Abstract
Apolipoprotein E, a 34 kDa protein, plays a key role in triglyceride and cholesterol metabolism. Of the three common isoforms (ApoE2, 3 and 4), only ApoE4 is a risk factor for Alzheimer’s Disease. All three isoforms of wild-type ApoE self-associate to form oligomers, a process that may have functional consequences. Although the C-terminal domain, residues 216–299, of ApoE is believed to mediate self-association, the specific residues involved in this process are not known. Here we report the use of hydrogen/deuterium exchange (H/DX) coupled with enzymatic digestion to identify those regions in the sequence of full-length apoE involved in oligomerization. For this determination, we compared the results of H/DX of the wild-type proteins and those of monomeric forms obtained by modifying four residues in the C-terminal domain. The three wild type and mutant isoforms show similar structures based on their similar H/DX kinetics and extents of exchange. Regions of the C-terminus (residues 230–270) of the ApoE isoforms show significant differences of deuterium uptake between oligomeric and monomeric forms, confirming that oligomerization occurs at these regions. To achieve single amino acid resolution, we examined the extents of H/DX by using electron transfer dissociation (ETD) fragmentation of peptides representing selected regions of both the monomeric and the oligomeric forms of ApoE4. From these experiments, we could identify the specific residues involved in ApoE oligomerization. In addition, our results verify that ApoE4 is composed of a compact structure at its N-terminal domain. Regions of C-terminal domain, however, appear to lack defined structure.
Apolipoprotein E (ApoE), a 299-residue, 34 kDa protein, a major lipid transporter in plasma and the central nervous system (1–3), is intimately involved in triglyceride and cholesterol metabolism (2). There are three major isoforms, ApoE2, ApoE3 and ApoE4, that differ only by single amino acid changes (3). ApoE3 has a cysteine at position 112 and an arginine at 158, whereas ApoE2 has cysteines, and ApoE4 has arginines at these two positions. These single amino acid differences cause significant differences in their biochemical and functional properties (2, 3). For example, ApoE4 is a major risk factor for Alzheimer’s disease and cardiovascular diseases, whereas ApoE2 serves a protective role (4).
Lipid-free ApoE consists of a 22 kDa N-terminal domain (1–191), which harbors the LDL receptor binding site, and a 10 kDa C-terminal domain (216–299), which has high affinity for lipids (5–7), linked by a protease-sensitive hinge region (3, 8). Structures of full length, wild-type ApoE are unknown, although x-ray crystallography of the N-terminal domain shows it is a monomer comprised of a four-helix bundle (9). The lack of structure of the wild-type proteins is probably due to a high tendency for them to aggregate at the high concentrations needed for crystallization (8, 10) and the likely presence of amphipathic helices in the full length protein (11, 12). Recently, however, an NMR structure of a monomeric mutant of ApoE3 was determined (13). Lipid-free ApoE exists predominantly as a tetramer at µM concentrations and dissociates to dimers and monomers at nM concentrations (1, 2, 10, 14–17). A monomeric form of ApoE3 at 150 µM can be obtained by making four or five substitutions in the C-terminal region without apparently affecting the overall secondary structure or function (18). This monomeric form is a reference point for our study to analyze the structure and oligomerization properties of ApoE isoforms.
We chose hydrogen/deuterium exchange (H/DX) coupled with mass spectrometry (MS) (19–22) as a structural tool to measure the relative rates of exchange of amide hydrogens with deuteriums from D2O and draw conclusions based on the measured m/z and their changes, rather than ion abundances. The experimental outcome allows inferences on the stability of the hydrogen bonding, the extent of which is inverse to relative solvent accessibilities. To achieve a high resolution comparison of the three isoforms of ApoE and their monomeric mutants, we coupled H/DX with enzymatic digestion and analyzed the exchanged peptides.
H/DX with digestion affords spatial resolution at the “peptide level” (i.e., to regions of the protein that are 6–10 amino-acid residues in length (23)) Although one can sometimes incorporate multiple enzymatic digestions to generate overlapping peptide fragments and improve the resolution to one or a few amide sites, we chose gas-phase fragmentation of the deuterated peptides released upon pepsin digestion because it has greater potential to achieve resolution at the amino-acid level. Fragmentation cannot be collisionally induced dissociation (CID) (collision-activated dissociation (CAD)) because it causes scrambling of H and D (24–29). To overcome this, we fragmented the peptides with electron-transfer dissociation (ETD) (30) under experimental conditions that we determined to be sufficiently “gentle” to avoid scrambling.
To insure minimal scrambling, we followed Jørgensen, who established a set of standard peptides as tests (several other methods were also established for the same purpose (31, 32)), using a bottom-up H/DX (23) to afford resolution at a single-amide site. The latter allowed us to improve the resolution to the single amide level so that we can identify those amides involved in self-association. This approach also affords an opportunity to continue the development of a structural model for the full-length, wild-type ApoE proteins.
EXPERIMENTAL PROCEDURES
a. Protein expression
Apolipoprotein E was expressed in E. coli and purified as described previously by Garai et al. (33). Briefly, ApoE proteins were expressed in Escherichia coli (Strain BL21, DE3 competent cells, Stratagene), which was grown in defined media to OD600 = 0.6. Mutations were introduced into the c-DNA of ApoE using the QuikChange site-directed mutagenesis kit (Stratagene). All sequences were verified using DNA sequencing. In all cases, the monomeric mutants contained the following four substitutions: F257A/W264R/L279Q and V287E.
b. Materials
Potassium chloride, HEPES hemisodium salt [N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid) hemisodium salt], tris(2-carboxyethyl)phosphine hydrochloride (TCEP), porcine pepsin, citric acid, sodium citrate, sodium chloride, and ethanolamine were purchased from Sigma-Aldrich (St. Louis, MO). The synthetic peptide HHHHHHIIKIIK used to test the extent of H/D scrambling was purchased from Genscript (Piscataway, NJ). D2O was from Cambridge Isotope Laboratories Inc (Andover, MA), and high purity guanidine hydrochloride was from Pierce (Rockford, IL).
c. Preparation of ApoE stock solution
ApoE, dissolved in 4 M GdnCl, 0.1% β-mercaptoethanol, was dialyzed against 10 mM HEPES and 150 mM KCl containing 100 µM TCEP disulfide reductant by using Slide-A-Lyzer Dialysis Cassettes 7000 MWCO (Pierce, CA) for 2 h. The buffer was exchanged for fresh buffer, and the solution was dialyzed overnight at 4 °C. The concentration of the final protein solution was established by absorbance at 280 nm (ε = 44950 cm−1M−1) (34). The protein solutions were stored at −80 °C as 50 µL aliquots.
d. H/D Exchange (H/DX) Protocol
The protein stock solution was first diluted in buffer (10 mM HEPES, 150 mM KCl, pH 7.4) to prepare a 4 µM final analytical concentration and equilibrated at 25 °C for 2 h. H/DX was initiated by diluting the protein solution 1:10 into D2O buffer (10 mM HEPES, 150 mM KCl, pH 7.4) at 25 °C. At varying times, the H/DX was quenched by adding sufficient 1 M HCl at 0 °C to give a final pH of 2.5.
