (m1 and m2) were stable (Figure 3B, C), this 3' region was not sufficient for the observed wild-type instability, leading us to conclude that both regions coordinate to destabilize miR-503.
Levels of miR-503 increase specifically during arrest in G1
Both the canonical miR-16 family members and miR-503 have previously been shown to repress mRNAs whose protein products are important for the G1-S transition and so overexpression of any of these miRNAs results in the accumulation of cells in G1 (Jiang et al., 2009; Linsley et al., 2007). Reasoning that cell cycle regulation of the miR-16 family might reflect the enrichment of its targets in genes promoting the G1-S transition, we decided to investigate the response of this miRNA family to various cell cycle perturbations more thoroughly.
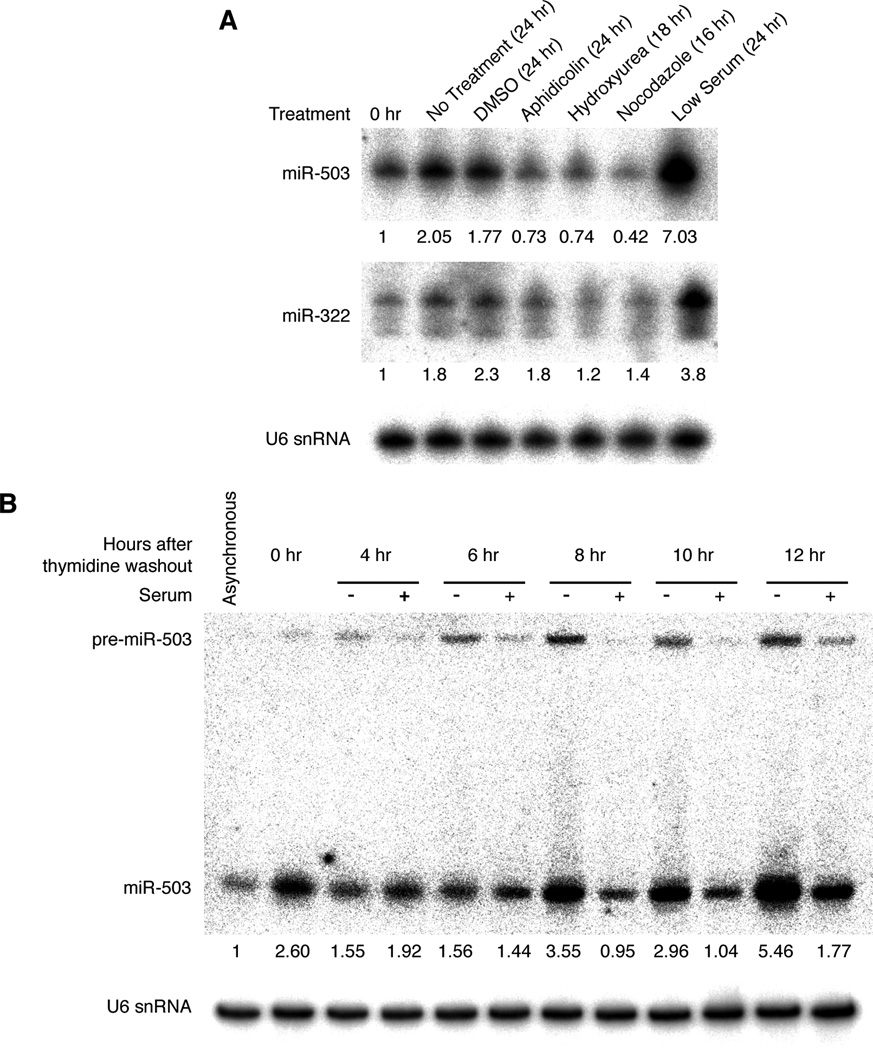
Levels of miR-503 increased in response to serum starvation (Figure 4A). Within 24 hours of serum withdrawal, a time period sufficient to arrest 95% of the cells, levels of miR-503 increased 7-fold. However, other types of cell cycle arresting agents failed to elicit an increase in miR-503 levels. For instance, treatment with either aphidicolin or hydroxyurea, drugs that arrest cells in S phase, had very little effect on miR-503 (Figure 4A). Similarly, the mitotic inhibitor, nocodazole, also failed to increase levels of miR-503. The increases in miR-503 observed in the no-treatment- and DMSO-control cells were likely due to the response of miR-503 to contact inhibition (see below). Consistent with a model of transcriptional control during such cell cycle changes, miR-322, expressed from the same miRNA cluster as miR-503 (Figure 1B), displayed similar responses to these agents (Figure 4A). Thus, the increases in miR-503 and miR-322 appeared to be dependent upon arrest in G1 rather than upon general cell cycle arrest. This specificity distinguished the miRNAs of the mir-322~503 cluster from other cell cycle–controlled miRNAs, such as the miR-34 family, which responded to various insults such as serum starvation and S phase arrest (Figure S4), and was consistent with the well-established role that the miR-16 miRNA family plays in the G1-S transition.
