Abstract
The molecular mechanisms by which microtubule-associated proteins (MAPs) regulate the dynamic properties of microtubules (MTs) are still poorly understood. Here, we review recent advances in our understanding of two conserved families of MAPs, the XMAP215/Dis1 and CLASP family of proteins. In vivo and in vitro studies show that XMAP215 proteins act as microtubule polymerases at MT plus ends to accelerate MT assembly, while CLASP proteins promote MT rescue and suppress MT catastrophe events. These are structurally related proteins that use conserved TOG domains to recruit tubulin dimers to MTs. We discuss models for how these proteins might use these individual tubulin dimers to regulate dynamic behaviors of MT plus ends.
Microtubule dynamics
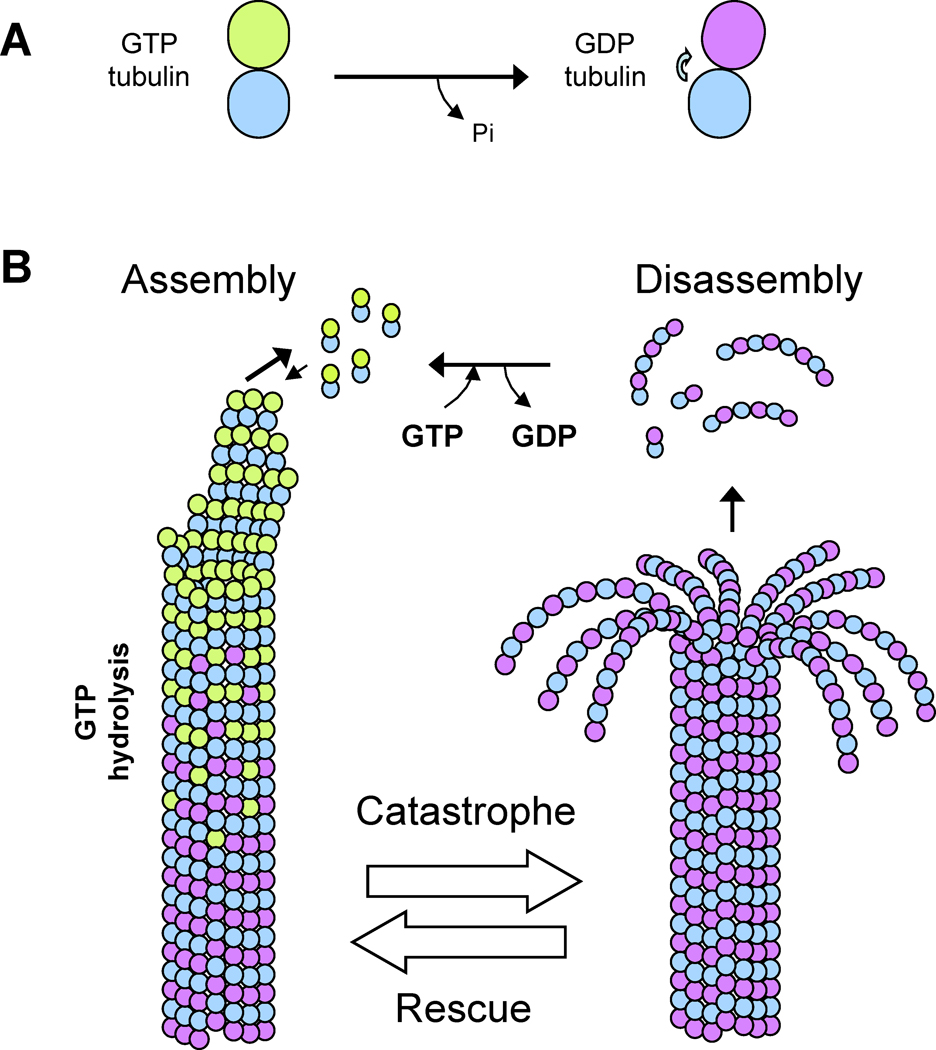
Microtubules (MTs) are dynamic protein polymers used to move and organize cellular components for processes such as cell division, membrane trafficking and cell morphogenesis. Alpha (α) and beta (β) tubulins form obligate αβ-heterodimers (tubulin dimers), which assemble in a head-to-tail fashion to form protofilaments with distinct polarity (Figure 1)[1, 2]. MTs are composed of approximately 13 protofilaments, which associate laterally in a parallel manner to produce a rigid, hollow 25 nm diameter tube (Figure 1)[3]. An essential property of MTs is their ability to grow and shrink at their ends. Transitions between these states are termed “catastrophe” and “rescue.” In a “catastrophe” event, the MT ceases growth and transitions into an explosive shrinkage phase in which protofilaments peel off from the MT plus end [1]. In a “rescue” event, a MT stops shrinkage and converts back to a state of MT assembly. These growth and shrinkage states are accompanied by dynamic conformational changes in the structure of the MT plus end. MT protofilaments at MT ends are straight structures during MT growth, and curved structures that peal outwards during MT shrinkage [4] (Figure 1). Cycles of GTP binding and hydrolysis in beta tubulin provide a conformational switch that helps to drive assembly and disassembly states in the polymer [3] (Figure 1). These dynamic states are highly sensitive to the local concentration of soluble dimer at the MT plus end [2, 5]. For instance, at low tubulin concentration, MTs uniformly shrink, while high tubulin concentrations promote MT assembly and rescue events, and inhibit catastrophe [5]. At physiological tubulin concentrations, pure MTs in vitro display “dynamic instability,” in which MTs in the same population exhibit assembly and disassembly, and undergo switch-like transitions stochastically [5].
Figure 1. Assembly and disassembly of dynamic microtubules (MTs).
A) Conformational change of αβ-tubulin accompanying GTP hydrolysis. In the GTP state (in β-tubulin in green), α and β tubulin monomer interfaces result in “straight” tubulin dimer. In the GDP state αβ-tubulin dimer interface is curved by 5 degrees (arrow), leading to a “bent” tubulin dimer.
B) Structural changes at MT plus ends. During MT assembly, MT plus ends form a sheet-like group of straight protofilaments. GTP-tubulin dimers (green) assemble on the ends, forming a cap of GTP tubulin. GTP hydrolysis over time converts GTP-tubulin in the lattice to GDP-tubulin (note that the extent of the GTP cap is not known). In the MT disassembly phase, GDP-tubulin protofilaments curl and peel off the MT plus ends. The transitions between growth and shrinkage states are termed catastrophe and rescue.
In vivo, a set of conserved MT regulatory proteins bind along the MT lattice or at MT ends and control almost every aspect of MT dynamic behavior [1]. These regulatory proteins modulate in space and time the dynamics and organization of MTs, creating MTs with distinct properties in certain regions of the cell or during certain cell cycle phases. Despite the extensive study, there is still little known about how regulators affect MT dynamics at molecular and structural levels. The molecular details of the MT plus-end itself are still poorly understood [2]. For instance, there are continuing debates in the field about what alternative MT lattice arrangements may exist at the plus end, and how GTP-GDP states of tubulin contribute to MT plus end regulation and structure [2, 6–8]. A recent study shows that the +TIP protein EB1/Mal3 binds in vitro to MTs containing GTPγS (a GTP analogue) but not to GDP or GMPCPP MTs, providing evidence that these EB proteins normally localize to the MT plus end by recognizing a conformation of tubulin dictated by its guanine nucleotide γ-phosphate binding site [9]. Although many proteins on MT plus ends have now been identified, how they may modulate the structure of an MT plus end and/or the state of the guanine nucleotide in tubulin is generally not known.
