Summary
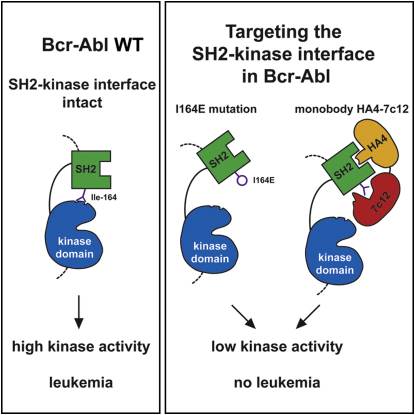
Chronic myelogenous leukemia (CML) is caused by the constitutively active tyrosine kinase Bcr-Abl and treated with the tyrosine kinase inhibitor (TKI) imatinib. However, emerging TKI resistance prevents complete cure. Therefore, alternative strategies targeting regulatory modules of Bcr-Abl in addition to the kinase active site are strongly desirable. Here, we show that an intramolecular interaction between the SH2 and kinase domains in Bcr-Abl is both necessary and sufficient for high catalytic activity of the enzyme. Disruption of this interface led to inhibition of downstream events critical for CML signaling and, importantly, completely abolished leukemia formation in mice. Furthermore, disruption of the SH2-kinase interface increased sensitivity of imatinib-resistant Bcr-Abl mutants to TKI inhibition. An engineered Abl SH2-binding fibronectin type III monobody inhibited Bcr-Abl kinase activity both in vitro and in primary CML cells, where it induced apoptosis. This work validates the SH2-kinase interface as an allosteric target for therapeutic intervention.
PaperFlick
Graphical Abstract
Highlights
► The SH2-kinase domain interface is necessary for high catalytic activity of Bcr-Abl ► This intramolecular interaction is critical for Bcr-Abl-dependent leukemogenesis ► Disrupting this interaction potentiates the effects of clinical kinase inhibitors ► Targeting of the SH2-kinase interface with a monobody inhibits Bcr-Abl allosterically
Intramolecular interaction between two domains of Bcr-Abl is essential for its oncogenic activity. Disrupting the interaction prevents leukemia, even in cases where Bcr-Abl has become resistant to existing kinase inhibitors.
Introduction
The deregulated, constitutively activated tyrosine kinase Bcr-Abl is expressed from the Philadelphia chromosome after the t(9;22) chromosomal translocation that leads to the fusion of the breakpoint cluster region (BCR) gene and the Abelson tyrosine kinase (ABL1) (Wong and Witte, 2004). The defining molecular event of chronic myelogenous leukemia (CML) in humans is the expression of Bcr-Abl, which is sufficient for the initiation and maintenance of CML-like disease in mouse models (Daley et al., 1990). Bcr-Abl activates a large number of signaling pathways that lead to uncontrolled proliferation, inhibition of apoptosis, and block of myeloid differentiation. Many of these pathways are thought to act in a redundant fashion, as only a few signaling components have thus been reported to be critical for Bcr-Abl-mediated oncogenic transformation. These included the transcription factors STAT5 and Myc, as well as the adaptor protein Gab2 (Nieborowska-Skorska et al., 1999; Hoelbl et al., 2010; Sattler et al., 2002).
Inhibition of Bcr-Abl tyrosine kinase activity by the highly specific Bcr-Abl inhibitor imatinib (Gleevec) leads to durable cytogenetic and molecular remissions in the majority of CML patients in the early chronic phase of the disease and is superior to previous therapies in advanced stage CML (Hochhaus et al., 2009; Deininger et al., 2005). The occurrence of imatinib resistance—mainly caused by point mutations in the Bcr-Abl kinase domain—leads to patient relapse, bears the risk of disease progression, and resulted in the development and rapid approval of the second-generation inhibitors nilotinib and dasatinib that target most imatinib-resistant Bcr-Abl variants (Shah and Sawyers, 2003; Quintás-Cardama et al., 2007). However, unsatisfactory responses in advanced disease stages, resistance, and problematic long-term tolerability of all three Bcr-Abl inhibitors remain major clinical problems (Jabbour et al., 2010). All approaches aimed at targeting the ATP-binding pocket of the Bcr-Abl kinase domain alone do not target the disease-initiating leukemic stem cells, and thus, patients are not cured from CML (Perrotti et al., 2010). Combination therapy of imatinib with drugs that target downstream signaling components of Bcr-Abl yielded promising results in preclinical studies (reviewed in Deininger et al., 2005). Still, these approaches have currently not been followed up further, as restoration of Bcr-Abl activity by resistance mutations appears to be dominant and override additive or synergistic inhibitory effects of the second drug (Deininger et al., 2005). For these reasons, approaches to target additional sites on Bcr-Abl itself in combination with the commonly targeted ATP-binding pocket may result in superior future therapeutic options.
Studies on the structure and dynamics of c-Abl/Bcr-Abl regulation have identified key regulatory mechanisms (reviewed in Hantschel and Superti-Furga, 2004). Binding of the N-terminal myristate moiety to a unique binding pocket in the c-Abl kinase domain was found to be critical for autoinhibition (Hantschel et al., 2003, Nagar et al., 2003). The myristate pocket was targeted by the non-ATP-competitive compound GNF-2/GNF-5 that led to inhibition of pan-TKI resistant Bcr-Abl variant T315I in a mouse model in combination with nilotinib (Zhang et al., 2010).
SH2 domains constitute one of the largest families of eukaryotic protein-protein interaction domains and bind phosphotyrosine moieties with a certain sequence specificity (Pawson et al., 2001). Aside from their well-described role in mediating intermolecular protein interactions, the SH2 domains in certain cytoplasmic tyrosine kinases, like c-Abl and Fes, were shown to activate the adjacent tyrosine kinase domain (Filippakopoulos et al., 2008). The ability of the Abl SH2 to stimulate kinase activity was dependent on the establishment of a tight interface between the SH2 domain and the N-terminal lobe of the kinase domain (Filippakopoulos et al., 2008; Nagar et al., 2006). Mutations in the SH2 domain that disrupt this SH2-kinase domain interface resulted in severe impairment of kinase activity. Thus, correct positioning of the SH2 and kinase domain modules appears to be critical for efficient activation of cytoplasmic tyrosine kinases (Filippakopoulos et al., 2008).
In the oncogenic fusion Bcr-Abl, the Bcr moiety contains important regulatory elements that contribute to constitutive activation and cellular transformation. The Grb2 docking site (Tyr177 in Bcr) and the coiled-coil oligomerization domain were shown to support Bcr-Abl leukemogenicity (McWhirter et al., 1993; Million and Van Etten, 2000). Therefore, whether or not SH2-mediated allosteric activation of the kinase domain is operable on the already highly activated Bcr-Abl fusion kinase or even affects its leukemogenicity were entirely unclear. Therefore, we set out to understand the functional role of the SH2-kinase domain interface in the context of Bcr-Abl.
The data presented here show that the SH2-kinase domain interface is critical for Bcr-Abl activity both in vitro and in cells. Disruption of this interface abolished the development of CML in a mouse bone marrow transplantation model and enhanced TKI sensitivity of nonmutated and TKI-resistant Bcr-Abl forms. Finally, an engineered high-affinity Abl SH2 domain-binding protein targeted to the SH2-kinase interface was found to strongly inhibit Bcr-Abl activity in vitro and to induce apoptosis in CML cell lines as well as in primary CML cells, demonstrating the potential druggability of this interface.
Results
Ile164 Is a Critical Residue for the Integrity of the Active Conformation of Bcr-Abl
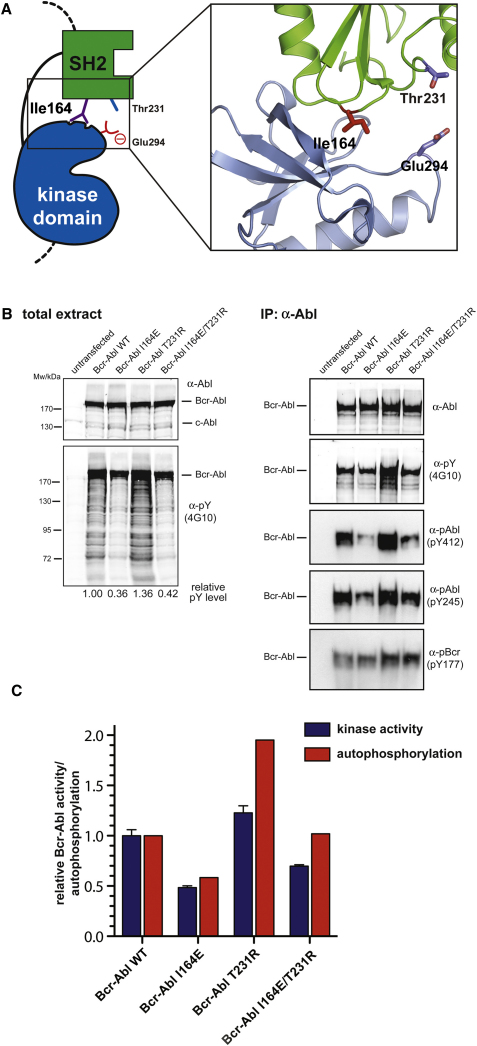
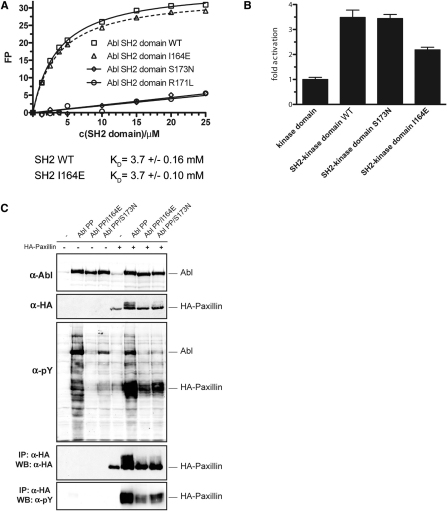
The SH2 domain is thought to act as an intramolecular allosteric activator of the tyrosine kinase domain in the protooncogenic kinases Fes and c-Abl (Filippakopoulos et al., 2008, 2009). Recently, a mutation of threonine 231 (Thr231) in the SH2 domain of the Bcr-Abl protein was identified in imatinib-treated patients and implicated in causing imatinib resistance (Sherbenou et al., 2010). The T231R mutation may stabilize the SH2-kinase domain interface by the formation of an additional ionic interaction with Glu294 in the N lobe of the kinase domain (Figure 1A and Sherbenou et al., 2010). In contrast, mutation of isoleucine 164 (Ile164), located in the αA-βB-loop of the Abl SH2 domain, to glutamate (I164E) may nullify the allosteric activation of the Abl kinase by the SH2 domain by disrupting the SH2-kinase domain interface (Filippakopoulos et al., 2008; Nagar et al., 2006 and Figure 1A). These two observations may suggest a possible role of the Bcr-Abl SH2-kinase domain interface for the activity of the oncoprotein and even in CML pathobiology.
Figure 1.
Structure-Function Analysis of the SH2-Kinase Domain Interface in Bcr-Abl
(A) Cartoon diagram of the active SH2-kinase domain unit of Abl. Critical residues involved in the SH2-kinase domain interface that are discussed in the text are highlighted.
(B) HEK293 cells were transiently transfected with Bcr-Abl WT, I164E, T231R, or I164E/T231R. Total protein extracts (left panels) and Abl immunoprecipitates (right panels) were analyzed by immunoblotting using the indicated antibodies.
(C) Abl immunoprecipitates were assayed for catalytic activity using an optimal Abl substrate peptide, and total autophosphorylation was quantified. Kinase activity and autophosphorylation of Bcr-Abl WT were set to 1. The bar graph shows averages ± standard deviation (SD) from two independent experiments done in triplicate.