Figure 4. miR-503 is rapidly up-regulated during arrest by serum starvation.
(A) miR-503 and miR-322 specifically respond to arrest by serum starvation. NIH3T3 cells were treated with various cell cycle inhibitors (DMSO, aphidicolin (5 µg/ml), hydroxyurea (2 mM), nocodazole (1 µg/ml) and serum starved) for the indicated time periods. Levels of miR-503 and miR-322 were determined by northern blotting (upper panel) and normalized with U6 snRNA (lower panel). (B) miR-503 is induced in response to low serum. After NIH3T3 were arrested in S phase by 2 mM thymidine, cells were released into the cell cycle in the presence or absence of serum, and samples were harvested at the indicated time points. Levels of miR-503 (upper panel) were determined by northern blotting and normalized to U6 snRNA (lower panel).
Nevertheless, an alternative explanation was formally possible. The specificity of the miR-322/503 response could reflect that the levels of these miRNAs are normally high during G1, and so the increase during serum starvation might instead result from a shift in cell cycle distribution, rather than a response to cell cycle arrest. In order to distinguish between these two possibilities, we arrested cells in S phase with thymidine and then synchronously released them into the cell cycle in media containing either normal or low serum. Those cells released into normal media proceeded through G1 and into the subsequent S phase (Figure S4). In contrast, upon release into media lacking serum, cells proceeded through the G2 and M phases but then, as expected, arrested in the subsequent G1. Importantly, cells released into normal media did not display a significant up-regulation of miR-503, which indicated that progression through G1 was insufficient to elicit this up-regulation (Figure 4B). Indeed, miR-503 increased only in samples lacking serum and did so rapidly as cells entered the subsequent G1. Concomitant with the increase in miR-503 was an increase in the levels of pre-miR-503, consistent with a model of increased production of the pre-miRNA in response to low serum.
The miR-16 family is up-regulated in response to arrest by contact inhibition
In order to address whether the up-regulation of miR-322 and miR-503 was specific to serum withdrawal or whether other types of arrest in G1 elicited such increases, we used contact inhibition to arrest NIH3T3 cells. After 5 days, over 95% of cells accumulated in G0 (Figure S5). Strikingly, on day 5, levels of miR-503 increased approximately 40-fold over those on day 0, when cells were growing asynchronously (Figure 5A). With the exception of miR-15b, other members of the extended miR-16 family also increased: miR-16 was up-regulated nearly 6-fold; miR-15a, 10.7-fold; miR-322, 32-fold. Consistent with a previous report that levels of miRNAs generally increase upon cellular contact (Hwang et al., 2009), let-7d increased 2.7-fold relative to total RNA, as approximated by U6 snRNA levels. However, such a change stood in contrast to the coordinated, large up-regulation of most members of the miR-16 family.
Figure 5. Contact inhibition elicits dramatic up-regulation of the miR-16 family.