For peptide-level H/DX experiments, the quenched solution was injected into an on-line pepsin-digestion device (Supplemental Figure 1) containing immobilized pepsin in a 1 mm diameter × 2 cm length guard column (Upchurch Scientific, WA), which was prepared as described previously (35). The quenched solution was injected into the pepsin column in 0.1% formic acid via an Agilent 1100 HPLC (Santa Clara, CA) operated at 50 µL/min flow rate for 3 min. The digested protein solution was trapped on a C18 guard column (1 mm diameter × 1.5 cm length, Optimize Technologies, Oregon City, OR). The peptide mixture was then eluted from the trap column and separated with a C18 analytical column (1 mm diameter × 5 cm length, Dionex, Bannockburn, IL) via an Agilent 1200 HPLC (Santa Clara, CA) with a gradient operated at 50 µL/min flow rate. Solvent A was water containing 0.1% formic acid, and solvent B was 80% acetonitrile, 20% water containing 0.1% formic acid. The gradient settings were: 5% to 15% solvent B in 0.3 min, 15% to 50% solvent B in 5.2 min, 50% to 100% solvent B in 0.5 min, and isocratic flow at 100% solvent B for 1.5 min, then returned to 5% solvent B in 0.1 min. All LC connection lines were immersed in a water-ice (0 °C) bath.
e. LC-ESI/MS Analysis with a Q-TOF Mass Spectrometer
Peptide-level H/DX results were acquired on a Maxis (Bruker, Bremen, Germany) quadrupole time-of-flight (Q-TOF) mass spectrometer. The settings were: capillary voltage, 3.8 kV; nebulizer gas, 0.4 bar; drying gas flow rate and temperature, 4.0 L/min and 180 °C, respectively; funnel RF, 400 V(pp).
f. Test of hydrogen scrambling
In the test procedure, described previously by Jørgensen and coworkers (30), 100 µM of synthetic peptide, HHHHHHIIKIIK (Supplemental Figure 2), was prepared in D2O and kept at 4 °C overnight to allow complete deuterium exchange. The solution was diluted 50-fold with cold H2O buffer (50% MeOH, 0.5 M acetic acid, pH 2.5) and frozen on dry ice to quench the exchange reaction. When needed for analysis, the peptide solution was manually thawed and injected into the ESI source via a pre-cooled 500 µL syringe (Hamilton, Reno, NV) operated at 50 µM/min flow rate.
Three instrumental conditions were chosen: (1) capillary temperature, 50 °C; capillary voltage, 20 V; tube lens, 50 V; (2) capillary temperature, 100 °C; capillary voltage, 50 V; tube lens, 50 V, and (3) capillary temperature, 200 °C; capillary voltage, 100 V; tube lens, 100 V. The active hydrogens of the test peptide were exchanged with deuterium and submitted to back exchange for approximately 1 min. The maximum number of deuteriums remaining in the peptide was approximately 4.5, which agreed with Jørgensen’s results under similar conditions (29). Upon fragmenting the peptide by ETD, we found the deuterium content for the c5 ion to be low (~0.6 D) under the first condition, indicating low H/D scrambling. As the capillary temperature and voltage were increased, increasing deuterium content was found in the N-terminal c5 ion (deuterium uptakes for the c ions are in Supplemental Figure 3), indicating that the first set of instrument parameters were optimum. Another advantage for tuning the experiment by using this standard peptide was that the solution flow rate and other instrument parameters were easily optimized to afford good peptide ion signals.
g. LC-ESI/MS/MS Analysis with an Orbitrap-ETD Mass Spectrometer
To confirm the sequence for each peptic peptide, the protein was digested as described previously and analyzed on a Thermo LTQ XL Orbitrap (Thermo Fisher, San Jose, CA). The settings were: spray voltage, 3.5 kV; sheath gas flow rate, 8 (arbitrary units); capillary temperature, 275 °C; capillary voltage, 35 V; and tube lens, 110 V. One full mass spectral acquisition triggered three scans of MS/MS (precursor ion activation to give CID) whereby the most abundant precursor ions were activated for sequencing. The peaks observed in the product-ion spectra (MS/MS) were centroided for each peptide during the acquisition.
H/DX-ETD MS was performed on a Thermo LTQ XL Orbitrap (Thermo Fisher, San Jose, CA), using the fluoranthene radical anion to cause ETD. One full mass-spectrum acquisition was used to trigger three scans of ETD. The parameters were: spray voltage, 3.5 kV; sheath gas flow rate, 8 (arbitrary units); capillary temperature, 50 °C; capillary voltage, 20 V; tube lens, 50 V; tandem MS selection threshold, 1000 counts; activation time, 100 ms for ETD. For ETD, the isolation width was 6.0 m/z, and the reaction time was “charge-state” dependent. A precursor mass list was generated based on the deuterated peptides’ centroid m/z, and MS/MS data were centroided during acquisition.
h. Mascot database search
Thermo RAW files were processed by using extract_msn (2007 version 4.0, Thermo Fisher, San Jose, CA) with a grouping tolerance of 0.8 Da, an intermediate scan setting of 1, and a minimum of 1 scan per group. The NCBI nonredundant database (version 20101004, restricted to human) was searched by using MASCOT 2.2.06 (Matrix Science, Oxford, U.K.) employing the following settings: enzyme, none; MS tolerance, 10 ppm; MS/MS tolerance, 0.8 Da; maximum number of missed cleavages, 3; peptide charge of 1+, 2+ and 3+; oxidation of methionine was set as variable modification.
i. Data analysis
For peptide-level H/DX, the uptake of deuterium for each peptide was measured as the average differences between the centroid masses of the deuterated peptide and the undeuterated peptide. The back-exchange was measured to be one deuterium loss per minute. No correction for back exchange was applied because the time between sample quench and the measurement with the mass spectrometer was less than 6 min, and all data were treated consistently. The centroid and width of the deuterium containing isotopic distribution for each peptide was analyzed using HX-Express software (36).
For H/DX-ETD, the centroid of the deuterium-containing isotopic distribution for each product ion was obtained through MagTran 1.03 (Amgen, Thousand Oaks, CA). A minimum of 10% relative ion abundance threshold was applied in the data processing. The deuterium uptake for nth amino acid was determined as the difference between the centroid of cn-1 and cn-2 ions or the difference between the centroid of zn+1 and zn ions. All experiments were performed in at least triplicate.
RESULTS AND DISCUSSION
a. Regions involved in ApoE self-association revealed by H/DX kinetics study
Apolipoprotein E isoforms exist predominantly as tetramers at µM concentrations and dissociate into dimers and monomers at nM analytical concentrations (17). The C-terminal domain seems to play the major role in this self-association (10). At analytical concentrations higher than µM, the protein tends to aggregate (17, 37), seriously hindering the elucidation of the full-length structure. Wang and coworkers (18) showed that a monomeric form of ApoE3 could be generated by substituting in the C-terminal region four to five bulky hydrophobic residues with either smaller hydrophobic or polar/charged residues. Although these substitutions prevent oligomerization, an understanding of the key residues involved in the self-association process and the differences between the ApoE isoforms remains to be achieved.