See also Figure S1.
We introduced the I164E and T231R single mutations and the I164E/T231R double mutation into Bcr-Abl. Upon expression in HEK293 cells, global tyrosine phosphorylation, Bcr-Abl autophosphorylation (on Tyr412 in the activation loop and on Tyr245 in the SH2-kinase linker), and in vitro tyrosine kinase activity were reduced in the I164E but increased in the T231R mutant. The I164E mutation was dominant over the T231R mutation, consistent with the proposed disrupting effect of the I164E mutation versus the stabilizing role of the T231R mutation (Figures 1B and 1C). In contrast, levels of phosphorylated Tyr177 in the Bcr part of Bcr-Abl, a site thought to be phosphorylated by Src kinases, were slightly higher in the T231R mutant. This might suggest higher Src activity in the presence of the T231R mutation (Figure 1B).
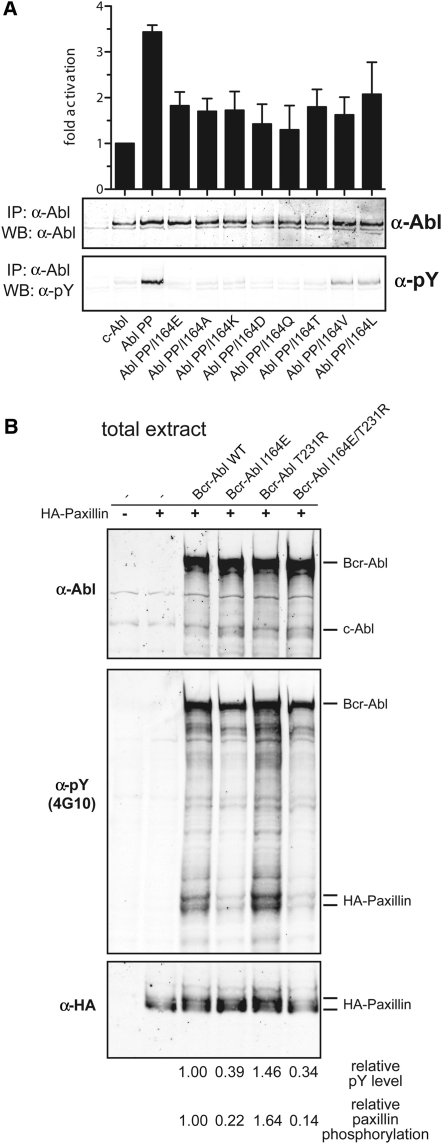
To study the structural role of Ile164 in more detail, we mutated Ile164 to different polar/charged amino acids (Glu, Gln, Thr, Asp, or Lys) or to Ala. All mutations led to a strong reduction of Abl autophosphorylation and in vitro kinase activity. Somewhat weaker effects were observed by mutating Ile164 to structurally related hydrophobic amino acids (Val or Leu) (Figure S1 available online).
Figure S1.
Effects of Mutations of Ile164 and/or Thr231 in the SH2-Kinase Interface on Abl Kinase Activity and Phosphorylation of the Abl Substrate Paxillin, Related to Figure 1
(A) The indicated mutations of Ile164 were introduced in the constitutive active mutant Abl PP and the resulting constructs were transiently expressed in HEK293 cells. Constructs were immunoprecipitated and subjected to in vitro kinase assays in the presence of an optimal Abl substrate peptide containing a single tyrosine phosphorylation site (upper panel) and western blotting using the indicated antibodies (lower panels). Mutation of Ile164 to all residues tested led to a significant reduction of in vitro kinase activity and tyrosine phosphorylation, indicating that the effect is not specifically restricted to the I164E mutation. Error bars represent SD.
(B) Bcr-Abl WT, Bcr-Abl I164E, Bcr-Abl T231R, and Bcr-Abl I164E/T231R constructs were coexpressed with HA-tagged paxillin in HEK293 cells and total protein extracts were analyzed by immunoblotting using the indicated antibodies.
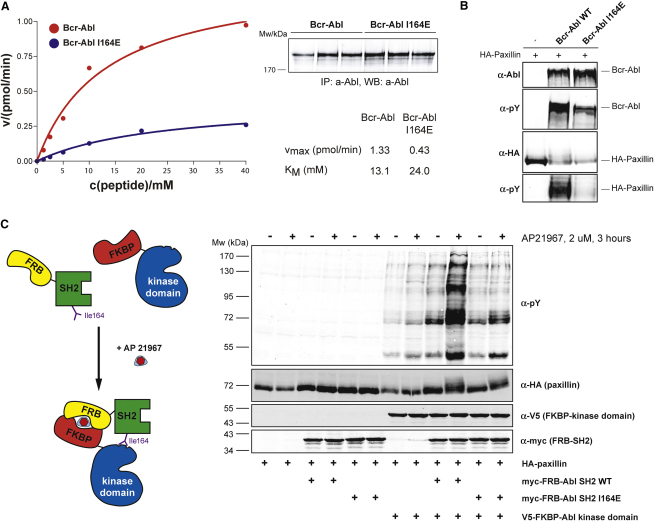
In enzyme-kinetic experiments, the Bcr-Abl I164E mutant protein displayed a >3-fold reduction in vmax compared to Bcr-Abl wild-type (WT) and a modest increase in Michaelis-Menten constant (KM) when assayed in vitro using an optimal Abl substrate peptide containing one single tyrosine (Figure 2A).
Figure 2.
The SH2-Kinase Domain Interface in Bcr-Abl Is Both Necessary and Sufficient for High Catalytic Activity of the Enzyme
(A) Immunoprecipitated Bcr-Abl WT and I164E proteins were assayed for catalytic activity in the presence of the indicated concentrations of an optimal Abl substrate peptide containing a single tyrosine phosphorylation site. Averages of three replicates are plotted. The KM and vmax values were calculated after fitting the data to the Michaelis-Menten equation.
(B) Bcr-Abl WT and I164E constructs were coexpressed with HA-tagged paxillin in HEK293 cells, and total protein extracts were analyzed by immunoblotting using the indicated antibodies.
(C) The indicated constructs were transiently transfected in HEK293 cells along with HA-paxillin. FRB-FKBP dimerization was induced by treating cells with AP21967. Total cell lysates were analyzed by immunoblotting using the indicated antibodies.
See also Figure S2.
Relationship between Phosphotyrosine Binding and Kinase Domain Binding of the Abl SH2 Domain
SH2 domain-containing tyrosine kinases have been proposed to use their SH2 domains to bind primed substrates to facilitate multisite (processive) phosphorylation of substrates with multiple tyrosine phosphorylation sites (Mayer et al., 1995). Upon cotransfection of the Bcr-Abl substrate paxillin with Bcr-Abl I164E, we observed a strong reduction in multisite phosphorylation of paxillin, whereas the T231R mutation had the opposite effect (Figures 2B and S1). These data indicate that disruption of the SH2-kinase domain interface also influences the cellular activity of Bcr-Abl.
As the canonical function of the SH2 domain is phosphotyrosine binding, we analyzed whether the I164E mutation would interfere with phosphopeptide-SH2 interactions (Pawson et al., 2001). Recombinant WT and I164E-mutated Abl SH2 domains displayed indistinguishable low micromolar binding constants for an optimal Abl SH2-domain phoshopeptide ligand (Figure S2). In contrast, Abl SH2 domains bearing the FLVRES mutants R171L or S173N were unable to bind tyrosine-phosphorylated peptides (Figure S2 and data not shown). Together these data show that the I164E mutant does not compromise phosphotyrosine binding or structural integrity of the SH2 domain. Nonetheless, in the context of full-length Abl kinase, the I164E mutant was as defective as the FLVRES mutant S173N in multisite phosphorylation of paxillin (Figure S2). This shows that aside from the intrinsic reduction in in vitro and cellular kinase activity, disruption of the SH2-kinase interface also interferes with multisite phosphorylation of Abl substrates, even though the phosphotyrosine peptide-binding properties of the SH2 domain are retained in Abl I164E. In addition, this indicates that the correct positioning of the SH2 domain may just be as important as the ability of the SH2 domain to bind primed tyrosine-phosphorylated substrates for further rounds of phosphorylation.
Figure S2.
The I164E Mutation Does Not Interfere with the Phosphotyrosine-Binding Capability of the Abl SH2 Domain, Related to Figure 2
(A) Fluorescence polarization binding assay of recombinant WT and mutant Abl SH2 domains to a fluorescently labeled tyrosine phosphorylated peptide (EPPVpYANLS). The recombinant Abl SH2 domain harboring the I164E mutation was found to bind a phosphorylated peptide with equal affinity as the wt Abl SH2 domain. In contrast, Abl SH2 domains bearing the FLVRES mutants R171L or S173N were unable to bind tyrosine phosphorylated peptides.
(B) HEK293 cells were transfected with expression constructs comprising the Abl kinase domain alone or the SH2-kinase domain module of c-Abl (Abl SH2-kinase domain WT, S173N, I164E). Constructs were immunoprecipitated and subjected to in vitro kinase assays in the presence of an optimal Abl substrate peptide containing a single tyrosine phosphorylation site. In line with previous results, the SH2 domain had a positive role on the kinase activity, as we observed higher kinase activity of the Abl SH2-kinase domain construct as compared to the Abl kinase domain alone, and introduction of the I164E reverted this effect. However, the phosphotyrosine-binding mutant S173N did not show significantly different kinase activity as compared to the WT Abl SH2-kinase domain construct. As the kinase substrate peptide used in this assay contains only one tyrosine residue, no (processive phosphorylation) effects dependent on the phosphotyrosine-binding capability of the Abl SH2 domain are observed. Together, these data rule out that the I164E mutation in the Abl SH2 domain leads to reduced kinase activity by interfering with phosphotyrosine binding of the SH2 domain. Furthermore, this experiment shows that the ability of the Abl SH2 domain to bind phosphotyrosine does not influence the in vitro kinase activity of the Abl kinase. Error bars represent SD.
(C) HEK293 cells were transiently transfected with the indicated c-Abl mutants in the presence or absence of a HA-tagged form of the Abl substrate paxillin. Total cell lysates were subjected to immunoblot analysis using antibodies against Abl, HA and phosphotyrosine. HA-tagged paxillin was immunoprecipitated from the same lysates and subjected to immunoblotting using antibodies against phosphotyrosine and HA. The Abl substrate paxillin contains multiple tyrosine residues that can serve as substrates for Abl. Thus, basal phosphorylation of one tyrosine residue might exert a positive feedback to Abl activity that is mediated by the Abl SH2 domain by binding the first phosphotyrosine and thereby positioning another unphosphorylated tyrosine close to the active site of the kinase. This phenomenon is referred to as processive phosphorylation and is dependent on the capability of the SH2 domain to bind to tyrosine-phosphorylated substrates. Paxillin was efficiently tyrosine phosphorylated by the constitutively active form of Abl (Abl PP) in a processive way (phosphorylation on multiple tyrosine residues as indicated by the shift of the HA-reactive bands to higher molecular weight species). Both Abl PP I164E and Abl PP S173N were able to phosphorylate paxillin at basal levels but neither of the mutants induced efficient processive phosphorylation of this substrate. This phenotype can be explained by the inability of the S173N mutant to bind to tyrosine-phosphorylated proteins, while in the case of the I164E mutant, this could be attributed to the lower overall activity of the kinase caused by the loss of the positive allosteric effect of the SH2 domain, which is independent of phosphotyrosine binding. In addition, this may indicate that for the recognition of Abl substrates with multiple phosphorylation sites the correct positioning of the SH2 domain appears to be of equal importance.