Here, we present a current understanding of two related families of MT regulatory proteins: XMAP215/Dis1 and Cytoplasmic Linker ASsociated Proteins CLASP. XMAP215/Dis1 proteins have been implicated primarily as an accelerator of MT assembly, while CLASP proteins are involved in promoting MT rescue and suppressing MT catastrophe. These MAPs, which are conserved from yeasts and plants to humans, have essential cellular functions in vivo, regulating MT dynamics in the mitotic spindle and interphase MT [10, 11]. Recent studies have shown that to regulate MT dynamics, these proteins use TOG domains to bind to soluble tubulin dimer [12–16]. How presentation of individual tubulin dimers to the MT plus end modulates MT growth or shrinkage remains an open and highly interesting question. We compare and contrast these structurally related proteins, and propose some working models for how these proteins function. The study of these TOG-domain proteins represents a promising avenue into understanding new mechanisms used to regulate MT polymers.
XMAP215/Dis1 proteins promote MT assembly
XMAP215/Dis1 proteins have conserved roles in promoting the assembly of MTs. These proteins include: S. cerevisae Stu2, S. pombe Alp14 and Dis1, A. thaliana MOR1, C. elegans Zyg9, D. melanogaster MiniSpindles (MSPS) and human ch-TOG [17–22]. XMAP215/Dis1 orthologs localize to MT plus ends, MT organizing centers (MTOCs), kinetochores and, to varying degrees, along MT lattices (Figure 2A,B). In vivo studies show that these proteins generally promote the rate of MT assembly. In general, loss of function leads to interphase MTs that are short and exhibit reduced growth rates and increased frequencies of MT catastrophe and pause events [17, 20, 21, 23–25]. As illustrated by the name of the Drosophila mutant “minispindles,” knockdown of these proteins often lead to small or abnormally organized spindles and short astral MTs [20, 26, 27]. At kinetochores, these proteins are needed for regulation of kinetochore-MT attachment [17, 28]. Stu2, for instance, has been shown to participate in the formation of MTs from the kinetochore to facilitate the attachment of kinetochores to the spindle [29].
Figure 2. Localization and activities of XMAP215/Dis and CLASP proteins.
A) Drosophila Msps (XMAP215/Dis1) on spindle MTs and poles.
B) Drosophila Msps on interphase MT plus ends (Msps, red, arrowhead; tubulin, green)
C) Human CLASP1 on anaphase spindle, at spindle midzone MTs and poles (arrows)(CLASP1, green; MT, red; DNA blue).
D) Human CLASP1 at kinetochore on a metaphase chromosome (CLASP green; DNA, blue; ACA centromere marker, red).
E) Human CLASP2 staining at interphase MT plus ends near plasma membrane (CLASP, red; MT, green).
F) Fission yeast CLASP Cls1p in clusters on interphase MT bundles near nuclear envelope (Cls1p, green; MT, red).
G) Human CLASP1 on the lattice of interphase MTs near the leading edge of the cell (arrowhead).
H) Schematic showing dynamic behavior of pure MTs (grey lines)
I) XMAP215 (green) at the MT plus end accelerates MT assembly and leads to formation of long MTs.
J) S. pombe CLASP, Cls1p (red) binds to the MT lattice and promotes local MT rescue, preventing MTs from shrinking completely.
Images in A–G are reproduced with permission from [40] [24, 48, 53, 61, 63].
XMAP215 was identified as a factor in Xenopus extracts that promotes MT assembly [30]. In vitro studies with recombinant XMAP215 show that these molecules bind directly to growing MT plus ends and accelerate MT plus end assembly roughly ten-fold (Figure 2 H,I)[14, 31]. Each XMAP215 molecule associates with the growing MT plus end only transiently and during this time, it helps to polymerize twenty-five tubulin dimers onto the MT end before disassociating [14]. Although XMAP215 was initially hypothesized to bind preformed tubulin oligomers and load them onto MT ends [32], several lines of evidence demonstrate that each XMAP215 can only bind one tublin dimer at a time. XMAP215 and Stu2 can also catalyze the reverse reaction, namely MT depolymerization, at low tubulin concentrations (added to MTs stabilized with a non-hydrolysable GTP analog, GMPCPP), and were also identified biochemically as MT depolymerases [33, 34]. Thus XMAP215 and its orthologs may act catalytically as MT polymerases that promote the reversible assembly of single tubulin dimers onto MT plus ends.
It remains to be tested whether all members of the XMAP215/Dis1 protein family act as polymerases or have other activities. For instance, S. pombe has two related proteins in this family: Alp14 and Dis1 [17, 35]. Alp14 appears to promote assembly of tubulin dimers at MT plus ends like XMAP215 (Al-Bassam and Chang, unpublished observations). Dis1 functions at kinetochores and spindles for chromosome segregation and is targeted to kinetochores by binding to the Ndc80 complex [36]. However, in addition, recent studies show that Dis1 is localized along the MT lattice in interphase cells, and appears to function in bundling MTs in the spindle and interphase arrays [37]. S.cerevisae Stu2 has been shown to interact with the EB1 and CLIP-170 orthologs, Bik1 and Bim1 [38]. In addition, many XMAP215/Dis1 proteins have been shown to function at MTOCs in a complex with Transforming Acidic coiled-coil (TACC) proteins [38–42]. TACC proteins, which have been identified as partners of XMAP215/Dis1 proteins in yeast to human cells, target XMAP215/Dis1 proteins to centrosomes or SPBs. At MTOCs, XMAP215-TACC complexes increase the number and length of centrosomal MTs during mitosis, possibly by stabilizing or anchoring MT minus ends [39, 42] [43]. These complexes are activated during mitosis by Aurora A kinase phosphorylation on TACC [42, 44, 45] and in yeast, by Ran-dependent nuclear import [46]. Whether XMAP215 functions as a MT plus end polymerase at the MTOC or contributes some other function, for instance at MT minus ends with TACC, remains to be characterized (Figure 3A).
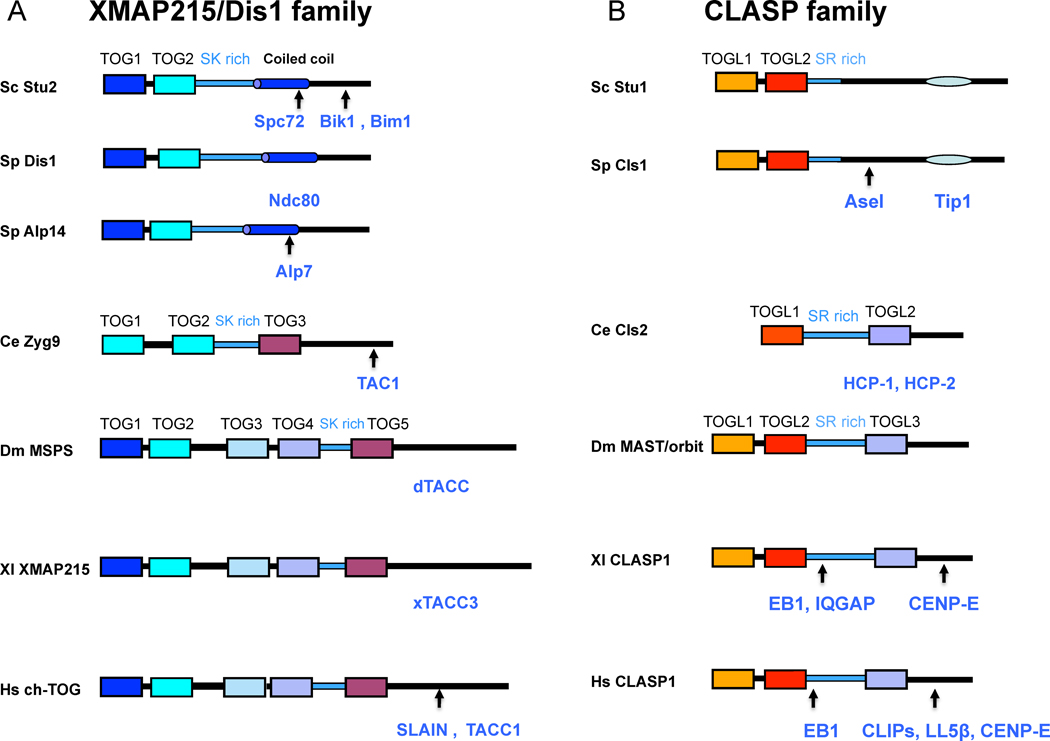
Figure 3. Domain organization of XMAP215/Dis1 and CLASP families from yeast, worms and mammals and their binding partners.