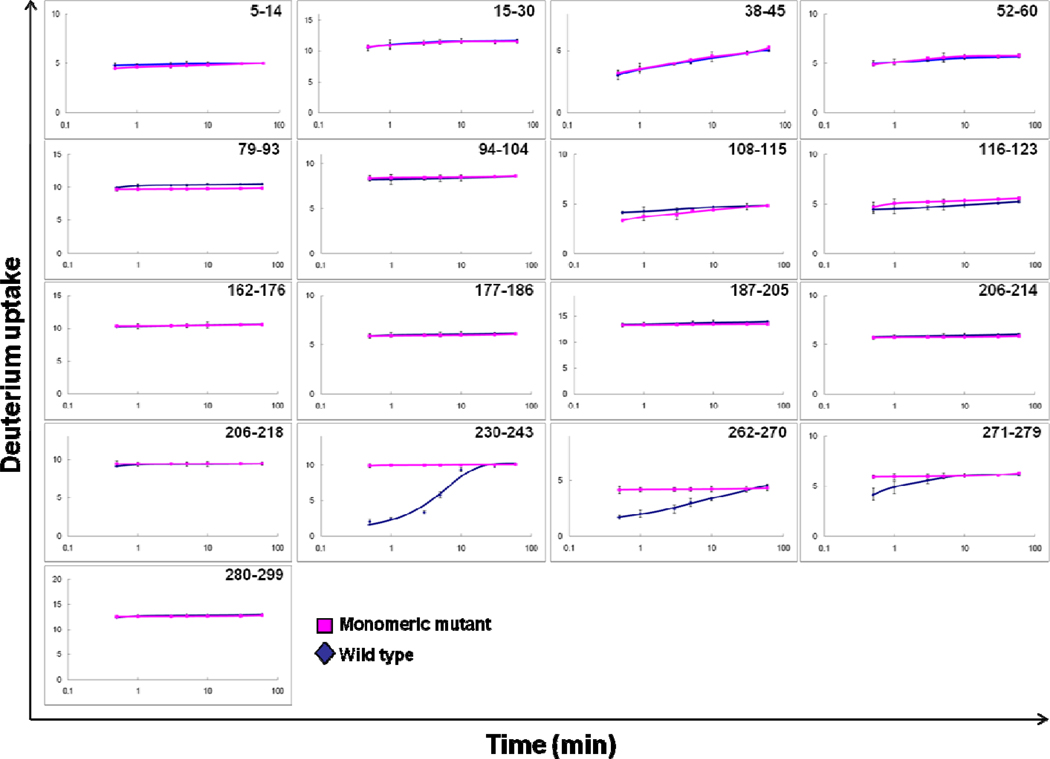
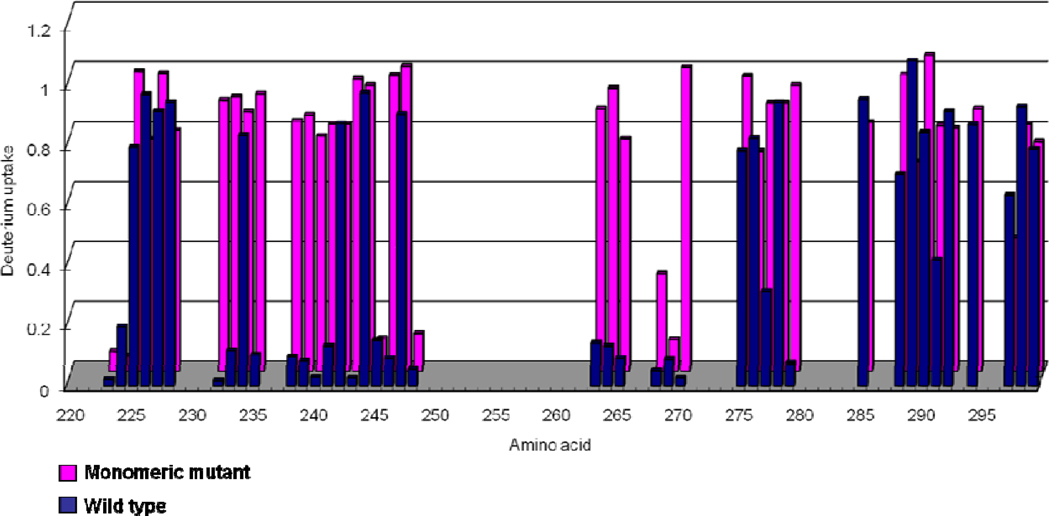
In this study, we performed peptide-level H/DX kinetic measurements at seven different exchange times (0.5, 1, 3, 5, 10, 30, 60 min) on all three isoforms of ApoE including the wild-type proteins and their monomeric mutants. Starting with protein solutions for which the analytical concentration was 4 µM (where ApoE exists predominantly as tetramer), the proteins were equilibrated at 25 °C for 2 h followed by introduction of D2O to initiate exchange and MS analysis to afford the kinetics of exchange at the peptide level for ApoE4 (Figure 1). Although the extents of H/DX for all regions of the wild-type proteins and the monomeric mutants located in the N-terminal domain are similar, as revealed by peptic peptides, significant differences between wild type and monomeric mutant occur in the C-terminal region for peptides 230–243 and 262–270, and small differences for peptide 271–279. These differences are most obvious at early exchange times (0.5–1 min). We suggest that this behavior is a consequence of the time-dependent dissociation of tetramer to monomer as discussed below. Interestingly, two substitution sites used to produce the monomeric mutant are located in two of these regions (W264R and L279Q) (18). Information for the peptide containing residue F257A (Peptide 257–261) was missing because the peptide signal intensity was low, and the signal dropped even more caused by further dispersion of signals by uptake of deuterium. Peptide 230–243 showed a dramatic decrease in deuterium uptake for this region of the wild type compared to the monomer. Overall, the regions that are affected by self-association are near the C-terminus, and this is consistent with previous findings that the C-terminal domain contains the residues important for ApoE oligomerization.
Figure 1.
Peptide-level H/DX kinetics of ApoE4. A comparison between wild type (blue) and monomeric mutant (pink) shows significant differences in H/DX only for peptides 230–243 and 262–270 and small difference for peptide 271–279.
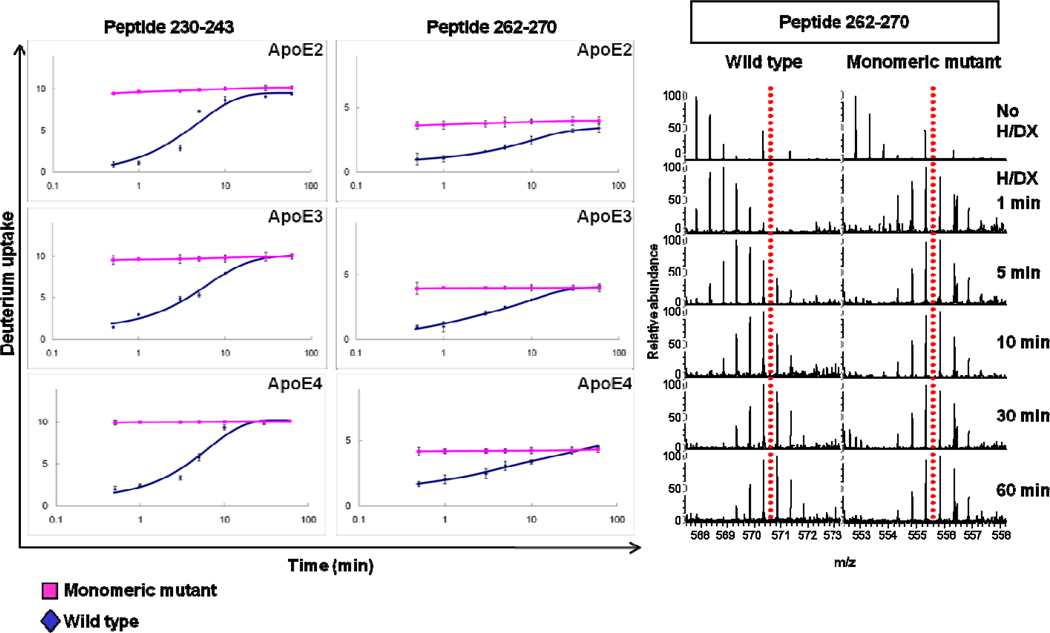
The deuterium uptake of the two regions represented by peptides 230–243 and 262–270 of all the wild-type isoforms are time-dependent as shown in Figure 2. This slow HD/X of the C-terminal peptide fragments (residues 230–243 and 262–270) of the wild-type ApoE isoforms may be consistent with the slow dissociation of the ApoE oligomers. Garai and Frieden (17) showed that association-dissociation of ApoE isoforms fit a monomer-dimer-tetramer model and that the rates for both association and dissociation at neutral pH are slow. The HD/X experiments were carried out by diluting ApoE from 4 µM to 400 nM analytical concentration in the D2O buffer; the result is dissociation of the tetramers to monomers. Although wild-type ApoE exists as a mixture of monomers, dimers and tetramers at 400 nM (17), deuterium uptake by the wild-type ApoE at this concentration reaches completion owing to the reversibility of the monomer-dimer-tetramer process. The dissociation rate constants for the isoforms, as measured by Garai and Frieden, are approximately 10−4 s−1 for dimer-monomer and 10−3 s−1 for tetramer-dimer conversion, the equilibrium constants for both processes are in the range of 107 M−1 (17). We fit the H/DX kinetic data with a fixed-rate-constant binning model by using MathCAD (Math-Soft. Inc. Cambridge, MA), as previously described (38, 39), whereby iterations were continued until the root mean square (RMS) was minimized. We chose three fixed rate-constant bins (k = 10, 1, 0.1 min−1); the latter bin on the basis of the time needed to effect nearly complete exchange (Table 1). According to the model, nearly all the amide hydrogens in these two regions of the wild-type ApoE exchange in 0.1 min−1 (1.6 × 10−3 s−1 rate-constant bin), whereas all amide hydrogens of the monomeric mutants undergo fast exchange rate (k = 10 min−1), indicating that the mutants are flexible in these two regions.
