Abstract
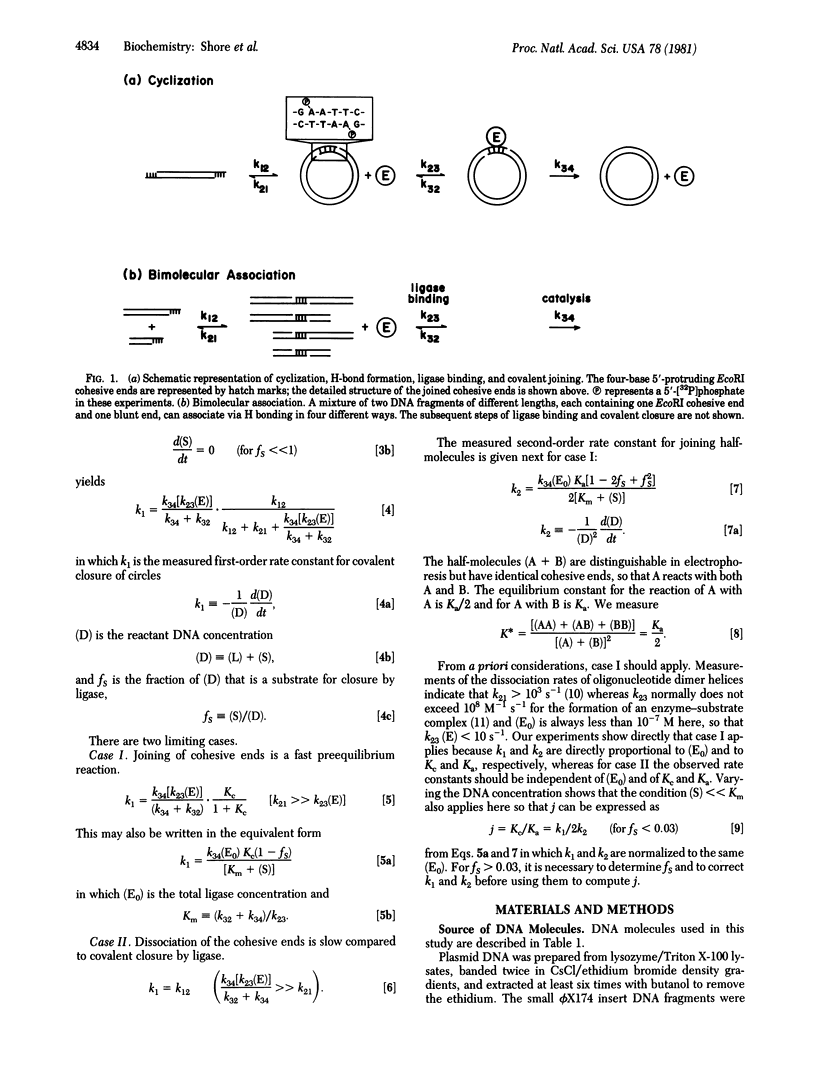
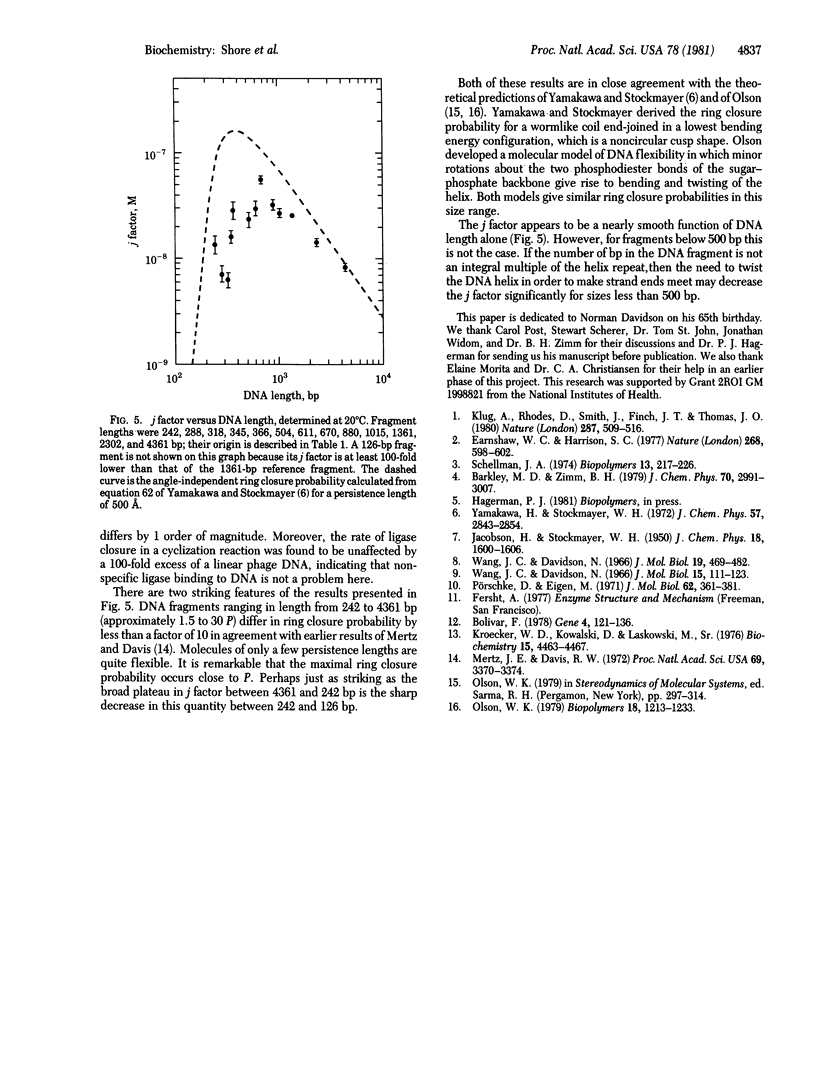
The ring closure probability, or j factor, has been measured for DNA restriction fragments of defined sequence bearing EcoRI cohesive ends and ranging in size from 126 to 4361 base pairs (bp). The j factor is defined as the ratio of the equilibrium constants for cyclization and for bimolecular association via the cohesive ends. The end-joining reactions are fast compared to covalent closure of the cohesive ends by T4 DNA ligase. The rate of ligase closure is shown to be proportional to the equilibrium fraction of DNA molecules with joined cohesive ends, both in cyclization and in bimolecular association reactions. The j factor changes by less than 10-fold between 242 and 4361 bp, whereas it decreases by more than 100-fold between 242 and 126 bp as the DNA reaches the size range of the persistence length (150 bp). As regards ring closure, short DNA fragments are surprisingly flexible. These data are in good agreement with predictions by others for the ring closure probability of a wormlike chain.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Harrison S. C. DNA arrangement in isometric phage heads. Nature. 1977 Aug 18;268(5621):598–602. doi: 10.1038/268598a0. [DOI] [PubMed] [Google Scholar]
- Klug A., Rhodes D., Smith J., Finch J. T., Thomas J. O. A low resolution structure for the histone core of the nucleosome. Nature. 1980 Oct 9;287(5782):509–516. doi: 10.1038/287509a0. [DOI] [PubMed] [Google Scholar]
