ABSTRACT
Carbapenem-resistant Enterobacteriaceae (CRE) have emerged as major causes of health care-associated infections worldwide. This diverse collection of organisms with various resistance mechanisms is associated with increased lengths of hospitalization, costs of care, morbidity, and mortality. The global spread of CRE has largely been attributed to dissemination of a dominant strain of Klebsiella pneumoniae producing a serine β-lactamase, termed K. pneumoniae carbapenemase (KPC). Here we report an outbreak of KPC-producing CRE infections in which the degree of horizontal transmission between strains and species of a promiscuous plasmid is unprecedented. Sixteen isolates, comprising 11 unique strains, 6 species, and 4 genera of bacteria, were obtained from 14 patients over the first 8 months of the outbreak. Of the 11 unique strains, 9 harbored the same highly promiscuous plasmid carrying the KPC gene blaKPC. The remaining strains harbored distinct blaKPC plasmids, one of which was carried in a strain of Klebsiella oxytoca coisolated from the index patient and the other generated from transposition of the blaKPC element Tn4401. All isolates could be genetically traced to the index patient. Molecular epidemiological investigation of the outbreak was aided by the adaptation of nested arbitrary PCR (ARB-PCR) for rapid plasmid identification. This detailed molecular genetic analysis, combined with traditional epidemiological investigation, provides insights into the highly fluid dynamics of drug resistance transmission during the outbreak.
IMPORTANCE
The ease of horizontal transmission of carbapenemase resistance plasmids across strains, species, and genera of bacteria observed in this study has several important public health and epidemiological implications. First, it has the potential to promote dissemination of carbapenem resistance to new populations of Enterobacteriaceae, including organisms of low virulence, leading to the establishment of reservoirs of carbapenem resistance genes in patients and/or the environment and of high virulence, raising the specter of untreatable community-associated infections. Second, recognition of plasmid-mediated outbreaks, such as those described here, is problematic because analysis of resistance plasmids from clinical isolates is laborious and technically challenging. Adaptation of nested arbitrary PCR (ARB-PCR) to investigate the plasmid outbreak facilitated our investigation, and the method may be broadly applicable to other outbreaks due to other conserved mobile genetic elements. Whether infection control measures that focus on preventing transmission of drug-resistant clones are effective in controlling dissemination of these elements is unknown.
Introduction
Enterobacteriaceae are responsible for a significant number of infections and death in the United States and worldwide each year, and the prevalence of antibiotic resistance in this family of bacteria continues to rise (1). One reason for this increase has been the dissemination of Klebsiella pneumoniae carbapenemase (KPC), a class A serine carbapenemase first isolated from K. pneumoniae in 1996 (2). blaKPC encodes KPC and is carried within the conserved Tn3 family transposon Tn4401 on transferable plasmids (3). A dominant strain of KPC-producing K. pneumoniae, multilocus sequence type (ST) 258, has disseminated throughout the United States and other parts of the world (4, 5). Institutional outbreaks of KPC-producing Enterobacteriaceae due to the spread of a single strain have also been reported for other species, including Enterobacter spp. and Serratia marcescens (6, 7). While these reports highlight the clonal nature of dissemination of carbapenem-resistant Enterobacteriaceae (CRE), several cases of horizontal transfer of blaKPC through transferable plasmids have also been described (7–9).
Tn4401, a 10-kb Tn3 family transposon, serves as the genetic support structure for blaKPC and is highly conserved. Tn4401 has been characterized in more than seven different plasmids, ranging 12 to 80 kb in size, each with a distinct insertion site (3, 10). While reassortment of the transposon has been found in an isolate from China (11), all other reports and deposited sequences in Tn4401 to GenBank to date have a largely conserved structure and sequence.
In August 2007 we identified the first known case of CRE at our institution (12), which prompted us to screen all clinical isolates of extended-spectrum β-lactamase (ESBL)-producing enterobacteriaceae for carbapenemase production. In contrast to other reported clusters, we observed a diversity of species and genera of CRE from the outset of the outbreak. In this article, we describe the molecular epidemiological characteristics of this heterogeneous outbreak of KPC-producing CRE.
RESULTS
Detection of KPC and in vitro antimicrobial susceptibility.