(A) The miR-16 family is up-regulated in response to contact inhibition. To arrest NIH3T3 cells by contact inhibition, cells were plated at low density and allowed to become confluent over five days. Samples were taken each day, and the expression of indicated miRNAs was determined by northern blotting with normalization to U6 snRNA. (B) Arrest by contact inhibition affects miRNA levels and composition. Profiles of miRNAs during contact inhibition were determined by Illumina sequencing. miRNA distributions within the day-0, day-3 and day-5 libraries are shown. The area of each chart reflects the number of total miRNA reads, relative to the day-5 library as determined by normalizing to let-7d. (C, D) The effect of confluency upon let-7 and miR-16 families. Fold change of (C) expressed let-7 and (D) expressed miR-16 family members upon contact inhibition. Read numbers were normalized to let-7d. (E) Scatter plot comparing the early response of each miRNA (day 0 to day 3) to the late response (day 3 to day 5). Red squares indicate miR-16 family members; purple triangles, let-7 family members.
In order to determine the effects of contact inhibition on miRNAs more completely, we profiled small RNAs from day-0, -3 and -5 samples by high-throughput sequencing. We first normalized reads to those from let-7d, which was affected only modestly by contact inhibition (Figure 5A), so as to control for the general increase in miRNA levels (Figure 5B). Other members of the let-7 family (let-7f, -7g, -7i and miR-98) increased during contact inhibition; however these members had the lowest expression in the family and so the percentage of reads derived from the let-7 family decreased nearly two-fold (Figure 5B, C).
In contrast to the let-7 family, many miRNAs increased dramatically during this arrest. In fact, some of the most striking examples were members of the miR-16 family, such as miR-503 and miR-322, which increased 20- and 10-fold, respectively, from day 0 and day 3 (Figure 5D). Consistent with our earlier observations, levels of most of the members of the miR-16 extended family increased rapidly upon contact inhibition with the majority of the up-regulation occurring within the first three days: relative to let-7d, the family increased almost four-fold by day 3 and nearly eight-fold by day 5 (Figure 5D). Generally, it appeared the majority of miRNAs that increased did so either early (between days 0 and 3), such as the miR-16, the miR-34 and the miR-17 families, or late (between days 3 to 5), such as the miR-199 family (Figure 5B, E).
miR-21 also displayed strong up-regulation (Figure 5B). This miRNA initially accounted for 3.9% of miRNA reads; yet, at the final time-point, miR-21 was the single most abundant miRNA, representing 18.6% of all miRNA reads. Such up-regulation during growth arrest was not anticipated because miR-21 has been reported to be up-regulated in numerous cancers (Chan et al., 2005; Volinia et al., 2006). On the other hand, hsa-miR-21, along with miRNAs of the hsa-mir-17~92 cluster and hsa-miR-424 (the human homologue of miR-322), is up-regulated during monocyte differentiation (Schmeier et al., 2009). Perhaps a similar transcriptional network is activated during arrest by contact inhibition.
Not all miRNA families increased (Figure 5B). For example, the miR-221 family, composed of miR-221 and miR-222, did not increase relative to let-7d during contact inhibition. Because the other miRNAs generally increased, the percentage of miRNA reads derived from these two miRNAs decreased three-fold. These miRNAs are known to increase as cells re-enter the cell cycle after arrest in G0, and this up-regulation facilitates proper resumption of the cell cycle through the targeting of negative regulators of cell cycle progression (p27 and p57) (Medina et al., 2008). Thus the decrease upon cell-cycle arrest was consistent with a role for these miRNAs in promoting cell cycle progression. Taken together, these results demonstrate that the miR-16 family is strongly up-regulated in response to cell cycle arrest in G0, though not in other stages of the cell cycle. These data suggest a potential role for this family, as well as miR-21, the miR-199 family and the miR-34 family, during arrest by contact inhibition.
