Abstract
Retinal mitochondria become dysfunctional and their DNA (mtDNA) is damaged in diabetes. Biogenesis of mitochondria DNA is tightly controlled by nuclear-mitochondrial transcriptional factors, and translocation of transcription factor A (TFAM) to the mitochondria is essential for transcription and replication. Our aim is to investigate the effect of diabetes on nuclear-mitochondrial communication in the retina, and its role in the development of retinopathy. Damage of mtDNA, copy number and biogenesis (PGC1, NRF1, TFAM) were analyzed in the retina from streptozotocin-diabetic wildtype (WT) and MnSOD transgenic (Tg) mice. Binding between TFAM and chaperone Hsp70 was quantified by co-immunoprecipitation. The key parameters were confirmed in isolated retinal endothelial cells, and in the retina from human donors with diabetic retinopathy. Diabetes in WT mice increased retinal mtDNA damage, and decreased copy number. The gene transcripts of PGC1, NRF1 and TFAM were increased, but mitochondrial accumulation of TFAM was significantly decreased, and the binding of Hsp70 and TFAM was subnormal compared to WT-non diabetic mice. However, Tg-diabetic mice were protected from retinal mtDNA damage, and alterations in mitochondrial biogenesis. In retinal endothelial cells, high glucose decreased the number of mitochondria, as demonstrated by MitoTracker green staining and by electron microscopy, and impaired the transcriptional factors. Similar alterations in biogenesis were observed in the donors with diabetic retinopathy. Thus, retinal mitochondria biogenesis is under the control of superoxide radicals, and is impaired in diabetes, possibly by decreased transport of TFAM to the mitochondria. Modulation of biogenesis by pharmaceutical or molecular means may provide a potential strategy to retard the development/progression of diabetic retinopathy.
Keywords: Diabetic Retinopathy, DNA damage, Mitochondria, Superoxide dismutase
Introduction
Retinopathy is one of the most severe complications of diabetes which affects almost 90% of the patients after 20 years of the disease (1). Loss of retinal capillary cells (both endothelial cells and pericytes) instigates the development of histopathology associated with diabetic retinopathy, and this cell loss precedes any histopathological changes in the retina (2–5). Oxidative stress is considered as one of the major contributing factors in the development of diabetic retinopathy (6–8), but the mechanism via which free radicals results in its development remains elusive.
Our previous studies have shown that oxidative stress has a major role in the pathogenesis of diabetic retinopathy; superoxide levels are elevated in the retina and its capillary cells, mitochondria are dysfunctional, and the activities of mitochondrial superoxide scavenging enzyme, MnSOD, and of complex III of the electron transport chain become subnormal (9–12). Furthermore, mitochondrial DNA (mtDNA) is damaged, the import of DNA repair enzymes to the mitochondria is decreased, and the expression of the genes encoded by mtDNA (important in electron transport chain) is subnormal (12, 13). Regulation of mitochondrial superoxide levels by MnSOD attenuates oxidative stress, mitochondrial dysfunction and mtDNA damage, and also prevents the development of diabetic retinopathy in mice (13, 14).
Transcription and replication of mtDNA is tightly regulated by nucleus-mitochondria signaling pathway initiated by peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC1). PGC1, a nuclear encoded protein, is induced by environmental signals, and this in turn targets other specific transcription factors (15), and among these, nuclear respiratory factor, NRF-1, is a key nuclear-encoded transcription factor that regulates the expression of a number nuclear-encoded genes important in mitochondrial function, including mitochondrial transcription factor A (TFAM) and cytochrome oxidase IV, Cox IV (16, 17). TFAM is responsible for stability, maintenance, and transcriptional control of mtDNA and its translocation to the mitochondria is important to initiate mtDNA transcription and replication (18). How diabetes affects mitochondria biogenesis in the retina remains to be explored.
Here we have investigated the effect of diabetes on mitochondria biogenesis in the retina, and the role of mitochondria biogenesis in the development of retinopathy. Since mtDNA is prone to increased oxidative damage, mitochondria biogenesis was examined in the retina obtained from diabetic mice in which mitochondrial superoxide levels are modulated by overexpression of MnSOD, the model that is protected from the development of diabetic retinopathy (11, 13). The retina is a complex structure with multiple layers and cells, mitochondria biogenesis was also investigated in the isolated retinal endothelial cells, the cells that are the target of histopathology characteristic of diabetic retinopathy, in which mitochondrial superoxide radicals are regulated either by pharmacological (MnSOD mimic) or genetic means (MnSOD overexpression). The key parameters of mitochondria biogenesis were also confirmed in the retina obtained from human donors with diabetic retinopathy.
Methods
Mice
A group of mice (8–10 weeks old), overexpressing MnSOD (Tg), and their wild type littermates (WT) were made diabetic by streptozotocin injection (55 mg/kg BW) for 5 consecutive days. The mice, which presented blood glucose levels above 250 mg/dl 3 days after the last injection, were considered as diabetic. Twelve months after initiation of the experiment mice (10 or more group) were sacrificed by an overdose of pentobarbital and the retina was isolated under a dissecting microscope. These mice are being routinely used in our laboratory, and phenotype details are provided in our previous publications (11, 14, 19). Diabetic mice had blood glucose values 4–5 times higher compared to their age-matched nondiabetic mice. However, the values among the two diabetic groups (WT and Tg) were not different from each other (420±93 and 396±94mg/dl respectively), suggesting that the severity of hyperglycemia was similar in both WT-diabetic and Tg-diabetic mice. Treatment of animals was carried out according to the National Institute of Health principals of laboratory animal care, the Association for Research in Vision and Ophthalmology resolution on the use of animals in research, and the institutional guidelines.
Retinal endothelial cells
Endothelial cells isolated from bovine retina (BRECs) were cultured on polystyrene culture plates coated with 0.1% gelatin in a humidified incubator at 37°C, in an atmosphere of 5% CO2 and 95% air, as routinely performed in our laboratory (10, 13, 19, 20). The cells from the 4th–6th passage were incubated in Dulbecco’s Modified Eagle Medium (DMEM) containing 2% heat inactivated fetal bovine serum, 10% Nu-serum, 50µg/ml heparin, 1µg/ml endothelial growth and antibiotic/anti-mycotic supplemented with 5mM glucose or 20mM glucose for four days in the presence or absence of a cell permeable MnSOD mimic, MnTBAP (200µM; Biomol, Plymouth Meeting, PA). The medium was replaced every other day. For transfection of cells with MnSOD, BRECs from 3rd–5th passage were incubated with 3µg of MnSOD plasmid and effectene transfection reagent (Qiagen, Valencia, CA) as previously described (13). At the end of the transfection, the cells were washed with phosphate-buffered saline (PBS) and incubated in 20mM glucose for four days. The transfection efficiency was verified by semi-quantitative PCR using MnSOD primers (Applied Biosystem, Foster City, CA), and the cells transfected with MnSOD had 70% increase in their MnSOD gene transcripts. Osmotic controls included the cells incubated in identical experimental conditions with 20mM mannitol instead of 20mM glucose. At the end of the experimental period, the cells were harvested by trypsin digestion and rinsed with ice-cold PBS to remove trypsin.
To investigate the effect of increased oxidative stress on mitochondria biogenesis, BRECs were incubated with 250µM H2O2 for 1 hour (20). The cells were quickly washed with DMEM and incubated in 5mM glucose and 20mM glucose for 4 additional days.
Human retina
Retina was isolated from human post-mortem eyes obtained from Midwest Eye Banks (Ann Arbor, MI). Donors with diabetic retinopathy were 47–75 years of age. For control, retina was obtained from age-matched donors with no history of diabetes (Table I).
Table I.
Patient characteristics
| Age | Type of diabetes |
Duration of diabetes (years) |
Retinopathy status |
Cause of death | Death- enucleation (hours) |
|
|---|---|---|---|---|---|---|
| Diabetic patients | ||||||
| 75 | T2DM | 30 | PDR | Myocardium infraction | 10 | |
| 54 | T2DM | 10 | PDR | Congestive heart failure | 6 | |
| 69 | T2DM | 10 | PDR | Respiratory failure | 6 | |
| 61 | T2DM | 10 | PDR | Myocardium infraction | 10 | |
| 59 | T2DM | 16 | PDR | Renal failure | 9 | |
| 47 | T1DM | 27 | PDR | Myocardium infraction | 4 | |
| Non-diabetic patients | ||||||
| 44 | - | Not applicable | - | Intracranial hemorrhage | 11 | |
| 77 | - | Not applicable | - | Myocardium infraction | 6 | |
| 72 | - | Not applicable | - | Unknown | 7 | |
| 55 | - | Not applicable | - | Subarachnoid hemorrhage | 10 |
Sample preparation
Retina or cells were sonicated in 30mM Tris-HCl, pH7.4 buffer containing 10mM EGTA, 5mM EDTA, 1% Triton X-100, 250mM sucrose, 1mM NaF, 1mM PMSF, 1mM Na3VO4 and protease inhibitors, and centrifuged at 700×g for 5 minutes to remove the cell debris.
