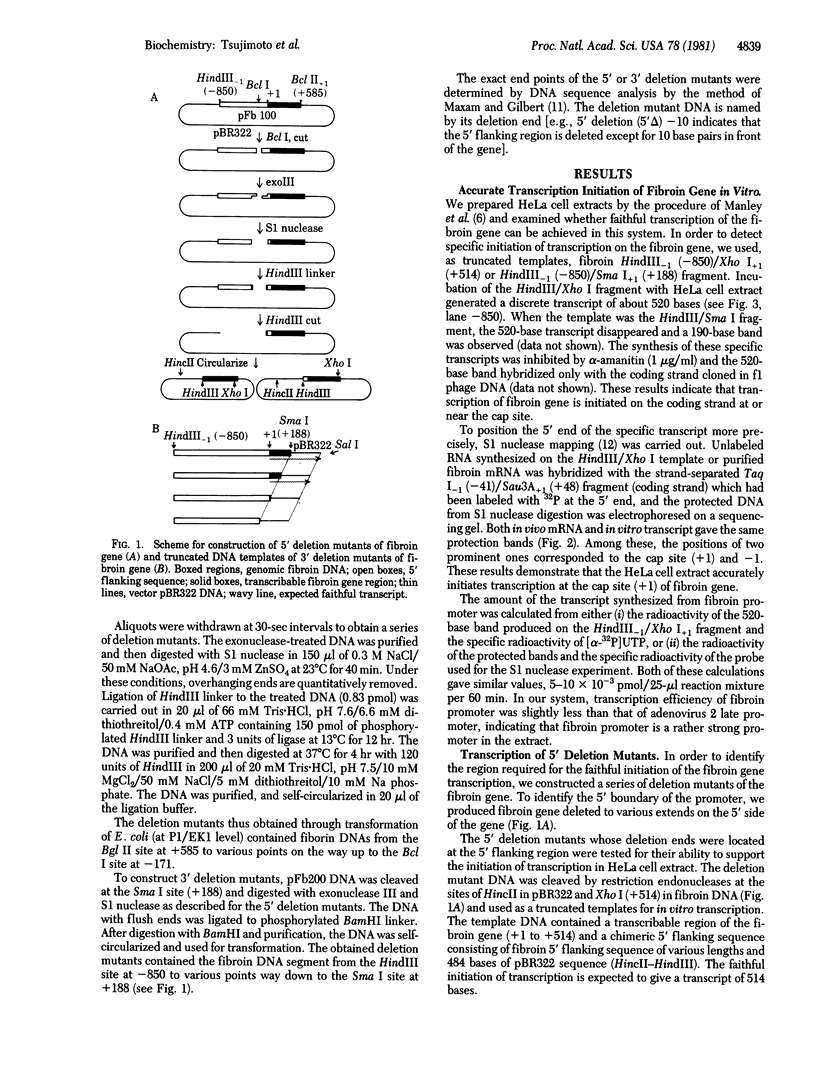
Abstract
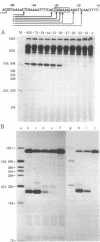
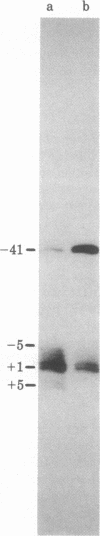
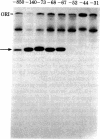
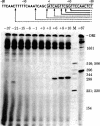
We have shown that the silk fibroin gene from Bombyx mori is faithfully transcribed in an in vitro transcription system of the HeLa cell extract prepared by the method of Manley et al. [Manley, J. L., Fire, A., Cano, A., Sharp, P. A. & Gefter, M. L. (1980) Proc. Natl. Acad. Sci. USA 77, 3855-3859]. Using this system and a series of deletion mutants of fibroin gene, we have assigned the promoter sequence of fibroin gene. The 5' boundary of the promoter is around nucleotide position -29, indicating that most of the T-A-T-A-A-A-A sequence (-30 to -24) is essential for the promoter function, where the transcription initiation point of fibroin gene is assigned as nucleotide position +1 [Tsuda, M., Ohshima, Y. & Suzuki, Y. (1979) Proc. Natl. Acad. Sci. USA 76, 4872-4876]. The 3' boundary is around nucleotide position +6. However, to support the efficient, faithful transcription, some additional (more than 26 but less than 41) nucleotides of nonspecific origin are required at the 5' side of -29. Functions ascribed to the promoter region are discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Identification of regulatory sequences in the prelude sequences of an H2A histone gene by the study of specific deletion mutants in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1432–1436. doi: 10.1073/pnas.77.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia T. D., Craig E. A., McCarthy B. J. Sequence of three copies of the gene for the major Drosophila heat shock induced protein and their flanking regions. Cell. 1980 Oct;21(3):669–679. doi: 10.1016/0092-8674(80)90430-4. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima Y., Suzuki Y. Cloning of the silk fibroin gene and its flanking sequences. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5363–5367. doi: 10.1073/pnas.74.12.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T., Sugimoto K., Sugisaki H., Takanami M. DNA regions essential for the function of a bacteriophage fd promoter. Nucleic Acids Res. 1977 Jul;4(7):2213–2222. doi: 10.1093/nar/4.7.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M., Ohshima Y., Suzuki Y. Assumed initiation site of fibroin gene transcription. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4872–4876. doi: 10.1073/pnas.76.10.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto Y., Suzuki Y. Structural analysis of the fibroin gene at the 5' end and its surrounding regions. Cell. 1979 Feb;16(2):425–436. doi: 10.1016/0092-8674(79)90018-7. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y., Suzuki Y. The DNA sequence of Bombyx mori fibroin gene including the 5' flanking, mRNA coding, entire intervening and fibroin protein coding regions. Cell. 1979 Oct;18(2):591–600. doi: 10.1016/0092-8674(79)90075-8. [DOI] [PubMed] [Google Scholar]
- Weil P. A., Luse D. S., Segall J., Roeder R. G. Selective and accurate initiation of transcription at the Ad2 major late promotor in a soluble system dependent on purified RNA polymerase II and DNA. Cell. 1979 Oct;18(2):469–484. doi: 10.1016/0092-8674(79)90065-5. [DOI] [PubMed] [Google Scholar]