Repression of targets of the miR-16 family is affected by cell cycle arrest and re-entry
Having observed the dynamic response of the miR-16 family in response to cell cycle arrest and re-entry, we wished to determine whether repression of targets was similarly affected by cell cycle changes. Analysis of array data profiling the mRNA changes eight hours after adding back serum to serum starved cells (Castellano et al., 2009) revealed that 105 of the 1,440 up-regulated genes (P < 0.005, at least 1.2-fold induction) were predicted targets of the extended miR-16 family. This fraction represented a 1.62-fold enrichment, which was statistically significant (p < 7 × 10−7, hypergeometric mean test). The enrichment remained significant (p < 0.001, computed using FAME (Ulitsky et al., 2010)) when we accounted for differences in the number of miRNA target sites overall (which was attributable to differences in 3' UTR length). These correlative results examining the changes of endogenous mRNAs are consistent with our model that due to the instabilities of several miR-16 family members, repression mediated by the miR-16 family is reduced during cell cycle entry.
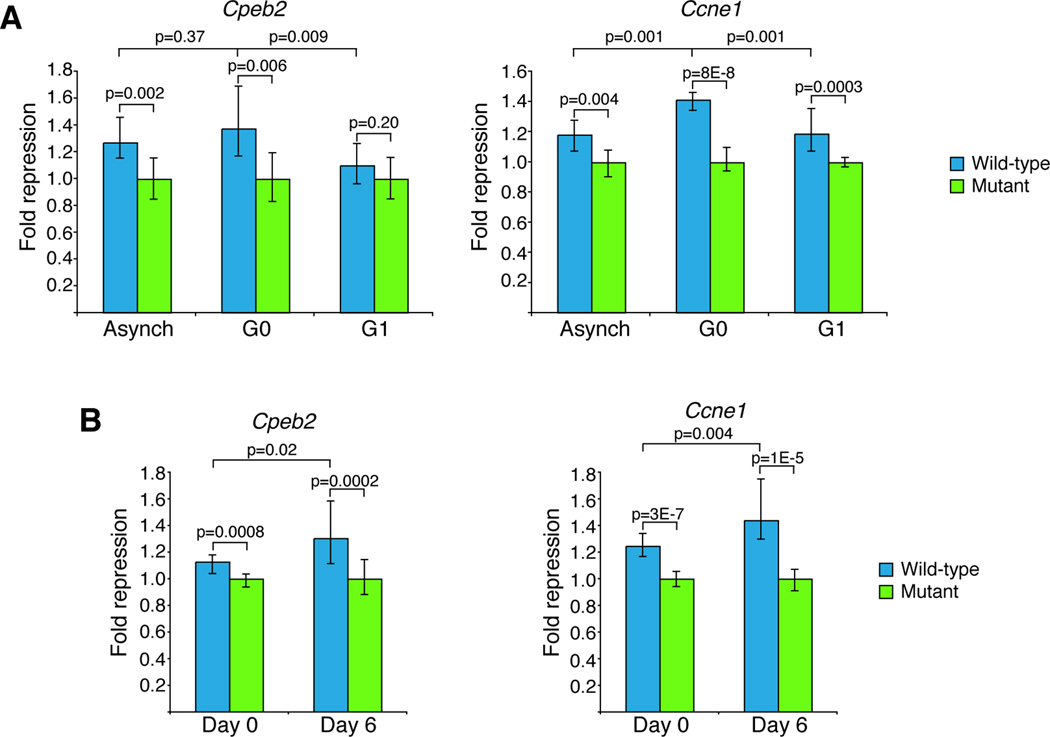
In order to address more directly whether repression by these miRNAs is indeed reduced during the G0-G1 transition, reporter constructs were generated with Renilla luciferase followed by either the 3' UTRs of various miR-16 family targets—specifically, Cpeb2, Cdc25a and Ccne1—or those 3' UTRs with the miRNA sites mutated. To ensure that repression was accurately monitored during transient cell cycle phases, the luciferase reporter was destabilized with a PEST-tag. As expected, statistically significant repression of all three constructs was observed in both asynchronous growth and during arrest by serum starvation (Figure 6A, Figure S6). 1.3- and 1.4-fold repression was observed for the Cpeb2 3' UTR during asynchronous growth and G0, respectively (p = 0.002, p = 0.006, Mann-Whitney U-test). Similarly, for the Cdc25a 3' UTR, the miR-16 family mediated 1.3- and 1.4-fold repression during asynchronous growth and G0 arrest (p = 2 × 10−5, p = 3 × 10−5). For the Ccne1 construct, repression increased from 1.2- to 1.4-fold during arrest by serum starvation (p = 0.004). Importantly, repression of all three targets was sensitive to cell cycle re-entry: for all three targets, release into the cell cycle by addition of serum significantly reduced repression (p = 0.009 for Cpeb2; p = 0.002 for Cdc25a; p = 0.0003 for Ccne1). For 3' UTRs of Cpeb2 and Cdc25a, no significant repression was observed during cell cycle re-entry (p = 0.20 and p = 0.28, respectively).
