ABSTRACT
Type IV secretion systems (T4SS) transfer DNA and/or proteins into recipient cells. Here we performed immunofluorescence deconvolution microscopy to localize the assembled T4SS by detection of its native components VirB1, VirB2, VirB4, VirB5, VirB7, VirB8, VirB9, VirB10, and VirB11 in the C58 nopaline strain of Agrobacterium tumefaciens, following induction of virulence (vir) gene expression. These different proteins represent T4SS components spanning the inner membrane, periplasm, or outer membrane. Native VirB2, VirB5, VirB7, and VirB8 were also localized in the A. tumefaciens octopine strain A348. Quantitative analyses of the localization of all the above Vir proteins in nopaline and octopine strains revealed multiple foci in single optical sections in over 80% and 70% of the bacterial cells, respectively. Green fluorescent protein (GFP)-VirB8 expression following vir induction was used to monitor bacterial binding to live host plant cells; bacteria bind predominantly along their lengths, with few bacteria binding via their poles or subpoles. vir-induced attachment-defective bacteria or bacteria without the Ti plasmid do not bind to plant cells. These data support a model where multiple vir-T4SS around the perimeter of the bacterium maximize effective contact with the host to facilitate efficient transfer of DNA and protein substrates.
IMPORTANCE
Transfer of DNA and/or proteins to host cells through multiprotein type IV secretion system (T4SS) complexes that span the bacterial cell envelope is critical to bacterial pathogenesis. Early reports suggested that T4SS components localized at the cell poles. Now, higher-resolution deconvolution fluorescence microscopy reveals that all structural components of the Agrobacterium tumefaciens vir-T4SS, as well as its transported protein substrates, localize to multiple foci around the cell perimeter. These results lead to a new model of A. tumefaciens attachment to a plant cell, where A. tumefaciens takes advantage of the multiple vir-T4SS along its length to make intimate lateral contact with plant cells and thereby effectively transfer DNA and/or proteins through the vir-T4SS. The T4SS of A. tumefaciens is among the best-studied T4SS, and the majority of its components are highly conserved in different pathogenic bacterial species. Thus, the results presented can be applied to a broad range of pathogens that utilize T4SS.
Introduction
Type IV secretion systems (T4SS) are multiprotein complexes used by Gram-negative and Gram-positive bacteria for transfer of DNA and/or proteins to other bacteria (1–3), plants (4), mammalian cells (5, 6), and yeast cells (7, 8). There are two major classes of T4SS. The first class comprises T4SS involved in conjugative transfer of plasmid DNA between bacteria (1, 9). The second class is involved in pathogenesis by transferring effector proteins into eukaryotic host cells or the extracellular milieu. Human pathogens such as Helicobacter pylori, Legionella pneumophila, Bordetella pertussis, and Rickettsia prowazekii require the T4SS for disease (10).
The canonical model for T4SS is the virulence (vir)-induced T4SS of the plant pathogen Agrobacterium tumefaciens. vir-T4SS delivers both DNA and proteins into plant cells, causing crown gall disease. Components of the vir-T4SS are encoded by the tumor-inducing plasmid (pTi). The vir-T4SS transports pTi-encoded single-stranded DNA (T strand) and at least four Vir proteins, VirD2, VirE2, VirE3, and VirF, into host cells. Insertion of the T strand into plant genomic DNA and its subsequent expression lead to the overproduction of T-DNA-encoded plant growth hormones, resulting in the tumorous phenotype. Crown gall tumors also produce opines, which are unusual amino acid-derived compounds (11) specifically catabolized by agrobacteria as a source of carbon and nitrogen, thus providing a selective advantage for their growth in the rhizosphere (12, 13). Strains of A. tumefaciens can be differentiated on the basis of the unique opine produced by the tumor, and each type of opine is specifically catabolized by the infecting strain (14). The two A. tumefaciens strains most extensively studied induce tumors that produce nopaline or octopine opines. The vir-T4SS components from these two strains are highly conserved (15).
The vir-T4SS is composed of 11 VirB proteins (VirB1 to -11) and VirD4. All VirBs and VirD4 are essential for maximal DNA and protein transport. The vir-T4SS components can be placed into three major groups. The first group consists of the T-pilus components and assembly factors VirB1 and VirB3. VirB2 is the major T-pilus structural component, while the minor component, VirB5, is localized at the T-pilus tip (16). The N-terminal portion of VirB1 has homology to lytic transglycosylases and cleaves the peptidoglycan (17) to facilitate the assembly of the vir-T4SS in the periplasmic space. The C terminus of VirB1, VirB1*, is secreted to the cell surface (18, 19) and is required for T-pilus formation (20). VirB3 is also required for T-pilus assembly (21). The second group consists of VirB6 to VirB10, which span the inner and outer membranes and periplasm, forming the translocation channel (22). Cryo-electron microscopy and crystal structures show that 14 copies each of VirB7, VirB9, and VirB10 form a 1.05-MDa “core complex” that connects the cytosol to the outer surface of the bacterium with a central core diameter of 76 Å (23, 24). The arrangement of VirB6 and VirB8 with respect to this core is unknown, but both proteins interact with the T strand (25). The third group consists of VirB4, VirB11, and VirD4, which have ATPase homology and ATP-binding motifs (26–29) and may energize assembly and/or substrate translocation. VirD4 is also the coupling protein that brings the T strand and its associated proteins to the vir-T4SS (30–32).
