Abstract
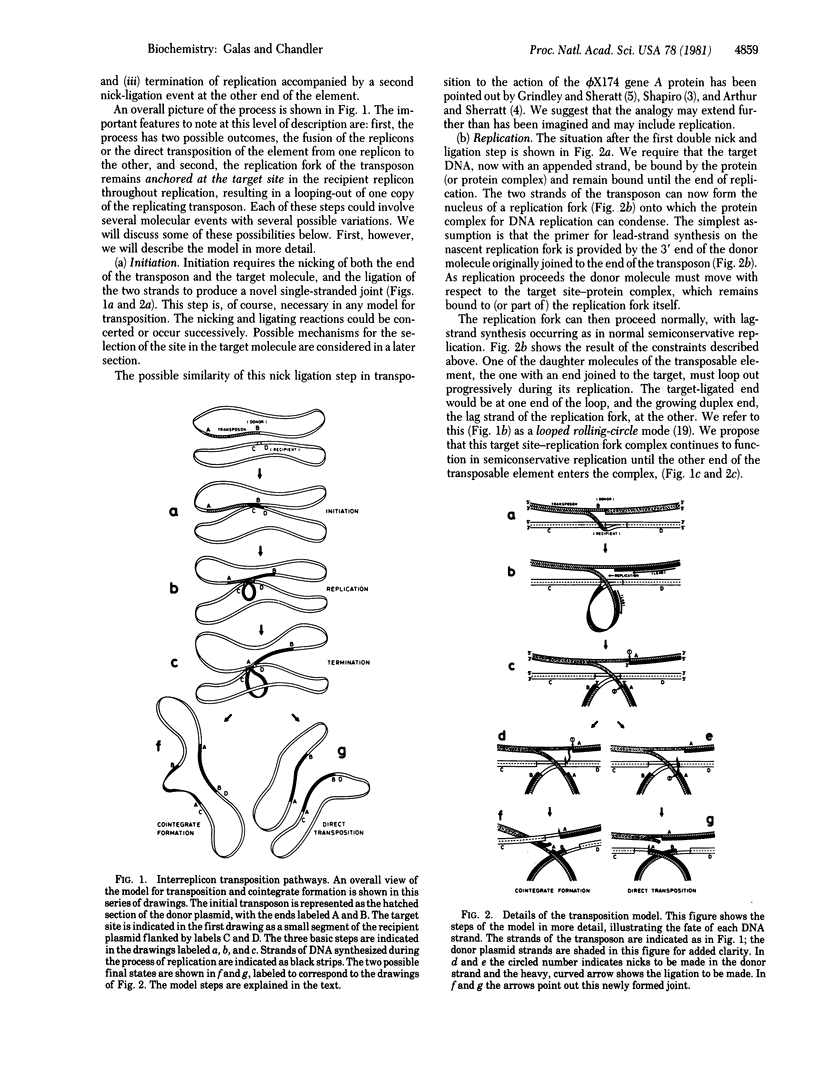
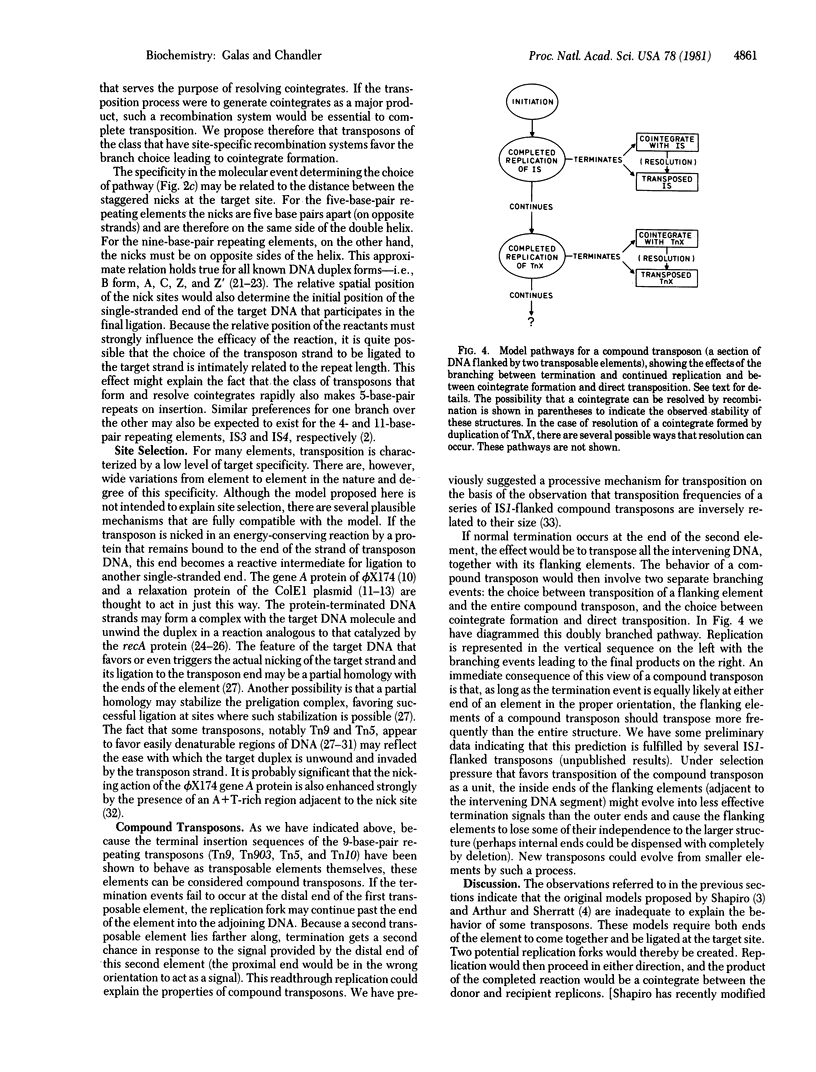
We present a model for transposition that allows a choice between cointegrate formation (replicon fusion) and direct transposition. We propose that initiation of the process occurs by invasion of the target DNA by a single-stranded end of the transposable element. This leads to nicking of one of the DNA strands of the target molecule and ligation of this strand to that of the invading transposon. Transposition then occurs in a processive way by replication of the element from the invading end into the target site in a looped rolling-circle mode similar to replication of phage phi X174 replicative form to viral strand. The choice between cointegrate formation and direct transposition occurs at the nick-ligation step, which terminates the process. We suggest that the choice is determined by the topology of the transposition enzymes and could be related to whether the element generates five- or nine-base-pair repeats in the target DNA on insertion.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Birdsall D. L., Leslie A. G., Ratliff R. L. Left-handed DNA helices. Nature. 1980 Feb 21;283(5749):743–745. doi: 10.1038/283743a0. [DOI] [PubMed] [Google Scholar]
- Arthur A., Sherratt D. Dissection of the transposition process: a transposon-encoded site-specific recombination system. Mol Gen Genet. 1979 Oct 1;175(3):267–274. doi: 10.1007/BF00397226. [DOI] [PubMed] [Google Scholar]
- Broome-Smith J. RecA independent, site-specific recombination between ColE1 or ColK and a miniplasmid they complement for mobilization and relaxation: implications for the mechanism of DNA transfer during mobilization. Plasmid. 1980 Jul;4(1):51–63. doi: 10.1016/0147-619x(80)90082-7. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Chaconas G., Harshey R. M., Sarvetnick N., Bukhari A. I. Mechanism of bacteriophage Mu DNA transposition. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):311–322. doi: 10.1101/sqb.1981.045.01.043. [DOI] [PubMed] [Google Scholar]
- Chandler M., Clerget M., Caro L. IS1-promoted events associated with drug-resistance plasmids. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):157–165. doi: 10.1101/sqb.1981.045.01.025. [DOI] [PubMed] [Google Scholar]
- Coelho A., Leach D., Maynard-Smith S., Symonds N. Transposition studies using a ColE1 derivative carrying bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):323–328. doi: 10.1101/sqb.1981.045.01.045. [DOI] [PubMed] [Google Scholar]
- Devos R., Contreras R., van Emmelo J., Fiers W. Identification of the translocatable element IS1 in a molecular chimera constructed with plasmid pBR322 DNA into which a bacteriophage MS2 DNA copy was inserted by the poly(dA).poly(dT) linker method. J Mol Biol. 1979 Mar 15;128(4):621–632. doi: 10.1016/0022-2836(79)90296-1. [DOI] [PubMed] [Google Scholar]
- Drew H., Takano T., Tanaka S., Itakura K., Dickerson R. E. High-salt d(CpGpCpG), a left-handed Z' DNA double helix. Nature. 1980 Aug 7;286(5773):567–573. doi: 10.1038/286567a0. [DOI] [PubMed] [Google Scholar]