Induced Tethering of the SH2 Domain to the Kinase Domain Activates Abl and Depends on Ile164
In order to assess its potential for pharmacological exploitation, we further investigated the molecular mechanism of the positive effect of the SH2 domain on kinase activity. We made use of the FKBP-FRB fusion protein system, in which the SH2 and kinase domains of Abl are expressed separately but can be induced to physically associate upon addition of rapamycin (Belshaw et al., 1996) (Figure 2C). Induced tethering of the Abl SH2 domain to the Abl kinase domain led to an unexpectedly strong increase in phosphorylation of cotransfected paxillin and in total cellular tyrosine phosphorylation (Figure 2C). The effect was reliant on the presence of both protein components and on the dimerization-inducing drug. Importantly, this effect was entirely dependent on the SH2-kinase interface, as mutation of Ile164 abolished the stimulatory effect. This provided conclusive evidence that docking of the SH2 domain is sufficient for the observed positive allosteric effect on kinase activity and could thus represent a targetable interaction.
The Bcr-Abl SH2-Kinase Interface Is Critical for the Transformation of Primary Murine Hematopoietic Cells and Leukemia Formation In Vivo
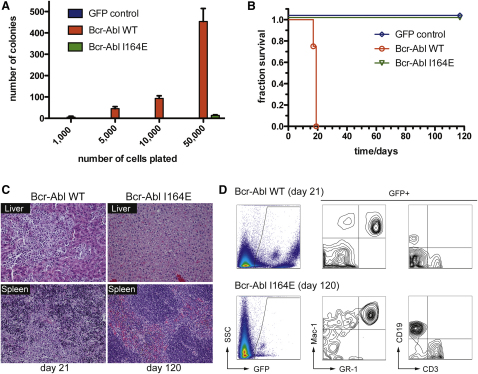
We next determined whether mutation of the SH2-kinase domain interface had an effect on oncogenic transformation and leukemogenicity. Primary murine bone marrow hematopoietic cells transduced with Bcr-Abl WT were able to form colonies in the absence of cytokines in semisolid media (Figure 3A). In contrast, this transforming capability was dramatically reduced in cells expressing Bcr-Abl I164E (Figure 3A).
Figure 3.
Bcr-Abl I164E Is Not Leukemogenic in a Bcr-Abl Mouse Bone Marrow Transplantation Model
(A) Indicated amounts of primary murine bone marrow cells expressing Bcr-Abl WT, Bcr-Abl I164E, or GFP were seeded in semisolid medium. Cytokine-independent colonies were scored after 8 days. Error bars represent SD.
(B) Equal amounts of primary murine Lin− c-Kit+ Sca-1+ cells expressing Bcr-Abl WT, Bcr-Abl I164E, or GFP were injected in lethally irradiated recipient mice (n = 4). Overall survival of transplanted mice was monitored over 120 days.
(C) Sections from liver (top panels) and spleen (bottom panels) of representative Bcr-Abl WT and I164E transplanted animals were stained with hematoxylin/eosin.
(D) Peripheral blood samples were prepared at indicated time points after transplantation and stained with the indicated antibodies and analyzed by FACS.
We then tested whether this reduction was also manifest in an in vivo model of Bcr-Abl-induced leukemia (Daley et al., 1990). Equal numbers of Bcr-Abl WT- and Bcr-Abl I164E-transduced hematopoietic stem cells were injected into lethally irradiated recipient mice. Mice transplanted with Bcr-Abl-expressing cells developed an aggressive myeloproliferative disorder, leading to death of all animals within 3 weeks (Figure 3B). These mice displayed massively infiltrated spleen and liver and loss of normal organ architecture (Figure 3C). In contrast, all mice transplanted with Bcr-Abl I164E-expressing cells remained alive for the 120 days of the study (Figure 3B). After this period, no obvious pathological alterations in the liver or spleen were detected despite confirmation of the presence of Bcr-Abl I164E-transduced cells in all lineages in peripheral blood, bone marrow, and spleen (Figures 3C and 3D and data not shown). On one hand, this indicates a lack of oncogenic properties of Bcr-Abl I164E that are necessary to induce a fatal leukemia, but it rules out a possible defect in engraftment and/or hematopoietic differentiation caused by the I164E mutation. Together, our results indicate a crucial role for the SH2-kinase domain interface in Bcr-Abl-mediated leukemic transformation.
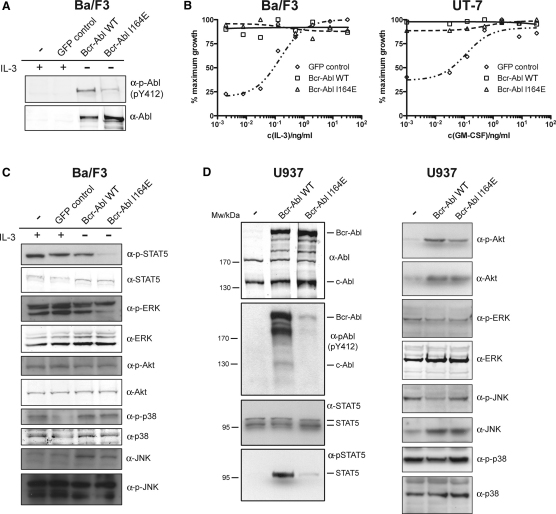
Bcr-Abl I164E Confers Growth-Factor Independence to Ba/F3 and UT-7 Cells despite Reduced Kinase Activity
To assess cellular effects and downstream signaling of the Bcr-Abl I164E mutation, we engineered the murine IL-3-dependent cell line Ba/F3 and the human GM-CSF-dependent cell line UT-7 to express Bcr-Abl WT and Bcr-Abl I164E. In both cell lines, we consistently observed higher expression levels of Bcr-Abl I164E as compared to Bcr-Abl WT, despite equal virus titers used (Figure 4A and data not shown). Nevertheless, global tyrosine phosphorylation was still lower in cells expressing Bcr-Abl I164E (Figure 4A). Single-cell clone pairs, initially selected for equal expression levels of Bcr-Abl WT and I164E, quickly showed increased Bcr-Abl I164E expression within a few passages (data not shown).
Figure 4.
Bcr-Abl I164E Renders Ba/F3 or UT-7 Cell Lines Factor Independent and Differentially Impacts Cellular Signaling Pathways
(A) Expression levels and cellular activity of Bcr-Abl WT and I164E transduced Ba/F3 cells.
(B) Proliferation of Bcr-Abl WT, I164E, or GFP control Ba/F3 cells or UT-7 cells was measured in the presence of the indicated concentrations of IL-3 or GM-CSF.
(C and D) Activation levels of selected components of different cellular signaling pathways by Bcr-Abl WT versus I164E were investigated by immunoblotting of total lysates from Ba/F3 (B) and U937 cells (C) stably expressing Bcr-Abl WT or Bcr-Abl I164E using the indicated antibodies.
See also Figure S3.
A widely used read-out of Bcr-Abl activity is the ability to confer cytokine-independent growth to Ba/F3 or UT-7 cells. Despite the complete lack of leukemogenic activity of Bcr-Abl I164E (see Figure 3), we observed that Bcr-Abl I164E was equally potent in transforming Ba/F3 or UT-7 cell lines to cytokine independence (Figure 4B). A possible explanation is that the reduced kinase activity of the I164E mutant may be compensated by upregulation of the Bcr-Abl mutant protein levels in order to allow cytokine-independent proliferation of hematopoietic cell lines.
Analysis of Downstream Signaling Pathways
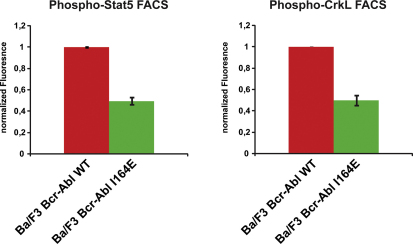
It is surprising that a single point mutation located outside of the tyrosine kinase domain and not affecting phosphotyrosine binding of the SH2 domain could have such a dramatic effect on the oncogenic activity of Bcr-Abl. Given this crucial function, we investigated the downstream signaling events that may be involved. We tested some major phosphorylation events in Ba/F3 cells, as well as in human U937 cells. As already observed in HEK293 cells, mutation of Ile164 led to a strong reduction of global tyrosine phosphorylation and phosphorylation of Tyr412 in the activation loop of Bcr-Abl (Figures 4A and 4D). Important signaling mediators downstream of Bcr-Abl include the phosphorylation and activation of STAT5, Akt, and Erk1/2 (Ren, 2005). We observed a strong reduction in tyrosine phosphorylation of STAT5 and CrkL in cells expressing Bcr-Abl I164E as compared to Bcr-Abl WT (Figures 4C, 4D, and S3). As activation of STAT5 is required for the induction and maintenance of CML by Bcr-Abl (Nieborowska-Skorska et al., 1999; Hoelbl et al., 2010), it is tempting to speculate that the inability of Bcr-Abl I164E to induce CML may mainly be caused by its inability to activate STAT5.
Figure S3.
Bcr-Abl I164E-Expressing Ba/F3 Cells Show Reduced Tyrosine Phosphorylation of Stat5 and CrkL, Related to Figure 4
Ba/F3 cells expressing Bcr-Abl WT or Bcr-Abl I164E were fixed and stained with antibodies against Stat5 phosphorylated on Tyr694 (BD Biosciences) (left panel) or CrkL phosphorylated on Tyr207 (Cell Signaling) (right panel) and analyzed by flow cytometry. Mean fluorescence intensities of samples from Bcr-Abl wt-expressing cells were normalized to 1. Error bars represent SD.
Surprisingly, despite the strong effect on Bcr-Abl activity, the I164E mutation had no effect on Erk1/2 and Akt phosphorylation or other MAPK or PI3K pathway members (Figures 4C and 4D and data not shown). This is in line with comparable levels of phosphorylated Tyr177 in Bcr-Abl, which was shown to be critical for PI3K and MAPK pathway activation (Figure 1B) (Sattler et al., 2002). Thus, interference with the SH2-kinase domain interface appears to result in a complex rewiring of the signaling network with specific downregulated events rather than general attenuation of signaling, suggesting that uncoupling of the SH2-kinase module may generate a not only quantitatively but also qualitatively impaired abnormal Bcr-Abl output. Furthermore, these data are consistent with different thresholds in Bcr-Abl activity that are required for full signaling output, with STAT5 activation being a critical event that is highly sensitive to disruption of the SH2-kinase domain interface.
Disruption of the SH2-Kinase Domain Interface Sensitizes Bcr-Abl WT and Drug-Resistant Forms to TKI Inhibition
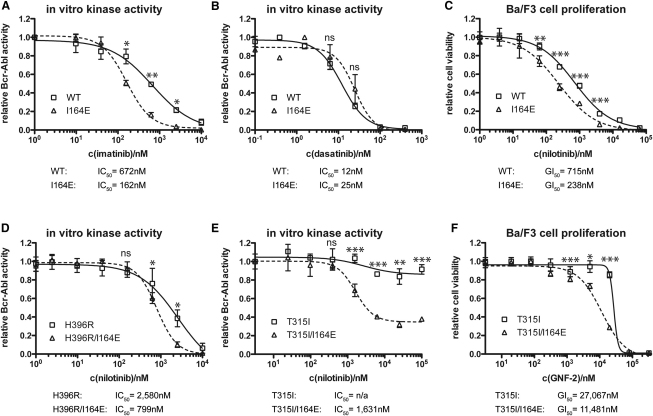
Pharmacological interference with the SH2-kinase domain interaction surface would be particularly attractive if it could alter the sensitivity of Bcr-Abl to existing TKIs. We tested the sensitivity of Bcr-Abl I164E toward the CML TKIs imatinib and dasatinib in in vitro kinase assays (Figures 5A and 5B). Whereas imatinib and nilotinib exclusively bind Bcr-Abl when its activation loop is not phosphorylated, binding of dasatinib requires an active conformation (Schindler et al., 2000; Vajpai et al., 2008). Therefore, different sensitivities to the two classes have been used by others and us to monitor different conformational states of the target enzymes (De Keersmaecker et al., 2008). We observed a 4-fold increase in sensitivity of Bcr-Abl I164E for imatinib, whereas no differences were observed in response to dasatinib (Figures 5A and 5B). In line with this, the sensitivity of Ba/F3 cells expressing Bcr-Abl I164E for nilotinib was increased 3-fold (Figure 5C). We next addressed whether disruption of the SH2-kinase domain interface also sensitized TKI-resistant mutants to nilotinib inhibition. Introduction of the I164E mutation in the imatinib-resistant Bcr-Abl clones H396R or E255K also increased the nilotinib sensitivity of these mutants 3-fold (Figure 5D and data not shown).