Approximately 280 members of the Enterobacteriaceae were screened for carbapenemase production by modified Hodge analysis during the study period. Sixteen isolates (5.7%) from 14 patients had positive phenotypic carbapenemase tests. Four genera and six species were represented: Enterobacter cloacae (n = 6), K. pneumoniae (n = 4), Klebsiella oxytoca (n = 3), Escherichia coli (n = 1), Enterobacter asburiae (n = 1), and Citrobacter freundii (n = 1) (Table 1). Two (14%) of 14 patients harbored clinical isolates of more than one CRE species. PCR analysis determined that all isolates carried blaKPC. According to 2009 Clinical and Laboratory Standards Institute (CLSI) breakpoints, most isolates were susceptible to imipenem (81%) by automated susceptibility testing, while 1 (6%) and 4 (25%) isolates were ertapenem susceptible and intermediate, respectively (Table 1). Disk diffusion did not accurately predict in vitro resistance; 15 (94%) isolates were imipenem or meropenem susceptible, while one isolate (6%) was intermediate (Table 1 and data not shown).
TABLE 1 .
Isolate description and carbapenem susceptibilitiesa
| Patient | Isolate | Species | Date of first isolationb |
Source(s) | MIC (µg/ml) determined by Vitek 2 |
Imipenem disk diffusion (mm) |
Pulsed- field group |
Sequence group |
Plasmid | Outcome | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Imipenem | Ertapenem | ||||||||||
| 1 | Kox1015 | Klebsiella oxytoca | 8/31/2007 | Blood, drain, BAL fluid |
4 | ≥8 | 16 | pfg-kox1 | sg2 | pUVA02 | Died |
| Kpn1016 |
Klebsiella
pneumoniae |
8/31/2007 | Drain, sputum |
≥16 | ≥8 | 16 | pfg-kpn1 | sg1 | pUVA01 | Died | |
| 2 | Kpn1017 |
Klebsiella pneumoniae |
10/05/2007 | Blood, sputum, urine |
8 | 4 | 17 | pfg-kpn1 | sg1 | pUVA01 | Died |
| 3 | Ecl1026 | Enterobacter cloacae | 11/30/2007 | Sputum, urine |
4 | ≥8 | 20 | pfg-ecl3 | sg1 | pUVA01 | Recovered |
| 4 | Ecl1027 | Enterobacter cloacae | 12/09/2007 | Urine | 2 | 4 | 19 | pfg-ecl2 | sg1 | pUVA01 | Recovered |
| 5 | Kox1028 | Klebsiella oxytoca | 1/15/2008 | Blood, sputum |
4 | ≥8 | 20 | pfg-kox1 | sg2 | pUVA02 | Died |
| 6 | Ecl1032 | Enterobacter cloacae | 1/24/2008 | Blood, ascites fluid |
≥16 | ≥8 | 14 | pfg-ecl4 | sg1 | pUVA01 | Recovered |
| 7 | Eco1036 | Escherichia coli | 2/7/2008 | BAL fluid | ≤1 | 1 | 19 | n/ac | sg3 | pUVA03 | Recovered |
| 8 | Ecl1034 | Enterobacter cloacae | 2/15/2008 | Blood, sputum |
2 | ≥8 | 18 | pfg-ecl1 | sg1 | pUVA01 | Died |
| 9 | Ecl1035 | Enterobacter cloacae | 2/20/2008 | Blood, urine |
4 | ≥8 | 18 | pfg-ecl1 | sg1 | pUVA01 | Died |
| 10 | Ecl1033 | Enterobacter cloacae | 2/20/2008 | BAL fluid, sputum |
2 | ≥8 | 17 | pfg-ecl1 | sg1 | pUVA01 | Recovered |
| 11 | Eas1043 |
Enterobacter
asburiae |
3/18/2008 | BAL fluid, sputum, wound |
4 | 4 | 19 | n/ac | sg1 | pUVA01 | Died |
| Kpn1041 |
Klebsiella
pneumoniae |
4/8/2008 | Blood, sputum |
2 | ≥8 | 21 | pfg-kpn2 | sg1 | pUVA01 | Died | |
| 12 | Cfr1047 | Citrobacter freundii | 3/21/2008 | Urine | ≤1 | 4 | 19 | n/ac | sg1 | pUVA01 | Died |
| 13 | Kox1039 | Klebsiella oxytoca | 3/31/2008 | Sputum, urine |
4 | ≥8 | 20 | pfg-kox1 | sg2 | pUVA02 | Recovered |
| 14 | Kpn1042 |
Klebsiella
pneumoniae |
4/6/2008 | Urine | ≤1 | ≥8 | 18 | pfg-kpn3 | sg1 | pUVA01 | Recovered |
Boldface values indicate intermediate or resistant susceptibilities by MIC testing. Patients 1 and 2 have been described previously (12).