To understand vir-T4SS function, it is critical to determine the localization pattern of the assembled vir-T4SS. Previously, we used deconvolution fluorescence microscopy to assess the localization of green fluorescent protein (GFP) fusions to structural components and substrates of the vir-T4SS. Due to the bulky nature of GFP, very few fusion proteins were functional; given that many T4SS proteins are multimeric or membrane spanning, GFP fusions are likely to interfere with complex assembly and function. Nevertheless, fusions of GFP to the cytoplasmic tails of VirD4 and VirB8 were functional, and these GFP fusion proteins localized in a helical/periodic pattern of multiple foci around the perimeter of the bacterial cell (33). However, as there are 12 vir-T4SS components, it is possible that there are subassemblies with different localization patterns. Thus, it is critical to determine the localization pattern of the T4SS using probes to all vir-T4SS components. Here, we performed immunofluorescence microscopy using antibodies to nine native Vir proteins residing in the inner membrane, periplasm, and outer membrane in the nopaline strain C58 and detected similar patterns of multiple foci supporting our initial findings. In addition, we extended our studies to the octopine strain A348, and the localization of its vir-T4SS is identical to that found in the nopaline strain. Fluorescent labeling of the vir-T4SS allowed us to examine the orientation of bacteria during binding to plant cells. The majority of bacteria bound laterally along their lengths to plant cells, and multiple vir-T4SS foci continued to be present during this binding. These data support a model where multiple vir-T4SS around the bacterium maximize effective contact with the host to facilitate efficient transfer of DNA and protein substrates.
RESULTS
Nopaline vir-T4SS membrane and periplasmic core structural components localize to multiple foci.
Here we use immunofluorescence followed by deconvolution microscopy to detect nine different structural components of the vir-T4SS. The use of antibodies allows localization of the native vir-T4SS. In our previous work, GFP fusions to VirB8 and VirD4 and to vir-T4SS substrate proteins VirD2, VirE2, and VirF localized to multiple foci in a helical/periodic pattern around the circumference of vir-induced cells (33). Importantly, these fusion proteins did not interfere with tumor formation, and GFP-VirB8 rescued a virB8 deletion (33). In contrast, GFP fusions to VirB4, VirB6, VirB7, VirB9, VirB10, and VirB11 exhibited dominant negative effects on tumor formation and did not form multiple foci, suggesting that the GFP fusion interfered with vir-T4SS complex assembly and/or function (33).
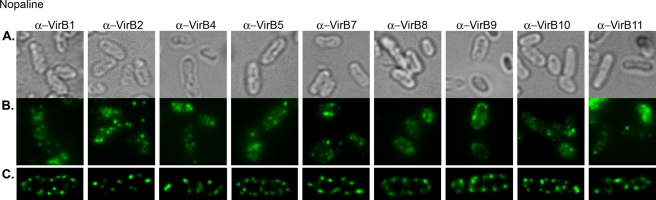
Immunofluorescent detection of nopaline strain vir-T4SS components VirB1, VirB2, VirB4, VirB5, VirB7, VirB8, VirB9, VirB10, and VirB11 revealed multiple foci (Fig. 1). Ten optical sections were taken for each bacterial cell and then deconvolved to a three-dimensional (3D) image to better resolve the multiple foci (Fig. 1C). The images recapitulate those seen with GFP-VirB8 fusions (33) and are suggestive of a periodic arrangement of foci. The numbers of foci in the panels in Fig. 1C vary between a minimum of 10 and a maximum of 19.
FIG 1 .
Immunofluorescent detection of VirB proteins in vir-induced A. tumefaciens nopaline strain (C58). vir-induced A. tumefaciens C58 was probed with primary antibodies to native VirB proteins (α-VirB1, α-VirB2, α-VirB4, α-VirB5, α-VirB7, α-VirB8, α-VirB9, α-VirB10, and α-VirB11) followed by fluorescent secondary antibodies (left to right). (A and B) Bright-field images (A) corresponding to fluorescent panels (B). (C) Maximum-intensity z projections of deconvolved z stacks of a representative cell.
We next analyzed thousands of individual cells to provide a quantitative assessment of the frequency of multiple foci detected by the nine different antibodies to native nopaline vir-T4SS components. As it is impractical to perform deconvolution microscopy of stacked optical images of thousands of cells, we instead counted foci in a single plane of focus. We can easily detect multiple foci, and we specifically assessed the frequency of cells exhibiting 3 or more fluorescent foci (Fig. 1B). Over 80% of the cells had 3 or more fluorescently labeled foci (Table 1). Most of the cells had between 5 and 10 foci, and a very small percentage of cells had only 3 or 4 foci. For example, the VirB8 antibody detected 3 or more foci in 90% of the fluorescently labeled cells, reinforcing the results seen with GFP-VirB8 in our earlier report (33). Antibodies to the T-pilus components VirB2 and VirB5 revealed that 92% of the labeled cells had 3 or more foci. In cells labeled with antibodies to the energetic components VirB4 and VirB11, 83 to 85% of the cells contained at least 3 foci. Antibodies to the core components VirB7, VirB9, and VirB10 showed that 85 to 91% of the labeled cells possessed 3 or more foci.
TABLE 1 .
Cellular localization of VirB proteins
| Strain | Antibody | No. of cells labeled | % of cells with ≥3 foci in one optical section |
|---|---|---|---|
| Nopaline | VirB1 | 504 | 94 |
| VirB2 | 600 | 92 | |
| VirB4 | 540 | 85 | |
| VirB5 | 450 | 92 | |
| VirB7 | 778 | 85 | |
| VirB8 | 343 | 90 | |
| VirB9 | 94 | 91 | |
| VirB10 | 1,160 | 89 | |
| VirB11 | 315 | 83 | |
| Octopine | VirB2 | 90 | 80 |
| VirB5 | 145 | 85 | |
| VirB7 | 159 | 72 | |
| VirB8 | 83 | 88 |
Each antibody labeled significantly more cells (P value, <0.05) of vir-induced cultures (+AS) than of noninduced cultures (−AS) (Table 2). The majority of antibodies labeled less than 7% of noninduced cells, signifying high specificity of the antibodies (Table 2). Antibodies to VirB1 and, to a lesser extent, antibodies to VirB7 were evidently less specific (Table 2). Nevertheless, antibodies to VirB1 and VirB7 detected 3 or more foci more frequently in vir-induced cells than in noninduced cells, indicating that many of the foci observed in vir-induced cells were due to specific antibody binding.