Figure 5.
Bcr-Abl I164E Sensitized WT and Imatinib-Resistant Bcr-Abl Forms to TKI Inhibition
(A, B, D, and E) Kinase activity of the indicated immunoprecipitated Bcr-Abl constructs was assayed in the presence of the indicated concentrations of imatinib (A), dasatinib (B), or nilotinib (D and E). For each dataset, the activities of the untreated samples were set to 1. Each data point in the graphs represents the averages ± SD from two independent experiments done in duplicate.
(C and F) Viability of Ba/F3 cells expressing Bcr-Abl WT, I164E, T315I, and T315I/I164E was measured in the presence of the indicated concentrations of nilotinib (C) or GNF-2 (F) and normalized to 1 for each dataset. For all panels, significance levels are indicated (ns: not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). Each data point in the graphs represents the averages ± SD from one representative experiment done in sextuplicate.
The T315I mutation in Bcr-Abl is the only mutation that is resistant to imatinib, nilotinib, and dasatinib. At concentrations that only marginally inhibited Bcr-Abl T315I, nilotinib caused a dramatic reduction in the in vitro tyrosine kinase activity in the Bcr-Abl T315I/I164E double mutant (Figure 5E). Finally, the I164E mutation also increased the sensitivity of Bcr-Abl T315I-expressing Ba/F3 cells toward the allosteric myristate-binding pocket inhibitor GNF-2 (Zhang et al., 2010 and Figure 5F). These results extend the recently proposed cooperativity between the myristate pocket and the ATP-binding site (Zhang et al., 2010) to the SH2-kinase domain interface and indicate that disruption of the SH2-kinase domain interface not only sensitizes Bcr-Abl WT to imatinib/nilotinib but could also be used to enhance inhibition of TKI-resistant Bcr-Abl mutants.
Targeting of the Bcr-Abl SH2-Kinase Interface using Monobodies
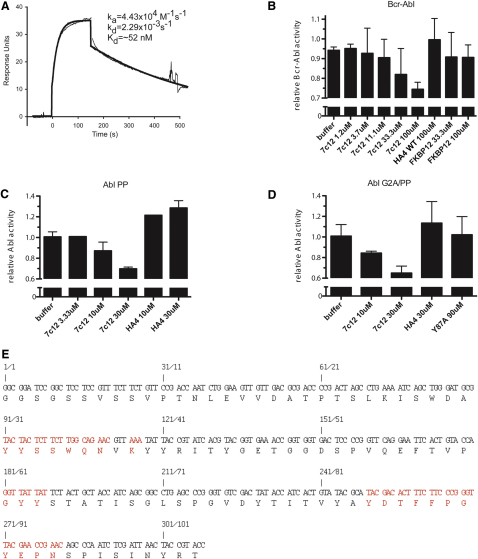
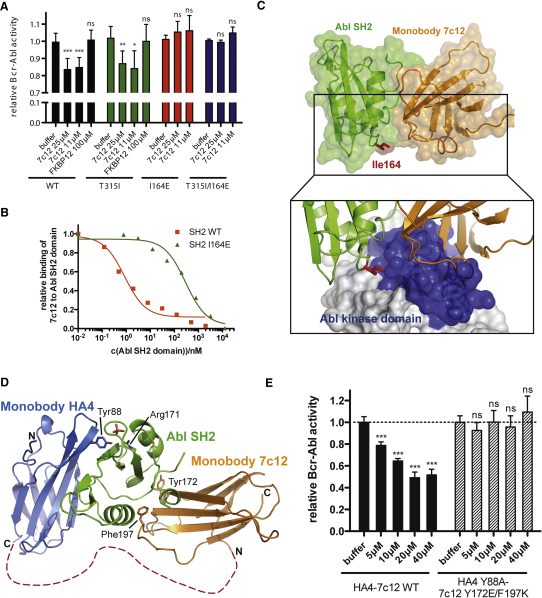
To test whether the SH2-kinase interface can be targeted using an inhibitor in trans, we generated single-domain binding proteins based on the fibronectin type III domain (FN3), termed monobodies, that target the Abl SH2 domain (Koide and Koide, 2007; Wojcik et al., 2010). We have previously described the Abl SH2-binding monobody HA4, which acts as a competitive inhibitor of phosphotyrosine binding (Wojcik et al., 2010). Using HA4 as a competitor in phage-disply library sorting, we identified an Abl SH2 monobody, designated 7c12, that bound to a distinct site. 7c12 binds the Abl SH2 domain with a dissociation constant (KD) of ∼50 nM (Figure S4 and data not shown). In in vitro kinase assays, 7c12 inhibited kinase activity of Bcr-Abl WT and T315I, but not of Bcr-Abl I164E or Bcr-Abl T315I/I164E (Figures 6A and S4). Although the effect was mild, it was statistically significant and did not occur with a recombinant control protein, which does not bind the Abl SH2 domain. This suggested a possible involvement of the Bcr-Abl SH2-kinase domain interface in 7c12 binding. In fact, the I164E mutation reduced binding of 7c12 by a factor of ∼400 (Figure 6B).
Figure S4.
Binding Parameters, Inhibition of Active Forms of Abl, Binding Properties, and Sequence of the SH2 Domain Monobody 7c12, Related to Figure 6
(A) Surface plasmon resonance trace for the Abl SH2 domain binding to immobilized 7c12. Parameters for the Langmuir fit of binding data are provided, and the black lines show the best fit.
(B–D) In vitro kinase activity of immunoprecipitated Bcr-Abl and the indicated constitutive active Abl mutant proteins (B, Bcr-Abl, C, Abl PP, D, Abl G2A-PP) was assayed in the presence of the indicated concentrations of 7c12, showing a dose-dependent inhibition of kinase activity of 7c12. FKBP12 or HA4 were used as control recombinant proteins.
(E) Nucleotide and amino acid sequence of 7c12. Positions that were randomized in the phage-display library that was used to select for Abl SH2 binders are indicated in red. Error bars represent SD.
Figure 6.
Targeting the SH2-Kinase Domain Interface with the Engineered Monobody Protein 7c12 Leads to Bcr-Abl Inhibition
(A) In vitro kinase activity of immunoprecipitated Bcr-Abl WT, I164E, T315I, and T315I/I164E proteins was assayed in the presence of the indicated concentrations of recombinant 7c12. FKBP12 was used as a control recombinant protein. Activity of each Bcr-Abl mutant-buffer control was normalized to 1. Significance levels are indicated (ns: not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). The bar graph shows averages ± SD from at least three independent experiments done in triplicate. See also Figure S4.
(B) Competition phage ELISA of the 7c12 monobody. Phages displaying the 7c12 monobody were bound to GST-Abl SH2 WT or I164E protein. Unbound phages were exposed to immobilized Abl SH2 domain and detected using an anti-phage antibody. Values are normalized to the intensity of the ELISA signal in the absence of a competitor.
(C) Crystal structure of the complex of the monobody 7c12-Abl SH2 domain complex (PDB entry 3T04). Ile164 is part of the interface and shown in red. Lower panel: Superposition of the 7c12-Abl SH2 complex and the active Abl structure. The surface of the Abl kinase domain is shown to highlight the incompatibility of simultaneous binding of 7c12 and the Abl kinase domain to the SH2 domain. See Table S1 and Figure S5 for details.
(D) Superposition of the 7c12-Abl SH2 domain complex (PDB entry 3T04) and the HA4-Abl SH2 complex (PDB entry 3K2M) crystal structures. HA4 and 7c12 are shown in blue and orange, respectively. The Gly-Ser linker between HA4 and 7c12 is indicated as a dotted red line. Arg171 in the Abl SH2 domain is shown to indicate the location of the phosphotyrosine-binding pocket. Residues Tyr88, Tyr172, and Phe197 were mutated to abolish binding to the Abl SH2 domain. See also Figure S5.
(E) In vitro kinase activity of immunoprecipitated Bcr-Abl WT protein was assayed in the presence of the indicated concentrations of recombinant HA4-7c12 WT or the nonbinding Y88A/Y172E/F197K mutant protein. Significance levels are indicated (ns: not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). The bar graph shows averages ± SD from one representative experiment done in triplicate. See also Figure S6.
Crystal Structure of the Abl SH2 Domain-7c12 Monobody Complex
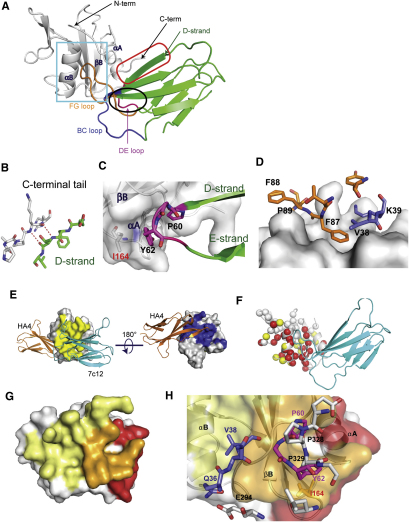
To elucidate the molecular details of the 7c12-Abl SH2 interaction, we determined the crystal structure of the complex at 2.1 Å resolution (PDB ID: 3T04; Table S1; Figures 6C and S5). The asymmetric unit consists of a single monobody-SH2 domain complex (Figures 6C and S5A). The 7c12 monobody and the Abl SH2 domain contribute nearly equally to the ∼1340 Å2 of surface area buried in the complex. The diversified loops contribute nearly 75% of the binding interface with each of the three loops making substantial contributions (91, 178, and 234 Å2 for the BC, DE, and FG loops, respectively). The D strand of the monobody scaffold also makes substantial contribution to the interface. It packs against the C-terminal tail of the SH2 domain through an intermolecular β sheet (Figure S5A, red box and S5B). Outside of these backbone-mediated interactions, the interface is dominated by hydrophobic contacts between monobody loop residues and two surface concavities on either side of β strand B of the SH2 domain. At the end of the monobody D strand, the DE loop forms a hairpin that packs against one of these concave surfaces that is formed by the βB strand and the αA helix of the Abl SH2 domain (Figure S5C). Pro60 and Tyr62 make most of the DE loop contacts, together burying more than 150 Å2 of surface area.
Figure S5.
Crystal Structure of the 7c12sm-Abl SH2 Domain Complex and Comparison to the Abl SH2-Kinase Domain Interface, Related to Figure 6
(A) The asymmetric unit of the 2.10 Å structure consists of a single copy of the 7c12sm-Abl SH2 complex. 7c12sm is a variant of 7c12 containing surface mutations that enhance solubility (see Extended Experimental Procedures). The Abl SH2 domain is shown as the white cartoon diagram. The N terminus and C terminus of the Abl SH2 domain are labeled, as are Abl SH2 helices αA and αB, and β strand B. 7c12sm is shown as a green cartoon diagram, with the BC, DE, and FG loops colored in blue, pink, and orange, respectively. The D strand of 7c12sm is indicated in green lettering and by the green arrow. An intramolecular interaction between 7c12sm strand D and the C-terminal tail of the SH2 domain is surrounded by the red oval, and shown in close-up in (B), where hydrogen bonds are shown as dotted red lines. The hairpin formed by strands D and E of 7c12sm, along with the DE loop, is circled in black, and shown in close-up in (C). Monobody residues are shown with black labels. I164 of the Abl SH2 domain is labeled in red and shown in stick form.