Figure 2.
Comparison of H/DX kinetics of peptide 230–243 and 262–270 among all three isoforms (ApoE2, ApoE3, and ApoE4). An example of the mass spectra of peptide 262–270 of ApoE4 as a function of H/DX time is shown on the right (the red, dotted line is a reference to guide the eye).
Table 1.
Numbers of amide hydrogen undergoing H/D exchange for wild-type (WT) and monomeric-mutant ApoE’s isoforms. The kinetic modeling used three fixed exchange rate constants and “binned” the number of amides with respect to the rate constants.
| Kinetic Fit | # of H's per fixed-rate bin | |||
|---|---|---|---|---|
| Peptide 230–243 | k (min−1) | 10 | 1 | 0.1 |
| WT | ApoE2 | 0 ± 0 | 2 ± 1 | 8 ± 1 |
| ApoE3 | 0 ± 0 | 3 ± 1 | 7 ± 1 | |
| ApoE4 | 1 ± 0 | 1 ± 0 | 8 ± 1 | |
| MM | ApoE2 | 9 ± 1 | 0 ± 0 | 0 ± 0 |
| ApoE3 | 10 ± 1 | 0 ± 0 | 0 ± 0 | |
| ApoE4 | 10 ± 1 | 0 ± 0 | 0 ± 0 | |
| Peptide 262–270 | k (min−1) | 10 | 1 | 0.1 |
| WT | ApoE2 | 1 ± 0 | 0 ± 0 | 2 ± 0 |
| ApoE3 | 0 ± 0 | 1 ± 0 | 3 ± 1 | |
| ApoE4 | 1 ± 0 | 1 ± 0 | 2 ± 1 | |
| MM | ApoE2 | 4 ± 1 | 0 ± 0 | 0 ± 0 |
| ApoE3 | 4 ± 1 | 0 ± 0 | 0 ± 0 | |
| ApoE4 | 4 ± 1 | 0 ± 0 | 0 ± 0 | |
The H/DX kinetics do not reveal the two-step behavior observed by Garai et al. (17) but instead are an integrated view of the various states of the protein. Given that the peptide-level H/DX kinetics for the wild-type ApoE’s C-terminal regions (~10−3 s−1) agrees well with the average dissociation constant determined for tetramer-dimer-monomer process (17), we suggest that the oligomeric proteins must first dissociate to monomers and then undergo H/DX in the two C-terminal regions.
ApoE2 and ApoE3 show similar HD/X behavior (Supplemental Figures 4 and 5). A comparison of the H/DX kinetics for monomeric mutant of all three isoforms (Supplemental Figure 6) reveals that some regions have minor differences among the three isoforms (peptide 15–30, 116–123, and 271–279) where ApoE4 uptakes slightly more deuterium than ApoE2 and ApoE3. Overall, the three isoforms must share similar structures and properties with respect to oligomerization. These properties as revealed by H/DX are also consistent with the differences found in our study using fast photochemical oxidation (FPOP) (40)
b. H/DX-ETD reveals residues involved in self-association of ApoE4
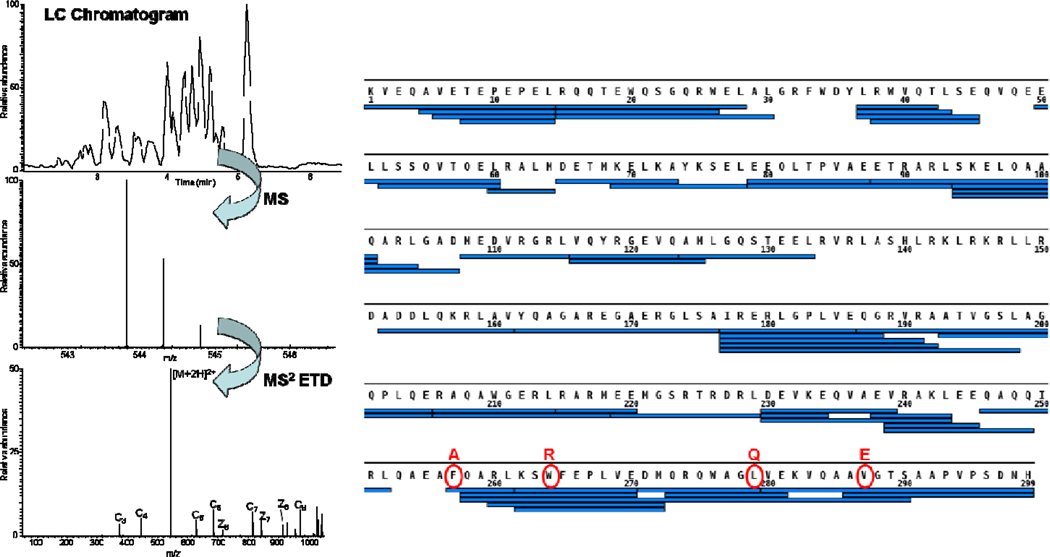
Peptide-level H/DX kinetics, as shown in this study, can reveal not only those regions that are involved in protein:protein interactions but also the dynamics of the interactions. To improve the resolution of the H/DX experiments, we chose a bottom-up strategy whereby, following H/DX of the protein and proteolysis, we subjected the peptide to ETD fragmentation (Figure 3). This approach is more appropriate than a top-down one because our ApoE samples contain several post-translational oxidations (Supplemental Figure 7) that disperse the signal and increase the difficulty of the top-down H/DX experiment. Furthermore, extensive sequence coverage for any 34 kDa protein is difficult to achieve in a top-down experiment.
Figure 3.
A diagram of the H/DX-ETD procedure. A peptide of interest produced from on-line pepsin digestion is subjected to ETD fragmentation. Peptide coverage (~89%) of ApoE4 by applying this strategy is shown in right (four substitution sites to achieve the MM are marked in red).