TABLE 2 .
Labeling specificity of VirB antibodies
| Strain | Antibodya | % labeled (total no. of cells) |
|
|---|---|---|---|
| Non-vir induced | vir induced | ||
| Nopaline | VirB1 | 46 (570) | 66 (761) |
| VirB2 | 4 (1,194) | 45 (1,341) | |
| VirB4 | 2 (752) | 66 (823) | |
| VirB5 | 7 (609) | 69 (654) | |
| VirB7 | 19 (593) | 56 (1,397) | |
| VirB8 | 4 (2,239) | 32 (1,064) | |
| VirB9 | 2 (487) | 8 (1,170) | |
| VirB10 | 2 (2,055) | 29 (3,970) | |
| VirB11 | 5 (265) | 59 (531) | |
| Octopine | VirB2 | 1 (1,380) | 15 (582) |
| VirB5 | 2 (541) | 63 (229) | |
| VirB7 | 20 (264) | 51 (312) | |
| VirB8 | 0 (555) | 8 (1,081) | |
P < 0.05 for all antibodies.
Octopine vir-T4SS structural components localize to multiple foci.
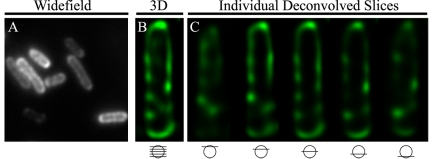
The vir-T4SS components are highly conserved among A. tumefaciens strains. For example, VirB8 shares 90% amino acid identity between nopaline and octopine strains (15). VirB8 is a bitopic membrane protein (34) that accommodates GFP fusions to its short cytoplasmic N-terminal tail without decreased functionality, as previously demonstrated in the nopaline strain (33). To test the localization of VirB8 in the octopine strain, we made a GFP fusion to the N terminus of octopine VirB8 and monitored its localization using deconvolution microscopy. Indeed, we detected multiple foci localized around the bacterial cell (Fig. 2), resembling the pattern of GFP-VirB8 fluorescence in the nopaline strain (33).
FIG 2 .
Localization of octopine VirB8 in a vir-induced A. tumefaciens octopine strain (A348). GFP-VirB8 in wild-type A. tumefaciens A348. (A) Wide-field image. (B) Maximum-intensity z projections of deconvolved z stacks of a representative cell. (C) Individual deconvolved slices from top to bottom of the cell in panel B.
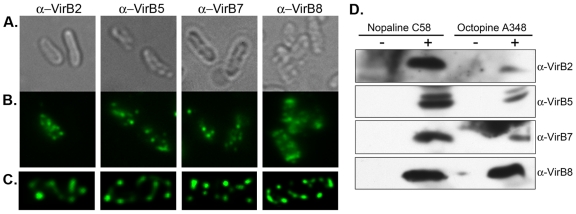
Antibodies to VirB2, VirB5, VirB7, and VirB8 from the nopaline strain detect vir-induced bands of the predicted molecular weights in extracts of the vir-induced octopine strain by Western blot analysis (Fig. 3D). Although antibody against nopaline VirB8 detects octopine VirB8 at similar levels, this antibody is not efficient in detecting VirB8 in whole cells by immunofluorescence (Table 2). In contrast, while antibodies against nopaline VirB2, VirB5, and VirB7 were less efficient in detecting their octopine homologs on Western blots, these antibodies were effective in detecting their respective proteins in whole cells by immunofluorescence microscopy. Such variation is expected due to the innate complexity of epitope conformations. Antibodies against nopaline T-pilus components VirB2 and VirB5 detected multiple foci in 80% and 85% of the labeled vir-induced octopine cells, respectively (Fig. 3; Table 1). Similarly, antibodies against the nopaline core components VirB7 and VirB8 detected multiple foci in 72% and 88% of the labeled vir-induced octopine cells, respectively (Fig. 3; Table 1). Nopaline antibodies against VirB2, VirB5, and VirB8 labeled less than 2% of the octopine cells under noninducing conditions, while VirB7 antibody labeled 20% of noninduced cells (Table 2). All antibodies used for immunofluorescence microscopy of the octopine strain showed significantly more labeling in vir-induced cells than in noninduced cells (P value, <0.05). In the vir-induced cells, over 70% of the labeled cells showed 3 or more foci (Table 1).
FIG 3 .
Detection of VirB proteins in A. tumefaciens A348. Antibodies from the nopaline strain C58 were used to detect native VirB proteins in the octopine strain A348. (A) Bright-field images corresponding to fluorescent panels in panel B. (B and C) Immunofluorescence detection with primary antibodies to native A348 VirB proteins (α-VirB2, α-VirB5, α-VirB7, and α-VirB8) followed by fluorescent secondary antibodies (left to right). (C) Maximum-intensity z projections of deconvolved z stacks of a representative cell. (D) A. tumefaciens bacteria from the nopaline and octopine strains were uninduced (−) or induced with 200 µM AS (+) for the expression of vir genes. AS-induced bands of the correct molecular weights were recognized by α-VirB2, α-VirB5, α-VirB7, and α-VirB8.
Do T4SS components colocalize?
Given the multiple foci observed for individual T4SS components, an obvious question is whether such components colocalize to the same foci. Biochemical studies indicate that VirB7-VirB9-VirB10 (24), VirB4-VirB11 (26), and VirB4-VirB8 and VirB5-VirB8-VirB10 (35) copurify. Yeast two-hybrid analyses confirm these interactions and further demonstrate that VirB8 interacts with VirB1, VirB4, and VirB11 and that VirB9 and VirB11 interact (36). Thus, eight core T4SS components are known to copurify/interact and thus colocalize.