Month/day/year.
n/a, not applicable.
Clinical outcomes.
All patients developed clinically significant infections with CRE (Table 1). All were adult inpatients with major comorbidities who had been hospitalized for a median of 31 days prior to the first isolation of CRE. A summary of the demographic and clinical characteristics are presented in Table 2. Cases were identified throughout the hospital: eight intensive care units, five medical or surgical wards, and one oncology care unit. Investigation of the outbreak revealed possible epidemiological links between most but not all of the cases. While 93% had received broad-spectrum antibiotics prior to infection with CRE, only 36% had received a carbapenem. All-cause 28-day mortality was 50%, and mortality among patients with bacteremia was 86% (Table 2).
TABLE 2 .
Demographics and clinical characteristics
| Characteristic | Value for case patientsa |
|---|---|
| Mean age, yr (range) | 60 (23–81) |
| Male | 8 (57) |
| Comorbid conditions | |
| Transplant recipient | 5 (36) |
| Diabetes mellitus | 4 (29) |
| Renal insufficiency | 4 (29) |
| Chronic heart disease | 3 (21) |
| Malignancy | 2 (14) |
| Health care-associated risk factors | |
| ICU stay > 48 h | 9 (64) |
| Any ICU stay | 11 (79) |
| Median length of stay prior to infection, days (range) |
31 (3–144) |
| Prior anti-Gram-negative antibiotic | 13 (93) |
| Beta-lactam/beta-lactamase inhibitor | 8 (57) |
| Fluoroquinolone | 7 (50) |
| Broad-spectrum cephalosporin | 5 (36) |
| Carbapenem | 5 (36) |
| Monobactam | 2 (14) |
| Clinical infection | |
| Bacteremia | 7 (50) |
| Urinary tract infection | 7 (50) |
| Pneumonia | 7 (50) |
| Intra-abdominal infection | 3 (21) |
| Mortality | 7 (50) |
| Bacteremia | 6 (86) |
n = 14. Data are no. (%) of patients unless otherwise indicated.
Genetic evaluation of Tn4401.
Bidirectional sequencing of blaKPC from isolates Kox1015, Kpn1016, and Eco1036 and alignment analyses revealed 100% identity to the blaKPC-2 allele. Using the primer sets listed in Table S1 in the supplemental material, PCR screening showed that all isolates carried blaKPC and all other putative coding elements of Tn4401 and had a genetic organization congruent with other previous reports (see Fig. S1) (3).
Several isoforms of Tn4401 have been characterized, differing by the presence of deletions in the intergenic region between the istB and blaKPC genes (3, 4). In this study, PCR amplification of the istB-blaKPC intergenic region in all 16 isolates produced amplicons consistent with a 757-bp product (see Fig. S1 in the supplemental material; also data not shown), demonstrating that none contained a large deletion in this nonconserved region of Tn4401.
Genetic relatedness.
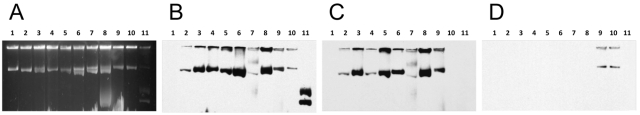
Pulsed-field gel electrophoresis (PFGE) was performed on species that were represented by more than one isolate. A striking degree of genetic diversity was observed among the E. cloacae and K. pneumoniae isolates: four pulsed-field groups (pfg’s) were identified among the six E. cloacae isolates, and three pfg’s were identified among the four K. pneumoniae isolates. In contrast, all three K. oxytoca isolates were of a single pfg (Fig. 1; see also Table 1).
FIG 1 .
Pulsed-field gel electrophoresis of outbreak isolates of the same species. (A) All three K. oxytoca isolates are a single strain. (B) Four K. pneumoniae isolates are three distinct strains. (C) Six E. cloacae isolates are four distinct strains.
Plasmid evaluation.