Bottom-up H/DX-ETD experiments were performed under the same conditions as for peptide-level H/DX experiments in which a 4 µM (analytical concentration) protein solution was equilibrated at 25 °C for 2 h followed by diluting the protein solution 1:10 into D2O buffer for 1 min, a time for which there is a large difference in H/DX between wild types and the monomeric mutants of ApoE. After quenching the exchange, the protein solution was submitted to on-line pepsin digestion, and the peptides were analyzed by MS and ETD whereby the selection of the precursor ion was directed by a precursor-mass list containing the peptides of interest. This list utilized the centroid m/z of the deuterium-containing isotopic distribution of the peptides of interest. ETD usually generates a product-ion spectrum of good signal-to-noise for those peptides that are doubly or more highly charged. Peptides that are +4 or higher, however, will show increased H/D scrambling due to the more energetic collisions they undergo upon acceleration in the ESI declustering field than do lower charged ions (30). Therefore, we selected doubly and triply charged peptides that had sufficient ion signal for ETD fragmentation.
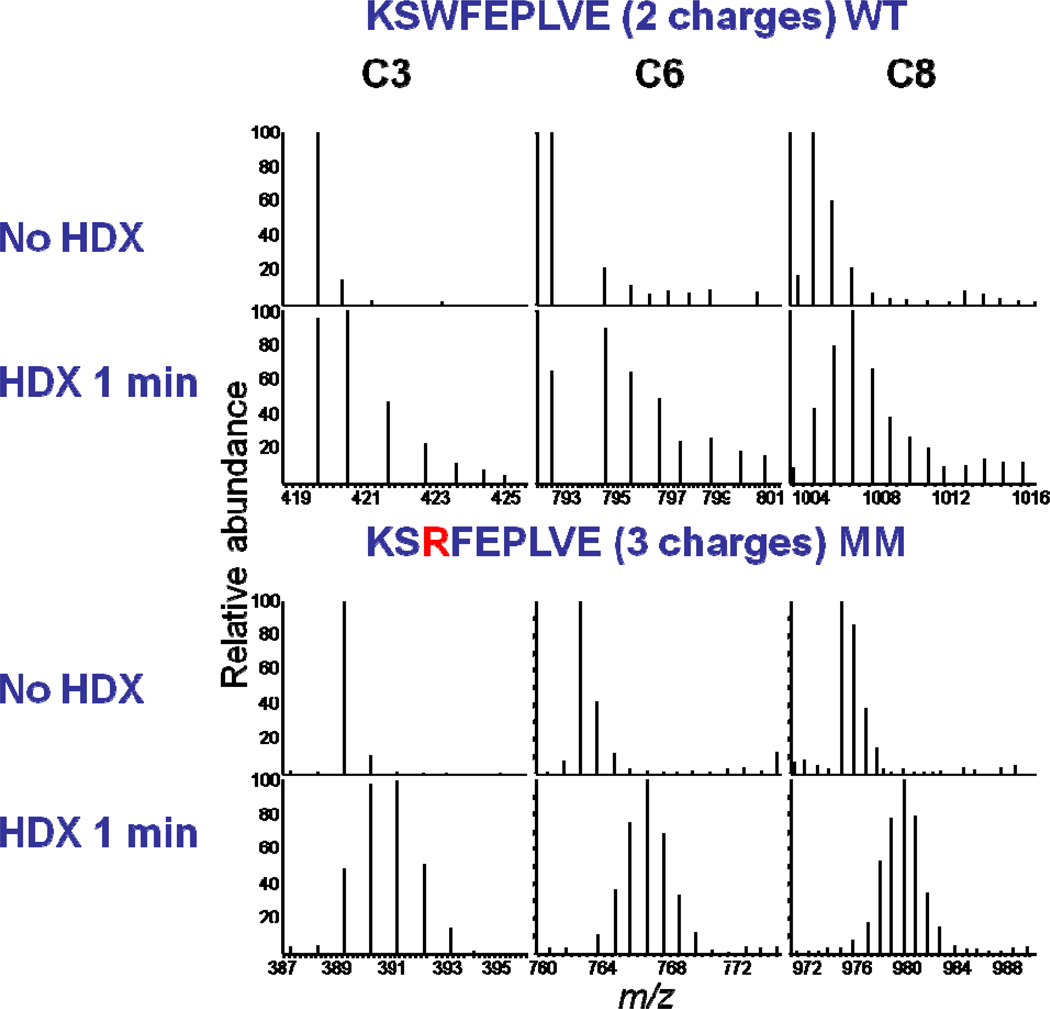
Although the c ions of peptides 206–214 show no significant differences in H/DX for wild type ApoE4 and monomeric mutant forms after 1 min (Supplemental Figure 8), the region 262–270 (Figure 4) undergoes significantly different H/DX. We measured the extent of deuteration at each amide site, using the difference of the isotopic-distribution centroid between two adjacent product ions (c or z ions). For example, the extent of deuterium on the nth amino acid is the difference between the centroid of cn-1 and cn-2 ions or between the centroid of zn+1 and zn ions. The outcome is a deuterium-uptake diagram for each amide of each residue of the ApoE4 wild type and its monomeric mutant (Supplemental Figure 9). We analyzed 132 out of 274 amides with exchangeable backbone hydrogens (excluding proline and the N-terminal amine hydrogens for each peptide), giving a coverage of ~ 50%. The coverage depends highly on the quality of the product-ion spectra; that quality was reduced when working under ESI conditions that minimize scrambling. No correction for back exchange was made because all experiments were treated consistently. Although three of four substitution sites of the ApoE4 monomeric mutant are located on C-terminal peptic peptides, which we selected for ETD fragmentation, these sites are located separately on different peptides; thus, the extents of back exchange should be similar for the same set of peptides from the two protein isoforms and will not affect conclusions about the relative differences of H/DX.
Figure 4.
ETD product-ion spectra of peptide 262–270 of ApoE4 after H/DX. The centroid of the distribution of c ions from monomeric mutant shows there are significant differences in the extent of H/DX between WT and MM for this region.
There are clearly no significant differences for all the amides examined in the N-terminal region of the wild type and the monomeric mutant form of ApoE4 (the complete extents of deuteration are in Supplemental Table 1). Not surprisingly, several amides in the C-terminal region underwent relatively low deuterium uptake (< 0.3 D) for the wild type ApoE4 whereas they exchanged more deuterium (> 0.7 D) for the monomeric mutant (Figure 5). The residues associated with these amides are: Val232, Lys233, Gln235, Glu238, Val239, Arg240, Ala241, Leu243, Gln246, Ser263, Trp264 (Arg in the monomeric mutant form), Phe265, Leu268, Glu270, and Leu279 (Gln in the monomeric mutant form). Interestingly, two of the substitution sites (Trp264Arg and Leu279Gln) are on the list, indicating the important roles played by the two residues in the self-association process. Several residues (e.g., Glu234 and Lys242) showed no significant differences in deuterium uptake between the wild-type and monomeric-mutant forms, indicating that these two residues are minimally affected by the self association of ApoE4. Weisgraber and coworkers (10), who studied the ApoE oligomerization by truncating various sites in the C-terminal region, found that residues within 267–299 are critical for tetramerization. Our study, using the full-length protein, show that three residues within this region are important in the ApoE self asoociation. Moreover, 12 additional residues, which are close to this region, also play a role in self association. A potential spinoff of this work is the use of H/DX results as a basis for designing future mutation studies in ApoE proteins.
Figure 5.