A) XMAP215/Dis1 proteins contain conserved TOG domains and SK rich domain. Domain organization of yeast orthologs S.cereviae Stu2, S.pombe Dis1 and Alp-14 with two TOG domains, C. elegans Zyg9 with three TOG domains and D. melanogaster MSPS, X. laevis XMAP215 and human ch-TOG with five TOG domains. All molecules contain regions with stretches of sequences rich in Serine, Glycine, Lysine (SK-rich domains). TOG domains are colored based on the conserved phylogenetic classes from sequence alignments (Figure 4): TOG1 class (Blue), TOG2 class (Cyan), TOG4 class (sky blue), TOG4 (purple), TOG5 class (maroon). Protein binding partners (Blue) described in the text, are shown below each protein, with arrows denoting approximate binding sites. An absence of an arrow denotes an interaction in which binding domains have not yet been mapped.
B) CLASP proteins contain conserved TOG-Like (TOGL) domains and SR-rich domains
Similar to A, domain organization of S. cerevisae Stu1 and S. pombe Cls1 with two TOGL domains, C. elegans Cls2 with two TOGL domains and D.melanogaster MAST/orbit, human and Xenpus CLASP1 with three TOGL domains. All molecules contain regions with stretches of sequences rich in Serine, Proline and Arginine (SR-rich domains). TOGL domains are also colored based on the conserved phylogenetic classes from sequence alignments (Figure 4): TOGL1 class (orange), TOGL2 class (red), TOGL3 class (purple). Protein binding partners (Blue), described in the text, are shown below each protein with arrows denoting approximate binding sites based on studies described in the text. An absence of an arrow denotes an interaction in which the interacting domain has not yet been mapped.
CLASP promotes MT rescue
CLASP proteins have been implicated in stabilizing subsets of MTs. CLASP family members include S. cerevisae Stu1, S. pombe Cls1, A. thailana CLASP, C. elegans Cls2, D. melanogaster MAST/orbit, and human CLASP1 and CLASP2 [47–52]. A prominent conserved function of CLASP is at the mitotic spindle [50, 53–55]. Loss of CLASP activity causes a variety of severe mitotic defects, often resulting in the collapse of spindles into a “monopolar spindle,” in which the chromosomes are located at the middle of the aster [54, 56]. CLASP localizes to the outer periphery of kinetochores, where it promotes the stability and growth of kinetochore MTs (Figure 2D) [54, 57]. In C.elegans, the CLASP ortholog Cls-2 is targeted to kinetochores via CENP-F-like proteins HCP-1/2 [58]. At the mammalian kinetochore, CLASP1 binds CENP-E and has been shown to be in a complex in early mitosis with the kinesin Kif2b to promote kinetochore movement and MT turnover, and in a separate complex in metaphase with astrin to promote MT stability [59, 60]. During anaphase, CLASP localizes to spindle midzone MTs, where it stabilizes these overlapping MTs and contributes to cytokinesis [53, 54, 56].(Figure 2C).
CLASPs mediate the selective stabilization of interphase MTs. In animal cells CLASPs are commonly seen on the growing MT plus ends (Figure 2E)[52], as well as on the MT lattice and other locations (see below). Knock down of mammalian CLASP2 causes a decrease in MT density at the periphery of the cell and a decrease in the frequencies of MT rescue and pauses, but little change in general MT growth or shrinkage rates [61]. CLASPs are implicated in the attachments of MT plus ends to cortex at the leading edge of fibroblasts [62](Figure 2E). In migrating epithelial cells, CLASP accumulates on the lattices of persistently growing MTs that extend into the leading edge of these migrating cells (Figure 2G) [63]. In Drosophila macrophages and neuronal growth cones, CLASP stabilizes MT bundles that contribute to directional migration, as well as cell-cell repulsion [64, 65] In plants, CLASP mediates the attachment of the MT lattice to the cortex [49]. CLASPs are also located at MTOC structures [52]. Notably, in migrating mammalian cells, a subset of CLASP molecules localizes to the Golgi apparatus and is responsible for organization of MTs emanating from the Golgi apparatus, which contributes to asymmetric distribution of MTs [66].
In addition to binding MT lattices directly, CLASPs are targeted to MT plus ends as well as to diverse cellular compartments through their associations with other proteins in their C-terminal halves, including ase1/PRC1 (MT bundles and spindle midzone), CENP-E and F (kinetochores), LL5β (patches at the plasma membrane), and GCC185 (Golgi apparatus)(Figure 3B) [48, 59, 62, 63, 66]. CLASP is recruited to MT plus ends through its interactions with the +TIP proteins EB1 and CLIP-170, which both contribute to plus end binding by binding to different regions of the molecule [52, 61, 67]. CLASP binds to EB proteins via an EB interaction domain (S-X-I-P motif) near its central MT lattice-binding domain [68]. In migrating epithelial cells, the localization of CLASP to MT plus ends and MT lattice is spatially regulated by GSK3β kinase, which phosphorylates CLASP at multiple sites near these domains. Differential phosphorylation at these sites may influence whether CLASP binds to MT lattice or plus ends [67].
The function of CLASP in promoting MT rescues is perhaps most clear in the fission yeast S. pombe [48]. The sole fission yeast CLASP ortholog cls1/peg1 localizes primarily on the MT lattice, at regions of MT anti-parallel bundling at spindles and in small clusters (of about 13 molecules) in the middle of interphase MT bundles through an interaction with the MT bundling protein, ase1/PRC1 (Figure 2F). Genetic studies show that cls1p is responsible for practically all the local rescue events within both the mitotic spindle and interphase MT bundles. In cls1 mutant cells, MT rescues are not observed, while other MT dynamic parameters are not affected. When overexpressed, cls1p accumulates along MTs and causes dramatically increased rescue frequencies [48].
In vitro studies with recombinant S. pombe cls1p show that it decreases the frequency of MT catastrophes, and increases the frequency of MT rescues [15](Figure 2J). It binds to the MT lattice directly without diffusing and does not track growing MT plus ends. MT rescue events occur at sites on the MT where Cls1 molecules are present at high concentration. Thus, these results together with genetic in vivo results show that cls1p is a MT rescue-promoting factor.
Although S. pombe cls1p is the only CLASP extensively characterized in vitro thus far, the proposed function of MT rescues is fully consistent with the effects seen with CLASPs in animal cells. If bound to the MT plus end, CLASP may promote sustained growth by causing repeated rescue events that immediately reverse catastrophes [54, 61]. At MTOCs and the trans-Golgi network, CLASPs may help to stabilize small segments of MTs by binding near the minus ends, thereby making them resistant to complete depolymerization every time they undergo catastrophe. Thus, CLASP may be regarded as a movable clamp that can locally promote rescue on specific portions of dynamic MTs.
TOG domains bind to soluble tubulin dimer
XMAP215/Dis1 and CLASP family proteins share the highest degree of conservation in their N-termini, which contain 250-residue sequence repeats, termed Tumor Overexpressed Gene (TOG) domains (named after the human ortholog ch-TOG [12]. In XMAP215/Dis1 proteins, the numbers of TOG domains vary from two TOG domains in yeast (Alp14, Dis1, Stu2), three TOG domains in worms (Zyg9), and five TOG domains in flies, plants and mammals (Dm MSPS, Xl XMAP215, Hs ch-TOG; Figure 3A). CLASP family proteins contain TOG-like (TOGL) domains which share weak sequence homology with XMAP215/Dis1 family TOG domains. Yeast CLASP orthologs (Sc Stu1, Sp Cls1) contain a minimum of two conserved TOGL domains, while fly and vertebrate CLASP orthologs (Ce Cls2, Dm MAST/orbit, Xl and Hs CLASP1) contain an additional TOGL domain in their C-terminal half, which cannot be identified in yeast CLASP orthologs [15, 69].