Figure 6. Repression of targets of the miR-16 family is sensitive to arrest in G0 and cell cycle re-entry.
(A) miR-16 family-mediated repression of reporters is affected by cell cycle re-entry. Reporters expressed unstable Renilla luciferase followed by the 3' UTR of various targets. After transfection, NIH3T3 cells were grown in the presence of serum to measure repression during asynchronous growth (asynch), arrested in the absence of serum (G0) or arrested by serum starvation and then released into the cell cycle by the addition of serum (G1). Fold repression was calculated relative to that of the mutant 3' UTR. Plotted are the normalized values, with error bars representing the third largest and third smallest values (n=12). (B) miR-16 family-mediated repression increases during arrest by contact inhibition. After transfection, NIH3T3 cells were grown asynchronously (day 0) or allowed to arrest by contact inhibition (day 6). Fold repression was calculated and depicted as in (A).
The increases in levels of miR-16 family members during contact inhibition suggested that their repression might also increase during this cell cycle arrest. To test this hypothesis, luciferase assays were performed in cells that had been grown for either one day, when cells were growing asynchronously, or six days, when cells had become fully confluent (Figure 6B). Repression of targets of the miR-16 family was sensitive to this cell cycle arrest, and, for reporters with the 3' UTRs of Cpeb2 and Ccne1, significantly increased (p=0.02 and p=0.004, respectively).
DISCUSSION
Here we have shown that the extended miR-16 family is subject to dynamic regulation in response to both G0 arrest and cell-cycle re-entry. Although the function of this family in repressing genes central for promoting the G1-S transition has been appreciated for several years, the inverse relationship—that of the cell cycle modulating this family of miRNAs—had not been described. These data moreover suggest a coherent regulatory mechanism whereby this family of miRNAs may reinforce arrest in G0. This study also raises several interesting questions both about post-transcriptional regulation of miRNAs and about the role of the miR-16 family in the cell cycle.
miRNA Stability
Although miRNAs generally seem to be stable (van Rooij et al., 2007), presumably through their association with Argonaute (Kai and Pasquinelli, 2010), recent work has highlighted the instability of some, specific animal miRNAs (Bail et al., 2010; Burns et al., 2011; Chatterjee and Grosshans, 2009; Hwang et al., 2007; Krol et al., 2010). In addition to these previously described miRNAs, we have now demonstrated that some members of the extended miR-16 family are constitutively unstable, and that this instability appears to be critical in enabling the rapid decrease in the family upon cell cycle re-entry.
How might such degradation occur? Our observation of the importance of both the seed region and the 3' end in mediating the instability of miR-503 hints suggestively to target-dependent degradation. In one decay mechanism, especially active in Drosophila melanogaster, miRNAs bound to highly complementary targets are tailed and trimmed (Ameres et al., 2010). However, this does not appear to be the primary mechanism of degradation, as judged by our analysis of untemplated nucleotides and 3' trimming on miR-503 and other family members (Figure S1J). Other potential decay mechanisms have also been described. In C. elegans, the 5'→3' exonuclease Xrn2 has been implicated in the degradation of mature miRNAs (Chatterjee and Grosshans, 2009), and in plants and animals, 3'→5' degradation has been implicated in miRNA degradation by, respectively, the small degrading nucleases (SDNs) or the cytoplasmic exosome, respectively (Bail et al., 2010; Ramachandran and Chen, 2008). However, knockdown of homologous enzymes did not stabilize miR-503 during G0–G1 transition (Table S3), and thus identification of the nucleases responsible for miR-503 degradation remains an important, outstanding question.