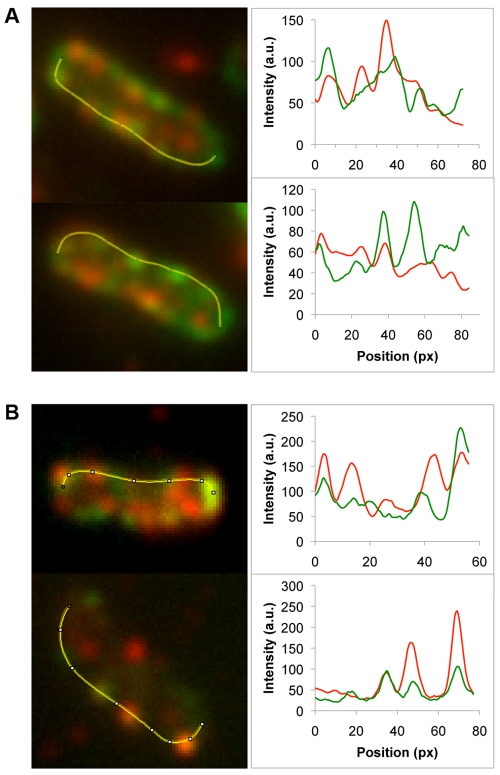
While the T pilus is assumed to localize at the outer limit of the T4SS channel, it is formally possible that the T pilus is a separate entity used only for making host cell contact. We therefore addressed whether a T4SS core component and the VirB2 pilus component colocalize. We detected the T4SS using a GFP-VirB8 fusion and used antibodies to detect VirB2 (red fluorescence). Figure 4A shows two representative cells. Strikingly, there are distinct red and green foci, suggesting that VirB2 and VirB8 do not colocalize; however, there are other orange or greenish yellow foci, suggesting partial colocalization. To the right of each fluorescent cell, we show quantification of the two fluorophores; these plots reinforce the idea that some foci result from colocalized red and green fluorescence and that other foci emit predominantly red or green fluorescence. These data suggest that VirB2 and VirB8 only sometimes colocalize.
FIG 4 .
Do T4SS components colocalize? (A) GFP-VirB8 and VirB2 colocalization; VirB2 is detected by primary antibodies to VirB2 and secondary antibodies with red fluorescence (AlexaFluor 546). (B) VirB colocalization with itself detected by GFP-VirB8 and primary antibodies to GFP followed by secondary antibodies with red fluorescence (AlexaFluor 546). Two independent cells are shown in panels A and B. To the right are fluorescent profiles through the regions indicated by the yellow lines in panels A and B; green or red curves indicate GFP or AlexaFluor 546 fluorescence intensity, respectively.
As a control for the above, we monitored whether this approach could accurately detect VirB8 colocalization with itself. We detected VirB8 as a GFP fusion and with antibodies to VirB8 (red fluorescence). The upper panel of Fig. 4B shows 3 possible colocalized foci and several foci that do not colocalize. The bottom panel and its tracing reveal excellent colocalization for 3 foci; however, the upper side of the same cell shows a clear red focus indicating no colocalization. Importantly, we expressed GFP-VirB8 in a virB8 deletion strain, so GFP fluorescence and VirB8 antibodies should detect the same molecule, without any wild-type VirB8 present to compete for antibody binding. We also tested for GFP-VirB8 colocalization using antibodies to VirB8 (red fluorescence) and antibodies to GFP (far-red fluorescence) and obtained similar results that only some foci colocalize (not shown). These data then lead to the unexpected conclusion that combining GFP fusions and immunofluorescence does not reliably result in colocalized foci under these conditions.
Thus, the data testing for VirB2-VirB8 and for VirB8-VirB8 colocalization are similar and do not allow us to conclude that colocalization occurs for all foci. We suggest two likely explanations. First, antibody detection is stochastic; following cell permeabilization and fixation, not all epitopes are in the correct conformation for detection. Second, fixation of cells may lead to unfolding of GFP and stochastic loss of GFP fluorescence.
The above data illustrate the shortcomings of immunofluorescence in resolving protein colocalization in the confines of bacterial cells. These results potentially illuminate why a previous report utilizing immunolocalization found several T4SS components localized to different sites around the bacterial cell (37).