We aligned the sequences of TOG/TOGL domains from diverse XMAP215/Dis1 and CLASP family orthologs (Figure S1). Phylogenetic analysis indicates that the XMAP215/Dis1 and CLASP families share common evolutionary origins in their N-termini (Figure 4A) and that their conserved TOG domains can be classified into distinct groups. For example, all TOG1 domains from XMAP215/Dis1 family proteins form a single class that is distinct from TOG2 domains.
Figure 4. XMAP215/Dis1 and CLASP proteins bind to a tubulin dimer to form a globular complex.
A) XMAP215/Dis1 molecules wrap around soluble tubulin dimers with their TOG domains to form globular complexes. Models for tubulin binding are shown on the left, and electron microscopy images are shown on the right (Stu2 or XMAP215 alone, above; tubulin complex, below). Yeast Stu2 is a homodimer with two sets of TOG domains that wrap around a single tubulin dimer, while Xenopus XMAP215 is a monomer with two internal “halves” each consisting of two TOG domains that interact with a single tubulin dimer. EM images of XMAP215 and Stu2 are reproduced with permission from [12, 14]. White Arrows denote the open conformation of XMAP215 and Stu2 molecules
B) S.pombe CLASP, Cls1 is a homodimer that wrap around soluble tubulin dimers with two sets of TOGL domains. Model for Cls1 dimer binding to the tubulin dimer is shown on the left; electron microscopy images are shown on the right (Cls1 alone, above; cls1-tubulin complex, below). EM of Cls1 were reproduced with permission from [15] White Arrows denote the open conformation of XMAP215 and Stu2 molecules
A key function of the XMAP215 TOG and CLASP TOGL domains is to bind to soluble tubulin dimers [12–15]. The TOG domains of XMAP215, Stu2 and cls1p have been shown to bind soluble (non-polymerized) tubulin dimers with high affinity, but do not bind the tubulin dimers polymerized in the MT lattice. This indicates that the TOG domains can recognize features of tubulin when tubulin is soluble, and not when it is incorporated into the MT lattice [14, 15].
One reason why the vertebrate XMAP215/Dis1 and CLASP proteins are larger and contain more TOG domains than the yeast orthologues is because vertebrate proteins are monomers while the yeast proteins function as dimers [12, 14]. The S. cerevisiae XMAP215 orthologue Stu2 is a homodimer, which contains two sets of two TOG domains; it binds soluble tubulin dimer with a stoichiometry of two Stu2 molecules to one tubulin dimer [12]. Interactions between a C-terminal 100 residue coiled-coil may be responsible for dimerization (Figure 3B). In contrast, XMAP215, containing five TOG domains, is a monomer which binds soluble tubulin dimers with a stoichiometry of one XMAP215 per tubulin dimer [14]. Thus, mammalian XMAP215/Dis1 orthologs contain two internal “halves”, each consisting of a pair of TOG domains (TOG1–2 and TOG 3–4). However, the role of the fifth TOG domain in mammalian orthologs remains unclear. S. pombe Cls1p resembles Stu2, as it also forms a homodimer that binds to a single tubulin dimer [15]. Although the oligomeric states of other CLASP orthologs have not been measured for purified recombinant proteins, fluorescence correlation spectroscopy suggests that mammalian CLASP2 is monomeric when diffuse in the cytoplasm [70]. The presence of a C-terminal TOGL domain suggests that the mammalian CLASPs may bind to tubulin as a monomer through two tubulin-binding halves of TOGL domains, similar to mammalian XMAP215/Dis1 (Figure 3B).
Images of purified negatively stained complexes by electron microscopy reveal how these related proteins clutch soluble tubulin dimers with their TOG domains [12, 14, 15]. In the absence of tubulin, both Stu2p and Cls1p dimers are elongated molecules with thin domains and flexible linkers. With addition of soluble tubulin dimer, the TOG domains of both proteins appear to “wrap” around the outer length of the tubulin dimer to form a globular complex (Figure 4A). The overall conformation of Cls1p and Stu2p tubulin complexes are highly similar (Figure 4). In both Cls1p-tubulin and Stu2p complexes, two identical sets of TOG domains (TOG1 and TOG2) contact a tubulin dimer. It is not yet known which TOG domain binds to which tubulin, or if each TOG in the pair binds to a different face of tubulin. Although XMAP215 molecules are monomers, they are similar to Stu2p in flexibility and wrap around a tubulin dimer to form a similar globular complex [14]. At this level, the striking similarity between the XMAP215 and CLASP proteins provides strong evidence that these are related proteins that use TOG domains to bind to tubulin dimers.
Crystal structures combined with mutational analyses define the TOG domain binding site for soluble tubulin dimer. Structures of three TOG domains from XMAP215/Dis1 proteins show that the TOG domains contain a highly conserved α-helical fold consisting of six conserved HEAT (Huntington, Elongation Factor 2, Phosphotase A2, TOR PI-3 kinase) repeats [13, 16]. Unlike other HEAT repeat proteins, which have curved solenoid-like structures, TOG domains form flat “paddles” with wide and thin edges. The wide edges of a TOG paddle are composed of HEAT repeat α-helices, while the narrow edges of the TOG paddle are composed of loops that connect those helices (Figure 5B, left panel). Two groups of residues are highly conserved in all TOG domains (Figure S1): i) Hydrophobic residues buried between neighboring HEAT repeats α-helices that stabilize the domain’s overall structure (Figure S1), ii) five short intra-HEAT repeat loops (T1 through T5) on one narrow edge, comprising the most conserved surface of the domain (Figure 5B and C). Mutations of the strictly conserved residues in these loops abolish binding to tubulin dimer, indicating that these five loops comprise the TOG domain tubulin-binding site [16].
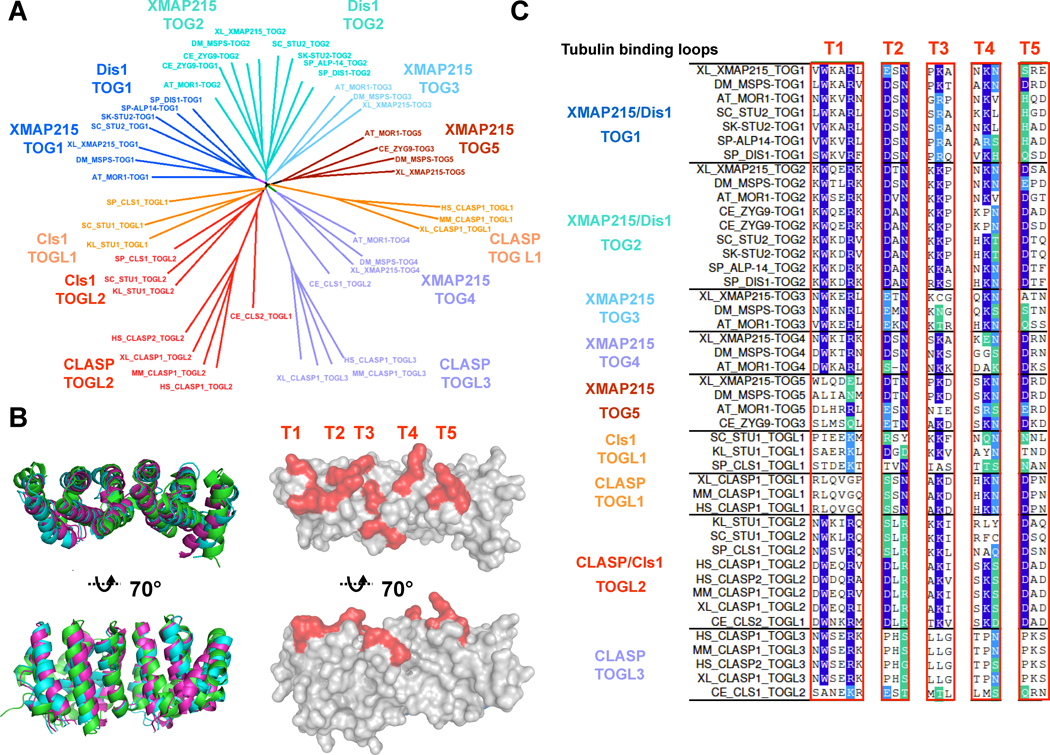
Figure 5. XMAP215/Dis1 TOG and CLASP TOGL domains and the tubulin dimer-binding interface.