The extent of deuterium uptake of various amino-acid residues in the C-terminal domain of ApoE4 (Residue 220–299). Several residues of wild type (blue) have lower level (<0.3D) of deuterium uptake compared to the monomeric mutant (pink). Note: The gaps correspond to missing information.
c. The structure of ApoE4
Before by mapping the H/DX-ETD results onto a structure of ApoE that contains the X-ray crystal structure of the N-terminal domain (PDB: 1GS9) (41), we wished to establish that a low extent of deuteration for a specific amide indicated low solvent accessibility and not extensive back exchange. Thus, we prepared a nearly 100% deuterium-exchanged ApoE4 monomeric mutant by diluting the protein solution into D2O buffer at 25 °C and exchanging for 24 h. After submitting the protein to on-line pepsin digestion and ETD-MS analysis (42), we found that the average percent back exchange was (13 ± 4)%. (Each deuterium uptake for a single amide was normalized to its corresponding D100% value, and the normalization was treated separately for each trial of experiments—see Supplemental table 1).
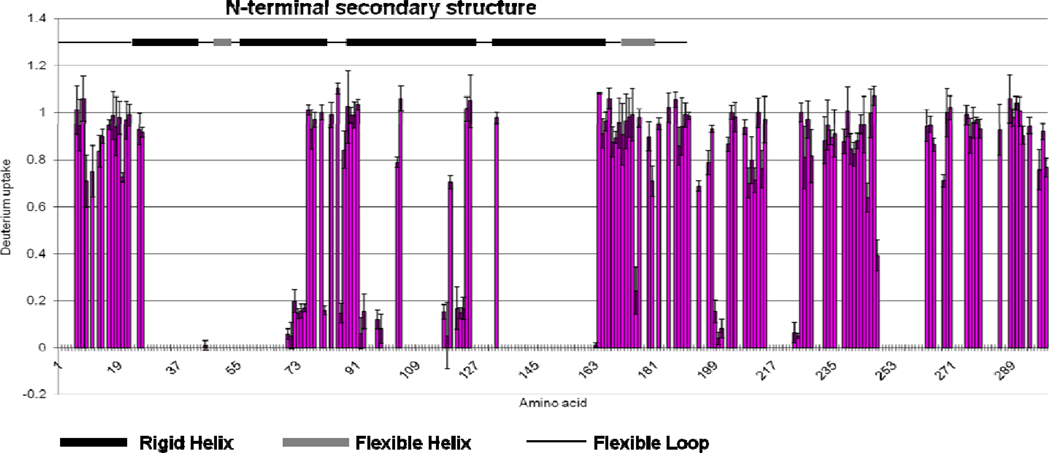
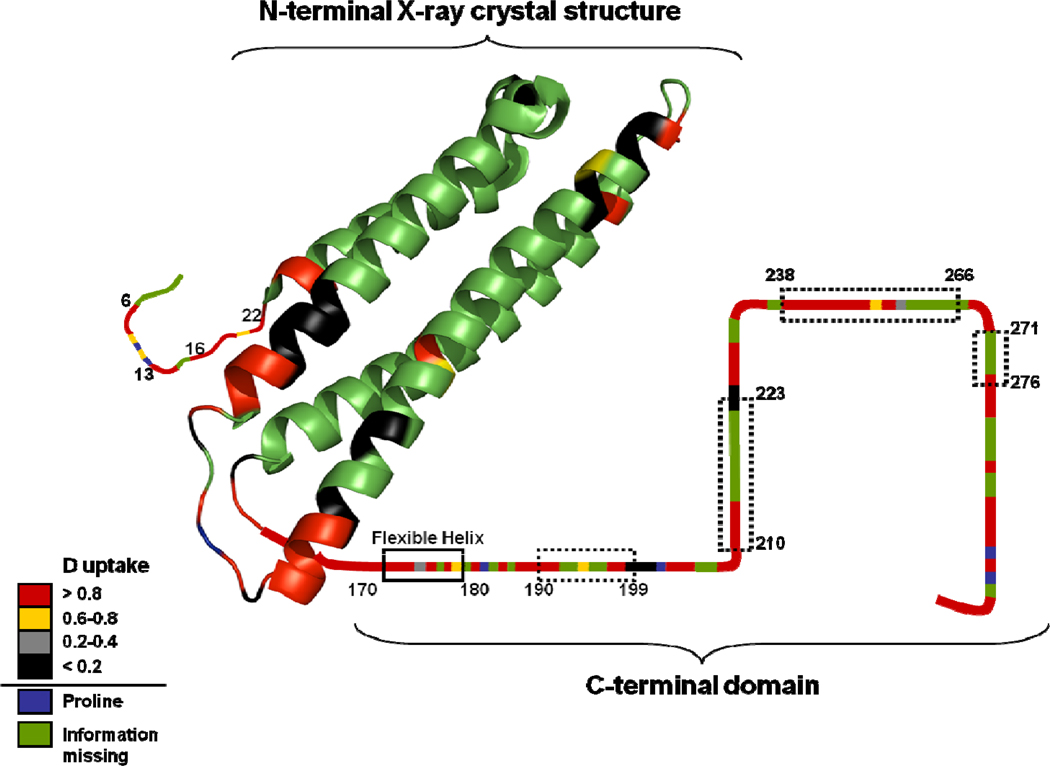
The deuterium uptake at a single amide of the ApoE4 monomeric mutant (Figure 6), show that the ApoE4 structure is primarily compact in the N-terminal region and more flexible in the C-terminal region, as previously assumed (43). Previous studies indicated that the C-terminal domain should contain amphipathic helices and several flexible loop regions (11, 12). Four possible helices, represented by peptide regions 190–199, 210–223, 238–266, and 271–276, exist in the C-terminal domain, as determined by Wang and coworkers (5). Their recent NMR structure of ApoE3 monomeric mutant shows that these helices are in the middle of long, unstructured regions (13). Our amide-level H/DX results, when mapped onto a schematic secondary structure of the C-terminal domain of ApoE (Figure 7), show that peptide regions 190–199 and 271–276 undergo extensive H/DX, consistent with being flexible helices. Although deuterium uptake information is missing for peptide regions 215–222 and 249–262, those regions adjoin those helices in C-terminal domain that also undergo extensive H/DX and should be flexible.
Figure 6.
The extent of deuterium uptake of each residue of ApoE4 monomeric mutant after correcting for back exchange (pink). Most of the residues in the C-terminus have high extent of deuterium uptake, indicating a flexible structure in this region. The extent of deuterium uptake in N-terminal region are consistent with the known secondary structure (top). (flexible loop, black line; flexible helix, gray box; rigid helix, black box). Note: The gaps correspond to missing information.
Figure 7.
Structure of ApoE4 on which residue-level H/DX of ApoE4 monomeric mutant is mapped. Left: X-ray crystal structure of N-terminal region of ApoE4 (PDB: 1GS9). Right: Proposed secondary structure of C-terminal region of ApoE (5). Possible helices are shown in dotted boxes. Different colors represent different levels of deuterium uptake for each residue (lower left).
In another peptide-level HD/X experiment, we treated the three ApoE’s isoforms with 0.5 M guanidine hydrochloride (GdnCl) (Supplemental Figure 10), a concentration at which the protein could lose some tertiary/quaternary structure while maintaining its secondary structure (43). The results show that this low concentration of denaturant has more impact on the C-terminal than the N-terminal region of ApoE. This is additional evidence that ApoE is composed of a flexible C-terminal region, making it susceptible to denaturation. Morrow and coworkers (44), who studied the denaturation of ApoE at various concentrations of GdnCl, showed that the regions nearby the C-terminus and in the presence of low concentration of GdnCl, unfold prior to those close to the N-terminal domain. Our results are consistent with their findings.