A. tumefaciens binds longitudinally to host cells.
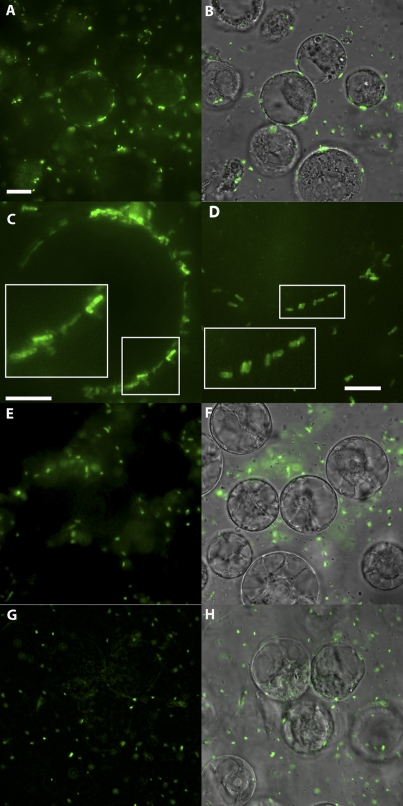
Attachment between the host cell and A. tumefaciens is necessary for the transfer of the vir-T4SS substrates and the T-strand complex. Several reports suggested that the vir-T4SS localizes to the cell poles (31, 32, 37–39). Earlier reports of a few A. tumefaciens bacteria attaching to a single plant cell via their poles lent support for polar attachment (40, 41). Recent reviews continue to suggest polar attachment (42, 43) based on these earlier reports. However, our present and previous findings (33) demonstrate that vir-T4SS components, including T pili, localize around the circumference of the cell, suggesting that there are multiple points of bacterial attachment to host cells. To test if attachment correlates with the localization of the vir-T4SS, we used GFP-VirB8 as a marker for the localization of the vir-T4SS following incubation of A. tumefaciens with plant cells. A. tumefaciens carrying a vir-inducible GFP-VirB8 plasmid in trans to pTi was induced for vir gene expression with acetosyringone (AS) prior to incubation with host cells. We used protoplasts of BY2 tobacco culture cells to mimic the wounded cells that A. tumefaciens naturally infects and to replicate conditions previously used to monitor attachment (40, 41). Bacteria were incubated with plant protoplasts for 3 h, and binding was monitored by fluorescence deconvolution microscopy. Most bacterial cells bind horizontally along their lengths to the surface of the plant cell (Fig. 5A and 5B). Higher-magnification images again reveal that most bacteria are attached along their length, with few bacteria attaching in a polar fashion (Fig. 5C and 5D); in these latter images, bacteria still contain multiple GFP-VirB8 foci, although their resolution is lower than that shown above (Fig. 1 to 3), as it is difficult to focus down on spherical plant cells without causing them to burst.
FIG 5 .
A. tumefaciens attachment to plant protoplasts. (A to D) Incubation of protoplasts with A. tumefaciens C58 expressing vir-inducible GFP-VirB8. (E and F) Incubation of protoplasts with A. tumefaciens C1 lacking the Ti plasmid but expressing GFP under the control of a lac-inducible promoter. (G and H) Incubation of protoplasts with A. tumefaciens attachment-deficient strain A1020 expressing vir-inducible GFP-VirB8. Magnifications: ×600 (A, B, E, F, G, and H) and ×1,000 (C and D). Panels A, C, D, E, and G are GFP epifluorescence. Panels B, F, and H are GFP epifluorescence and bright-field microscopy combined. Bar, 10 µm (A, B, and E to H) or 5 µm (C and D). The insets in panels C and D are shown magnified 2-fold.
We performed two control assays to demonstrate that the observed horizontal attachment is biologically relevant. First, we monitored attachment of A. tumefaciens lacking the Ti plasmid but expressing an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible GFP. Figure 5E and F show no bacteria binding to the protoplasts. Second, we placed vir-inducible GFP-VirB8 into attachment-deficient A. tumefaciens strain A1020 (41), and the cells were grown and induced for vir gene expression exactly as the wild-type strain was. Figure 5G and H show excellent GFP-VirB8 fluorescence in vir-induced A1020, yet none of the cells are attached to the protoplasts.
Thus, wild-type A. tumefaciens strains carrying the Ti plasmid bind along their length to plant protoplasts, and A. tumefaciens strains lacking the Ti plasmid or defective in plant cell attachment do not bind. The images in Fig. 5 are representative of independent experiments, each monitoring hundreds of protoplasts.
DISCUSSION
Immunofluorescence deconvolution microscopy revealed that the A. tumefaciens vir-T4SS is arranged in multiple foci around the circumference of the bacterial cell, supporting our initial findings with GFP fusions to vir-T4SS components VirB8 and VirD4 and substrates VirD2, VirE2, and VirF (33). Here, we determined the localization of the vir-T4SS by monitoring nine of its components (VirB1, VirB2, VirB4, VirB5, VirB7, VirB8, VirB9, VirB10, and VirB11) in the nopaline strain of A. tumefaciens. Over 80% of the fluorescently labeled cells in the nopaline strain showed multiple foci around the bacterial cell. Previously, GFP fusions to the ATPases, VirB4 and VirB11, and core components VirB7, VirB9, and VirB10 interfered with tumor formation; therefore, their inability to form multiple GFP foci was not biologically significant (33). In this report, we used antibodies against these native proteins to perform immunofluorescence microscopy and were now able to determine their localization as multiple foci. The spacing of the vir-T4SS foci suggests that they are arranged in a regular pattern. Indeed, an independent quantitative study to theoretically and experimentally assess the pattern of vir-T4SS localization supports the idea that vir-T4SS are periodically arranged (T. Cameron, M. Roper, and P. Zambryski, unpublished data).
We also extended our localization studies to include VirB1 and T-pilus components VirB2 and VirB5, and all showed similar multifocal localization patterns in whole cells. We did not detect these proteins in extracellular T pili. Thus, our antibodies detected VirB1, VirB2, and VirB5 as native membrane-associated proteins following the mild fixation used for immunofluorescence and also when denatured on Western blots. Different proteins may change conformation in response to chemical treatments and alter the affinity of antigenic epitopes for the primary antibody. Indeed, following harsh chemical fixation and detection via immunogold electron microscopy, the VirB5 antibody detected denatured VirB5 at T-pilus tips, but under these conditions, the VirB2 antibody does not detect cell-bound T pili (16); these authors used VirB2 and VirB5 antibodies from our lab for their studies. Here, antibodies to VirB1 also labeled a fraction of non-vir-induced cells. This nonspecific binding may be due to the recognition of endogenous (non-vir) lytic transglycosylases or other epitopes with similarity to the C terminus of the lytic transglycosylase domain of VirB1, the antibody recognition site.