A) Phylogenetic tree based on sequence alignment of TOG and TOGL domains: Separate XMAP215/Dis1 family TOG domains and CLASP family TOGL domains from S. cereviase, S. Kluyveri, S .pombe, C. elegans, D. melangaster, A. thaliana, X. laevis, mouse, and human orthologs were aligned using MUSCLE alignment server (further described in Figure S1,[78]). Distance matrices for aligned sequences were used to calculate a phylogenetic tree to classify TOG/TOGL sequences. The tree shows that aligned sequences of TOG and TOGL domains (shown in figure S1) are grouped in conserved phylogenetic classes based on TOG domain position in the protein. This analysis indicates that XMAP215/Dis1 TOG and CLASP family TOGL domain sequences have a common origin. The degree of separation between the different classes describes further divergence in the sequences. The colors of the different branches of the phylogenetic tree are consistent with the coloring scheme of TOG/TOGL domains in Figure 3. The classes are highlighted as follows: XMAP215/Dis1 TOG1 (aqua blue), XMAP215/Dis1 TOG2 (cyan), XMAP215 TOG3 (light blue), XMAP215 TOG4 (purple), XMAP215 TOG5 (brown), Cls1 TOGL1 and CLASP TOGL1 (beige), CLASP/Cls1 TOGL2 (orange), CLASP TOGL3 (purple).
B) Structures of TOG domains and mutational analyses reveal the site for tubulin dimer binding. Top panel: structures of three TOG domains from Zyg9, MSPS, and Stu2 TOG domains show that TOG domains have a flat paddle shape with six conserved HEAT repeats [13, 16]). The overlaid structures are shown narrow side of the paddle overlooking the tubulin binding loops, and the wide side of the paddle overlooking the HEAT repeat helices. Lower panel: sequence conservation and mutational analyses show that TOG domains bind tubulin dimers using intra-HEAT repeat Loops T1–T5 (shown in red) in similar views as described in the upper panel.
C) Detailed sequences of the tubulin binding loops in XMAP215/Dis1 TOG domain and CLASP TOGL domain classes. These aligned tubulin-binding loops describe the amino acid variations in the tubulin-binding sites of TOG and TOGL domains. All the tubulin-binding loop (T1–T5) sequences contain strictly conserved residues (purple), moderately conserved residues (blue), and weakly conserved residues (either in cyan or not colored). Although strictly conserved residues are maintained throughout TOG domains, there are class-conserved residues that differ between classes. For example, XMAP215/Dis1 TOG1 and TOG2 classes differ in many residues neighboring strictly conserved residues within T1 and T2; those variations are maintained in each class. Some classes such as XMAP215 TOG3, XMAP215 TOG4, CLASP TOGL1 and CLASP TOGL3 contain divergent variations in strictly conserved residues, suggesting they may have a weaker affinity for binding tubulin dimer.
Sequence comparisons in the tubulin-binding loops within the TOG domains begin to explain functional differences between TOG domains. Some residues in these tubulin-binding loops are strictly conserved across all classes (Figure 5C, purple), while others are conserved only among different classes. These class-specific variations may modulate the affinity of each TOG domain for tubulin. For instance, in mammalian XMAP215/Dis1 orthologs, the composition of class-conserved residues in these tubulin-binding loops suggests that TOG1 and TOG2 bind with high affinity, while TOG3, TOG4 and TOG5 domains bind with lower affinity.
The affinity of TOG domains for tubulin dimer is critical for function. A recent study found a direct correlation between XMAP215 affinity for tubulin dimer and its MT polymerase activity [71]. Roles of each class of TOG domains in the XMAP215 MT polymerase activity were examined by mutating strictly conserved residues in tubulin binding loops. Inactivation of all five XMAP215 TOG domains resulted in full loss of its MT polymerase. Inactivating the high affinity TOG1–TOG2 domains led to a 75% reduction in both tubulin affinity and MT polymerase activity, while inactivating the low affinity TOG3–TOG4 domains decreased both tubulin binding affinity and MT polymerase by 25%. Although a shortened XMAP215 with just a high-affinity site (TOG1–TOG2) is functional, its MT polymerase activity is still lower than that of full length XMAP215. Thus, both weak and strong tubulin binding sites are likely required for full activity. In comparison, yeast XMAP215 orthologs like Dis1 or Stu2 are predicted to have two identical high affinity sites (TOG1–TOG2) in these homodimeric molecules. It remains to be seen how yeast and vertebrate XMAP215 orthologs compare in their tubulin dimer affinities and MT polymerase activities.
It is very likely that CLASP TOGL domains bind tubulin dimers in a similar manner through analogous residues in the tubulin binding loops. Mutations of the strictly conserved residues in TOGL1 and TOGL2 domains of S. pombe CLASP, Cls1p, cause loss of tubulin dimer binding without affecting MT lattice binding [15]. Multiple mutations in these loops cause a complete loss of Cls1p MT rescue promoting activity in vitro and in vivo. CLASP TOGL domains show a higher degree of variation in their tubulin binding loops than the XMAP15/Dis1 TOG domains (Figure 5C). Some differences in the strictly conserved residues of TOGL1 suggest a weakened affinity for tubulin dimer (Figure 5C). Although Cls1 TOGL domains form stable complexes with tubulin similar to XMAP215 or Stu2, the detailed on and off-rates have not been measured [12, 15]. Differences in TOGL2 tubulin binding loops (such as T2 of TOGL2) are strictly conserved across CLASP orthologs. It remains to be tested if differences in key residues in the TOG/TOGL domains of the CLASP and XMAP215/Dis1 proteins cause different affinities of tubulin binding, and if these affinities underlie differences in protein function.
Binding to the MT lattice
Another key activity of XMAP215/Dis1 and CLASP families is binding to MT lattices. In vivo, multiple interactions with MTs and association with other MAPs may contribute further to XMAP215/Dis1 and CLASP localization to MTs [48, 61, 67]. Although TOG domains were originally suggested to bind MT lattices, there is little evidence that TOG domains themselves interact directly with MT lattices [14, 15]. Domain analysis suggests that direct MT lattice binding activities are mediated primarily by stretches of positively charged residues immediately C-terminal to the TOG domains in both XMAP215/Dis1 and CLASP families [14, 15]. Although these regions are predicted to be mostly unstructured and less conserved in exact sequence, they retain specific features across many species. In XMAP215/Dis1 proteins, MT lattice binding domains consist of stretches of Serine, Glycine, and Lysine residues (termed S/K rich domains), while in CLASP family MT binding domains consist of stretches of Serine, Proline, and Arginine residues (termed S/R rich domains) [12, 15]. As with other MAPs (such as Ndc80 and Dam/DASH kinetochore complexes), these positively charged domains may bind MT lattices by interacting with negatively charged residues at the C-terminus of tubulins [72–75]. In XMAP215, swapping its C-terminus and SK domain with three copies of generic poly-lysine MT binding sequence from Kinesin-3 (Kif1a/unc104), forms an active MT polymerase [71]. Thus the MT lattice binding domains C-terminal to the TOG domains are primarily used for MT localization and are not specifically required for XMAP215/Dis1 polymerase activity.