Although the instability of miR-503 does not appear to be regulated by the cell cycle, other cellular contexts might eventually be shown to stabilize this miRNA, in which case it would be regarded as a differentially stabilized miRNA. In this regard, a new example of a potentially differentially stabilized miRNA comes from a comparison of our results to those of Krol et al. (2010). miR-16 was relatively stable during the cell cycle, whereas in mouse retinas this miRNA is unstable.
miR-16 Family, Targets and the Cell Cycle
Although various external stimuli globally affect miRNAs, the miR-16 family seems to be unique in its specific, cell-cycle-dependent regulation. Like the miR-34 family (He et al., 2007), cell cycle arrest can elicit up-regulation of this family—primarily through activation of the mir-322~503 cluster, but unlike the miR-34 family, this up-regulation occurs only when cells are arrested in G1. These results are consistent with the known role of this family in targeting multiple genes involved in the G1–S transition and thus in repressing progression through this transition (Bandi et al., 2009; Jiang et al., 2009; Linsley et al., 2007). Since changes in the cell cycle (both arrest by contact inhibition and cell cycle re-entry after serum addition) modulate repression mediated by the miR-16 family, we suggest that the cell cycle regulation of the miR-16 family could serve to reinforce the G0-arrested state and might thus be important in various developmental processes. For instance, arrest in G1 is important for the differentiation of myoblasts into myotubes, and a recent report has highlighted that, during myogenesis, the mir-322~503 cluster is up-regulated (Sarkar et al., 2010). Perhaps such a response reflects this coherent relationship between the cell cycle and these miRNAs.
Sensitivity to the levels of these miRNAs might help further explain the connection between the miR-16 family and tumorigenesis. Deletion of the mir-15a~16-1 locus is associated with B-CLL (Calin et al., 2002), and deletion of this region in the mouse is sufficient to partly phenocopy the disease (Klein et al., 2010). If the observed sensitivity of repression of targets (many of which are involved in the G1-S transition and growth-factor signaling) to levels of the miR-16 family were to correspond to a sensitivity of cell cycle progression, even a two-fold reduction in miRNA levels would be sufficient to cause phenotypic changes. Accordingly, even partial down-regulation of this family—through either deletion of the 13q14 region or other mechanisms—might thus alleviate some repression of cell cycle progression upon, for instance, cell-cell contact. Investigation into the relationship between the levels of the miR-16 family, the cell cycle and repression of targets will likely be a promising avenue to address these issues.
MATERIALS AND METHODS
Cell culture
NIH3T3 (ATCC), 293 (ATCC) and Tet-Off 3T3/MEF cells (Clontech) were cultured as described by the manufacturer. Phoenix cells were a gift from Michael Hemann (MIT, Cambridge, USA) and cultured in DMEM supplemented with 10% fetal bovine serum (FBS, Clontech) and penicillin/streptomycin (P/S). To generate stable Tet-Off 3T3 lines expressing bicistronic miRNA cassettes or NIH3T3 lines stably expressing shRNAs, retroviral particles were generated. Phoenix cells, used as the packaging cell line, were transfected with Lipofectamine 2000 (Invitrogen) and TRE plasmids (see Table S3). 24 hours after transfection, media was replaced with DMEM supplemented with 10% FBS and P/S. Viral particles were harvested 48 hours later. 24 hours after infection, the media was replaced with complete, fresh media; after a further 24 hours, puromycin (2 µg/ml) was added. Selected, polyclonal populations were obtained after 2 days. To repress transcription of the bicistronic construct, cells were grown as recommended in the presence of 1 µg/ml doxycycline (Clontech). Before transcriptional shut-off experiments, cells were grown in the absence of doxycycline for at least 24 hours to ensure robust expression of the bicistronic construct.