- Eisenberg S., Denhardt D. T. Structure of nascent phiX174 replicative form: evidence for discontinuous DNA replication. Proc Natl Acad Sci U S A. 1974 Mar;71(3):984–988. doi: 10.1073/pnas.71.3.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N. V. Structure of deletion derivatives of a recombinant plasmid containing the transposable element Tn9 in the spacer sequence of Xenopus laevis 5S DNA. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1287–1292. doi: 10.1101/sqb.1979.043.01.146. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Calos M. P., Miller J. H. Sequence analysis of Tn9 insertions in the lacZ gene. J Mol Biol. 1980 Nov 25;144(1):19–41. doi: 10.1016/0022-2836(80)90213-2. [DOI] [PubMed] [Google Scholar]
- Gill R., Heffron F., Dougan G., Falkow S. Analysis of sequences transposed by complementation of two classes of transposition-deficient mutants of Tn3. J Bacteriol. 1978 Nov;136(2):742–756. doi: 10.1128/jb.136.2.742-756.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Joyce C. M. Analysis of the structure and function of the kanamycin-resistance transposon Tn903. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):125–133. doi: 10.1101/sqb.1981.045.01.021. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Sherratt D. J. Sequence analysis at IS1 insertion sites: models for transposition. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1257–1261. doi: 10.1101/sqb.1979.043.01.142. [DOI] [PubMed] [Google Scholar]
- Harshey R. M., Bukhari A. I. A mechanism of DNA transposition. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1090–1094. doi: 10.1073/pnas.78.2.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., Bedinger P., Champoux J. J., Falkow S. Deletions affecting the transposition of an antibiotic resistance gene. Proc Natl Acad Sci U S A. 1977 Feb;74(2):702–706. doi: 10.1073/pnas.74.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Hu S., Otsubo E., Davidson N., Saedler H. Electron microscope heteroduplex studies of sequence relations among bacterial plasmids: identification and mapping of the insertion sequences IS1 and IS2 in F and R plasmids. J Bacteriol. 1975 May;122(2):764–775. doi: 10.1128/jb.122.2.764-775.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S., Arber W. On the role of IS1 in the formation of hybrids between the bacteriophage P1 and the R plasmid NR1. Mol Gen Genet. 1980 Jan;177(2):261–270. doi: 10.1007/BF00267437. [DOI] [PubMed] [Google Scholar]
- Kamp D., Kahmann R. Two pathways in bacteriophage Mu transposition? Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):329–336. doi: 10.1101/sqb.1981.045.01.046. [DOI] [PubMed] [Google Scholar]