Several authors have observed the localization of vir-T4SS components at the bacterial cell poles (31, 32, 37–39). These authors used standard wide-field fluorescence microscopy, where the low signal-to-noise ratio may have obscured other foci present on the cell. Standard wide-field fluorescence microscopy is limited, as it does not give any information in the z dimension. In addition, the earlier reports of polar localization of GFP fusions to VirD4, VirE2, VirC1, and VirB6 may result from overexpression of these proteins from the multicopy IncP plasmid. When overexpressed, these proteins would potentially accumulate at the poles in aggregates, as seen in Escherichia coli (44). For example, in the report of polar VirC1, the bacterial cells were misshapen with large bulges at the poles (39). In contrast, our studies with GFP fusions to Vir proteins did not use multicopy plasmids (33 and this study), and the bacterial cells have the expected rod-shaped morphology.
Notably, multiple foci along the bacterial cell also have been observed for homologs of the vir-T4SS proteins, such as the VirB4 homolog TrhC (45, 46) and the VirD4 coupling protein homolog TraG (47) from the IncH1 conjugative transfer system of E. coli. Immunofluorescence studies showed that TrhC and TraG localized at discrete foci around the circumference of the cell membrane.
We previously suggested that the multiple T4SS around the bacterial cell are arranged in a helical pattern (33) by analogy to bacterial cytoskeletal proteins, such as MreB, proposed to form helical arrays (48). Two recent reports now characterize MreB localization during cell wall synthesis and suggest that MreB and cell elongation machinery move circumferentially around the cell perpendicular to the length of the cell (49, 50). As A. tumefaciens does not contain an MreB homolog (51), we cannot make direct comparison with these new data and the localization of vir-T4SS. It is formally possible that an underlying helical filament is responsible for the anchoring of the vir-T4SS. Nevertheless, to be cautious in our interpretation of the data, we suggest that a more accurate description is that the A. tumefaciens vir-T4SS localizes in a periodic pattern around the bacterial circumference.
Here we show that A. tumefaciens attaches to plant protoplast cells predominantly by making lateral contacts along their sides with few bacteria also attaching via their poles or subpoles. These data suggest that several vir-T4SS localized around the bacterial cell may contact the host cell for the transfer of the T strand and the Vir effector proteins. Several previous studies that addressed A. tumefaciens binding to plant cells suggested polar attachment (40, 41). There are two significant differences between the earlier reports and the data presented here. First, the early reports detected few bacteria binding to plant cells. In contrast, the use of GFP facilitates the detection of numerous bacteria binding to plant cells along their lengths. Second and more importantly, we used vir-induced A. tumefaciens to monitor attachment. vir-induced A. tumefaciens cells are likely primed and ready to attach. Under our conditions, agrobacteria are induced for 48 h at 19°C; this time frame results in maximal labeling of the T4SS (33). We further suggest that A. tumefaciens attachment is likely a stepwise dynamic process. Pole- and subpole-attached A. tumefaciens observed here or previously may be in transition to become laterally attached or in transition from detaching from the host cell following transfer of DNA and proteins.
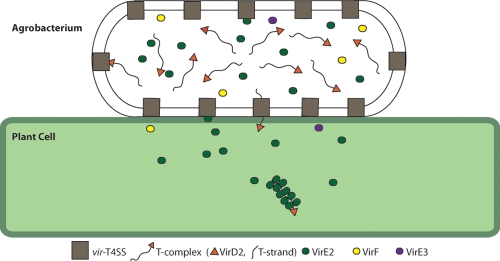
The arrangement of multiple vir-T4SS around the bacterial circumference should maximize effective contact between bacteria and host cells. Multiple vir-T4SS complexes and lateral attachment points may be required to facilitate efficient transfer of T strands and Vir effector proteins into the host cell (Fig. 6). It is currently estimated that each induced bacterial cell produces 50 T strands (39). To coat each T strand would require approximately 30 molecules of VirE2 per kilobase of T-DNA (52, 53). Therefore, thousands of VirE2 molecules need to be delivered into the host cytoplasm to prevent T-strand degradation. In addition to the numerous VirE2 molecules, VirE3 and VirF proteins also must be transported through the vir-T4SS. Thus, multiple vir-T4SS would be advantageous to the bacterial cell to expedite transport of the vast quantities of proteins and T strands as shown in the model in Fig. 6.
FIG 6 .
Model for the localization of vir-T4SS and lateral attachment of A. tumefaciens to a plant cell. It may be advantageous for A. tumefaciens to assemble multiple vir-T4SS and attach laterally to the host cell to facilitate efficient transfer of the tens of T complexes and thousands of Vir-effector proteins transported through the vir-T4SS.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Wild-type A. tumefaciens strain C58 contains nopaline pTiC58, and strain A348 contains octopine pTiA6. Strain A1020 carries a Tn5 insertion at the chvB gene and is attachment deficient (41). For induction of the vir system, an overnight culture was diluted to an optical density at 600 nm (OD600) of 0.1 in minimal AB medium, pH 5.5, and grown for 5 h at 19°C (18). Cultures were then plated on AB agar plates with 200 µM acetosyringone (AS) for 2 days at 19°C.
GFP was amplified using iProof DNA polymerase (Bio-Rad, Inc.) according to the manufacturer’s instructions. Primers incorporated an NdeI restriction site at the 5′ end of the coding sequence and an SpeI restriction site at the 3′ end. The amplified product was digested with NdeI and SpeI and ligated to similarly digested pSRKGm (54) to create pJZ210. This strain was induced with 10 mM IPTG at 19°C for 48 h prior to incubation with BY2 protoplasts.
Western analysis.
Proteins from 108 cells were loaded into each lane. After protein separation, gels were transferred to Immobilon-P polyvinylidene difluoride 0.45-µm membranes and analyzed by standard methods for Western blotting.