The MT lattice binding behaviors of XMAP215/Dis1 and CLASP families are quite different, reflecting their different localization patterns in vivo. XMAP215 molecules bind MT lattices with a very low affinity (<1 µM), resulting in molecules that undergo cycles of binding, release, and rebinding that lead to rapid diffusion (0.2 µm/s) along the MT lattice [14, 71]. How XMAP215 localizes on the MT plus end is not known, and we speculate that the molecules diffuse on the MT and then accumulate at the plus end by recognizing some additional feature of the plus end This behavior of XMAP215 may allow it to diffuse to and accumulate at the MT plus end, and is similar to other MT-binding proteins that interact with MT lattices via positive charge, such as kinetochore associated Ndc80 and Dam/DASH complexes [74, 75]. Although XMAP215 recognizes MT plus ends in vitro, in vivo, the accumulation XMAP21 on MT plus ends may be enhanced by interactions with other +TIPs such as EB1. Indeed, SLAIN has been identified as a bridge between EB1 and ch-TOG, the human XMAP215 ortholog, which may serve to enhance plus end binding [76]
In contrast, S. pombe CLASP (Cls1p) binds MT lattices with very high affinity: Cls1-molecules remain attached to specific sites along MT lattices without dissociating for minutes, and exhibit a low diffusion coefficient (0.0015 µm/s), over 100-fold slower than most MAPs [15]. Tubulin dimer binding via TOG domains and MT lattice binding via SR or SK domains are not mutually exclusive, and hence XMAP215 and Cls1p can bind to both MTs and tubulin dimer simultaneously, bringing their tubulin dimers to the MT lattice. In vivo, multiple interactions with MTs and association with other MAPs and regulatory proteins may contribute to the affinity of XMAP215/Dis1 and CLASP on MTs and other intracellular sites [48, 61, 67]. For instance, FRAP experiments show that at least of a subset of CLASP2 in fibroblasts are stable in cortical regions [70]. However, FRAP of CLASP2 specifically on the lattice of MTs in the lamella of eptithelial cells show very rapid dynamics [63], suggesting that the affinity of this protein for MTs is modulated in vivo.
Models for XMAP215 and CLASP mechanisms
We propose working models for how XMAP215 and CLASP families function to regulate MT dynamics. XMAP215 functions as a MT polymerase, which accelerates MT polymerization at the growing MT plus end. XMAP215 molecules may diffuse on the MT due to the low affinity of their SK domains, and accumulate on the MT plus by recognizing some feature at MT plus ends. We propose two models for how XMAP215 promotes tubulin assembly while persisting at growing MT plus ends Figure 6A). Model 1: XMAP215 molecules may repeatedly bind and release individual tubulin dimers that are immediately incorporated into the MT plus end. Model 2: XMAP215 may structurally stabilize the MT plus end in a growth phase by stabilizing a polymerized tubulin intermediate at the growing MT plus end. It may thus prevent the plus end from undergoing small depolymerization events that are observed even in growing MTs [2, 77]. In this view, the tubulin dimer held by the TOG domains may be used as an element to stabilize the newly formed MT lattice, and it may or may not be released into the lattice (Figure 5A, model 2).
Figure 6. Models for XMAP215/Dis1 and CLASP mechanisms.
A) XMAP215/Dis1 family proteins are MT polymerases. Upper panel: XMAP215 binds tubulin dimers with TOG domains and the MT lattice with its SK domain (left). XMAP215-tubulin complexes bind and diffuse along MT lattices. In the absence of XMAP215-tubulin MT assembly is slow. At MT plus ends, XMAP215-tubulin complexes accumulate at the MT plus end accelerates MT assembly (right). Lower panel shows two models for how XMAP215-tubulin complexes increase MT assembly rate: Model 1, XMAP215 TOG domains cycles to load its bound tubulin dimer at growing MT ends. It promotes multiple cycles of tubulin dimer binding, accompanied by conformational change to release tubulin at MT plus ends. Model 2, XMAP215 stabilizes the assembly conformation of a microtubule by binding and stabilizing polymerized-tubulin conformation (yellow) with its TOG domains. The bound tubulin may be a soluble tubulin dimer, or a dimer located at specific site on the MT, such as the very MT end or seam. The conformation of the tubulin dimer or its nucleotide state while bound to XMAP215 is not known and is shown as GDP.
B) CLASP family proteins promote MT rescues and inhibit MT catastrophes. Upper panel: CLASP binds tubulin dimer with its TOGL domains and binds MT lattices with high affinity with its SR-rich domain (left). CLASP high affinity leads to sites of high concentration along MTs. When a dynamic MT undergoes catastrophe, MT disassembly occurs until the plus end reaches such a site of high local CLASP concentration (middle). There, CLASP locally promotes rescue events, in which MT depolymerization halts and MT assembly reinitiates (right). Lower panel shows two models of how CLASP molecules induce MT rescue: Model 1, CLASP molecules releases their loaded tubulin dimer into the MT plus end, to reinitiate polymerization. In this model, CLASP molecules act as local polymerase, similar to model 1 of XMAP215. Model 2, CLASP molecules utilize their loaded tubulin to prevent MT disassembly and restore MT to the assembly phase. During this activity, CLASP molecules may or may not release their bound tubulin. CLASP-tubulin complexes may halt depolymerization, while MT assembly may be reinitiated by tubulin dimers polymerizing from solution.
We propose that CLASP is a MT rescue-promoting factor. CLASP binds tightly on the MT lattice, pre-loaded with a tubulin dimer. When the MT undergoes a catastrophe, the MT plus end depolymerizes until it reaches a site on the MT lattice where CLASP molecules are present at high concentration. These CLASP molecules somehow reverse MT disassembly and mediate rescue. We envision two mechanisms by which CLASP might use its loaded tubulin dimer to accomplish rescue (Figure 6B). Model 1: CLASP may act as a MT polymerase that acts only locally on a depolymerizing MT end, possibly by releasing its bound tubulin dimer to polymerize into the depolymerizing MT lattice and restarting MT assembly. In this model, CLASP is similar to XMAP215 in that it increases local tubulin dimer concentration (Figure 6B, model I). Model 2: CLASP molecules may halt rapid MT disassembly by locally stabilizing the depolymerizing MT lattice from shrinking, possibly by preventing protofilament curling (Figure 5B, model II). CLASP may use its bound tubulin dimer to transiently stabilize the lattice, and may not need to release it. A variation of model 2 is that CLASP and its bound tubulin may induce some alternative MT lattice conformation that makes it resistant to MT depolymerization.
In these models, we note that both CLASP and XMAP215 may share similar functions, either as local polymerases or as local stabilizers. One key difference distinguishing these models is whether the TOG domains release the loaded tubulin dimer into the MT lattice.
Concluding remarks
We have just begun to understand how XMAP215/Dis1 and CLASP family proteins utilize tubulin dimers to regulate the dynamic states of MTs. Through analyses of representative proteins in each class, it is clear that these are related protein families that have distinct roles in MT regulation; XMAP215/Dis1 family proteins promote MT assembly, while CLASP family proteins promote MT rescues and inhibit catastrophes. A key feature of both of these protein families is that they function by binding to soluble tubulin dimers via TOG domains. There are many questions still remaining about how TOG domains function and how the tubulin dimer is used in MT regulation. A detailed understanding of the interaction between TOG domains and the tubulin dimer will be critical to dissect the mechanism for how these proteins regulate dynamic MTs. What is the structural basis for why TOG domains bind to the soluble tubulin dimer and not to tubulin dimers polymerized in the lattice? Which TOG domains bind to which tubulin in the dimer, and do a pair of TOG domains bind to different faces on a single tubulin molecule? Some models predict that the TOG domain releases the tubulin dimer at MT plus ends. Do TOG domains release tubulin into the MT lattice? If so, what signals might allow these proteins to “sense” being at a MT plus end and cause the hand off of the tubulin dimer to the MT end at the right time and place? Although TOG domains bind to both GTP-tubulin and GDP-tubulin dimers in vitro (Al-Bassam, unpublished observation), whether CLASP and XMAP215 proteins function by somehow promoting or stabilizing the GTP state of tubulin during MT polymerization remains an open question. XMAP215/Dis1 TOG domains and CLASP TOGL domains may affect the incorporation of tubulin dimers by stabilizing a polymerizing conformation of tubulin at MT plus ends.