The structure of the N-teminal domain of ApoE was determined by X-ray crystallography and NMR to be a four helix-bundle (5, 9, 41). Our results, when compared to the X-ray crystal structure of N-terminal domain (Figure 7), show that amides in the regions of residues 6–25, 76–91, 124–125, and 165–191 undergo extensive deuterium uptake, consistent with their location at or near the flexible-loop regions and as part of the flexible-helix region (170–180). The amides in regions 70–75 and 92–103, which are located in a rigid helix, undergo low extents of deuteration, in accord with the strong H-bonding in that region of the protein.
CONCLUSIONS
The observation that ApoE has a high tendency to aggregate has hindered determination of the full-length structures of the three wild-type isoforms. We are approaching structure analysis of the three isoforms and their monomeric mutants by chemical footprinting (40) including H/DX coupled with MS. Unlike other covalent protein footprinting methods, H/DX is general, marking all the amino acids except proline and covering nearly the entire protein backbone provided the proteolysis and LC/MS efficiencies are high.
Our H/DX results not only confirm the role of the C-terminal domain in the oligomerization process but also reveal the dissociation rate of oligomers to give monomers. We found that the three wild-type isoforms largely share oligomerization properties. A bottom-up H/DX-ETD experiment also located deuterium on lipid-free ApoE4 including its wild type and monomeric mutant form and afforded information on approximately 50% of the amides. We obtained clear, single-amide-level H/DX results for ApoE4, identified the residues involved in self-association, and showed they are located in the C-terminal domain (i.e., residues 232–272).
The region of apoE that shows differential H/DX between the wild-type protein and the monomeric mutant includes the residues 244–272, which harbor the lipid-binding site (45) and are involved in Aβ binding, with implications for understanding Alzheirmer’s Disease (45). In addition, residues 241–272 mediate the toxic effects of ApoE4 in neuroblastoma cells (46). Thus, the residues 232–272 in the C-terminal domain seem to be involved in the multiple processes that are crucial for biological function and that mediate toxicity. Additional studies along these lines should increase our understanding of ApoE interactions with itself and with various ligands, an understanding that will be difficult to obtain by NMR and X-ray crystallography. Furthermore, H/DX and other footprinting strategies should not only provide information for the design of other ApoE monomeric mutants that contain a minimum number of mutation sites but also be a means for evaluating their equivalency.
Supplementary Material
ACKNOWLEDGMENT
Funding: Funding was provided by the National Center for Research Resources (NCRR) of the National Institutes of Health (grant no. 2P41RR000954) to MLG.
We thank Dr. Jun Zhang for assistance with the H/DX plate form and Berevan Baban for technical assistance.
Abbreviations
- ApoE
Apolipoprotein E
- ESI
electrospray ionization
- ETD
electron-transfer dissociation
- H/DX
hydrogen/deuterium exchange
- HPLC
high performance liquid chromatography
- Q-TOF
quadrupole time-of-flight
Footnotes
SUPPORTING INFORMATION AVAILABLE
Supplemental figures are included in the Supporting Information section. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Yokoyama S, Kawai Y, Tajima S, Yamamoto A. Behavior of human apolipoprotein E in aqueous solutions and at interfaces. J. Biol. Chem. 1985;260:16375–16382. [PubMed] [Google Scholar]
- 2.Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends in Biochemical Sciences. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Weisgraber KH, Anfinsen CB, J. T. E. F. M. R., David SE. Advances in Protein Chemistry. Academic Press; 1994. Apolipoprotein E: Structure-Function Relationships; pp. 249–302. [DOI] [PubMed] [Google Scholar]
- 4.Herz J, Beffert U. Apolipoprotein E receptors: linking brain development and alzheimer's disease. Nat. Rev. Neurosci. 2000;1:51–58. doi: 10.1038/35036221. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Chen J, Wang J. A complete backbone spectral assignment of lipid-free human apolipoprotein E (apoE) Biomolecular NMR Assignments. 2008;2:207–210. doi: 10.1007/s12104-008-9122-8. [DOI] [PubMed] [Google Scholar]
- 6.Raussens V, Drury J, Forte TM, Choy N, Goormaghtigh E, Ruysschaert J-M, Narayanaswami V. Orientation and mode of lipid-binding interaction of human apolipoprotein E C-terminal domain. Biochem. J. 2005;387:747–754. doi: 10.1042/BJ20041536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wetterau JR, Aggerbeck LP, Rall SC, Weisgraber KH. Human apolipoprotein E3 in aqueous solution. I. Evidence for two structural domains. J. Biol. Chem. 1988;263:6240–6248. [PubMed] [Google Scholar]
- 8.Vasudevan S, Sojitrawala R, Zhao W, Cui C, Xu C, Fan D, Newhouse Y, Balestra R, Jerome WG, Weisgraber K, Li Q, Wang J. A Monomeric, Biologically Active, Full-Length Human Apolipoprotein E. Biochemistry. 2007;46:10722–10732. doi: 10.1021/bi700672v. [DOI] [PubMed] [Google Scholar]
- 9.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 10.Westerlund JA, Weisgraber KH. Discrete carboxyl-terminal segments of apolipoprotein E mediate lipoprotein association and protein oligomerization. J. Biol. Chem. 1993;268:15745–15750. [PubMed] [Google Scholar]
- 11.Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, Anantharamaiah GM. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 1992;33:141–166. [PubMed] [Google Scholar]
- 12.Choy N, Raussens V, Narayanaswami V. Inter-molecular Coiled-coil Formation in Human Apolipoprotein E C-terminal Domain. J. Mol. Biol. 2003;334:527–539. doi: 10.1016/j.jmb.2003.09.059. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Li Q, Wang J. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc. Nat. Acad. Sci. 2011 doi: 10.1073/pnas.1106420108. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou C-Y, Lin Y-L, Huang Y-C, Sheu S-Y, Lin T-H, Tsay H-J, Chang G-G, Shiao M-S. Structural Variation in Human Apolipoprotein E3 and E4: Secondary Structure, Tertiary Structure, and Size Distribution. Biophys. J. 2005;88:455–466. doi: 10.1529/biophysj.104.046813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perugini MA, Schuck P, Howlett GJ. Self-association of Human Apolipoprotein E3 and E4 in the Presence and Absence of Phospholipid. J. Biol. Chem. 2000;275:36758–36765. doi: 10.1074/jbc.M005565200. [DOI] [PubMed] [Google Scholar]
- 16.Aggerbeck LP, Wetterau JR, Weisgraber KH, Wu CS, Lindgren FT. Human apolipoprotein E3 in aqueous solution. II. Properties of the amino- and carboxyl-terminal domains. J. Biol. Chem. 1988;263:6249–6258. [PubMed] [Google Scholar]