Immunofluorescence microscopy.
vir-induced cells were fixed in 2.67% paraformaldehyde and 0.01% gluteraldehyde for 15 minutes at room temperature. Cells were washed once in GTE (50 mM glucose, 25 mM Tris, pH 8, 10 mM EDTA), pelleted, resuspended in GTE, and stored overnight at 4°C. Cells were washed in GTE and permeabilized by treatment with 2 mg/ml of lysozyme and 5 mM EDTA for 45 min at room temperature. Primary antibodies were diluted in 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS): anti-VirB1, anti-VirB2, anti-VirB4, anti-VirB5, and anti-VirB7 were diluted 1:50; anti-VirB8 was diluted 1:200; anti-VirB9 was diluted 1:50; anti-VirB10 and anti-VirB11 were diluted 1:100; and chicken anti-GFP was diluted 1:100 (Aves Labs). Antibodies were added individually to each sample and incubated at 37°C for 60 min. The cells were then washed twice with PBS containing 0.05% Tween 20. The secondary antibodies were diluted in PBS: AlexaFluor 488 and AlexaFluor 546 goat anti-rabbit IgG were diluted 1:250 and AlexaFluor 647 goat anti-chicken IgG was diluted 1:100. Secondary antibodies were added as appropriate, and the samples were incubated at 37°C for 60 min. Cells were washed three times in PBS containing 0.05% Tween 20 and once in PBS. Cells were resuspended in PBS and imaged with an Applied Precision Deltavision Spectris DV4 deconvolution microscope as previously described (33). To assess colocalization, representative dually labeled cells were chosen from single wide-field images for analysis. Fluorescence profiles were measured in ImageJ (http://rsbweb.nih.gov/ij) using the RGB Profiles Tool macro.
A. tumefaciens interaction with host cells.
A. tumefaciens C58 containing GFP-VirB8 in trans to pTi was grown in AB minimal medium with appropriate antibiotics and induced with 200 µM AS for 48 hours as described above. Bacteria were then scraped off plates and resuspended in AB medium. BY2 tobacco protoplasts were made by treating the culture cells with protoplast enzyme solution (1% cellulase, 0.01% pectolyase, and 0.4 M d-mannitol) at pH 5.5 for 150 minutes. BY2 protoplasts were collected by centrifugation at 900 × g for 5 minutes. BY2 protoplasts were washed twice with ice-cold 0.4 M mannitol and resuspended in protoplast medium (66 mM calcium chloride, 7 mM sodium acetate, 247 mM d-mannitol, pH 5.8). A. tumefaciens was incubated with BY2 protoplasts for 3 h at 19°C. Plant cells were imaged with an Applied Precision Deltavision Spectris DV4 deconvolution microscope.
ACKNOWLEDGMENTS
We thank Steve Ruzin and Denise Schichnes of the College of Natural Resources Biological Imaging Facility for advice.
This work was supported by a predoctoral fellowship to J.A. (NIH grant F31GM089088) and an NSF grant (MCB-0343566) to P.Z.
Footnotes
Citation Aguilar J, Cameron TA, Zupan J, Zambryski P. 2011. Membrane and core periplasmic Agrobacterium tumefaciens virulence type IV secretion system components localize to multiple sites around the bacterial perimeter during lateral attachment to plant cells. mBio 2(6):e00218-11. doi:10.1128/mBio.00218-11.
REFERENCES
- 1. Llosa M, Gomis-Rüth FX, Coll M, de la Cruz F. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1–8 [DOI] [PubMed] [Google Scholar]
- 2. Wallden K, Rivera-Calzada A, Waksman G. 2010. Type IV secretion systems: versatility and diversity in function. Cell. Microbiol. 12:1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fronzes R, Christie PJ, Waksman G. 2009. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7:703–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zupan J, Muth TR, Draper O, Zambryski P. 2000. The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J. 23:11–28 [DOI] [PubMed] [Google Scholar]
- 5. Waters VL. 2001. Conjugation between bacterial and mammalian cells. Nat. Genet. 29:375–376 [DOI] [PubMed] [Google Scholar]
- 6. Schröder G, Schuelein R, Quebatte M, Dehio C. 2011. Conjugative DNA transfer into human cells by the VirB/VirD4 type IV secretion system of the bacterial pathogen Bartonella henselae. Proc. Natl. Acad. Sci. U. S. A. 108:14643–14648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas PJ. 1995. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 14:3206–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Piers KL, Heath JD, Liang X, Stephens KM, Nester EW. 1996. Agrobacterium tumefaciens-mediated transformation of yeast. Proc. Natl. Acad. Sci. U. S. A. 93:1613–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Juhas M, Crook DW, Hood DW. 2008. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 10:2377–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Llosa M, Roy C, Dehio C. 2009. Bacterial type IV secretion systems in human disease. Mol. Microbiol. 73:141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dessaux Y, Petit A, Tempe J. 1993. Chemistry and biochemistry of opines, chemical mediators of parasitism. Phytochemistry 34:31–38 [Google Scholar]
- 12. Gelvin SB, Thomashow MF, McPherson JC, Gordon MP, Nester EW. 1982. Sizes and map positions of several plasmid-DNA-encoded transcripts in octopine-type crown gall tumors. Proc. Natl. Acad. Sci. U. S. A. 79:76–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ooms G, et al. 1982. T-DNA organization in homogeneous and heterogeneous octopine-type crown gall tissues of Nicotiana tabacum. Cell 30:589–597 [DOI] [PubMed] [Google Scholar]
- 14. Montoya AL, Chilton MD, Gordon MP, Sciaky D, Nester EW. 1977. Octopine and nopaline metabolism in Agrobacterium tumefaciens and crown gall tumor cells: role of plasmid genes. J. Bacteriol. 129:101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aly KA, Baron C. 2007. The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens. Microbiology 153:3766–3775 [DOI] [PubMed] [Google Scholar]
- 17. Zahrl D, et al. 2005. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology 151:3455–3467 [DOI] [PubMed] [Google Scholar]