To identify the causative KPC-encoding plasmid during the course of the investigation, we adapted the nested arbitrary PCR (ARB-PCR) method to identify the DNA sequence flanking the Tn4401 insertion site. A schema of this method is presented in Fig S2 in the supplemental material. This technique has previously been used in genetic studies of bacterial virulence to rapidly identify the insertion site of transposon chromosomal disruptions (13, 14).
Tn4401-flanking DNA sequence was obtained from 15 isolates; one isolate, Ecl1032, could not be analyzed by the ARB-PCR method despite repeated attempts. Sequence lengths were on average 561 bp (range, 249 to 703 bp). Flanking sequences of 11 isolates were determined to be of the same sequence group (sg), sg1, suggesting that the isolates carried the same KPC-encoding plasmid. The Tn4401 insertion site for sg1 was in the transposase gene tnpA (99.6% identity over 509 bp; E value, 5e−202; GenBank accession no. FJ410927). The three related K. oxytoca isolates were of the same sequence group, sg2, with a Tn4401 insertion site in a novel open reading frame. The solitary E. coli isolate has a unique sequence group, sg3, characterized by a Tn4401 insertion site in a gene encoding a MobA-like relaxase (97.9% identity over 390 bp; GenBank accession no. AY589571). In all cases, the Tn4401 insertion sites identified by these sequence groups were distinct from corresponding transposon insertion sites found in previously described blaKPC-containing plasmids.
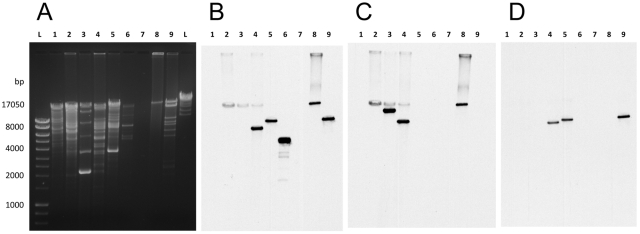
To verify the findings obtained by ARB-PCR, Southern blot analysis of plasmid DNA from isolates representing each pfg was performed (Fig. 2). Gel electrophoresis demonstrated that pUVA01, pUVA02, and pUVA03 were of 54, 80, and 22-kb, respectively (12; data not shown), and Southern hybridization confirmed that all isolates harbored blaKPC plasmids (Fig. 2A and B). Additional Southern hybridization confirmed that all sg1 isolates (but not the sg2 or sg3 isolates) and Ecl1032 hybridized to the sg1-specific probe, while only the K. oxytoca and C. freundii isolates hybridized to the sg2-specific probe (Fig. 2C and D).
FIG 2 .
Plasmid electrophoresis and Southern blot analysis of all unique outbreak clones. (A) Plasmid DNA electrophoresis resolved on a 0.6% agarose gel stained with ethidium bromide. Southern blot of gel A with a blaKPC-specific probe (B), sg1-specific probe (C), or sg2-specific probe (D). Lanes: 1, Kpn1014 (negative control); 2 to 4, K. pneumoniae; 5 to 7, E. cloacae; 8, E. asburiae; 9, C. freundii; 10, K. oxytoca; 11, E. coli. Supercoiled and relaxed blaKPC-hybridizing plasmid DNA was obtained from E. coli strain Eco1036.
Functional carbapenemase encoded by plasmid-borne blaKPC was confirmed by the generation of the pUVA01 and pUVA02 transformants AMGH01-T and AMGH07-T, respectively. The transformants had positive modified Hodge tests and were confirmed by PCR to carry blaKPC, all other elements of Tn4401, and the original sg designation. Interestingly, AMGH01-T had significantly reduced carbapenem MICs compared to those of Kpn1016, whereas carbapenem MICs for AMGH07-T were comparable those for Kox1015 (Table 2).
Plasmid DNA of the transformants and representative CRE isolates was subjected to restriction digestion with KpnI and Southern blot analysis using blaKPC, sg1, and sg2 hybridization probes (Fig. 3). For plasmid digests of the sg1 isolates Kpn1016, Ecl1027, Cfr1047, and AMGH01-T, the blaKPC and sg1 probes hybridized to a 20-kb fragment. Similarly, for digests of the sg2 isolate K. oxytoca Kox1028 and AMGH07-T, the blaKPC and sg2 probes hybridized to a 10-kb fragment. Plasmid digest of the single sg3 isolate, E. coli Eco1036, hybridized to the blaKPC probe but neither the sg1 nor the sg2 probe. These results confirmed that sequence groups determined by ARB-PCR correctly identified the resistance plasmid carried by the CRE strain.