One prime tool in the future studies of the XMAP215 and CLASP families of proteins will be to exploit the differences and similarities between the two families. Differences in TOG/TOGL tubulin binding and SK/SR domain MT lattice association are likely to underlie the functional differences between these proteins. For instance, variations of the TOG domain tubulin binding loop sequences may be responsible for different kinetics of tubulin binding and release. The further study of these intriguing proteins will provide critical understanding into how MT polymers are regulated in cells and provide new conceptual insights into ways MTs are regulated at a molecular level.
Supplementary Material
Sequence alignment of TOG and TOGL domains from many orthologs of XMAP215/Dis1 and CLASP proteins were performed using the MUSCLE server [78]. The TOG domain sequences were identified based on sequence conservations and structural prediction. The TOG domains and TOGL domains are numbered according to their position along the gene sequence. Above the aligned sequences are the structural boundaries of TOG domains based on the TOG domain structures of Zyg9 TOG3, MSPS TOG2 and Stu2 TOG2 [16] [13] Using MUSCLE pairwise sequence alignments, we calculated distance matrices and those were plotted into a phylogenetic tree (shown in Figure 4A) using the Neighbor Joining method [79]. XMAP215 and CLASP family TOG and TOGL domains formed phylogenetic classes based on their position in the protein (Figure 4A). The aligned sequences of classes are shown highlighted here, as follows: XMAP215/Dis1 TOG1 (aqua blue), XMAP215/Dis1 TOG2 (cyan), XMAP215 TOG3 (light blue), XMAP215 TOG4 (purple), XMAP215 TOG5 (brown), Cls1 TOGL1 and CLASP TOGL1 (beige), CLASP/Cls1 TOGL2 (orange), CLASP TOGL3 (purple). Sequences analyzed includes the species initials: XL_XMAP215 (Xenopus laevis), DM_MSPS (Drosophila melanogaster), AT_MOR1 (Arabidopsis thaliana), CE_Zyg9, CE_Cls2 (Caenorhabditis elegans), SC_STU2, SC_Stu1 (Sacchromyces cerevisae), SK_Stu2, SK_Stu1 (Sacchromyces kluyveri), SP_Alp14, SP_Dis1, SP_Cls1 (Schizoscchromyces pombe), HS_CLASP1, HS_CLASP2 (Human), MM_CLASP1, MM_CLASP2 (Mus musculus).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 2.Howard J, Hyman AA. Growth, fluctuation and switching at microtubule plus ends. Nat Rev Mol Cell Biol. 2009;10:569–574. doi: 10.1038/nrm2713. [DOI] [PubMed] [Google Scholar]
- 3.Downing KH, Nogales E. Tubulin structure: insights into microtubule properties and functions. Curr Opin Struct Biol. 1998;8:785–791. doi: 10.1016/s0959-440x(98)80099-7. [DOI] [PubMed] [Google Scholar]
- 4.Mandelkow EM, et al. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 6.Akhmanova A, Steinmetz MO. Microtubule End Binding: EBs Sense the Guanine Nucleotide State. Curr Biol. 2011;21:R283–R285. doi: 10.1016/j.cub.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 7.Sandblad L, et al. The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell. 2006;127:1415–1424. doi: 10.1016/j.cell.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 8.Nogales E, et al. Tubulin rings: which way do they curve? Curr Opin Struct Biol. 2003;13:256–261. doi: 10.1016/s0959-440x(03)00029-0. [DOI] [PubMed] [Google Scholar]
- 9.Maurer SP, et al. GTPgammaS microtubules mimic the growing microtubule end structure recognized by end-binding proteins (EBs) Proc Natl Acad Sci U S A. 2011;108:3988–3993. doi: 10.1073/pnas.1014758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gard DL, et al. MAPping the eukaryotic tree of life: structure, function, and evolution of the MAP215/Dis1 family of microtubule-associated proteins. Int Rev Cytol. 2004;239:179–272. doi: 10.1016/S0074-7696(04)39004-2. [DOI] [PubMed] [Google Scholar]
- 11.Bratman SV, Chang F. Mechanisms for maintaining microtubule bundles. Trends Cell Biol. 2008;18:580–586. doi: 10.1016/j.tcb.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Al-Bassam J, et al. Stu2p binds tubulin and undergoes an open-to-closed conformational change. J Cell Biol. 2006;172:1009–1022. doi: 10.1083/jcb.200511010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brouhard GJ, et al. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Bassam J, et al. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev Cell. 2010;19:245–258. doi: 10.1016/j.devcel.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Bassam J, et al. Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure. 2007;15:355–362. doi: 10.1016/j.str.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Garcia MA, et al. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 2001;20:3389–3401. doi: 10.1093/emboj/20.13.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whittington AT, et al. MOR1 is essential for organizing cortical microtubules in plants. Nature. 2001;411:610–613. doi: 10.1038/35079128. [DOI] [PubMed] [Google Scholar]
- 19.Charrasse S, et al. The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J Cell Sci. 1998;111(Pt 10):1371–1383. doi: 10.1242/jcs.111.10.1371. [DOI] [PubMed] [Google Scholar]
- 20.Cullen CF, et al. mini spindles: A gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J Cell Biol. 1999;146:1005–1018. doi: 10.1083/jcb.146.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang PJ, Huffaker TC. Stu2p: A microtubule-binding protein that is an essential component of the yeast spindle pole body. J Cell Biol. 1997;139:1271–1280. doi: 10.1083/jcb.139.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rockmill B, Fogel S. DIS1: a yeast gene required for proper meiotic chromosome disjunction. Genetics. 1988;119:261–272. doi: 10.1093/genetics/119.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tournebize R, et al. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat Cell Biol. 2000;2:13–19. doi: 10.1038/71330. [DOI] [PubMed] [Google Scholar]
- 24.Brittle AL, Ohkura H. Mini spindles, the XMAP215 homologue, suppresses pausing of interphase microtubules in Drosophila. EMBO J. 2005;24:1387–1396. doi: 10.1038/sj.emboj.7600629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawamura E, Wasteneys GO. MOR1, the Arabidopsis thaliana homologue of Xenopus MAP215, promotes rapid growth and shrinkage, and suppresses the pausing of microtubules in vivo. J Cell Sci. 2008;121:4114–4123. doi: 10.1242/jcs.039065. [DOI] [PubMed] [Google Scholar]
- 26.Gergely F, et al. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 2003;17:336–341. doi: 10.1101/gad.245603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cassimeris L, Morabito J. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol Biol Cell. 2004;15:1580–1590. doi: 10.1091/mbc.E03-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka K, et al. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- 29.Kitamura E, et al. Kinetochores generate microtubules with distal plus ends: their roles and limited lifetime in mitosis. Dev Cell. 2010;18:248–259. doi: 10.1016/j.devcel.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gard DL, Kirschner MW. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J Cell Biol. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinoshita K, et al. Reconstitution of physiological microtubule dynamics using purified components. Science. 2001;294:1340–1343. doi: 10.1126/science.1064629. [DOI] [PubMed] [Google Scholar]
- 32.Cassimeris L, et al. XMAP215 is a long thin molecule that does not increase microtubule stiffness. J Cell Sci. 2001;114:3025–3033. doi: 10.1242/jcs.114.16.3025. [DOI] [PubMed] [Google Scholar]
- 33.van Breugel M, et al. Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus end-binding microtubule destabilizer. J Cell Biol. 2003;161:359–369. doi: 10.1083/jcb.200211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shirasu-Hiza M, et al. Identification of XMAP215 as a microtubule-destabilizing factor in Xenopus egg extract by biochemical purification. J Cell Biol. 2003;161:349–358. doi: 10.1083/jcb.200211095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohkura H, et al. Dis1/TOG universal microtubule adaptors - one MAP for all? J Cell Sci. 2001;114:3805–3812. doi: 10.1242/jcs.114.21.3805. [DOI] [PubMed] [Google Scholar]
- 36.Hsu KS, Toda T. Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Curr Biol. 2011;21:214–220. doi: 10.1016/j.cub.2010.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roque H, et al. The fission yeast XMAP215 homolog Dis1p is involved in microtubule bundle organization. PLoS One. 2010;5:e14201. doi: 10.1371/journal.pone.0014201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen XP, et al. The yeast spindle pole body component Spc72p interacts with Stu2p and is required for proper microtubule assembly. J Cell Biol. 1998;141:1169–1179. doi: 10.1083/jcb.141.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee MJ, et al. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat Cell Biol. 2001;3:643–649. doi: 10.1038/35083033. [DOI] [PubMed] [Google Scholar]