Mitochondria were isolated from retina or from BRECs using a Mitochondria Isolation kit from Invitrogen (Carlsbad, CA), as routinely used in our laboratory (12). Samples were homogenized in ice-cold mitochondrial isolation buffer A with protease inhibitor using a Dounce homogenizer, and mitochondrial isolation buffer C was added to the homogenate. Samples were centrifuged at 700×g for 5 minutes, and the resultant supernatant was centrifuged at 12,000×g for 15 minutes at 4°C. The pellet containing mitochondrial fraction was washed in PBS and resuspended with mitochondrial isolation buffer. Protein was determined by the Bicinchoninic Acid protein assay (Sigma-Aldrich, St Louis, MO, USA).
Gene Expression
Total RNA was extracted from the retina or BRECs with Trizol reagent (Invitrogen), and cDNA was synthesized by High Capacity cDNA Reverse Transcription Kit (Applied Biosystem). To evaluate the effect of mtDNA damage on the proteins encoded by this DNA, transcript abundance of subunit 6 (ND6) of complex I and cytochrome B (Cyt b) of complex III of the electron transport chain were quantified by semi-quantitative PCR using ND6 and Cyt b gene-specific primers. Relative amplification was quantified by normalizing the gene specific band intensity to that of β-actin.
Gene expression of PGC1, NRF1 and TFAM in mouse retina was quantified by real time RT-PCR using the Sybr Green assay to gene specific primers (Table II), and in BRECs and human retina by real time RT-PCR using TaqMan assay (Applied Biosystems). For mouse, β-actin was used as an internal standard, and 18s rRNA was used for BRECs and human. Change in mRNA abundance was calculated using the ddCt method as routinely used by us (21).
Table II.
Primers for the target genes
| Target | Sequence | |
|---|---|---|
| Mouse DNA | ||
| mtDNA long | forward | 5’-GCCAGCCTGACCCATAGCCATAATAT-3’ |
| reverse | 5’-GAGAGATTTTATGGGTGTAATGCGG-3’ | |
| mtDNA short | forward | 5’-CCCAGCTACTACCATCATTCAAGT-3' |
| reverse | 5’-GATGGTTTGGGAGATTGGTTGATGT-3’ | |
| nDNA long | forward | 5'-TTGAGACTGTGATTGGCAATGCCT-3' |
| reverse | 5'-CCTTTAATGCCCATCCCGGACT-3' | |
| nDNA short | forward | 5'- AGCCACAGATCCTATTGCCATGC-3' |
| reverse | 5'-TGTTGCTTGGTAAACACAGAGGGAAA-3' | |
| Cyt b | forward | 5’-GCAACCTTGACCCGATTCTTCGC-3’ |
| reverse | 5’- TGAACGATTGCTAGGGCCGCG-3’ | |
| COII | forward | 5’-ATTGCCCTCCCCTCTCTACGCA-3’ |
| reverse | 5’-CGTAGCTTCAGTATCATTGGTGCCC-3’ | |
| Mouse RNA | ||
| ND6 | forward | 5’-TGGTTGGTTGTCTTGGGTTGGCA-3’ |
| reverse | 5’-CCGCTACCCAATCCCTCCCT-3’ | |
| Cyt b | forward | 5’-ACCCGCCCCATCCAACATCTCAT-3’ |
| reverse | 5’-TTGAGGCTCCGTTTGCGTGT-3’ | |
| PGC1 | forward | 5’- AGACGGATTGCCCTCATTTG-3’ |
| reverse | 5’-CCGTCAGGCATGGAGGAA-3’ | |
| NRF1 | forward | 5’-TTCCAGTCTCTGTGGACAAAATGA-3’ |
| reverse | 5’-CGACCTGTGGAATACTTGAGCAT-3’ | |
| TFAM | forward | 5’- GCACCCTGCAGAGTGTTCAA-3’ |
| reverse | 5’- CGCCCAGGCCTCTACCTT-3’ | |
| Human DNA | ||
| mtDNA long | forward | 5'-TCT AAG CCT CCT TAT TCG AGC CGA-3' |
| reverse | 5'-TTT CAT CAT GCG GAG ATG TTG GAT GG-3' | |
| mtDNA short | forward | 5'-CCC CAC AAA CCC CAT TAC TAA ACC CA-3' |
| reverse | 5'-TTT CAT CAT GCG GAG ATG TTG GAT GG-3' | |
| nDNA long | forward | 5-GCA CTG GCT TAG GAG TTG GAC T-3 |
| reverse | 5-CGA GTAAGA GAC CAT TGT GGC AG-3 | |
| nDNA short | forward | 5'-TCCAGGTAGGGGCAGGATTCAGG-3' |
| reverse | 5'-CCCAACGTGATCGCCTTTCTCCC-3' | |
| Cyt b | forward | 5'-TCACCAGACGCCTCAACCGC-3' |
| reverse | 5'-GCCTCGCCCGATGTGTAGGA-3' | |
| COII | forward | 5'-GGCACATGCAGCGCAAGTAGG-3' |
| reverse | 5'-GGCGGGCAGGATAGTTCAGACG-3' | |
| Human RNA | ||
| ND6 | forward | 5'-GACCTCAACCCCTGACCCCCA-3' |
| reverse | 5'-GCGGTGTGGTCGGGTGTGTTAT-3' | |
| Cyt b | forward | 5'-TCACCAGACGCCTCAACCGC-3' |
| reverse | 5'-GCCTCGCCCGATGTGTAGGA-3' |
Mitochondria DNA damage
Since DNA damage prevents the progression of polymerase along the DNA template, this can be exploited using extended length PCR. DNA from mouse retina or human retina was isolated using DNeasy kit (Qiagen, Valencia, CA), and quantified using the Quant-iT dsDNA assay (Invitrogen). Genome specific (mitochondrial and nuclear) quantitative extended length PCR was performed with the GeneAmp XL PCR kit (Applied Biosystems) according to the method routinely employed in our laboratory (12, 13) using sequence specific primers (Table II). Relative amplification was quantified by normalizing the intensity of the long PCR product to the short PCR product (mtDNA=13.4kb/210bp and nDNA=13.4kb/195bp), and reduction in relative amplification was indicative of an increase in DNA damage (22).
Mitochondrial copy number and mass
Mitochondrial copy number was measured by real time RT-PCR. DNA primers were designed to detect Cyt b and cytochrome C oxidase subunit II (COII) as markers for mitochondrial DNA, and β-actin for nuclear DNA (Table II). PCR reaction was carried out using 15ng of gDNA, 200nM of each primer and SYBR green PCR master mix (Applied Biosystem). To amplify mtDNA and nDNA products, conditions included 10 minute at 95°C followed by 40 cycles of 15 second at 95°C and 60 second at 60°C. Relative values for mitochondrial DNA product (Cyt b and COII), and nuclear DNA product (β-actin or 18s rRNA) within each sample were used to obtain a ratio of mtDNA to nDNA.
In retinal endothelial cells, mitochondria mass was determined by using MitoTracker green (Molecular Probes), a mitochondrial-selective membrane potential-independent dye. The cells grown on 12mm diameter cover slips coated with 0.1% gelatin were incubated with 5mM or 20mM glucose for 4 days. At the end of the incubation, the cells were incubated with 200nM MitoTracker green, for 30 minutes. The green fluorescence intensity was examined under a Zeiss ApoTome using 40× magnification (Carl Zeiss Inc, Chicago, IL, was counted (23).
Mitochondria number was also quantified using electron microscopy. The cells were fixed in 2% glutaraldehyde in 100mM cacodylate, washed with 7% sucrose and embedded in 2% agar. This was followed by incubation in 1% osmium oxide, dehydration with graded ethanol solutions, and embedding in 812 resin (Electron Microscope Sciences, Hatfield, PA). Ultrathin sections were cut and stained with uranyl acetate and lead citrate, and imaged using transmission emission microscope (Model 400; Phillips, Eindhoven, The Netherlands). Mitochondria number was counted manually in a blind manner by calculating the number of mitochondria over total area of the cell (analyzed by Image J software) in at least 8–10 images/sample.
Protein expression
Protein (30–60µg, mitochondria or homogenate) was separated on a 4–16% SDS-PAGE, transferred to a nitrocellulose membrane and blocked with 5% non-fat milk for 1 hour. The membranes were incubated with antibodies against PGC1, NRF1 and TFAM (Santa Cruz Biotechnology) overnight at 4°C. Cox IV (Molecular Probe, Carlsbad, CA) was used as loading control for mitochondria and β-actin for homogenate.
Mitochondrial functional integrity
Citrate synthase activity was measured in a total volume of 100µl reaction mixture containing 15µg isolated retinal mitochondrial protein, 50mM Tris-HCl (pH 8.0), 100µM 5, 5-dithiobis (2-nitrobenzoic acid) (DTNB) and 300µM acetyl coenzyme A. The rate of the reduction of DTNB, initiated by oxaloacetate (500µM), was followed for 3 minutes at 412nm, and the enzyme activity was calculated using an extinction coefficient of E 13.6mM/cm.