(D) Close-up of the interactions highlighted in the cyan box in (A). Selected 7c12sm BC and FG loop residues are shown as stick models colored as in (A) and labeled in black. The SH2 domain is shown as a white surface model.
(E) Orthogonal views of HA4-Abl SH2 and 7c12sm-Abl SH2 complexes superimposed according to the coordinates of the SH2 domain common to both structures. HA4 is shown as the orange cartoon model, 7c12sm as the cyan cartoon model. The Abl SH2 domain is shown as a white surface model, with additional coloring as follows: HA4 interface residues (defined as Abl SH2 domain residues within 5 Å of a monobody) are colored in blue and 7c12sm interface residues are colored in yellow.
(F) NMR epitope mapping of the 7c12sm interaction interface. 7c12sm is shown as cyan. Abl SH2 is shown as the white cartoon model, with individual residues as colored spheres. Red, yellow, and white spheres indicate the Cα atom positions for residues whose NMR signals are strongly affected, weakly affected, and not affected, respectively, by the binding of 7c12sm.
(G) Abl SH2 is shown as a transparent white surface model with additional coloring as follows: red: surfaces contacted by the kinase domain; yellow: surfaces contacted by 7c12sm; orange: surfaces contacted by both 7c12sm and the kinase domain in their respective structures.
(H) Close-up of (G) emphasizing the interface contacted by both the kinase domain and 7c12sm. The SH2 domain is also shown as a ribbon diagram beneath the transparent surface. Selected residues are highlighted as stick models. Kinase domain residues are shown with carbon atoms in white, nitrogen in blue, and oxygen in red and are labeled in black. 7c12-BC loop Q36 and V38 are labeled in blue, DE loop P60 and Y62 are in pink. Abl SH2 I164 is labeled in red, and shown as orange sticks. The αA, αB, and βB secondary structure elements of the Abl SH2 domain are labeled in black.
The second surface concavity is bounded on one side by β strand B of the Abl SH2 domain, on the other by α helix B, and from above and below by the N-terminal and C-terminal tails of the Abl SH2 domain (Figure S5A, cyan box). Phe87 of the 7c12 FG loop penetrates deep into the center of this pocket, burying 133 Å2 of surface area on its own. Other residues in the FG loop (Phe88, Pro89), Val38, and the aliphatic portion of Lys39 of the BC loop form a ring of hydrophobic contacts at the periphery of this surface pocket (Figure S5D). Together, these residues contribute nearly 300 Å2 to total monobody surface area burial.
The interface observed in the crystal structure is also consistent with the strong negative effect of the I164E mutant on 7c12-SH2 interaction (Figure 5B). Ile164 is located at the edge of the 7c12-binding interface and contributes 29 Å2 of surface to the interface. It makes hydrophobic interactions with Tyr62 of the DE loop (Figure S5C). These features rationalize the ∼400-fold reduction in binding affinity by the I164E mutation (Figure 6B).
In order to further confirm the authenticity of the interaction interface observed in the crystal structure, we used nuclear magnetic resonance (NMR) spectroscopy to conduct epitope mapping. The Abl SH2 backbone amide resonances most greatly affected by the presence of 7c12, as measured in 15N HSQC experiments, are in good agreement with portions of the SH2 domain that are contacted by 7c12 in the crystal structure (Figure S5F), suggesting that the interface observed in the crystal structure corresponds to the actual solution-phase interaction. To further validate the crystal structure using an independent experimental approach, we mutated each of the loop regions individually back to the template sequence and measured the binding affinity of these mutants. In each case, the mutant-Abl SH2 domain interaction was at least 100 times weaker than the wild-type 7c12-Abl SH2 interaction (data not shown). These results are consistent with the crystal structure, in which each loop makes a significant contribution to the 7c12-Abl SH2 interaction. Together, these studies provide significant validation of the observed crystal structure interface.
Comparison of the 7c12-Abl SH2 Interface to the Abl SH2-Kinase Domain Interface
We next compared the Abl SH2-kinase domain interface with the 7c12-Abl SH2 interface (Figures 6C and S5). The size of the SH2-kinase domain interface is ∼1030 Å2, ∼75% of that of the 7c12-Abl SH2 interface. More than half of the total surface area of the SH2-kinase interface overlaps with the 7c12-SH2 domain interface centered on β strand B of the Abl SH2 domain (Figures S5G and S5H). This significant overlap in the 7c12-Abl SH2 and Abl SH2-kinase interfaces strongly suggests that the binding of the SH2 domain to the kinase domain and to 7c12 are mutually exclusive (Figures 6C, S5G, and S5H). The incompatibility of simultaneous binding of the SH2 to both the kinase domain and 7c12, therefore, likely explains the observed inhibition of Bcr-Abl kinase activity, as full 7c12 binding requires the disruption of the SH2-kinase domain interface. Given the theoretical difficulty of competing intermolecularly with an ∼1100 Å2 intramolecular protein-protein interaction and the partial occlusion of the 7c12-binding site on the SH2 domain by the kinase domain, we consider the degree of inhibition that is achieved by the 7c12 monobody to be symptomatic and strongly indicative of a specific vulnerability.
Improved Targeting of the SH2 Interface with a Tandem Fusion of the HA4 and 7c12 Monobodies
Despite the clear demonstration of the general feasibility of targeting the SH2-kinase interface using 7c12, we aimed at improving its biological potency. Superposition of the structure of the previously characterized HA4 monobody bound to the Abl SH2 domain (Wojcik et al., 2010) with the 7c12-Abl SH2 structure reveals that the two monobodies bind on opposite faces of the SH2 domain with no overlap (Figures 6D and S5E). This arrangement is consistent with the selection scheme employed for the generation of 7c12, in which HA4 was used as a competitor, and with sandwich-ELISA data indicating that HA4 and 7c12 can bind simultaneously to the same Abl SH2 domain molecule (data not shown). HA4 binds with low nanomolar affinity to the phosphotyrosine-binding pocket of the Abl SH2 domain, which is not part of an intramolecular interaction in Bcr-Abl and is able to effectively outcompete phosphotyrosine ligand binding (Wojcik et al., 2010). For these reasons, we attempted to link HA4 and 7c12 in tandem, which should increase the local concentration of 7c12 in proximity to the SH2-kinase domain interface and further enhance the specificity toward the Abl SH2 domain. A 19 amino acid Gly-Ser linker was used to bridge the C terminus of HA4 to the N terminus of 7c12, based on structural modeling (Figure 6D). As negative controls, we introduced point mutations in both parts of the HA4-7c12 tandem monobody. For HA4, we used the Y88A (Y87A in the original numbering scheme) mutation that was previously shown to reduce binding to the Abl SH2 domain >1000-fold (Wojcik et al., 2010). We also designed a double point mutation (Y172E/F179K; Y62E/F87K in the original numbering scheme, Figure S5) for the 7c12 part, which is located at structurally central positions of the 7c12-Abl SH2 interface and showed a strong reduction in binding in pull-down experiments (data not shown). We tested the HA4-7c12 tandem monobody in Bcr-Abl in vitro kinase assays. We observed a strong inhibition of Bcr-Abl activity with low micromolar concentrations of the HA4-7c12 tandem monobody, whereas the nonbinding Y88A/Y172E/F179K mutant failed to inhibit Bcr-Abl activity (Figure 6E). Importantly, the degree of inhibition of Bcr-Abl activity achieved by HA4-7c12 was comparable to that upon introduction of the I164E mutant (Figures 1C and 2A). HA4-7c12 was also able to inhibit Bcr-Abl T231R, although to a lesser degree than Bcr-Abl WT (Figure S6). This indicates that the tandem monobody may lead to a complete disruption of the Bcr-Abl kinase domain interface.
Figure S6.
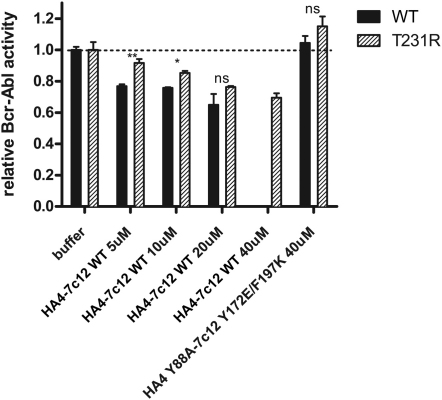
The T231R Mutation in the SH2-Kinase Interface Partially Blocks Inhibition of Bcr-Abl Activity by the HA4-7c12 Tandem Monobody, Related to Figure 6
In vitro kinase activity of immunoprecipitated Bcr-Abl WT and T231R proteins was assayed in the presence of the indicated concentrations of recombinant HA4-7c12. The nonbinding mutant HA4 Y88A-7c12 Y173E/F197K was used as a control. Activity of each Bcr-Abl mutant-buffer control was normalized to 1. Significance levels are indicated (ns: not significant, ∗p < 0.05, ∗∗p < 0.01). Error bars represent SD.
The HA4-7c12 Tandem Monobody Strongly Inhibits Cellular Bcr-Abl Activity and Induces Apoptosis in CML Cells
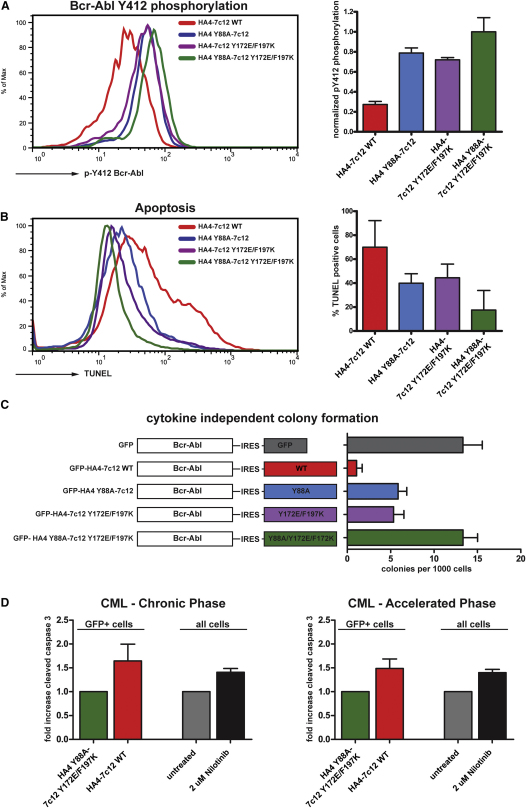
Finally, we tested whether the HA4-7c12 tandem monobody would also inhibit Bcr-Abl activity in CML cells. Transient expression of a HA4-7c12-GFP fusion protein in the Bcr-Abl-positive CML cell line K562 led to a strong reduction of activation loop (Tyr412) phosphorylation of Bcr-Abl (Figure 7A). This effect was dependent on both HA4 and 7c12 moieties of the tandem monobody, as expression of point mutants that abolish binding of either HA4 (Y88A) or 7c12 (Y172E/F197K) or both to the Abl SH2 domain did not show this effect (Figure 7A).
Figure 7.