FIG 3 .
Southern blot analysis of KpnI-digested plasmid DNA of representative outbreak clones and resistance plasmid transformants. (A) Restriction digests of plasmid DNA electrophoresis resolved on a 0.6% agarose gel stained with ethidium bromide. (B to D) Southern blot of gel A with a blaKPC-specific probe (B), sg1-specific probe (C), or sg2-specific probe (D). Lanes: L, ladder; 1, Kpn1014 (negative control); 2, Kpn1016; 3, Ecl1027; 4, Cfr1047; 5, Kox1028; 6, Eco1036; 7, blank; 8, AMGH01-T; 9, AMGH07-T.
Southern blot hybridization of Cfr1047 with the blaKPC probe also revealed a second, lower-molecular-weight fragment, suggesting the presence of two plasmids with the KPC gene (Fig 3). Furthermore, a second band in both Ecl1027 and Cfr1047 hybridized to the sg1 but not the blaKPC probe. Together, these results demonstrate that the clonal K. oxytoca isolates harbor a single KPC resistance plasmid, pUVA02, the single E. coli isolate carries a unique blaKPC-containing plasmid, pUVA03, and the remaining 12 isolates (representing 9 [82%] of the 11 unique outbreak strains) carry a highly promiscuous KPC resistance plasmid, pUVA01. Southern blotting also demonstrated that Cfr1047 harbors a second uncharacterized blaKPC plasmid and that Ecl1027 and Cfr1047 harbor plasmids that share homology with the Tn4401 plasmids identified by ARB-PCR. These results illustrate the diverse nature of assortments of resistance plasmids found in multidrug-resistant members of the Enterobacteriaceae (15).
Combining the molecular epidemiology of strain and resistance plasmid typing with classic epidemiological evaluation of CRE infection provides valuable insight into the dynamics of blaKPC dissemination during this outbreak (Fig. 4). Transmission of carbapenem resistance occurred primarily through horizontal interstrain or interspecies transfer of the Tn4401-bearing plasmid pUVA01 and, to a lesser extent, transmission of isogenic strains between patients. The spread of blaKPC correlated well with epidemiological risk factors, including the patient’s geographical location, health care team, and time period of possible exposure (Table 3; see also Fig. S3 in the supplemental material). We hypothesize that pUVA03 acquired Tn4401 via mobilization from pUVA01 or pUVA02, given the congruity of the transposon structure and the patient’s epidemiological risk factors. Consequently, all CRE infections appear to be linked to the index patient.
FIG 4 .
Hypothesized possible route(s) of spread of blaKPC between patients based on plasmid profiles and epidemiological risk for CRE acquisition. Each number denotes a patient. Shape and color denote the plasmid type: blue circle, pUVA01; red triangle, pUVA02; black diamond, pUVA03. Solid arrows denote a possible route of spread based on shared plasmids and epidemiological risk factors. Dashed arrows denote a possible route of spread without a shared plasmid type. Note that arrows do not necessarily signify direct patient-to-patient transmission; the spread of CRE may have occurred to a source that shared the same epidemiological space as the infected patient.
TABLE 3 .
Transformed strains
| Strain | Plasmid | MHT resulta | MIC (µg/ml) determined by Vitek 2 |
|
|---|---|---|---|---|
| Imipenem | Ertapenem | |||
| AMGH01-T | pUVA01 | + | ≤1 | ≤0.5 |
| AMGH07-T | pUVA02 | + | 4 | ≥8 |
MHT, modified Hodge test.
DISCUSSION
While sometimes difficult to recognize, plasmids have previously been linked to outbreaks of drug-resistant Gram-negative bacteria. In 1985, O’Brien and colleagues reported an outbreak of plasmid-mediated gentamicin resistance, which moved to six species of the Enterobacteriaceae over 8 years in geographically diverse areas of the United States and Venezuela (16). Outbreaks of enterobacteriaceae resistant to sulfonamides, β-lactams hydrolyzed by ESBL, aminoglycosides, and fluoroquinolones have also been reported (17–21). Most recently, a multispecies outbreak of plasmid-mediated VIM-1 metallo-carbapenemase has been reported in Spain (22), and A/C-type plasmid carriage of the New Delhi metallo-β-lactamase gene blaNDM-1 has emerged in Enterobacteriaceae in India and the United Kingdom (22). While movement of blaKPC-containing plasmids between Enterobacteriaceae has been described (7, 8, 23, 24), the degree of horizontal transmission between strains and species observed in this outbreak is unprecedented.