Interactions between TFAM and HSP70
Retina (200µg protein) was incubated overnight at 4°C with 1µg of Hsp70 antibody (Santa Cruz Biotechnology) followed by 1 hour with 20µl of protein A and G Plus agarose immunoprecipitation beads reagent (prewashed and suspended in lysis buffer). The beads were washed four times with lysis buffer, and the proteins were separated by SDS-PAGE. The membranes were immunoblotted with anti-TFAM antibody (Santa Cruz Biotechnology).
Statistical analysis
Data are expressed as mean ±standard deviation. Statistical analysis was carried out using SPSS software. Shapiro-Wilk test were used to test for normal distribution of the data. For variables with normal distribution ANOVA followed by Bonferrone test were applied, while Kruskal-Wallis test followed by Mann-Whitney U-test were used for data which did not present normal distribution. *p value <0.05 was considered statistically significant.
Results
Effect of diabetes on mitochondria DNA damage and biogenesis in mouse retina
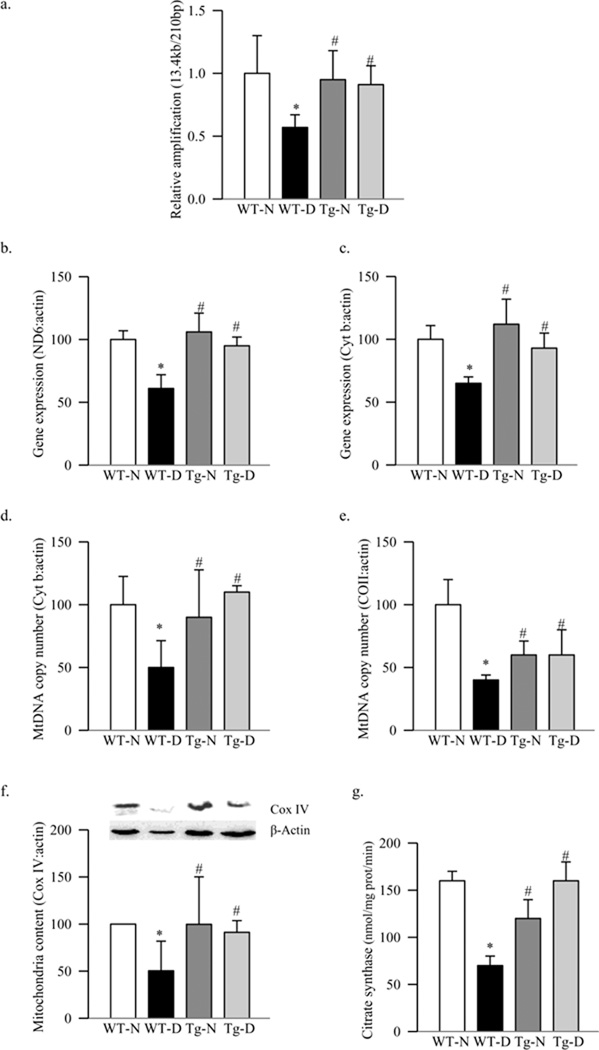
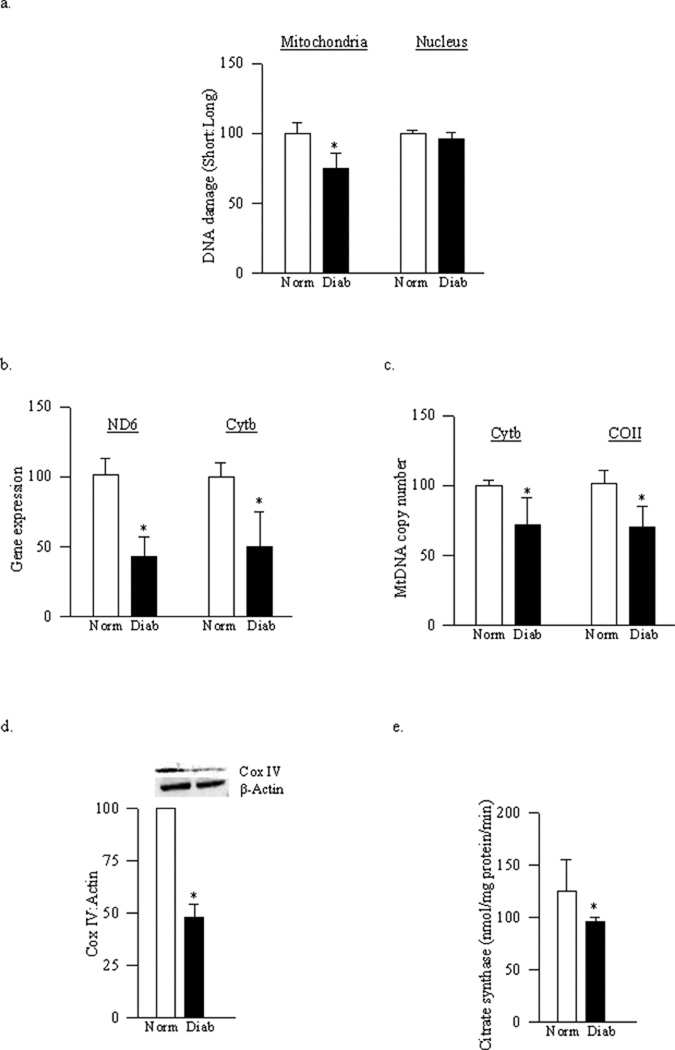
Mitochondrial DNA damage in the retina induced by diabetes was evaluated using extended length PCR with genome specific primers. Similar to the results obtained from rat retina (12), mouse retinal mtDNA was damaged in diabetes as confirmed by over 40% decrease in its amplification, and gene expressions of mtDNA encoded retinal proteins, ND6 and Cyt b, were decreased by 35–40% compared to the values obtained from age-matched WT-normal mice (Figure 1a–1c).
Figure 1.
Effect of diabetes on mitochondrial DNA damage and functional integrity. (a) DNA damage of retinal mitochondria was assessed using mitochondrial specific primers for long (13.4kb) and short (210bp) products. The relative amplification was calculated by normalizing the intensity 13.4kb and 210bp product within each sample. Transcript abundance of mitochondrial-encoded genes was assessed in DNase-treated RNA using conventional RT-PCR for (b) ND6 and (c) Cyt b using β-actin as housekeeping protein. MtDNA copy number was assessed in retinal DNA by real time RT-PCR using mitochondrial (COII and Cyt b) or nuclear specific (β-actin) mouse primers. Gene expressions of (d) Cyt b and (e) COII, adjusted to β-actin, were calculated within each sample. (f) Protein expression of Cox IV was measured in the homogenate and adjusted to the expression of β-actin in each sample (g) Citrate synthase activity was assessed in the isolated mitochondria following the reduction of DTNB. Values are represented as mean±SD obtained from 5–7 mice each in the four experimental groups: wild type diabetic (WT-D) and normal (WT-N), and MnSOD-Tg diabetic mice (Tg-D) normal (Tg-N) mice. *p< 0.05 compared to the values obtained from the WT-N group and #p< 0.05 compared to the values obtained from the WT-D group.
Mitochondria copy number, quantified by the ratio between mitochondrial genes (Cyt b or COII) and nuclear gene (β-actin), was decreased by about 50–60% in the retina from WT-diabetic mice compared to the WT-normal mice (Figures 1d–e). To further confirm the effect of diabetes on mitochondria biogenesis, the expression of the mitochondrial-specific protein, Cox IV, was determined. As shown in figure 1f, the expression of Cox IV, relative to that of β-actin, was decreased by about 50% in diabetes, suggesting decreased mitochondria biogenesis. In the same retina, mitochondrial function was also decreased as evidenced by 50% decrease in the activity of citrate synthase (Figure 1g).