- 17.Garai K, Frieden C. The Association Dissociation Behavior of the ApoE Proteins: Kinetic and Equilibrium Studies. Biochemistry. 2010;49:9533–9541. doi: 10.1021/bi101407m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan D, Li Q, Korando L, Jerome WG, Wang J. A Monomeric Human Apolipoprotein E Carboxyl-Terminal Domain. Biochemistry. 2004;43:5055–5064. doi: 10.1021/bi035958w. [DOI] [PubMed] [Google Scholar]
- 19.Pan J, Han J, Borchers CH, Konermann L. Conformer-Specific Hydrogen Exchange Analysis of Ab(1-42) Oligomers by Top-Down Electron Capture Dissociation Mass Spectrometry. Anal. Chem. 2011;83:5386–5393. doi: 10.1021/ac200906v. [DOI] [PubMed] [Google Scholar]
- 20.Chalmers MJ, Busby SA, Pascal BD, West GM, Griffin PR. Differential hydrogen/deuterium exchange mass spectrometry analysis of protein-ligand interactions. Expert Rev. Proteomics. 2011;8:43–59. doi: 10.1586/epr.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engen JR. Analysis of Protein Conformation and Dynamics by Hydrogen/Deuterium Exchange MS. Anal. Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Pan H, Smith DL. Hydrogen Exchange-Mass Spectrometry. Mol. Cell. Proteomics. 2002;1:132–138. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 23.Rand KD, Zehl M, Jensen ON, Jorgensen TJD. Protein Hydrogen Exchange Measured at Single-Residue Resolution by Electron Transfer Dissociation Mass Spectrometry. Anal. Chem. 2009;81:5577–5584. doi: 10.1021/ac9008447. [DOI] [PubMed] [Google Scholar]
- 24.Jorgensen TJD, Gardsvoll H, Ploug M, Roepstorff P. Intramolecular Migration of Amide Hydrogens in Protonated Peptides upon Collisional Activation. J. Am. Chem. Soc. 2005;127:2785–2793. doi: 10.1021/ja043789c. [DOI] [PubMed] [Google Scholar]
- 25.Demmers JAA, Rijkers DTS, Haverkamp J, Killian JA, Heck AJR. Factors Affecting Gas-Phase Deuterium Scrambling in Peptide Ions and Their Implications for Protein Structure Determination. J. Am. Chem. Soc. 2002;124:11191–11198. doi: 10.1021/ja0125927. [DOI] [PubMed] [Google Scholar]
- 26.Hoerner JK, Xiao H, Dobo A, Kaltashov IA. Is There Hydrogen Scrambling in the Gas Phase? Energetic and Structural Determinants of Proton Mobility within Protein Ions. J. Am. Chem. Soc. 2004;126:7709–7717. doi: 10.1021/ja049513m. [DOI] [PubMed] [Google Scholar]
- 27.Ferguson PL, Pan J, Wilson DJ, Dempsey B, Lajoie G, Shilton B, Konermann L. Hydrogen/Deuterium Scrambling during Quadrupole Time-of-Flight MS/MS Analysis of a Zinc-Binding Protein Domain. Anal. Chem. 2006;79:153–160. doi: 10.1021/ac061261f. [DOI] [PubMed] [Google Scholar]
- 28.Jorgensen TJD, Bache N, Roepstorff P, Gardsvoll H, Ploug M. Collisional Activation by MALDI Tandem Time-of-flight Mass Spectrometry Induces Intramolecular Migration of Amide Hydrogens in Protonated Peptides. Mol. Cell. Proteomics. 2005;4:1910–1919. doi: 10.1074/mcp.M500163-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Rand KD, Jorgensen TJD. Development of a Peptide Probe for the Occurrence of Hydrogen (1H/2H) Scrambling upon Gas-Phase Fragmentation. Anal. Chem. 2007;79:8686–8693. doi: 10.1021/ac0710782. [DOI] [PubMed] [Google Scholar]
- 30.Zehl M, Rand KD, Jensen ON, Jorgensen TJD. Electron Transfer Dissociation Facilitates the Measurement of Deuterium Incorporation into Selectively Labeled Peptides with Single Residue Resolution. J. Am. Chem. Soc. 2008;130:17453–17459. doi: 10.1021/ja805573h. [DOI] [PubMed] [Google Scholar]
- 31.Rand KD, Zehl M, Jensen ON, Jorgensen TJD. Loss of Ammonia during Electron-Transfer Dissociation of Deuterated Peptides as an Inherent Gauge of Gas-Phase Hydrogen Scrambling. Anal. Chem. 2010;82:9755–9762. doi: 10.1021/ac101889b. [DOI] [PubMed] [Google Scholar]
- 32.Hamuro Y, Tomasso JC, Coales SJ. A Simple Test To Detect Hydrogen/Deuterium Scrambling during Gas-Phase Peptide Fragmentation. Anal. Chem. 2008;80:6785–6790. doi: 10.1021/ac800645f. [DOI] [PubMed] [Google Scholar]
- 33.Garai K, Mustafi SM, Baban B, Frieden C. Structural differences between apolipoprotein E3 and E4 as measured by 19F NMR. Protein Science. 2010;19:66–74. doi: 10.1002/pro.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatters DM, Budamagunta MS, Voss JC, Weisgraber KH. Modulation of Apolipoprotein E Structure by Domain Interaction. J. Biol. Chem. 2005;280:34288–34295. doi: 10.1074/jbc.M506044200. [DOI] [PubMed] [Google Scholar]
- 35.Busby SA, Chalmers MJ, Griffin PR. Improving digestion efficiency under H/D exchange conditions with activated pepsinogen coupled columns. Int. J. Mass Spectrom. 2007;259:130–139. [Google Scholar]
- 36.Weis D, Engen J, Kass I. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J. Am. Soc. Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 37.Garai K, Baban B, Frieden C. Self-Association and Stability of the ApoE Isoforms at Low pH: Implications for ApoE-lipid Interactions. Biochemistry. 2011;50:6356–6364. doi: 10.1021/bi2006702. [DOI] [PubMed] [Google Scholar]
- 38.Zhu MM, Chitta R, Gross ML. PLIMSTEX: a novel mass spectrometric method for the quantification of protein-ligand interactions in solution. Int. J. Mass Spectrom. 2005;240:213–220. [Google Scholar]
- 39.Zhu MM, Rempel DL, Gross ML. Modeling data from titration, amide H/D exchange, and mass spectrometry to obtain protein-ligand binding constants. J. Am. Soc. Mass Spectrom. 2004;15:388–397. doi: 10.1016/j.jasms.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Gau B, Garai K, Frieden C, Gross ML. Mass Spectrometry-based Protein Footprinting Characterizes the Structures of Oligomeric Apolipoprotein E2, E3, and E4. Biochemistry. 2011 doi: 10.1021/bi200911c. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters-Libeu CA, Weisgraber KH, Segelke BW, Rupp B, Capila I, Hernáiz MJ, LeBrun LA, Linhardt RJ. Interaction of the N-Terminal Domain of Apolipoprotein E4 with Heparin. Biochemistry. 2001;40:2826–2834. doi: 10.1021/bi002417n. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: A new tool for protein structure elucidation. Protein Science. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel AB, Khumsupan P, Narayanaswami V. Pyrene Fluorescence Analysis Offers New Insights into the Conformation of the Lipoprotein-Binding Domain of Human Apolipoprotein E. Biochemistry. 2010;49:1766–1775. doi: 10.1021/bi901902e. [DOI] [PubMed] [Google Scholar]
- 44.Morrow JA, Segall ML, Lund-Katz S, Phillips MC, Knapp M, Rupp B, Weisgraber KH. Differences in Stability among the Human Apolipoprotein E Isoforms Determined by the Amino-Terminal Domain. Biochemistry. 2000;39:11657–11666. doi: 10.1021/bi000099m. [DOI] [PubMed] [Google Scholar]
- 45.Mahley RW, Rall SC. APOLIPOPROTEIN E: Far More Than a Lipid Transport Protein. Annual Review of Genomics and Human Genetics. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 46.Chang S, Ma Tr, Miranda RD, Balestra ME, Mahley RW, Huang Y. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc. Natl. Acad. Sci. 2005;102:18694–18699. doi: 10.1073/pnas.0508254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.