Why did the outbreak of CRE occur in such a wide assortment of strains in our institution? One possibility is that the plasmid-mediated nature of the outbreak may indicate that pUVA01 is more mobile than other blaKPC plasmids. This is supported by the observation that pUVA01 moved between a wide array of strains over a short period of time, often in the absence of carbapenem pressure. In contrast, horizontal transmission of pUVA02 or pUVA03 was not observed during the study period. Clearly, facile transfer of blaKPC plasmids adds an additional layer of complexity to the epidemiology of this rapidly spreading resistance mechanism. A second possibility is that by screening all possible ESBL-producing Enterobacteriaceae, we may have captured non- or weakly lactose-fermenting KPC-producing organisms that may have been overlooked in a targeted surveillance and evaluation program, as well as organisms with low carbapenem MICs that would otherwise have been considered carbapenem susceptible. Regardless of the etiology, we predict that the diverse nature of this outbreak will render detection and control of CRE spread even more problematic if it becomes increasingly widespread.
One important aspect of this investigation was the use of ARB-PCR to rapidly identify causative KPC resistance plasmids during the investigation. This technology allowed us to quickly recognize the high degree of genetic relatedness among the outbreak isolates at the level of plasmid carriage—relationships missed by traditional species- and strain-based analysis. Investigations of so-called “plasmid outbreaks” are difficult for all but sophisticated molecular epidemiology labs. Analysis of large resistance plasmids from clinical isolates of the Enterobacteriaceae, such as the ones observed in this study, can be laborious and technically challenging. ARB-PCR allows rapid determination of DNA sequences that flank a conserved element, which can be used as the molecular signature for a transmissible plasmid. Given its simplicity, ARB-PCR should be translatable to not only transposons but also resistance genes and other conserved components of mobile DNA elements. ARB-PCR does have several limitations. A product may not able to be generated from all samples, as was seen in this study. However, we were able to obtain products in all but one of our isolates without optimization of the protocol conditions or primer sets. Another limitation is that only one product will be generated from an isolate, even if it contains more than one plasmid with blaKPC, as was found with isolate Cfr1047. Finally, generation of flanking sequence does not ensure that the product necessarily has a plasmid origin; transfer of drug resistance to a recipient bacterial strain and demonstration of a plasmid-encoded drug-resistant determinant are required to prove the presence of a resistance plasmid in the donor strain.
The ease of horizontal transmission of carbapenem resistance observed in this study has serious public health and epidemiological implications. Dissemination of blaKPC through mobile genetic elements could allow carbapenem resistance to move to the community, as has been observed with the worldwide emergence of community-onset CTX-M-15 β-lactamase-producing E. coli (25–28). Additionally, transmission of blaKPC to enterobacteriaceae of reduced or no virulence could lead to the establishment of a reservoir of carbapenem resistance genes in patients and/or the environment. Understanding the dynamics of blaKPC horizontal gene transfer will be crucial to limiting the spread of CRE, since epidemiological and containment efforts must focus not only on the spread of multidrug-resistant strains but also on the associated resistance plasmids and transposons.
MATERIALS AND METHODS
Patients and bacterial isolates.
This study was conducted at the University of Virginia (UVA) Medical Center, a 619-bed tertiary care hospital in central Virginia, from 1 September 2007 to 30 April 2008. Following the detection of the first case of CRE at UVA (12), all clinical Enterobacteriaceae isolates identified as possible extended-spectrum β-lactamase (ESBL) producers by automated testing (Vitek 2; bioMérieux, Durham, NC) were prospectively screened for carbapenemase production using the direct carbapenemase and modified Hodge tests (29, 30). Species identification and antibiotic susceptibility testing were performed using the Vitek 2 system with the AST-GN18 card (bioMérieux). Imipenem susceptibility was determined by the Kirby-Bauer disk-diffusion and broth microdilution methods in accordance with CLSI guidelines (31). Patient characteristics, antibiotic exposure, and outcomes were obtained from clinical records. For this study, epidemiological risk factors for acquisition of CRE were defined as hospitalization on the same floor as a patient with CRE infection or receipt of care from a health care team concurrently caring for a patient with CRE infection. Infection was defined as isolation of CRE from a clinical specimen which necessitated treatment. Mortality was defined as death from any cause within 28 days of the latest positive clinical culture with CRE. The study protocol was approved by the UVA Institutional Review Board for Health Sciences Research (no. 13558).