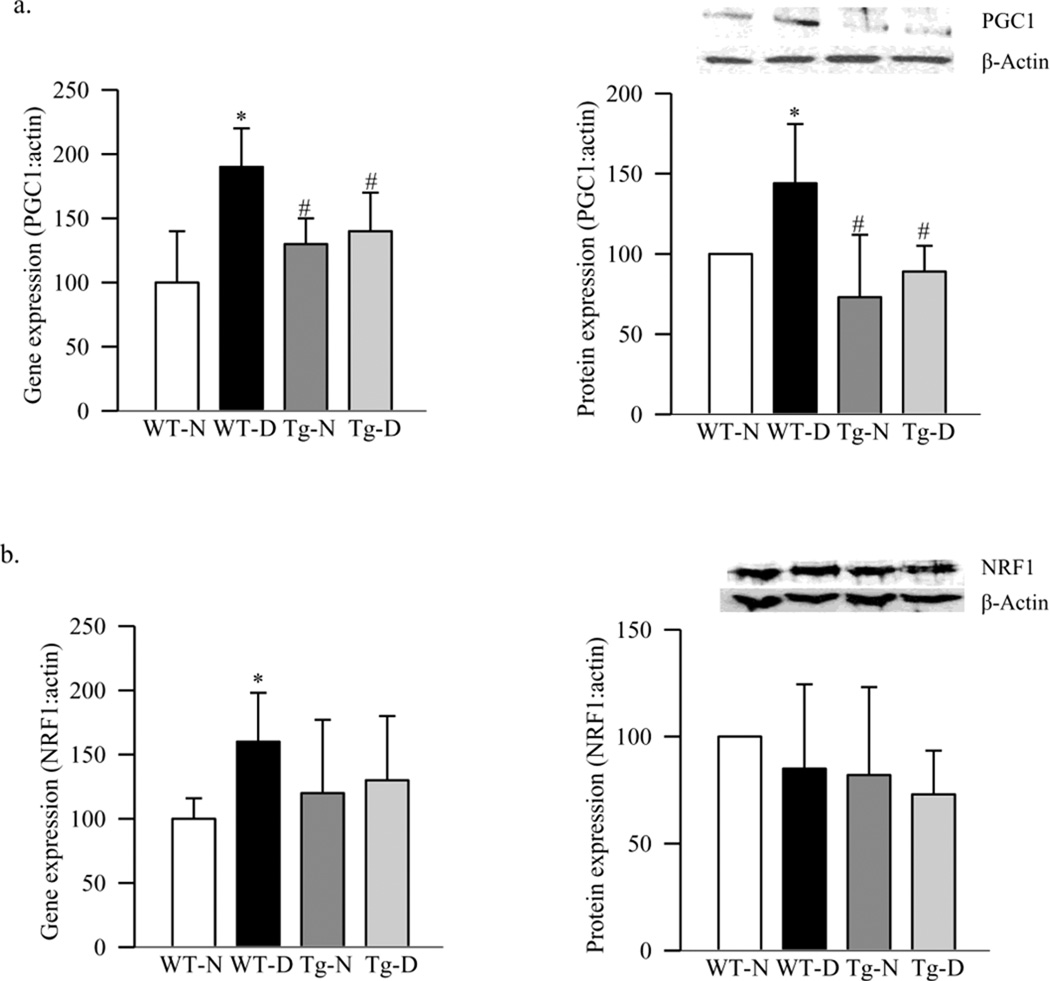
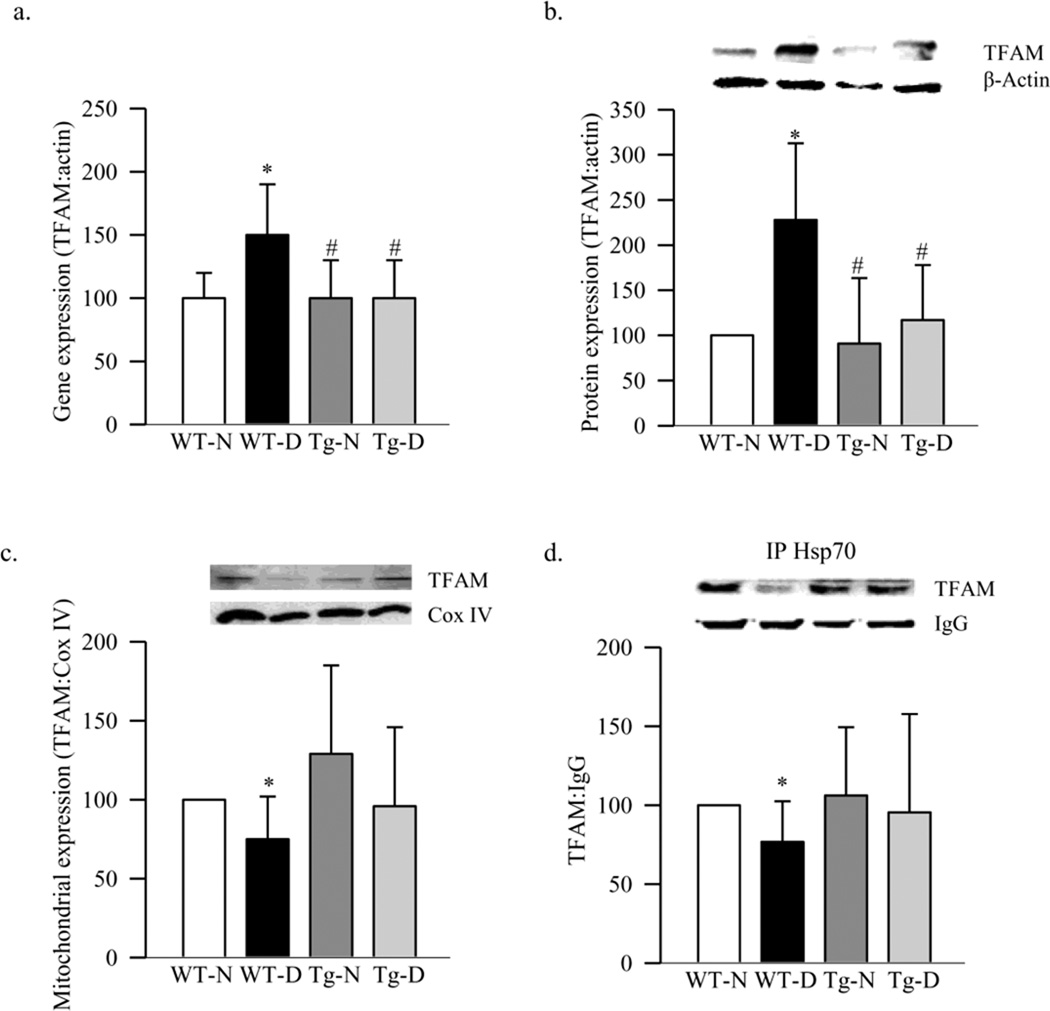
To address if the decrease in retinal mitochondria was due to impaired biogenesis, expressions of PGC1 and NRF1 were evaluated. Diabetes in WT mice increased the gene expressions of PGC1 by 80%, and its protein expression was increased by ~45% compared to the values obtained from WT-normal mice (Figure 2a). In the same retina, although the gene expression of NRF1 was increased by 60%, diabetes had no effect on its protein expression (Figure 2b). The gene expression of TFAM, a NRF1-regulated transcription factor important in the transcription and replication, was increased by 50% in diabetes (Figure 3a), and this was accompanied by a significant increase in its overall protein expression (Figure 3b). Since TFAM translocates to the mitochondria for its action, we quantified its expression in the mitochondria. As shown in figure 3c, protein accumulation of TFAM in the mitochondria was significantly decreased, suggesting an impaired subcellular translocation system. Translocation of TFAM to the mitochondria requires binding with chaperone proteins (18), and figure 3d shows that diabetes decreased interaction of retinal TFAM with Hsp70 by 25% compared to the results obtained from WT-normal mice.
Figure 2.
Effect of diabetes in PGC1 and NRF1 expression. RNA was quantified by real-time RT-PCR and protein expression by western blotting technique using β-actin as internal control; (a) PGC1 and (b) NRF1. Values are represented as mean±SD obtained from 5–7 animals in each group *p< 0.05 compared to the WT-N, and #p< 0.05 to WT-D.
Figure 3.
Effect of diabetes on TFAM and its binding with Hsp70. (a) Gene expression of TFAM was quantified by real time PCR using β-actin as internal standard. Protein expression of TFAM was measured in the (b) retinal homogenate and (c) mitochondria fraction by western blot technique using β-actin and Cox IV, respectively as loading controls. (d) For TFAM-Hsp70 interactions, Hsp70 was immunopreciptated in the retinal homogenate, and TFAM was detected in the immunoprecipitate by western blot. Values are represented as mean±SD from 4–7 animals in each group. *p< 0.05 compared to WT-N, and #p< 0.05 to WT-D
Effect of overexpression of MnSOD on mitochondria biogenesis
Since mtDNA is one of the major targets of superoxide (24–26), effect of inhibition of superoxide accumulation on retinal mtDNA and its biogenesis was investigated in mice overexpressing MnSOD. Our previous study has shown that the retina of Tg-diabetic mice is protected from increased superoxide levels, mitochondrial dysfunction and DNA damage, impairment in mitochondrial complex III activity, and the development of retinopathy (11, 13). Here we show that MnSOD overexpression protected diabetes-induced retinal mtDNA damage, and the gene expressions of ND6 and Cyt b, the proteins encoded by mtDNA, were significantly higher compared to the values obtained from WT-diabetic mice (Figures 1a–c). In the same Tg-diabetic mice, mtDNA copy number, mitochondria content and citrate synthase activity were also significantly higher (Figures 1d–g). This was accompanied by decreased transcripts of PGC1, NRF1 and TFAM, and increased accumulation of TFAM in the mitochondria compared to the values obtained from WT-diabetic mice, and these values were not different (p>0.05) from those obtained from Tg-normal mice (Figures 2a&b and 3a–c). Decreased interactions of TFAM with Hsp70, observed in WT-diabetic mice, were also ameliorated in Tg-diabetic mice (Figure 3d).