To arrest cells by serum starvation, cells were first plated at low density (about 12% confluency) and then washed, 24 hours later, 3 times with 1xPBS. DMEM supplemented with P/S and 0.5% donor calf serum (DCS) or FBS (for NIH3T3 cells or Tet-Off 3T3 cells, respectively) was then added. To release cells from arrest, after 48 hours, DMEM media, containing P/S and 20% DCS or FBS, was added. For other drugs, NIH3T3 cells were treated as follows: aphidicolin (5 µg/ml) was added for 24 hours; hydroxyurea (2 mM) was added for 18 hours; nocodazole (1 µg/ml) was added for 16 hours; thymidine (2 mM) was added for 14–16 hours. Arrest was confirmed by FACS analysis (see below). For contact inhibition experiments, NIH3T3 cells were plated at low density, and samples harvested at corresponding time points; media was exchanged daily.
Plasmids
See supplemental materials and methods for details of plasmid constructions. Plasmids have been deposited in Addgene.
Northern blotting
RNA was isolated using Tri-reagent (Ambion), according to manufacturer’s instructions. miRNA northern blots were performed as previously described (Drinnenberg et al., 2009): 2.5–10 µg of total RNA were loaded, and cross-linking to the membrane was mediated by carbodiimide (Pall et al., 2007). For a complete list of probes, see table S2.
FACS analysis
Cells were harvested at appropriate time points and fixed with 75% ice-cold ethanol for at least 30 minutes. Cells were then stained with propidium iodide (50 µg/ml) and RNase A (40 U/ml). DNA content was measured on a FACS Calibur HTS (Benton Dickinson). The percentage of diploid cells in G1, S and G2 was modeled by ModFitLT V3.1 software.
Luciferase assays
NIH3T3 cells were plated in 24-well plates 24 hours prior to transfection at 6×104 cells/well. Cells were transfected using Lipofectamine 2000 and Opti-MEM with 100 ng Renilla luciferase reporter plasmid and, when applicable, 20 ng firefly luciferase control reporter plasmid pISO (Grimson et al., 2007) per well. Media (e.g. containing 10% DCS or 0.5% DCS) replaced the transfection media after 12 hours, and cells were harvested at various time-points. Luciferase activities were measured using dual-luciferase assays, as described by the manufacturer (Promega). Renilla activity was normalized to firefly activity to control for transfection efficiency. Statistical significance was calculated using the Mann-Whitney U test.
Small RNA sequencing and analysis
Small RNAs were cloned from total RNA as described previously (Chiang et al.) and sequenced using the Illumina SBS platform. After removing adaptor sequences, 16–30 nucleotide reads were mapped to the mouse genome, allowing no mismatches and recovering all hits to miRNA loci (miRBase ver 14) (Griffiths-Jones et al., 2008). Reads were first separated into those having come from the mature or star arm of the pre-miRNA hairpin. Then, when necessary, to account for the ability of multiple loci to generate the same mature miRNA, reads were annotated and pooled as corresponding to various mature miRNAs (see Tables S1–2). To determine the addition of untemplated nucleotides, non-genome matching reads were progressively trimmed from the 3' end and queried for their ability to match miRNA loci, until a prefix length of 16 nucleotides was reached. When necessary, untemplated tails were then manually inspected to ensure that sequencing errors did not lead to erroneous classification of tailed species.
Microarray analysis
We obtained processed microarray data of gene expression following release from serum starvation from GEO (accession GSE14829). Expression profiles of probes corresponding to the same gene were averaged. We compared the 0 hr and 8 hr time points from the wild-type samples and selected genes up-regulated by at least 20% and with t-test p-value lower than 0.005. Those were compared with TargetScan 5.1 predicted targets of miR-16 family, miR-503 and the union of these targets. Correction of the p-value for differences in the overall number of microRNAs target sites in 3' UTRs of the up-regulated genes (e.g., due to differences in 3’ UTR length) was done using FAME as described (Ulitsky et al., 2010).