Bacteria, media, and reagents.
Strains and oligonucleotides are listed in Table 1 (see also Table S1 in the supplemental material). Enzymes were obtained from New England Biolabs (Ipswich, MA). Strains were routinely grown at 37°C on Luria-Bertani (LB) broth or agar supplemented with freshly prepared 0.1-µg/ml meropenem (AstraZeneca, Wilmington, DE). All strains were stored at −75°C in LB broth containing 15% glycerol. DNA sequencing was performed by the UVA Biomolecular Research Facility (Charlottesville, VA).
Genetic confirmation of blaKPC and Tn4401.
PCR amplification of blaKPC was performed on all isolates as previously reported (2), and blaKPC allele identification was performed by bidirectional DNA sequencing for a representative strain of each strain type. Confirmation of additional elements of the Tn4401 transposon was performed by PCR using the primer sets listed in Table S1 in the supplemental material. For each sample, whole-cell lysate was prepared as follows: an individual bacterial colony was suspended in 100 µl sterile water, boiled at 95°C for 10 min, and centrifuged at 16,100 × g for 1 min to sediment cell debris, and 2 µl of the supernatant was used as template DNA for PCR. PCR experiments were carried out using the following conditions: 3 min of initial denaturation at 95°C, and 30 cycles of 30 s of denaturation at 95°C, 30 s of annealing at 57°C, a 45-s extension at 72°C, and a final extension of 5 min at 72°C with positive and negative K. pneumoniae controls for each run.
PFGE.
Genotypic analysis was performed by PFGE of XbaI-digested total DNA with the Chef mapper system (Bio-Rad, Hercules, CA) using conditions outlined in the PulseNet protocol for the Enterobacteriaceae (32). Dendrogram construction was performed using the Dice coefficient and unweighted-pair group method using average linkages (UPGMA) clustering (BioNumerics 5.10; Applied Maths, Austin, TX). For this study, isolates of the same species were considered to share a common pulsed-field group (pfg) if they had a Dice similarity coefficient greater than 0.90.
Rapid determination of Tn4401 insertion site: ARB-PCR.
DNA sequences flanking the Tn4401 plasmid insertion site were determined using the nested arbitrary PCR (ARB-PCR) method (13, 14). The method entails two sequential rounds of PCR amplification using the transposon-specific primer AM14 and the arbitrary primer Arb1.1 in the first round and the nested primers AM11 and Arb2.1 in the second round, followed by DNA sequencing using primer AM13 (see Table S1 in the supplemental material). K. pneumoniae Kpn1014, which lacks a KPC resistance plasmid, and sterile double-distilled water (ddH2O) served as negative controls for both rounds of PCR amplification and DNA sequencing. Each isolate underwent ARB-PCR and DNA sequencing twice, and the resultant Tn4401 insertion site sequences were aligned using the ClustalW software program to assess the degree of identity. Isolates carrying plasmids with DNA sequences flanking the Tn4401 integrant that had a BLAST E value of ≤1e−50 and ≥95% identity by pairwise analysis and the same transposon insertion site, as determined by visual inspection, were considered to be of the same sequence group (sg). Unidirectional sequencing of the region flanking Tn4401 (~1.5 to 2.4 kb) was performed on distinctive plasmids.
Plasmid characterization.
To differentiate between plasmids isolated in the study, primer sets targeting the sequence groups were used for PCR analysis (see Table S1 in the supplemental material).