- 40.Cullen CF, Ohkura H. Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat Cell Biol. 2001;3:637–642. doi: 10.1038/35083025. [DOI] [PubMed] [Google Scholar]
- 41.Sato M, et al. Interdependency of fission yeast Alp14/TOG and coiled coil protein Alp7 in microtubule localization and bipolar spindle formation. Mol Biol Cell. 2004;15:1609–1622. doi: 10.1091/mbc.E03-11-0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinoshita K, et al. Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J Cell Biol. 2005;170:1047–1055. doi: 10.1083/jcb.200503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bellanger JM, Gonczy P. TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Curr Biol. 2003;13:1488–1498. doi: 10.1016/s0960-9822(03)00582-7. [DOI] [PubMed] [Google Scholar]
- 44.Barros TP, et al. Aurora A activates D-TACC-Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J Cell Biol. 2005;170:1039–1046. doi: 10.1083/jcb.200504097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peset I, et al. Function and regulation of Maskin, a TACC family protein, in microtubule growth during mitosis. J Cell Biol. 2005;170:1057–1066. doi: 10.1083/jcb.200504037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato M, Toda T. Alp7/TACC is a crucial target in Ran-GTPase-dependent spindle formation in fission yeast. Nature. 2007;447:334–337. doi: 10.1038/nature05773. [DOI] [PubMed] [Google Scholar]
- 47.Pasqualone D, Huffaker TC. STU1, a suppressor of a beta-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J Cell Biol. 1994;127:1973–1984. doi: 10.1083/jcb.127.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bratman SV, Chang F. Stabilization of overlapping microtubules by fission yeast CLASP. Dev Cell. 2007;13:812–827. doi: 10.1016/j.devcel.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ambrose JC, Wasteneys GO. CLASP modulates microtubule-cortex interaction during self-organization of acentrosomal microtubules. Mol Biol Cell. 2008;19:4730–4737. doi: 10.1091/mbc.E08-06-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dumont J, et al. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat Cell Biol. 2010;12:894–901. doi: 10.1038/ncb2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathe E, et al. Orbit/Mast, the CLASP orthologue of Drosophila, is required for asymmetric stem cell and cystocyte divisions and development of the polarised microtubule network that interconnects oocyte and nurse cells during oogenesis. Development. 2003;130:901–915. doi: 10.1242/dev.00315. [DOI] [PubMed] [Google Scholar]
- 52.Akhmanova A, et al. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- 53.Maiato H, et al. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 2003;113:891–904. doi: 10.1016/s0092-8674(03)00465-3. [DOI] [PubMed] [Google Scholar]
- 54.Maiato H, et al. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hannak E, Heald R. Xorbit/CLASP links dynamic microtubules to chromosomes in the Xenopus meiotic spindle. J Cell Biol. 2006;172:19–25. doi: 10.1083/jcb.200508180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inoue YH, et al. Mutations in orbit/mast reveal that the central spindle is comprised of two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J Cell Biol. 2004;166:49–60. doi: 10.1083/jcb.200402052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheeseman IM, et al. The CENP-F-like proteins HCP-1 and HCP-2 target CLASP to kinetochores to mediate chromosome segregation. Curr Biol. 2005;15:771–777. doi: 10.1016/j.cub.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 59.Maffini S, et al. Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr Biol. 2009;19:1566–1572. doi: 10.1016/j.cub.2009.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manning AL, et al. CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. EMBO J. 2010;29:3531–3543. doi: 10.1038/emboj.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mimori-Kiyosue Y, et al. Mammalian CLASPs are required for mitotic spindle organization and kinetochore alignment. Genes Cells. 2006;11:845–857. doi: 10.1111/j.1365-2443.2006.00990.x. [DOI] [PubMed] [Google Scholar]
- 62.Lansbergen G, et al. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev Cell. 2006;11:21–32. doi: 10.1016/j.devcel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 63.Wittmann T, Waterman-Storer CM. Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3beta in migrating epithelial cells. J Cell Biol. 2005;169:929–939. doi: 10.1083/jcb.200412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stramer B, et al. Clasp-mediated microtubule bundling regulates persistent motility and contact repulsion in Drosophila macrophages in vivo. J Cell Biol. 2010;189:681–689. doi: 10.1083/jcb.200912134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee H, et al. The microtubule plus end tracking protein Orbit/MAST/CLASP acts downstream of the tyrosine kinase Abl in mediating axon guidance. Neuron. 2004;42:913–926. doi: 10.1016/j.neuron.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 66.Efimov A, et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar P, et al. GSK3beta phosphorylation modulates CLASP-microtubule association and lamella microtubule attachment. J Cell Biol. 2009;184:895–908. doi: 10.1083/jcb.200901042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Honnappa S, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 69.Slep KC. Structural and mechanistic insights into microtubule end-binding proteins. Curr Opin Cell Biol. 2010;22:88–95. doi: 10.1016/j.ceb.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 70.Drabek K, et al. Role of CLASP2 in microtubule stabilization and the regulation of persistent motility. Curr Biol. 2006;16:2259–2264. doi: 10.1016/j.cub.2006.09.065. [DOI] [PubMed] [Google Scholar]
- 71.Widlund PO, et al. XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc Natl Acad Sci U S A. 2011;108:2741–2746. doi: 10.1073/pnas.1016498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang HW, et al. Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat Struct Mol Biol. 2007;14:721–726. doi: 10.1038/nsmb1274. [DOI] [PubMed] [Google Scholar]
- 73.Wei RR, et al. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 74.Westermann S, et al. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- 75.Powers AF, et al. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–875. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van der Vaart B, et al. SLAIN2 links microtubule plus end-tracking proteins and controls microtubule growth in interphase. J Cell Biol. 2011 doi: 10.1083/jcb.201012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schek HT, 3rd, et al. Microtubule assembly dynamics at the nanoscale. Curr Biol. 2007;17:1445–1455. doi: 10.1016/j.cub.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of TOG and TOGL domains from many orthologs of XMAP215/Dis1 and CLASP proteins were performed using the MUSCLE server [78]. The TOG domain sequences were identified based on sequence conservations and structural prediction. The TOG domains and TOGL domains are numbered according to their position along the gene sequence. Above the aligned sequences are the structural boundaries of TOG domains based on the TOG domain structures of Zyg9 TOG3, MSPS TOG2 and Stu2 TOG2 [16] [13] Using MUSCLE pairwise sequence alignments, we calculated distance matrices and those were plotted into a phylogenetic tree (shown in Figure 4A) using the Neighbor Joining method [79]. XMAP215 and CLASP family TOG and TOGL domains formed phylogenetic classes based on their position in the protein (Figure 4A). The aligned sequences of classes are shown highlighted here, as follows: XMAP215/Dis1 TOG1 (aqua blue), XMAP215/Dis1 TOG2 (cyan), XMAP215 TOG3 (light blue), XMAP215 TOG4 (purple), XMAP215 TOG5 (brown), Cls1 TOGL1 and CLASP TOGL1 (beige), CLASP/Cls1 TOGL2 (orange), CLASP TOGL3 (purple). Sequences analyzed includes the species initials: XL_XMAP215 (Xenopus laevis), DM_MSPS (Drosophila melanogaster), AT_MOR1 (Arabidopsis thaliana), CE_Zyg9, CE_Cls2 (Caenorhabditis elegans), SC_STU2, SC_Stu1 (Sacchromyces cerevisae), SK_Stu2, SK_Stu1 (Sacchromyces kluyveri), SP_Alp14, SP_Dis1, SP_Cls1 (Schizoscchromyces pombe), HS_CLASP1, HS_CLASP2 (Human), MM_CLASP1, MM_CLASP2 (Mus musculus).