Mitochondrial biogenesis in retinal endothelial cells and regulation by MnSOD
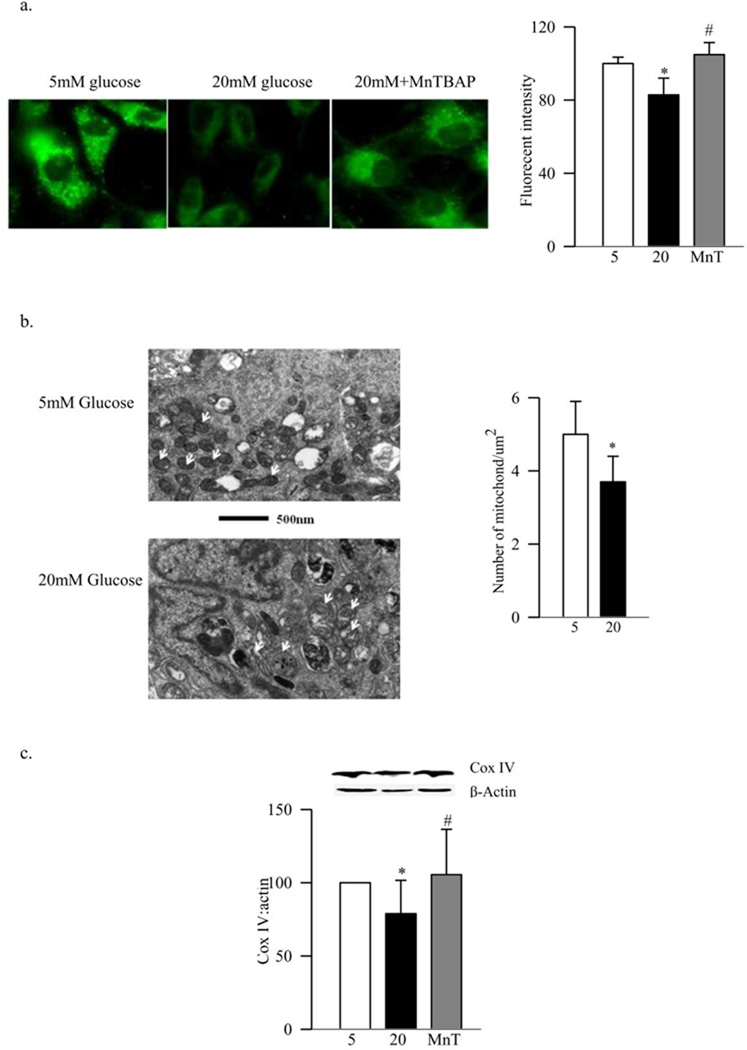
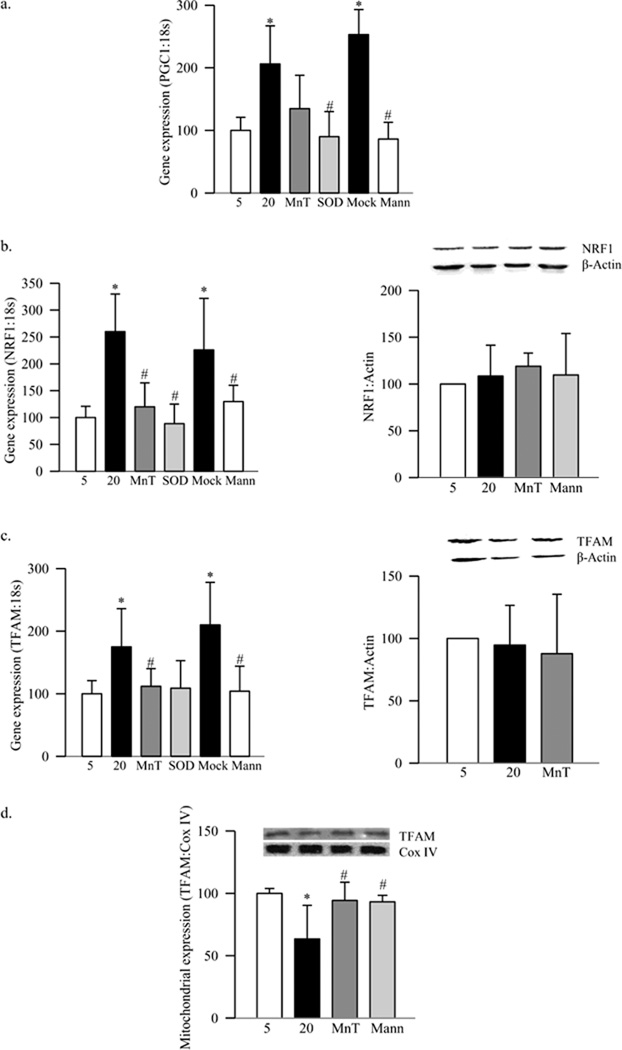
Our previous work has shown that high glucose exposure damages mtDNA in retinal endothelial cells (13). Consistent with this, high glucose decreased the fluorescent intensity of MitoTracker green, a dye that is insensitive to oxidative stress and membrane potential (27) (Figure 4a). Electron micrograph showed 25% less mitochondria in the cells exposed to high glucose compared to the cells exposed to normal glucose (Figure 4b), and the expression of Cox IV was decreased by over 20% (Figure 4c). In the same cells, gene expressions of PGC1 and NRF1 were increased by over 100% compared to the values obtained from cells incubated in normal glucose without any change in the protein expression of NRF1 (Figures 5a&b). Furthermore, gene expression of TFAM was increased by 70%, but its mitochondrial accumulation was decreased by 40% despite no change in its protein expression (Figure 5c–d). In contrast, incubation of BRECs in 20mM mannitol (instead of 20mM glucose) did not increase the gene expression of PGC1, NRF1 and TFAM, and the mitochondrial accumulation of TFAM remained normal, suggesting that the effects observed under high glucose conditions were not due to the increased osmolarity.
Figure 4.
Effect of high glucose on mitochondria mass and Cox IV expression. Mitochondria number was quantified (a) using MitoTracker Green. The cells were stained with MitoTracker Green, and examined under a Zeiss ApoTome using 40× magnification, and the fluorescence was quantified using Image J software. Each measurement was made in triplicate using 3–4 different cell preparations. (b) TEM was performed on ultrathin sections of the cells embedded in 812 resin. Mitochondria number was counted manually in a blind manner by calculating the number of mitochondria over total area of the cell in at least 8–10 images/sample. Each group had 3–4 different samples. (c) The expression of Cox IV was determined in the cell homogenate by western blot technique, and adjusted to that of β-actin. Each measurement was made in duplicate in 3–5 preparations. The values are represented as mean±SD. 5=5mM glucose, 20=20mM glucose, MnT=20mM glucose+200µM MnTBAP
Figure 5.
Effect of high glucose on mitochondria biogenesis in retinal endothelial cells. RNA isolated from retinal endothelial cells was assessed by real-time RT-PCR for (a) PGC1 (b) NRF1 and (c) TFAM. The values were normalized to 18s rRNA in each sample. Percentage relative to 5mM glucose treatment was calculated using the ddCt method. Protein expression was determined in the cell homogenate by western blot technique using β-actin as loading control. (d) Mitochondrial accumulation of TFAM was assessed by quantifying its expression in the isolated mitochondria using Cox IV as loading control. Each measurement was made in duplicate in at least 3 different cell preparations, and the values are represented as mean±SD. 5=5mM glucose, 20=20mM glucose, MnT=20mM glucose+200µM MnTBAP, SOD=cells transfected with MnSOD, Mock=cells treated with the transfection reagent alone and Mann=20mM mannitol. *p< 0.05 compared to the values from 5mM glucose, and #p< 0.05 to 20mM glucose.
Inhibition of glucose-induced increase in superoxide levels by MnSOD (either by its mimic or by its plasmid) prevented glucose-induced decrease in mitochondria biogenesis. MnTBAB inhibited glucose-induced decrease in the fluorescent intensity of MitoTracker green and expression of Cox IV (Figure 4a&c). Furthermore, increases in gene expressions of the transcriptional factors, PGC1, NRF1 and TFAM, observed in high glucose conditions, were ameliorated by both MnTBAP and also by overexpression of MnSOD (Figures 5a–c). Regulation of superoxide also inhibited glucose-induced increased TFAM gene expression and its decreased mitochondrial accumulation (Figure 5c&d). The values obtained from the cells incubated with transfection reagent alone (without MnSOD plasmid), and exposed to high glucose (Mock), were similar to those obtained from the untransfected cells exposed to high glucose, but were significantly different from the untransfected cells exposed to normal glucose (Figures 5).