- 18. Llosa M, Zupan J, Baron C, Zambryski P. 2000. The N- and C-terminal portions of the Agrobacterium VirB1 protein independently enhance tumorigenesis. J. Bacteriol. 182:3437–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baron C, Llosa M, Zhou S, Zambryski PC. 1997. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1. J. Bacteriol. 179:1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zupan J, Hackworth CA, Aguilar J, Ward D, Zambryski P. 2007. VirB1* promotes T-pilus formation in the vir-type IV secretion system of Agrobacterium tumefaciens. J. Bacteriol. 189:6551–6563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones AL, Shirasu K, Kado CI. 1994. The product of the virB4 gene of Agrobacterium tumefaciens promotes accumulation of VirB3 protein. J. Bacteriol. 176:5255–5261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Das A, Xie YH. 2000. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182:758–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chandran V, et al. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462:1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fronzes R, et al. 2009. Structure of a type IV secretion system core complex. Science 323:266–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cascales E, Christie PJ. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Atmakuri K, Cascales E, Christie PJ. 2004. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 54:1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gomis-Rüth FX, et al. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637–641 [DOI] [PubMed] [Google Scholar]
- 28. Savvides SN, et al. 2003. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 22:1969–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yeo HJ, Savvides SN, Herr AB, Lanka E, Waksman G. 2000. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol. Cell 6:1461–1472 [DOI] [PubMed] [Google Scholar]
- 30. Cascales E, Atmakuri K, Liu Z, Binns AN, Christie PJ. 2005. Agrobacterium tumefaciens oncogenic suppressors inhibit T-DNA and VirE2 protein substrate binding to the VirD4 coupling protein. Mol. Microbiol. 58:565–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Atmakuri K, Ding Z, Christie PJ. 2003. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol. Microbiol. 49:1699–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kumar RB, Das A. 2002. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol. Microbiol. 43:1523–1532 [DOI] [PubMed] [Google Scholar]
- 33. Aguilar J, Zupan J, Cameron TA, Zambryski PC. 2010. Agrobacterium type IV secretion system and its substrates form helical arrays around the circumference of virulence-induced cells. Proc. Natl. Acad. Sci. U. S. A. 107:3758–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thorstenson YR, Zambryski PC. 1994. The essential virulence protein VirB8 localizes to the inner membrane of Agrobacterium tumefaciens. J. Bacteriol. 176:1711–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yuan Q, et al. 2005. Identification of the VirB4-VirB8-VirB5-VirB2 pilus assembly sequence of type IV secretion systems. J. Biol. Chem. 280:26349–26359 [DOI] [PubMed] [Google Scholar]
- 36. Ward DV, Draper O, Zupan JR, Zambryski PC. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. U. S. A. 99:11493–11500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Judd PK, Kumar RB, Das A. 2005. Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc. Natl. Acad. Sci. U. S. A. 102:11498–11503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Judd PK, Kumar RB, Das A. 2005. The type IV secretion apparatus protein VirB6 of Agrobacterium tumefaciens localizes to a cell pole. Mol. Microbiol. 55:115–124 [DOI] [PubMed] [Google Scholar]
- 39. Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ. 2007. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J. 26:2540–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matthysse AG. 1987. Characterization of nonattaching mutants of Agrobacterium tumefaciens. J. Bacteriol. 169:313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Binns AN. 1991. Transformation of wall deficient cultured tobacco protoplasts by Agrobacterium tumefaciens. Plant Physiol. 96:498–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Merritt PM, Danhorn T, Fuqua C. 2007. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J. Bacteriol. 189:8005–8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tomlinson AD, Fuqua C. 2009. Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens. Curr. Opin. Microbiol. 12:708–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bardy SL, Maddock JR. 2007. Polar explorations: recent insights into the polarity of bacterial proteins. Curr. Opin. Microbiol. 10:617–623 [DOI] [PubMed] [Google Scholar]
- 45. Gilmour MW, Taylor DE. 2004. A subassembly of R27-encoded transfer proteins is dependent on TrhC nucleoside triphosphate-binding motifs for function but not formation. J. Bacteriol. 186:1606–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gilmour MW, Lawley TD, Rooker MM, Newnham PJ, Taylor DE. 2001. Cellular location and temperature-dependent assembly of IncHI1 plasmid R27-encoded TrhC-associated conjugative transfer protein complexes. Mol. Microbiol. 42:705–715 [DOI] [PubMed] [Google Scholar]
- 47. Gunton JE, Gilmour MW, Alonso G, Taylor DE. 2005. Subcellular localization and functional domains of the coupling protein, TraG, from IncHI1 plasmid R27. Microbiology 151:3549–3561 [DOI] [PubMed] [Google Scholar]
- 48. Shih YL, Rothfield L. 2006. The bacterial cytoskeleton. Microbiol. Mol. Biol. Rev. 70:729–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Domínguez-Escobar J, et al. 2011. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333:225–228 [DOI] [PubMed] [Google Scholar]
- 50. Garner EC, et al. 2011. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333:222–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Daniel RA, Errington J. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767–776 [DOI] [PubMed] [Google Scholar]
- 52. Zupan JR, Citovsky V, Zambryski P. 1996. Agrobacterium VirE2 protein mediates nuclear uptake of single-stranded DNA in plant cells. Proc. Natl. Acad. Sci. U. S. A. 93:2392–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zambryski PC. 1992. Chronicles from the Agrobacterium-plant cell DNA transfer story. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43:465–490 [Google Scholar]
- 54. Khan SR, Gaines J, Roop RM, II, Farrand SK. 2008. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl. Environ. Microbiol. 74:5053–5062 [DOI] [PMC free article] [PubMed] [Google Scholar]