- McEntee K., Weinstock G. M., Lehman I. R. recA protein-catalyzed strand assimilation: stimulation by Escherichia coli single-stranded DNA-binding protein. Proc Natl Acad Sci U S A. 1980 Feb;77(2):857–861. doi: 10.1073/pnas.77.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J., Iida S., Arber W. Does the insertion element IS1 transpose preferentially into A+T-rich DNA segments? Mol Gen Genet. 1980;178(2):471–473. doi: 10.1007/BF00270502. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Calos M. P., Galas D., Hofer M., Büchel D. E., Müller-Hill B. Genetic analysis of transpositions in the lac region of Escherichia coli. J Mol Biol. 1980 Nov 25;144(1):1–18. doi: 10.1016/0022-2836(80)90212-0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Zenilman M., Ohtsubo H. Plasmids containing insertion elements are potential transposons. Proc Natl Acad Sci U S A. 1980 Feb;77(2):750–754. doi: 10.1073/pnas.77.2.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo H., Ohmori H., Ohtsubo E. Nucleotide-sequence analysis of Tn3 (ap): implications for insertion and deletion. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1269–1277. doi: 10.1101/sqb.1979.043.01.144. [DOI] [PubMed] [Google Scholar]
- Ptashne K., Cohen S. N. Occurrence of insertion sequence (IS) regions on plasmid deoxyribonucleic acid as direct and inverted nucleotide sequence duplications. J Bacteriol. 1975 May;122(2):776–781. doi: 10.1128/jb.122.2.776-781.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Helical periodicity of DNA determined by enzyme digestion. Nature. 1980 Aug 7;286(5773):573–578. doi: 10.1038/286573a0. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt D., Arthur A., Burke M. Transposon-specified, site-specific recombination systems. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):275–281. doi: 10.1101/sqb.1981.045.01.040. [DOI] [PubMed] [Google Scholar]
- Shibata T., DasGupta C., Cunningham R. P., Radding C. M. Homologous pairing in genetic recombination: formation of D loops by combined action of recA protein and a helix-destabilizing protein. Proc Natl Acad Sci U S A. 1980 May;77(5):2606–2610. doi: 10.1073/pnas.77.5.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starlinger P. IS elements and transposons. Plasmid. 1980 May;3(3):241–259. doi: 10.1016/0147-619x(80)90039-6. [DOI] [PubMed] [Google Scholar]
- Warren G. J., Clark A. J. Sequence-specific recombination of plasmid ColE1. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6724–6728. doi: 10.1073/pnas.77.11.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. J., Twigg A. J., Sherratt D. J. ColE1 plasmid mobility and relaxation complex. Nature. 1978 Jul 20;274(5668):259–261. doi: 10.1038/274259a0. [DOI] [PubMed] [Google Scholar]
- Wilson J. H. Nick-free formation of reciprocal heteroduplexes: a simple solution to the topological problem. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3641–3645. doi: 10.1073/pnas.76.8.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mansfeld A. D., Langeveld S. A., Baas P. D., Jansz H. S., van der Marel G. A., Veeneman G. H., van Boom J. H. Recognition sequence of bacteriophage phi X174 gene A protein--an initiator of DNA replication. Nature. 1980 Dec 11;288(5791):561–566. doi: 10.1038/288561a0. [DOI] [PubMed] [Google Scholar]