Accession numbers
Illumina sequencing is available on the GEO, accession number GSE31225.
HIGHLIGHTS.
The miR-16 family increases during cell-cycle exit and decreases during re-entry.
The changes in miRNA levels can change the amount of target repression.
The constitutive instability of the miRNAs facilitates their rapid decrease.
The determinants of miR-503 instability map to its 5' and 3' end regions.
Supplementary Material
ACKNOWLEDGEMENTS
We thank past and present members of the Bartel laboratory for helpful discussions, especially V. Auyeung, A. Grimson, C. Jan, G. Lafkas, D. Shechner and I. Ulitsky. We also especially thank I. Ulitsky for performing microarray analysis and T. DiCesare for graphical assitance. This work has been supported by the NIH (grant GM067031 to D.P.B.). O.S.R. was supported by a Ruth L. Kirschstein National Research Service Award (GM088872). S-J.H. was supported by the Canandian Institutes of Health Research. D.P.B. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bail S, Swerdel M, Liu H, Jiao X, Goff LA, Hart RP, Kiledjian M. Differential regulation of microRNA stability. RNA. 2010;16:1032–1039. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, Kappeler A, Brunner T, Vassella E. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553–5559. doi: 10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- Burns DM, D'Ambrogio A, Nottrott S, Richter JD. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature. 2011;473:105–108. doi: 10.1038/nature09908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Cannell IG, Kong YW, Johnston SJ, Chen ML, Collins HM, Dobbyn HC, Elia A, Kress TR, Dickens M, Clemens MJ, et al. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc Natl Acad Sci U S A. 2010;107:5375–5380. doi: 10.1073/pnas.0910015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano E, Guerrero C, Nunez A, De Las Rivas J, Santos E. Serum-dependent transcriptional networks identify distinct functional roles for H-Ras and N-Ras during initial stages of the cell cycle. Genome Biol. 2009;10:R123. doi: 10.1186/gb-2009-10-11-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. Cell-cell contact globally activates microRNA biogenesis. Proc Natl Acad Sci U S A. 2009;106:7016–7021. doi: 10.1073/pnas.0811523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Feng MG, Mo YY. Systematic validation of predicted microRNAs for cyclin D1. BMC Cancer. 2009;9:194. doi: 10.1186/1471-2407-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai ZS, Pasquinelli AE. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol. 2010;17:5–10. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, Dalla-Favera R. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schubeler D, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Chen L, Xu Y, Li R, Du X. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem Biophys Res Commun. 2010;400:236–240. doi: 10.1016/j.bbrc.2010.08.046. [DOI] [PubMed] [Google Scholar]
- Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasa BS, Srikantan S, Masuda K, Abdelmohsen K, Kuwano Y, Yang X, Martindale JL, Rinker-Schaeffer CW, Gorospe M. Increased MKK4 abundance with replicative senescence is linked to the joint reduction of multiple microRNAs. Sci Signal. 2009;2:ra69. doi: 10.1126/scisignal.2000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, Croce CM, Stein GS. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773–2780. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Dey BK, Dutta A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by downregulation of Cdc25A. Mol Biol Cell. 2010;21:2138–2149. doi: 10.1091/mbc.E10-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeier S, MacPherson CR, Essack M, Kaur M, Schaefer U, Suzuki H, Hayashizaki Y, Bajic VB. Deciphering the transcriptional circuitry of microRNA genes expressed during human monocytic differentiation. BMC Genomics. 2009;10:595. doi: 10.1186/1471-2164-10-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Laurent LC, Shamir R. Towards computational prediction of microRNA function and activity. Nucleic Acids Res. 2010;38:e160. doi: 10.1093/nar/gkq570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.