The Tandem Monobody HA4-7c12 Strongly Inhibits Cellular Bcr-Abl Kinase Activity and Induces Apoptosis in CML Cell Lines and Primary Cells
(A) K562 cells were transiently transfected with the indicated HA4-7c12-GFP fusion constructs, and Bcr-Abl Tyr412 phosphorylation was measured using flow cytometry. Left panel: Histograms of Bcr-Abl pY412 fluorescence intensities of the GFP-positive fraction of cells transfected with the indicated constructs. Right panel: Quantitation of Bcr-Abl pY412 fluorescence. The intensities were normalized to K562 cells treated with nilotinib (complete inhibition—arbitrarily set to 0; Figure S5) and cells transfected with the nonbinding mutant HA4 Y88A-7c12 Y172E/F197K (no inhibition—arbitrarily set to 1) (n = 3).
(B) K562 cells treated as described in (C) were analyzed for apoptosis using TUNEL staining. Left panel: Histograms of TUNEL fluorescence of the GFP-positive fraction of cells transfected with the indicated constructs. Right panel: Quantitation of TUNEL-positive cells (n = 3). The intensities were normalized to K562 cells treated with nilotinib (arbitrarily set to 100%; Figure S7C) and mock-transfected cells (arbitrarily set to 0%) (n = 3).
(C) Primary murine bone marrow cells were transduced with the indicated retroviral constructs. GFP-positive cells were seeded in semisolid medium, and cytokine-independent colonies were scored.
(D) Primary cells from CML patients from chronic phase (left panel) or accelerated phase (right panel) were transduced with lentiviruses encoding HA4-7c12 WT or HA4 Y88A-7c12 Y172E/F197K. Parallel cultures from the same patients were treated with nilotinib for 5 days. Apoptosis was measured after 5 days via cleaved caspase staining by FACS. Levels of apoptosis in the GFP-positive fraction of cells transduced with HA4 Y88A-7c12 Y172E/F197K or in untreated cells were normalized to 1. p values: Chronic phase (five patients): 0.0007; accelerated phase (two patients): 0.011; one-way ANOVA).
In all panels, error bars represent SD. See Table S2 and Figure S7 for details.
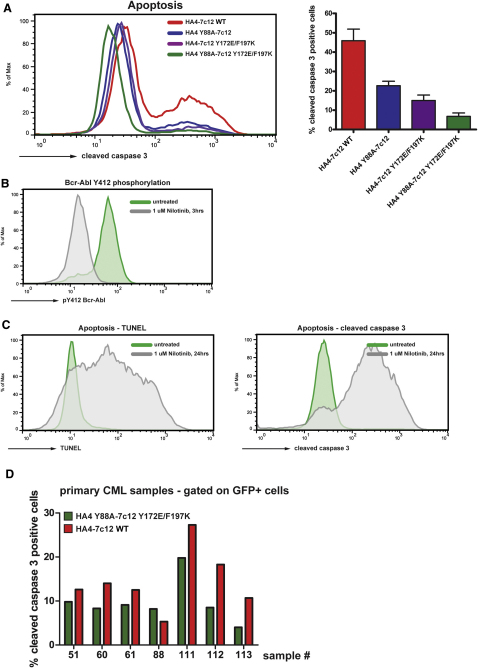
We next tested whether the HA4-7c12-induced decrease in Bcr-Abl activity with HA4-7c12 would result in cell death. Indeed, K562 cells expressing the HA4-7c12 tandem monobody showed high amounts of apoptosis, as measured by TUNEL and cleaved caspase 3 staining (Figures 7B and S7A). Again, this effect was not evident when the nonbinding mutant was used.
Figure S7.
The Tandem Monobody HA4-7c12 Induces Apoptosis in CML Cell Lines and Primary Cells from CML Patients, Related to Figure 7
(A) K562 cells were transiently transfected with the indicated HA4-7c12-GFP fusion constructs and the levels of cleaved caspase 3 were measured 48 hr later using flow cytometry. Left panel: Histograms of fluorescence intensities of the GFP-positive fraction of cells transfected with the indicated constructs. Right panel: Quantitation of cleaved caspase-3-positive cells. The percentages were normalized to K562 cells treated with 3 μM Nilotinib for 24 hr (arbitrarily set to 100%, see C, right panel) and mock-transfected cells (no apoptosis—arbitrarily set to 0%) (n = 3). Error bars represent SD.
(B) K562 cells were treated with 1 μM nilotinib for 3 hr or left untreated and Bcr-Abl activation loop phosphorylation (P-Tyr412) was measured by flow cytometry.
(C) K562 cells were treated with 1 μM nilotinib for 24 hr or left untreated and apoptosis was measured by flow cytometry using TUNEL (left panel) or cleaved caspase 3 staining (right panel) according the manufacturer's instructions.
(D) HA4-7c12 WT and the nonbinding mutant HA4 Y88A-7c12 Y173E/F197K were expressed in primary cells from CML patients (see Table S2 for patient characteristics) using lentiviral constructs expressing GFP as a marker. Seven days later, cells were fixed and analyzed for cleaved caspase 3-positive cells by FACS. Shown are percentages of apoptotic cells from the GFP-positive population.
Next, we asked whether HA4-7c12 would inhibit Bcr-Abl-induced transformation of primary murine bone marrow cells. In contrast to the nonbinding mutants, bicistronic expression of Bcr-Abl and GFP-HA4-7c12 WT severely impaired cytokine-independent colony formation in semisolid media (Figure 7C).
Finally, we tested whether HA4-7c12 would induce apoptosis in primary bone marrow or peripheral blood cells from CML patients at different stages of the disease (Table S2). Lentiviral expression of HA4-7c12 induced an ∼65% increase in apoptotic cells over the nonbinding mutant HA4 Y88A-7c12-Y172E/F197K in cells from patients in chronic phase (Figure 7D, left panel). This increase was comparable to the induction of apoptosis upon treatment of the same cultures with 2 μM nilotinib for 5 days (41%). Similar results were obtained when cells from patients in the accelerated phase of the disease were used (Figure 7D, right panel). Here, nilotinib treatment led to a 40% increase whereas expression of HA4-7c12 induced a 49% increase in apoptosis.
Together, these data show that Bcr-Abl activity can be inhibited by targeting the SH2-kinase interface using FN3-based monobodies in CML cell lines, as well as in primary cells from CML patients.
Discussion
In this study, we have identified the SH2-kinase domain interface as a critical structural feature that is important for the adoption and maintenance of the active conformation of Bcr-Abl. We show that disruption of this interface completely abolishes Bcr-Abl leukemogenicity. Therefore, the SH2-kinase domain interface in Bcr-Abl represents an attractive “druggable” target in addition to its ATP- and myristate-binding sites. As a molecular tool, we developed the FN3-based monobody 7c12 and the tandem monobody HA4-7c12 that showed strong inhibition of Bcr-Abl activity in vitro and induced apoptosis in CML cells. The HA4-7c12 fusion is an example of the use of two FN3 monobodies in tandem to achieve a higher level of target inhibition. Although we show that lentiviral delivery of the HA4-7c12 cDNA was able to induce apoptosis in primary cells from CML patients, difficulties with intracellular delivery of the HA4-7c12 protein will most probably limit or preclude the use of monobodies as drug-like molecules in clinical applications. In contrast, the exquisite affinity and specificity of HA4-7c12 for Bcr-Abl suggest general utility of monobodies as target validation tools for preclinical studies.
Recent data confirmed evidence for a positive role of the SH2 domain on the adjacent tyrosine kinase domain in selected cytoplasmic tyrosine kinases (Filippakopoulos et al., 2008; Mikkola and Gahmberg, 2010; Joseph et al., 2007), originally identified in the v-Fps/Fes oncoprotein (Sadowski et al., 1986). Despite this conserved allosteric regulatory property of the SH2 domain, the surfaces involved in establishing the interface between the SH2 domain and the tyrosine kinase domain N lobe appear to be diverse in terms of biophysical properties, size, and affinity among different tyrosine kinases. The underlying structural mechanism responsible for the activation of the Abl kinase domain by its SH2 domain is still elusive and should be the subject of an independent study. However, the I164E mutation disrupts the SH2-kinase domain interface while leaving the core function of the SH2 domain—binding of phosphotyrosine ligands—unaffected. Previous studies have attempted to address the role of the SH2 domain for oncogenic transformation and leukemogenicity of Bcr-Abl either by mutating the conserved Arg171 in the phosphotyrosine-binding pocket or by deleting the entire domain (Ilaria and Van Etten, 1995; Roumiantsev et al., 2001). However, the widely used mutation of Arg171 is associated with a loss of the integrity of the SH2 domain (Figure S1 and data not shown). Therefore, as in the case of deletion of the entire domain, both the phosphotyrosine-binding function as well as the maintenance of the SH2-kinase domain interface are likely to be compromised upon introduction of the Arg171 mutation. Thus, the observed effects on transformation and leukemogenicity likely reflect a combination of multiple effects caused by interference with the protein architecture.
As interference with the SH2-kinase domain interface leads to a dramatic reduction in Bcr-Abl activity, one would expect a uniform decrease in the activation of the major pathways that are activated by Bcr-Abl. Instead, we observed an almost exclusive loss of STAT5 activation. STAT5 is a central component that is required for the induction and maintenance of CML induced by Bcr-Abl (Nieborowska-Skorska et al., 1999; Hoelbl et al., 2010). Therefore, it is tempting to speculate that the inability of Bcr-Abl I164E to induce CML is caused by its inability to activate STAT5. It has been shown that even short-term blockage of Bcr-Abl action resulted in severe and complete inhibition of STAT5 activity and target gene expression (Shah et al., 2008; Hantschel et al., 2008). In contrast, other pathways, including the Ras-MAPK or the PI3K-Akt pathways, were not compromised strongly. This may be the reason that Bcr-Abl I164E, although not leukemogenic, may confer cytokine independence to Ba/F3 or UT-7 cells.
Together, our work shows that interfering with the SH2-kinase domain interface can allosterically inhibit Bcr-Abl activity. It should be feasible to develop a small molecule that interferes with this SH2-kinase domain interaction. Such allosteric modulators could be of either antagonistic or agonistic nature depending on the targeted surface and binding mode. This may represent an attractive option for patients with chronic phase CML that only achieve a suboptimal response or are resistant to TKI therapy, as well as for patients with advanced stage CML or Ph-positive acute lymphoblastic leukemia.
Experimental Procedures
Cells
Cell lines or primary cells were transduced with the indicated constructs using pMSCV-IRES-GFP-based retroviral vectors for 72 hr in the presence of IL-3, IL-6, SCF, and 7 μg/ml polybrene. For transient monobody expression, 1 × 106 K562 cells were transfected with 5 μg of expression vectors encoding GFP fusion constructs of HA4-7c12 and mutants thereof using the Nucleofector Kit V (Amaxa), applying program T-016. Samples were harvested 48 hr later. Primary CML cells were isolated from bone marrow or peripheral blood samples of patients with Ph-chromosome-positive CML at time of diagnosis. All patients gave written informed consent before blood or bone marrow was obtained. Cells were transduced with concentrated lentiviral supernatants via spinoculation (1000 × g, 90 min) at a multiplicity of infection (moi) of 20 and cultivated in RPMI 1640 medium plus 10% FCS and 100 ng/ml human Interleukin 3 (PeproTech). The study was approved by the Institutional Review Board (IRB) of the Medical University of Vienna.
Primary Cell Transformation Assay and Bone Marrow Transplantation
GFP-positive cells were isolated using fluorescence-activated cell sorting (FACS) and seeded in cytokine-free methylcellulose, and colonies were scored 8 days later. For bone marrow transplantation studies, transduced cells were injected into lethally irradiated (10 Gy) WT recipients via the tail vein. Peripheral blood samples of animals were regularly scored for the presence of GFP-Bcr-Abl-positive cells. Upon signs of sickness, mice were sacrificed and hematopoietic organs were analyzed for leukemic cells by FACS using fluorescently labeled antibodies against GR-1, Mac-1, CD19, and CD3. Tissues were fixed in paraformaldehyde prior to sectioning and staining with hematoxylin/eosin.