- Kroeker W. D., Kowalski D., Laskowski M., Sr Mung bean nuclease I. Terminally directed hydrolysis of native DNA. Biochemistry. 1976 Oct 5;15(20):4463–4467. doi: 10.1021/bi00665a020. [DOI] [PubMed] [Google Scholar]
- Mertz J. E., Davis R. W. Cleavage of DNA by R 1 restriction endonuclease generates cohesive ends. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3370–3374. doi: 10.1073/pnas.69.11.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson W. K. The flexible DNA double helix. I. Average dimensions and distribution functions. Biopolymers. 1979 May;18(5):1213–1233. doi: 10.1002/bip.1979.360180514. [DOI] [PubMed] [Google Scholar]
- Pörschke D., Eigen M. Co-operative non-enzymic base recognition. 3. Kinetics of the helix-coil transition of the oligoribouridylic--oligoriboadenylic acid system and of oligoriboadenylic acid alone at acidic pH. J Mol Biol. 1971 Dec 14;62(2):361–381. doi: 10.1016/0022-2836(71)90433-5. [DOI] [PubMed] [Google Scholar]
- Schellman J. A. Flexibility of DNA. Biopolymers. 1974 Jan;13(1):217–226. doi: 10.1002/bip.1974.360130115. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Davidson N. On the probability of ring closure of lambda DNA. J Mol Biol. 1966 Aug;19(2):469–482. doi: 10.1016/s0022-2836(66)80017-7. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Davidson N. Thermodynamic and kinetic studies on the interconversion between the linear and circular forms of phage lambda DNA. J Mol Biol. 1966 Jan;15(1):111–123. doi: 10.1016/s0022-2836(66)80213-9. [DOI] [PubMed] [Google Scholar]