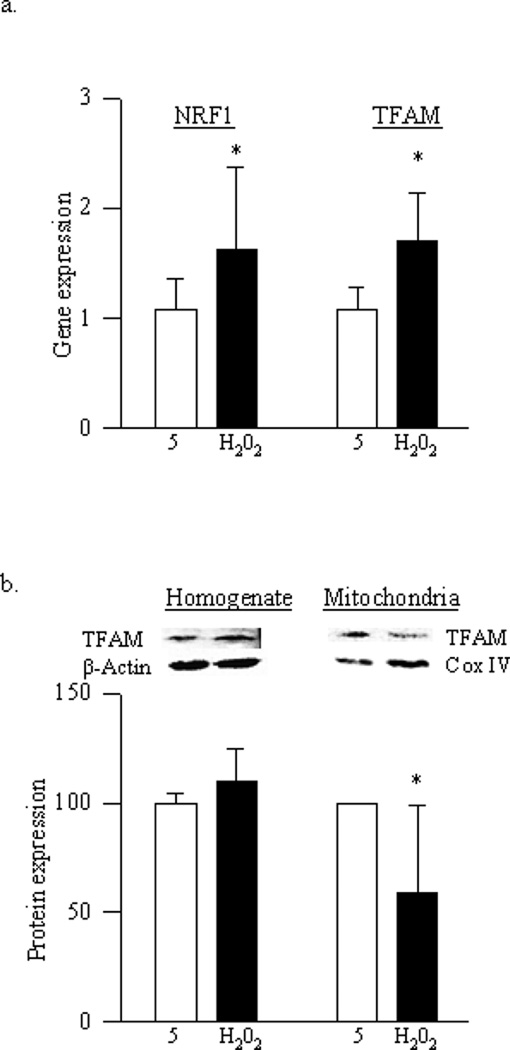
As with high glucose, exposure of cells to H2O2 elevated gene expressions of NRF1 and TFAM, however, mitochondrial accumulation of TFAM remained compromised (Figure 6a&b), suggesting that these transcriptional factors are under the control of oxidative stress.
Figure 6.
Effect of H2O2 on mitochondria biogenesis: BRECs were incubated with 250µM H2O2 for 1 hour, and after washing the cells with DMEM, they were incubated in 5mM glucose for 4 additional days. (a) Gene expression of NRF1 and TFAM was quantified by real time PCR. (b) Protein expression of TFAM was quantified in the cell homogenate and also in the mitochondrial fraction by western blot technique using β-actin (homogenate) and Cox IV (mitochondria) as loading control. Each measurement was made in duplicate in at least 3–4 different cell preparations. 5=5mM glucose and H2O2=cells incubated with H2O2; *p<0.05 compared to 5mM glucose.
Human retina
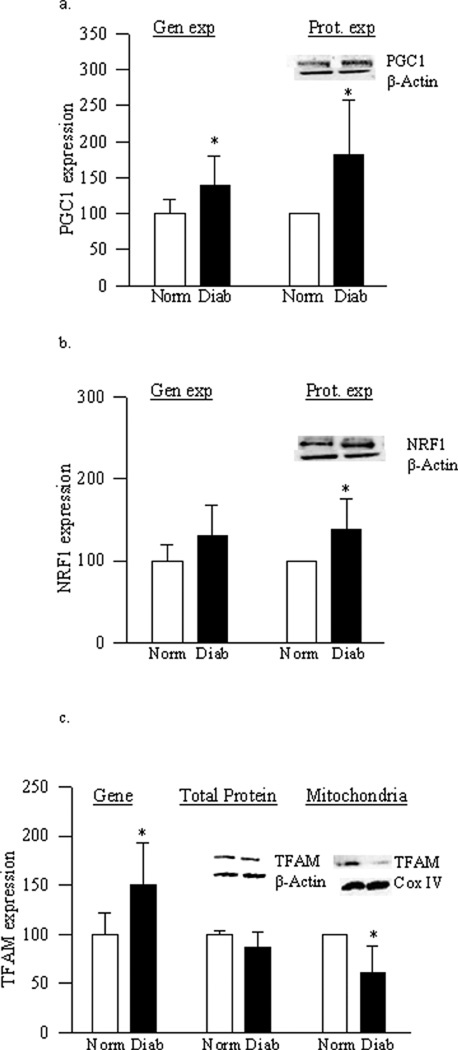
Retinal mtDNA was damaged by 25% in the donors with diabetic retinopathy (Figure 7a), and the gene expression of the proteins encoded by mtDNA, ND6 and Cyt b, were also decreased by 50–60% compared to the values from non-diabetic donors (Figure 7b). This was accompanied by significant decrease ratios of Cyt b/COII to β-actin (Figure 7c), and decreased expression of Cox IV (Figure 7d), suggesting decrease in the copy numbers. In addition, citrate synthase activity was also subnormal (Figure 7e). As with diabetic mice and retinal capillary cells, mitochondria biogenesis was also impaired in the retina of donors with diabetic retinopathy. The gene expressions of retinal PGC1, NRF1 and TFAM in the retina of donors with diabetic retinopathy were increased, but the overall protein expression of TFAM was similar in the donors with diabetic retinopathy and age-matched non diabetic donors (Figures 8a–c). Consistent with the results from mouse retina and from BRECs, TFAM accumulation in the mitochondria remained subnormal compared to the values from the retina obtained from age-matched normal donors (Figure 8c).
Figure 7.
Retinal mtDNA damage and mitochondrial functional integrity in donors with diabetic retinopathy. (a) Mitochondrial DNA damage was assessed using mitochondrial and nucleus specific primers for long (13.4kb) and short (210bp) products. (b) Transcript abundance of mitochondria-encoded genes ND6 and Cyt b was assessed in DNase-treated RNA by real time RT-PCR using 18s rRNA as housekeeping gene. (c) Mitochondria copy number was assessed in retinal DNA using mitochondrial (COII and Cyt b) or nuclear specific (β-actin) human primers by real time RT-PCR, and the ratios of COII or Cyt b, with β-actin were calculated within each sample. (d) The expression of Cox IV was determined in the retinal homogenate by western blot technique and was adjusted to that of β-actin. (e) Citrate synthase activity was assessed in the isolated mitochondria fraction by following the reduction of DTNB. Values are represented as mean±SD obtained from non diabetic donors (Norm), and donors with diabetic retinopathy (Diab). *p< 0.05 compared to non diabetic donors
Figure 8.
Mitochondria biogenesis in donors with diabetic retinopathy. Gene transcript of (a) PGC1, (b) NRF1 and (c) TFAM was quantified by real-time RT-PCR, and protein expression by western blotting technique using β-actin as internal control. TFAM mitochondrial accumulation was quantified in the mitochondria fraction, and Cox IV was used as a loading control. Results are represented as mean±SD. *p< 0.05 compared to non diabetic donors.
Discussion
Mitochondrial superoxide production is considered as a single unifying mechanism for diabetic complications (7). In the pathogenesis of diabetic retinopathy, superoxide levels are elevated in the retina and its capillary cells, mitochondria become dysfunctional, the activity of complex III of the electron transport chain becomes subnormal, and superoxide scavenging enzyme is decreased. Furthermore, mtDNA is damaged, and regulation of superoxide radicals by overexpression of MnSOD prevents mitochondrial dysfunction and the development of retinopathy in diabetic mice (11–13, 28). Since mtDNA maintenance depends on coordinated expression of nucleus and mitochondria genes responsible for mitochondria biogenesis, we provide a mechanism responsible for mtDNA damage and dysfunction. Our results show that diabetes decreases mitochondria copy number and mass in the retina, and their functional integrity is compromised. To overcome DNA damage and protect the integrity, transcription factors associated with nucleus-mitochondria signaling pathway responsible for mitochondria biogenesis are increased. However, transport of TFAM, which is the main activator of mitochondrial transcription, from nucleus to the mitochondria remains compromised, resulting in decreased mitochondria copy number and the proteins encoded by this DNA. One of the possible reasons for decreased accumulation of TFAM appears to be impaired interactions with the key chaperone Hsp70. Inhibition of superoxide radicals by overexpression of MnSOD, which prevents apoptosis of retinal capillary cells and the development of diabetic retinopathy (11), ameliorates abnormalities of retinal mitochondria biogenesis, suggesting that mitochondria biogenesis is under the control of superoxide and is an important component in the development of diabetic retinopathy. Similar high glucose-induced impaired biogenesis in the mitochondria of retinal endothelial cells, the site of histopathology associated with diabetic retinopathy, and also in the retina from donors with diabetic retinopathy, further supports the crucial role of mitochondria biogenesis in diabetic retinopathy.
Our results show that mitochondria copy number is decreased and the functional integrity is impaired in the retina and its capillary cells in diabetes. Regulation of mitochondrial biogenesis is considered essential for proper cellular functioning, and with aging, the copy number decreases (29). Decreased mtDNA copy number is reported in other chronic diseases, such as multiple sclerosis, type 2 diabetes and cardiomyopathy (30, 31). Since mitochondria biogenesis directly controls the copy number (30), and as presented here, this biogenesis machinery is impaired in diabetes. The possibility that the decreased retinal mitochondria copy number in diabetes, however, could be due to DNA mutation, cannot be ruled out.