Kinase Assays
Immunoprecipitation of Bcr-Abl/c-Abl protein and Abl in vitro kinase assay were carried out as described previously (Hantschel et al., 2003). Immunoprecipitated Bcr-Abl proteins were resuspended in kinase assay buffer containing the indicated concentrations of tyrosine kinase inhibitor or recombinant monobody protein. A peptide with the preferred Abl substrate sequence carrying an N-terminal biotin (biotin-GGEAIYAAPFKK-amide) was used as substrate. The terminated reaction was spotted onto a SAM2 Biotin Capture membrane (Promega) and further treated according to the instructions of the manufacturer. The relative amount of immunoprecipitated Abl protein was determined by immunoblotting and subsequent relative quantification using the Li-Cor Odyssey system.
Induced Tethering of the Abl SH2 Domain to the Abl Kinase Domain
For the experiments described in Figure 2C, we used the Regulated Heterodimerization Kit from Ariad Inc. FRB was fused to the N terminus of the SH2 domain and FKBP12 to the N terminus of the kinase domain and transiently transfected in HEK293 cells along with HA-Paxillin. Forty-eight hours after transfection, we induced dimerization of FRB-SH2 domains with FKBP-kinase domain constructs by treating cells with 2 μM of AP21967 for 3 hr.
Monobody Selection and Preparation
To identify monbobodies to the kinase-interaction surface of the Abl SH2 domain, a phage-display library was sorted using biotinylated Abl SH2 as a target in the presence of a 5-fold excess of the monobody HA4, which binds to the phosphopeptide interface of the SH2 domain (Wojcik et al., 2010). Otherwise, selection, cloning, and expression of the 7c12 monobody were carried out as described previously (Koide and Koide, 2007, Wojcik et al., 2010). Proteins were produced with an N-terminal His10-tag and purified using a Ni–Sepharose column (GE Lifesciences) to apparent homogeneity. The His-tag moiety was removed by protease cleavage prior to crystallization. For the HA4-7c12 tandem monobody, the sequences of HA4 and 7c12 were linked by a (Gly-Ser)-linker.
Intracellular Flow Cytometry
Cells were fixed in 3.2% paraformaldehyde and stored in Methanol, followed by staining with antibodies recognizing Stat5 phosphorylated on Tyr697 (BD Biosciences), CrkL phosphorylated on Tyr207 (Cell Signaling), or Abl phosphorylated on Tyr412 (Cell Signaling) followed by a PE-conjugated goat anti-rabbit secondary antibody (Imgenex). For analysis of apoptosis, cells were stained with antibodies against cleaved caspase 3 (Cell Signaling) or subjected to TUNEL followed by staining with a PE-conjugated anti-BrdU antibody according to the manufacturer's instructions (BioVision).
Extended Experimental Procedures.
Kinase Inhibitors
Dasatinib, nilotinib, and imatinib were synthesized by WuXi PharmaTech (Shanghai, China). GNF-2 was purchased from Sigma Aldrich.
Immunoblotting
Samples were analyzed by standard procedures using the following antibodies: anti-Abl (Ab-3, Oncogene Science), anti-phospho-tyrosine (4G10, Millipore), anti-phospho-Abl (Tyr245, Cell Signaling Technology), anti-phospho-Abl (Tyr412, Cell Signaling Technology), anti-phospho-STAT5 (pY696, Cell Signaling Technology), anti-STAT5 (Cell Signaling Technology), and anti-HA (HA11, Covance). Secondary antibodies were either labeled with AlexaFluor 680-labeled goat anti-mouse IgG (Molecular Probes) or IRDye800 goat anti-rabbit IgG (Rockland) and detected using the Li-Cor Odyssey system. Alternatively, peroxidase-labeled anti-mouse/anti-rabbit-HRP antibodies (AP Biotech) were used.
Fluorescence Polarization SH2-Binding Assay
Peptide preparation and the determination of SH2 domain binding affinities by fluorescence polarization were preformed as previously described in Filippakopoulos et al. (2008).
Cloning of FKBP-Abl Kinase Domain and FRB-Abl SH2 Domain Fusion Constructs
The DNA sequence encompassing amino acid residues 248 to 534 of human c-Abl 1b was amplified from pSGT-human c-Abl1b (Barilá and Superti-Furga, 1998) by PCR and was fused to the C terminus of the PCR-amplified sequence of FKBP12 from plasmid pC4EN-F1 (Ariad). The resulting FKBP12-Abl kinase domain fusion construct was shuttled to pcDNA3.1-N-V5-TEV (Invitrogen) using gateway cloning. The FRB-Abl-SH2 construct was generated in analogy, amplifying the DNA encompassing amino acids 143 to 239 of the human c-Abl 1b sequence and fusing it C-terminal to the FRB sequence from plasmid pC4-RHE (Ariad). The FRB-Abl-SH2 fusion was expressed with 6 N-terminal myc epitopes. No additional linkers were used.
Surface Plasmon Resonance
Measurements were taken in 10 mM Na2PO4, pH 7.4, 150 mM NaCl, 0.005% Tween 20, and 50 μM EDTA with a BIAcore2000 instrument at 298 K. Purified 7c12 was immobilized via a His-tag to a nickel–NTA sensor chip (BIAcore). The Abl SH2 domain (concentration: 500 nM) was then flowed over the sensor chip at a rate of 50 μl/min and the association and dissociation kinetics were monitored. Data was fit to a 1:1 Langmuir binding model using a single kinetic trace with the BIAevaluation software.
Protein Expression, Modification, and Purification
The 7c12 monobody as isolated from a phage-display library was not sufficiently soluble for crystallization trials. In order to increase solubility, we introduced four mutations to the FN3 scaffold (A18D, L25K, N51D, and S72K) on the surface of the monobody opposite the diversified loops where mutations are expected to have little effect on binding. Together, these mutations dramatically enhanced the soluble expression yield. In particular, the S72K mutation seemed to have a particularly large effect. This mutation was, in fact, a reversion mutation that restored a wild-type lysine residue that had been mutated to a serine residue as part of a surface-entropy-reduction strategy aimed at enhancing the ability of the FN3 scaffold to form crystal contacts (Garrard et al., 2001). The new protein, termed 7c12sm, containing all four mutations retained its ability to bind the Abl SH2 domain. Proteins were produced with an N-terminal His10-tag and purified using a Ni–Sepharose column (GE Lifesciences) to apparent homogeneity as described previously (Koide et al., 2007). Point mutations to enhance the solubility of the 7c12 monobody were made using Kunkel mutagenesis (Kunkel et al., 1991). The His-tag moiety was removed by protease cleavage prior to crystallization.
Crystallization, Data Collection, and Structure Determination
The 7c12 monobody as isolated from a phage-display library was not sufficiently soluble for crystallization trials. In order to increase solubility, we introduced four mutations to the FN3 scaffold (A18D, L25K, N51D, and S72K) on the surface of the monobody opposite the diversified loops where mutations are expected to have little effect on binding. Together, these mutations dramatically enhanced the soluble expression yield. In particular, the S72K mutation seemed to have a particularly large effect. This mutation, was, in fact, a reversion mutation that restored a wild-type lysine residue that had been mutated to a serine residue as part of a surface-entropy-reduction strategy aimed at enhancing the ability of the FN3 scaffold to form crystal contacts (Garrard et al., 2001). The new protein containing all four mutations retained its ability to bind the Abl SH2 domain.
The HA4-Abl SH2 domain complex was purified with a Superdex 75 column (GE Lifesciences) in 10 mM Tris-HCl pH 8.0, 50 mM NaCl, 10 mM Na2SO4. The complex was concentrated to ∼7.5 mg/ml and crystallized in 0.2M Mg(NO3)2, 100 mM LiCl 20% PEG 3350 pH 6.0 by the hanging-drop vapor-diffusion method. Glycerol (20%) was used as a cryoprotectant. X-ray diffraction data were collected at the Advanced Photon Source beamline 24 ID-E (Argonne National Laboratory) at a wavelength of 0.97917 Å and a temperature of 100 K. Data collection and structure determination statistics are given in Table S1. Diffraction data were processed and scaled with the HKL2000 package (Otwinowski and Minor, 1997). The structures were solved by molecular replacement using the MOLREP program in the CCP4 program suite (CCP4, 1994; Potterton et al., 2003). A multicopy search was performed with the Abl SH2 domain and the FN3 scaffold, without the loop regions, as the search models (PDB IDs 2ABL and 1FNA, respectively). Rigid-body refinement was carried out using REFMAC5 in the CCP4 program suite. TLS (translation/libration/screw) groups were defined using the TLSMD server (Painter and Merritt, 2006), and TLS refinement, B-factor refinement, bulk solvent parameters, final positional refinement, and the search for and refinement of water molecules was carried out using REFMAC5. Model building and evaluation were carried out using the Coot program, and molecular graphics were generated using PyMOL (DeLano, 2002). The final structure had 100% of residues within allowed Ramachandran regions, and 97% in favored regions as measured by MOLPROBITY (Davis et al., 2007). Surface area calculations were performed using the PROTORP protein–protein interaction server (Reynolds et al., 2009). For details, see also Table S1.
Culture of Primary CML Cells
After isolation, cells were washed and stored in liquid nitrogen until used. After thawing, cells were resuspended in RPMI 1640 medium plus 10% FCS supplemented with 1,500 U/ml DNase (Sigma) and centrifuged. Then, cells were resuspended in RPMI 1640 medium plus 10% FCS and 150 U/ml DNase for 30 min (37°C). Thereafter, cells were centrifuged and resuspended in RPMI 1640 medium plus FCS without DNase in the presence of 100 ng/ml human Interleukin 3 (PeproTech).
Acknowledgments
This work was supported by the Austrian Academy of Sciences, in part by grants from the Austrian Science Fund (FWF) (#P18737 to O.H., I.K., and G.S.-F. and #P22282 to F.G.), a Terry Fox Programme Project grant from the Canadian Cancer Society and CIHR (#MOP-6849 to T.P.), an EMBO short-term fellowship (#ASTF 293.00-2009 to J.W.), the National Institutes of Health (R01-GM72688 and R21-CA132700 to S.K. and T32GM07281 to J.W.), and the University of Chicago Cancer Research Center (to S.K.). Part of the work was conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award RR-15301 from the National Center for Research Resources at NIH. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. We wish to thank all members of the participating laboratories for continuous support and discussions, D. Printz (CCRI Vienna) for expert FACS sorting, V. Komnenovic (IMBA Vienna) for help with histology, H. Pickersgill for help with manuscript preparation, and K. De Keersmaecker and T. Buerckstuemmer for critical reading of the manuscript. P.V. received a research grant and honorarium from Novartis and a research grant and honorarium from BMS. This work is dedicated to the memories of Maggie Pawson and Hartmut Beug.
Published: October 13, 2011
Footnotes
Supplemental Information includes Extended Experimental Procedures, seven figures, and two tables and can be found with this article online at doi:10.1016/j.cell.2011.08.046.
Contributor Information
Oliver Hantschel, Email: oliver.hantschel@epfl.ch.
Giulio Superti-Furga, Email: gsuperti@cemm.oeaw.ac.at.