Mitochondria biogenesis is a tightly regulated process which is controlled by the nuclear genome (32). Regulation of mitochondrial biogenesis is initiated by a transcription factor coactivator, PGC1, which promotes the transcription of NRF1, and binds to NRF1 to increase the transcription of enzymes necessary for electron transport and ATP synthesis (16, 17). Others have shown a tissue-specific effect of diabetes on the mitochondrial biogenesis pathway; while chronic hyperglycemia decreases PGC1, NRF1 and TFAM in skeletal muscle, hepatocytes and heart (33–35), these factors are increased in brain (35, 36). Our results clearly show that, despite decreased mitochondria copy number in diabetes, the transcripts of retinal PGC1 and NRF1 are significantly increased, and suggest that in order to overcome mitochondrial dysfunction in diabetes, the retina starts to increase the transcripts of PGC1 and NRF1, but due to dysfunctional transport mechanism, the biogenesis remains impaired.
NRF1, in its inert form, is localized in the cytosol, but when activated, it translocates to the nucleus to coordinate the expression of several nucleus-encoded mitochondrial proteins (37, 38). Here we show that diabetic environment induces increase in retinal NRF1 gene transcripts, and this could be important in initiating the expression of other nucleus-encoded mitochondrial protein. In support, despite continued damaged retinal mtDNA in diabetes, the gene transcripts of the DNA repair enzymes remain significantly elevated (12).
TFAM is a transcription factor important in mitochondria biogenesis as it initiates transcription of mtDNA (18), and in addition to the DNA packaging function, it plays a direct role in replicating mtDNA (39). TFAM is also a key regulator of mtDNA copy number, which is controlled by directly binding and stabilizing of mtDNA. Its levels are closely associated with the mtDNA content and with the proteins encoded by mtDNA. Depletion of TFAM has been linked with decreased mtDNA, mtDNA-encoded proteins and oxidative phosphorylation capacity (39–42). Our results demonstrate that in diabetic conditions, despite increased gene expression of TFAM in the retina and its capillary cells, mitochondria copy number is decreased. TFAM protein is synthesized in the nucleus, but the major site of its action is the mitochondria (32, 40), and we show that its abundance in the mitochondria is subnormal. This suggests that despite the cell’s efforts to produce more TFAM to overcome impairments in mitochondria biogenesis experienced in diabetic milieu, it fails to reach its target-the mitochondria, resulting in a compromised mitochondria biogenesis and decreased copy number.
Since TFAM is nuclear encoded, and has be translocated to the mitochondria via translocase of the outer membrane and of the inner membrane complexes (TOM-TIM complexes), to understand the failure of mitochondrial accumulation of TFAM, we investigated its transport mechanism to the mitochondria (43). Interactions between the proteins incoming to the mitochondria and Hsps are critical in translocation reactions, and chaperone protein Hsp70 docking to the TOM receptor is important in pre-protein recognition (44, 45). Decreased binding between TFAM and Hsp70 suggests that there could be a failure of the transporter system to interact efficiently with TFAM to transport it to the mitochondria, the site of its action. The role of other members of the chaperon family and transporters, including TIM/TOM complex, in decreased mitochondria biogenesis in diabetes, however, cannot be ruled out.
Retina and its capillary cells experience increased oxidative damage in diabetes, and antioxidant defense mechanism is impaired (2–4, 7, 8). Retinal mitochondria become dysfunctional and start to leak cytochrome c into the cytosol, and superoxide levels are elevated (4, 8). Our results using H2O2 to induce oxidative stress clearly show that, despite increased gene expression of TFAM, its mitochondrial accumulation remains compromised, suggesting that the transport of TFAM to the mitochondria is under the direct control of superoxide. MnSOD overexpression prevents diabetes-induced decrease in copy number and functional integrity of the mitochondria. In addition, mitochondria biogenesis machinery is also protected, the transcripts of PGC1, NRF1 and TFAM remain similar to WT-non diabetic and Tg-non diabetic groups, and mitochondrial accumulation of TFAM becomes normal, suggesting that mitochondria biogenesis is under the control of mitochondrial superoxide production. In support, regulation of mitochondrial superoxide generation by overexpression of MnSOD or by its mimic protects increased accumulation of superoxide, mitochondrial dysfunction, mtDNA damage and also the development of retinopathy in diabetic mice suggesting a major role of mitochondrial superoxide in the development of diabetic retinopathy (11, 13).
In the pathogenesis of diabetic retinopathy, the microvasculature of the retina is the site of pathology associated with the disease. Because of the complexity of the retinal structure with multiple layers and cell types, key experiments were confirmed in isolated retinal endothelial cells, the major targets of the histopathology characteristic of diabetic retinopathy (46). Similar abnormalities in mitochondria biogenesis, with decreased mitochondria copy number and overall number of mitochondria in high glucose conditions, and protection of these abnormalities by overexpression of MnSOD in these cells further confirms the role of mitochondria biogenesis in the development of diabetic retinopathy.
Effect of diabetes on mitochondria biogenesis was also confirmed in the retina from diabetic donors with documented retinopathy. Our recent studies have shown that the similar H-Ras signaling cascade to activate matrix metalloproteinase-9 is operational in the retinal microvessels obtained from donors with diabetic retinopathy (47). Similar mtDNA damage, decrease in copy number, impairment in the functional integrity inhibition, and decreased mitochondrial accumulation of TFAM despite increased gene expressions of the transcriptional factors in the retina from donors with diabetic retinopathy further supports the importance of mitochondria biogenesis in the development of diabetic retinopathy.
We recognize that the mitochondria mass is regulated by the dynamic interplay between biogenesis and the rate of removal by autophagy, and autophagy is also regulated by mitochondrial ROS (48, 49). The contribution of autophagy in regulating mitochondrial copy number in the pathogenesis of diabetic retinopathy is unclear and is the subject of a further study.
In summary, here we show for the first time that in the pathogenesis of diabetic retinopathy, retinal mitochondria biogenesis is impaired and the number of mitochondria is decreased, and impaired transport of the transcription factor important for initiation of transcription of mtDNA, TFAM, to the mitochondria appears to be one of the possible mechanisms. Mitochondria biogenesis machinery is under the control of superoxide, and has an important role in the development of diabetic retinopathy. Modulation of mitochondria biogenesis by pharmaceutical or molecular means may provide a potential strategy to retard the development/progression of diabetic retinopathy.
Highlights.
In the pathogenesis of diabetic retinopathy, retinal mitochondria biogenesis is impaired and the number of mitochondria is decreased.
Decreased transport of the transcription factor important for initiation of transcription of mtDNA, TFAM, to the mitochondria appears to be responsible for impaired mitochondria biogenesis.
Regulation of oxidative stress protects the retina from diabetes-induced abnormalities in mitochondria biogenesis.