Supplemental Information
References
- Belshaw P.J., Ho S.N., Crabtree G.R., Schreiber S.L. Controlling protein association and subcellular localization with a synthetic ligand that induces heterodimerization of proteins. Proc. Natl. Acad. Sci. USA. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley G.Q., Van Etten R.A., Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- De Keersmaecker K., Versele M., Cools J., Superti-Furga G., Hantschel O. Intrinsic differences between the catalytic properties of the oncogenic NUP214-ABL1 and BCR-ABL1 fusion protein kinases. Leukemia. 2008;22:2208–2216. doi: 10.1038/leu.2008.242. [DOI] [PubMed] [Google Scholar]
- Deininger M., Buchdunger E., Druker B.J. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005;105:2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P., Kofler M., Hantschel O., Gish G.D., Grebien F., Salah E., Neudecker P., Kay L.E., Turk B.E., Superti-Furga G. Structural coupling of SH2-kinase domains links Fes and Abl substrate recognition and kinase activation. Cell. 2008;134:793–803. doi: 10.1016/j.cell.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P., Müller S., Knapp S. SH2 domains: modulators of nonreceptor tyrosine kinase activity. Curr. Opin. Struct. Biol. 2009;19:643–649. doi: 10.1016/j.sbi.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantschel O., Superti-Furga G. Regulation of the c-Abl and Bcr-Abl tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2004;5:33–44. doi: 10.1038/nrm1280. [DOI] [PubMed] [Google Scholar]
- Hantschel O., Nagar B., Guettler S., Kretzschmar J., Dorey K., Kuriyan J., Superti-Furga G. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell. 2003;112:845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Hantschel O., Gstoettenbauer A., Colinge J., Kaupe I., Bilban M., Burkard T.R., Valent P., Superti-Furga G. The chemokine interleukin-8 and the surface activation protein CD69 are markers for Bcr-Abl activity in chronic myeloid leukemia. Mol. Oncol. 2008;2:272–281. doi: 10.1016/j.molonc.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochhaus A., O'Brien S.G., Guilhot F., Druker B.J., Branford S., Foroni L., Goldman J.M., Müller M.C., Radich J.P., Rudoltz M., IRIS Investigators Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia. 2009;23:1054–1061. doi: 10.1038/leu.2009.38. [DOI] [PubMed] [Google Scholar]
- Hoelbl A., Schuster C., Kovacic B., Zhu B., Wickre M., Hoelzl M.A., Fajmann S., Grebien F., Warsch W., Stengl G. Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia. EMBO Mol Med. 2010;2:98–110. doi: 10.1002/emmm.201000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilaria R.L., Jr., Van Etten R.A. The SH2 domain of P210BCR/ABL is not required for the transformation of hematopoietic factor-dependent cells. Blood. 1995;86:3897–3904. [PubMed] [Google Scholar]
- Jabbour E., Hochhaus A., Cortes J., La Rosée P., Kantarjian H.M. Choosing the best treatment strategy for chronic myeloid leukemia patients resistant to imatinib: weighing the efficacy and safety of individual drugs with BCR-ABL mutations and patient history. Leukemia. 2010;24:6–12. doi: 10.1038/leu.2009.193. [DOI] [PubMed] [Google Scholar]
- Joseph R.E., Min L., Xu R., Musselman E.D., Andreotti A.H. A remote substrate docking mechanism for the tec family tyrosine kinases. Biochemistry. 2007;46:5595–5603. doi: 10.1021/bi700127c. [DOI] [PubMed] [Google Scholar]
- Koide A., Koide S. Monobodies: antibody mimics based on the scaffold of the fibronectin type III domain. Methods Mol. Biol. 2007;352:95–109. doi: 10.1385/1-59745-187-8:95. [DOI] [PubMed] [Google Scholar]
- Mayer B.J., Hirai H., Sakai R. Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr. Biol. 1995;5:296–305. doi: 10.1016/s0960-9822(95)00060-1. [DOI] [PubMed] [Google Scholar]
- McWhirter J.R., Galasso D.L., Wang J.Y. A coiled-coil oligomerization domain of Bcr is essential for the transforming function of Bcr-Abl oncoproteins. Mol. Cell. Biol. 1993;13:7587–7595. doi: 10.1128/mcb.13.12.7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola E.T., Gahmberg C.G. Hydrophobic interaction between the SH2 domain and the kinase domain is required for the activation of Csk. J. Mol. Biol. 2010;399:618–627. doi: 10.1016/j.jmb.2010.04.045. [DOI] [PubMed] [Google Scholar]
- Million R.P., Van Etten R.A. The Grb2 binding site is required for the induction of chronic myeloid leukemia-like disease in mice by the Bcr/Abl tyrosine kinase. Blood. 2000;96:664–670. [PubMed] [Google Scholar]
- Nagar B., Hantschel O., Young M.A., Scheffzek K., Veach D., Bornmann W., Clarkson B., Superti-Furga G., Kuriyan J. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- Nagar B., Hantschel O., Seeliger M., Davies J.M., Weis W.I., Superti-Furga G., Kuriyan J. Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Mol. Cell. 2006;21:787–798. doi: 10.1016/j.molcel.2006.01.035. [DOI] [PubMed] [Google Scholar]
- Nieborowska-Skorska M., Wasik M.A., Slupianek A., Salomoni P., Kitamura T., Calabretta B., Skorski T. Signal transducer and activator of transcription (STAT)5 activation by BCR/ABL is dependent on intact Src homology (SH)3 and SH2 domains of BCR/ABL and is required for leukemogenesis. J. Exp. Med. 1999;189:1229–1242. doi: 10.1084/jem.189.8.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T., Gish G.D., Nash P. SH2 domains, interaction modules and cellular wiring. Trends Cell Biol. 2001;11:504–511. doi: 10.1016/s0962-8924(01)02154-7. [DOI] [PubMed] [Google Scholar]
- Perrotti D., Jamieson C., Goldman J., Skorski T. Chronic myeloid leukemia: mechanisms of blastic transformation. J. Clin. Invest. 2010;120:2254–2264. doi: 10.1172/JCI41246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintás-Cardama A., Kantarjian H., Cortes J. Flying under the radar: the new wave of BCR-ABL inhibitors. Nat. Rev. Drug Discov. 2007;6:834–848. doi: 10.1038/nrd2324. [DOI] [PubMed] [Google Scholar]
- Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat. Rev. Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- Roumiantsev S., de Aos I.E., Varticovski L., Ilaria R.L., Van Etten R.A. The src homology 2 domain of Bcr/Abl is required for efficient induction of chronic myeloid leukemia-like disease in mice but not for lymphoid leukemogenesis or activation of phosphatidylinositol 3-kinase. Blood. 2001;97:4–13. doi: 10.1182/blood.v97.1.4. [DOI] [PubMed] [Google Scholar]
- Sadowski I., Stone J.C., Pawson T. A noncatalytic domain conserved among cytoplasmic protein-tyrosine kinases modifies the kinase function and transforming activity of Fujinami sarcoma virus P130gag-fps. Mol. Cell. Biol. 1986;6:4396–4408. doi: 10.1128/mcb.6.12.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M., Mohi M.G., Pride Y.B., Quinnan L.R., Malouf N.A., Podar K., Gesbert F., Iwasaki H., Li S., Van Etten R.A. Critical role for Gab2 in transformation by BCR/ABL. Cancer Cell. 2002;1:479–492. doi: 10.1016/s1535-6108(02)00074-0. [DOI] [PubMed] [Google Scholar]
- Schindler T., Bornmann W., Pellicena P., Miller W.T., Clarkson B., Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- Shah N.P., Sawyers C.L. Mechanisms of resistance to STI571 in Philadelphia chromosome-associated leukemias. Oncogene. 2003;22:7389–7395. doi: 10.1038/sj.onc.1206942. [DOI] [PubMed] [Google Scholar]
- Shah N.P., Kasap C., Weier C., Balbas M., Nicoll J.M., Bleickardt E., Nicaise C., Sawyers C.L. Transient potent BCR-ABL inhibition is sufficient to commit chronic myeloid leukemia cells irreversibly to apoptosis. Cancer Cell. 2008;14:485–493. doi: 10.1016/j.ccr.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Sherbenou D.W., Hantschel O., Kaupe I., Willis S., Bumm T., Turaga L.P., Lange T., Dao K.-H., Press R.D., Druker B.J. BCR-ABL SH3-SH2 domain mutations in chronic myeloid leukemia patients on imatinib. Blood. 2010;116:3278–3285. doi: 10.1182/blood-2008-10-183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajpai N., Strauss A., Fendrich G., Cowan-Jacob S.W., Manley P.W., Grzesiek S., Jahnke W. Solution conformations and dynamics of ABL kinase-inhibitor complexes determined by NMR substantiate the different binding modes of imatinib/nilotinib and dasatinib. J. Biol. Chem. 2008;283:18292–18302. doi: 10.1074/jbc.M801337200. [DOI] [PubMed] [Google Scholar]
- Wojcik J., Hantschel O., Grebien F., Kaupe I., Bennett K.L., Barkinge J., Jones R.B., Koide A., Superti-Furga G., Koide S. A potent and highly specific FN3 monobody inhibitor of the Abl SH2 domain. Nat. Struct. Mol. Biol. 2010;17:519–527. doi: 10.1038/nsmb.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S., Witte O.N. The BCR-ABL story: bench to bedside and back. Annu. Rev. Immunol. 2004;22:247–306. doi: 10.1146/annurev.immunol.22.012703.104753. [DOI] [PubMed] [Google Scholar]
- Zhang J., Adrián F.J., Jahnke W., Cowan-Jacob S.W., Li A.G., Iacob R.E., Sim T., Powers J., Dierks C., Sun F. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature. 2010;463:501–506. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- Collaborative Computational Project, Number 4. (1994). The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763. [DOI] [PubMed]
- Barilá, D., and Superti-Furga, G. (1998). An intramolecular SH3-domain interaction regulates c-Abl activity. Nat. Genet. 18, 280–282. [DOI] [PubMed]
- Davis, I.W., Leaver-Fay, A., Chen, V.B., Block, J.N., Kapral, G.J., Wang, X., Murray, L.W., Arendall, W.B., 3rd, Snoeyink, J., Richardson, J.S., and Richardson, D.C. (2007). MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35 (Web Server issue), W375–W383. [DOI] [PMC free article] [PubMed]
- DeLano, W.L. (2002). The PyMOL Molecular Graphics System. http://pymol.sourceforge.net.
- Filippakopoulos, P., Kofler, M., Hantschel, O., Gish, G.D., Grebien, F., Salah, E., Neudecker, P., Kay, L.E., Turk, B.E., Superti-Furga, G., et al. (2008). Structural coupling of SH2-kinase domains links Fes and Abl substrate recognition and kinase activation. Cell 134, 793–803. [DOI] [PMC free article] [PubMed]
- Garrard, S.M., Longenecker, K.L., Lewis, M.E., Sheffield, P.J., and Derewenda, Z.S. (2001). Expression, purification, and crystallization of the RGS-like domain from the Rho nucleotide exchange factor, PDZ-RhoGEF, using the surface entropy reduction approach. Protein Expr. Purif. 21, 412–416. [DOI] [PubMed]
- Koide, A., Gilbreth, R.N., Esaki, K., Tereshko, V., and Koide, S. (2007). High-affinity single-domain binding proteins with a binary-code interface. Proc. Natl. Acad. Sci. USA 104, 6632–6637. [DOI] [PMC free article] [PubMed]
- Kunkel, T.A., Bebenek, K., and McClary, J. (1991). Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 204, 125–139. [DOI] [PubMed]
- Lawrence, M.S., Phillips, K.J., and Liu, D.R. (2007). Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 129, 10110–10112. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z., and Minor, W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Painter, J., and Merritt, E.A. (2006). Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450. [DOI] [PubMed]
- Potterton, E., Briggs, P., Turkenburg, M., and Dodson, E. (2003). A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137. [DOI] [PubMed]
- Reynolds, C., Damerell, D., and Jones, S. (2009). ProtorP: a protein-protein interaction analysis server. Bioinformatics 25, 413–414. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.