Acknowledgements
Authors thank Gulrez Mahmood, Doug Putt, Yakov Shamailov and Loan Dang for technical help. This study was supported in part by grants to R.A.K. from the National Institutes of Health, the Juvenile Diabetes Research Foundation, the Thomas Foundation, Research to Prevent Blindness and Midwest Eye Banks, and to A.F.X.G. from the National Center for Research Resources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Frank RN. Diabetic retinopathy. N Engl J Med. 2004;350:48–58. doi: 10.1056/NEJMra021678. [DOI] [PubMed] [Google Scholar]
- 2.Mizutani M, Kern TS, Lorenzi M. Accelerated death of retinal microvascular cells in human and experimental diabetic retinopathy. J Clin Invest. 1996;97:2883–2890. doi: 10.1172/JCI118746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barber AJ, Lieth E, Khin SA, Antonetti DA, Buchanan AG, Gardner TW. Neural apoptosis in the retina during experimental and human diabetes. Early onset and effect of insulin. J Clin Invest. 1998;102:783–791. doi: 10.1172/JCI2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kern TS, Tang J, Mizutani M, Kowluru R, Nagraj R, Lorenzi M. Response of capillary cell death to aminoguanidine predicts the development of retinopathy: Comparison of diabetes and galactosemia. Invest Ophthalmol Vis Sci. 2000;41:3972–3978. [PubMed] [Google Scholar]
- 5.Kern TS, Engerman RL. Pharmacological inhibition of diabetic retinopathy: aminoguanidine and aspirin. Diabetes. 2001;50:1636–1642. doi: 10.2337/diabetes.50.7.1636. [DOI] [PubMed] [Google Scholar]
- 6.Kowluru RA, Tang J, Kern TS. Abnormalities of retinal metabolism in diabetes and experimental galactosemia. VII. Effect of long-term administration of antioxidants on the development of retinopathy. Diabetes. 2001;50:1938–1942. doi: 10.2337/diabetes.50.8.1938. [DOI] [PubMed] [Google Scholar]
- 7.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54:1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- 8.Santos JM, Mohammad G, Zhong Q, Kowluru RA. Diabetic retinopathy, superoxide damage and antioxidant. Curr Pharm Biotechnol. 2011;12:352–361. doi: 10.2174/138920111794480507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowluru RA. Diabetic retinopathy: Mitochondrial dysfunction and retinal capillary cell death. Antioxidants & Redox Signaling. 2005;7:1581–1587. doi: 10.1089/ars.2005.7.1581. [DOI] [PubMed] [Google Scholar]
- 10.Kowluru RA, Atasi L, Ho YS. Role of mitochondrial superoxide dismutase in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2006;47:1594–1599. doi: 10.1167/iovs.05-1276. [DOI] [PubMed] [Google Scholar]
- 11.Kanwar M, Chan PS, Kern TS, Kowluru RA. Oxidative damage in the retinal mitochondria of diabetic mice: possible protection by superoxide dismutase. Invest Ophthalmol Vis Sci. 2007;48:3805–3811. doi: 10.1167/iovs.06-1280. [DOI] [PubMed] [Google Scholar]
- 12.Madsen-Bouterse SA, Mohammad G, Kanwar M, Kowluru RA. Role of mitochondrial DNA damage in the development of diabetic retinopathy, and the metabolic memory phenomenon associated with its progression. Antioxidant & Redox Signaling. 2010;13:797–805. doi: 10.1089/ars.2009.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madsen-Bouterse S, Zhong Q, Mohammad G, Ho YS, Kowluru RA. Oxidative damage of mitochondrial DNA in diabetes, and its protection by manganese superoxide dismutase. Free Rad Research. 2010;44:313–321. doi: 10.3109/10715760903494168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowluru RA, Kowluru V, Ho YS, Xiong Y. Overexpression of mitochondrial superoxide dismutase in mice protects the retina from diabetes-induced oxidative stress. Free Rad Biol Med. 2006;41:1191–1196. doi: 10.1016/j.freeradbiomed.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 16.Dhar SS, S. O, Wong-Riley MT. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem. 2008;283:3120–3120. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen JQ, Cammarata PR, Baines CP, Yager JD. Regulation of mitochondrial respiratory chain biogenesis by estrogens/estrogen receptors and physiological, pathological and pharmacological implications. Biochim Biophys Acta. 2010;1793:1540–1570. doi: 10.1016/j.bbamcr.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scarpulla RC. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann N Y Acad Sci. 2008;1147:321–334. doi: 10.1196/annals.1427.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohammad G, Kowluru RA. Matrix metalloproteinase-2 in the development of diabetic retinopathy and mitochondrial dysfunction. Lab Invest. 2010;90:1365–1372. doi: 10.1038/labinvest.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowluru V, Kowluru RA. Increased oxidative stress in diabetes regulates activation of a small molecular weight G-protein, H-Ras, in the retina. Mol Vis. 2007;13:602–610. [PMC free article] [PubMed] [Google Scholar]
- 21.Kowluru RA, Kanwar M, Chan PS, Zhang JP. Inhibition of retinopathy and retinal metabolic abnormalities in diabetic rats with AREDS-based micronutrients. Arch Ophthalmol. 2008;126:1266–1272. doi: 10.1001/archopht.126.9.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayala-Torres S, Chen Y, Svoboda T, Rosenblatt J, Van Houten B. Analysis of gene-specific DNA damage and repair using quantitative polymerase chain reaction. Methods. 2000;22:135–147. doi: 10.1006/meth.2000.1054. [DOI] [PubMed] [Google Scholar]
- 23.Parra V, Eisner V, Chiong M, Criollo A, Moraga F, Garcia A, Härtel S, Jaimovich E, Zorzano A, Hidalgo C, Lavandero S. Changes in mitochondrial dynamics during ceramide-induced cardiomyocyte early apoptosis. Cardiovasc Res. 2008;77:387–397. doi: 10.1093/cvr/cvm029. [DOI] [PubMed] [Google Scholar]
- 24.Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci U S A. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarrett SG, Lin H, Godley BF, Boulton ME. Mitochondrial DNA damage and its potential role in retinal degeneration. Prog Retin Eye Res. 2008;27:596–607. doi: 10.1016/j.preteyeres.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Jarrett SG, Albon J, Boulton M. The contribution of DNA repair and antioxidants in determining cell type-specific resistance to oxidative stress. Free Radic Res. 2006;40:1155–1165. doi: 10.1080/10715760600876613. [DOI] [PubMed] [Google Scholar]
- 27.Pendergrass W, Wolf N, Poot M. Cytometry Part A. 2004 October;Volume 61A(Issue 2):162–169. doi: 10.1002/cyto.a.20033. [DOI] [PubMed] [Google Scholar]
- 28.Kowluru RA, Abbas SN. Diabetes-induced mitochondrial dysfunction in the retina. Inves Ophthalmol Vis Sci. 2003;44:5327–5334. doi: 10.1167/iovs.03-0353. [DOI] [PubMed] [Google Scholar]
- 29.Laderman KA, Penny JR, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-dependent functional alterations of mitochondrial DNA (mtDNA) from human fibroblasts transferred into mtDNA-less cells. J Biol Chem. 1996;271:15891–15897. doi: 10.1074/jbc.271.27.15891. [DOI] [PubMed] [Google Scholar]
- 30.Clay Montier LL, Deng JJ, Bai Y. Number matters: control of mammalian mitochondrial DNA copy number. J. Genet Genomics. 2009;36:125–131. doi: 10.1016/S1673-8527(08)60099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian W, Van Houten B. Alterations in bioenergetics due to changes in mitochondrial DNA copy number. Methods. 2010;51:452–457. doi: 10.1016/j.ymeth.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 33.Palmeira CM, Rolo AP, Berthiaume J, Bjork JA, Wallace KB. Hyperglycemia decreases mitochondrial function: the regulatory role of mitochondrial biogenesis. Toxicol Appl Pharmacol. 2007;225:214–220. doi: 10.1016/j.taap.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 34.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bugger H, Chen D, Riehle C, Soto J, Theobald HA, Hu XX, et al. Tissue-specific remodeling of the mitochondrial proteome in type 1 diabetic akita mice. Diabetes. 2009;58:1986–1997. doi: 10.2337/db09-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edwards JL, Quattrini A, Lentz SI, Figueroa-Romero C, Cerri F, Backus C, et al. Diabetes regulates mitochondrial biogenesis and fission in mouse neurons. Diabetologia. 2010;160:160–169. doi: 10.1007/s00125-009-1553-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piantadosi CA, Suliman HB. Mitochondrial transcription factor A induction by redox activation of nuclear respiratory factor 1. J Biol Chem. 2006;281 doi: 10.1074/jbc.M508805200. 324-233. [DOI] [PubMed] [Google Scholar]
- 38.Suliman HB, Carraway MS, Tatro LG, Piantadosi CA. A new activating role for CO in cardiac mitochondrial biogenesis. J Cell Sci. 2006;120:299–308. doi: 10.1242/jcs.03318. [DOI] [PubMed] [Google Scholar]
- 39.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 40.Mazzanti R, Giulivi C. Coordination of nuclear- and mitochondrial-DNA encoded proteins in cancer and normal colon tissues. Biochim Biophys Acta. 2006;1757:618–623. doi: 10.1016/j.bbabio.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Wang J, Wilhelmsson H, Hansson A, Thoren P, Duffy J, et al. Genetic modification of survival in tissue-specific knockout mice with mitochondrial cardiomyopathy. Proc Natl Acad Sci U S A. 2000;97:3467–3472. doi: 10.1073/pnas.97.7.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mihara K, Omura T. Cytoplasmic chaperones in precursor targeting to mitochondria: the role of MSF and hsp 70. Trends Cell Biol. 1996;6:104–108. doi: 10.1016/0962-8924(96)81000-2. [DOI] [PubMed] [Google Scholar]
- 44.Voos W, Röttgers K. Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim Biophys Acta. 2002;159:51–62. doi: 10.1016/s0167-4889(02)00264-1. [DOI] [PubMed] [Google Scholar]
- 45.Young JC, Hoogenraad NJ, Hartl FU. Chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- 46.Kern TS, Kowluru R, Engerman RL. Dog and rat models of diabetic retinopathy. In: Shafrir E, editor. Lessons from Animal Diabetes. London: Smith-Gordon; 1996. pp. 395–408. [Google Scholar]
- 47.Mohammad G, Kowluru RA. Novel role of mitochondrial matrix metalloproteinase-2 in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2011;52:3832–3841. doi: 10.1167/iovs.10-6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17:422–427. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 49.Romanello V, Sandri M. Mitochondrial biogenesis and fragmentation as regulators of muscle protein degradation. Curr Hypertens Rep. 2010;12:433–439. doi: 10.1007/s11906-010-0157-8. [DOI] [PubMed] [Google Scholar]