Southern blot hybridization using sg-specific amplicons as probes was performed to confirm sg designations. Plasmid DNA was isolated using the Qiagen Plasmid Midi kit (Qiagen) using 60°C prewarmed eluting buffer. Uncut plasmid DNA was subjected to electrophoresis in a 0.6% UltraPure DNA-grade agarose (Bio-Rad) gel at 70 V for 20 h at 4°C in 0.5× Tris-borate-EDTA buffer. Migration distances of the DNA were compared to reference plasmids of E. coli strain V517. For restriction site analysis, plasmid DNA was digested with KpnI at 37°C overnight and subjected to electrophoresis in a 0.7% agarose gel at 80 V for 4 h in Tris-acetate-EDTA buffer. For Southern blot hybridization, plasmid DNA was immobilized to an Amersham Hybond N+ nylon membrane (GE Healthcare, Piscataway, NJ) via capillary transfer and cross-linked in a UV cross-linker (Agilent Technologies, Santa Clara, CA). Membranes were sequentially hybridized with PCR amplicon probes generated for blaKPC, sg1, and sg2 (see Table S1 in the supplemental material) labeled using the ECL Direct nucleic acid labeling and detection system (GE Healthcare) in accordance with the manufacturer’s instructions.
Transfer of plasmid and carbapenemase phenotype.
In order to determine if carbapenem resistance was attributable to carriage of the Tn4401 element, plasmid DNA extracts from Kox1015 (for pUVA02) and Kpn1016 (for pUVA01) were transferred into E. coli GeneHog (Invitrogen) by electroporation. Transformants were selected on LB medium supplemented with meropenem. Phenotypic carbapenemase resistance of the transformants was determined by the modified Hodge test (29). Clinical isolates and transformants were verified to carry resistance plasmids via PCR analyses and Southern blot hybridization.
SUPPLEMENTAL MATERIAL
Organization of the Tn4401 transposon. (A) Schematic representation of the genetic organization of transposon Tn4401 (~10 kb) with designated open reading frames, including blaKPC. Gray triangles with horizontal lines depict primer sets and PCR products of elements spanning all open reading frames; sizes of products are listed below. Adapted from the work of Naas et al. (3). (B) Representative electrophoresis gel with 100-bp ladder from a representative CRE isolate (Cfr1047) demonstrating the presence of all predicted products numbered in panel A: L, ladder; 1, tnpR-tnpA; 2, tnpA-istA; 3, istA-istB; 4, istB-blaKPC; 5, blaKPC-tnpA*. Asterisks indicate the tnpA gene of ISKpn6. Download Figure S1, PDF file, 0.1 MB.
Schematic of the ARB-PCR method, used to determine the Tn4401 insertion site. Round 1 PCR is performed using the intertransposon outward-facing primer AM14 and arbitrary primer Arb1.1 with a fixed “tail” (black square), at 10 times the concentration. Round 2 PCR enriches for the region flanking the transposon by using the nested intertransposon outward-facing primer AM11 and primer Arb2.1 at 10 times the concentration, which anneals to the fixed tail of Arb1.1. Unidirectional sequencing is performed using a third nested intertransposon primer. Download Figure S2, PDF file, 0.1 MB.
Representations of outbreak isolates or plasmid carriage during the investigation. (A) Representation of the outbreak by species and strain of the CRE isolate through the outbreak. (B) Representation of the outbreak by the blaKPC-containing plasmid of the CRE isolate through the outbreak. Download Figure S3, PDF file, 0.6 MB.
Primers used in the study.
ACKNOWLEDGMENTS
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
This work was supported by a Howard Hughes Medical Institute Early Career Award to C.D.S. and the UVA Biodefense Research Training and Career Development Program (NIH grant T32AI055432).
We thank Kate Barowski for providing technical assistance and Barry Farr, Jean Patel, and Kyle Enfield for comments on the project and the manuscript.
W.M.S. and C.D.S. have received research funding from Wyeth. All other authors declare that they have no conflicts of interests.
Footnotes
Citation Mathers AJ, et al. 2011. Molecular dissection of an outbreak of carbapenem-resistant enterobacteriaceae reveals intergenus KPC carbapenemase transmission through a promiscuous plasmid. mBio 2(6):e00204-11. doi:10.1128/mBio.00204-11.
REFERENCES
- 1. Rhomberg PR, Deshpande LM, Kirby JT, Jones RN. 2007. Activity of meropenem as serine carbapenemases evolve in US Medical Centers: monitoring report from the MYSTIC Program (2006). Diagn. Microbiol. Infect. Dis. 59:425–432 [DOI] [PubMed] [Google Scholar]
- 2. Yigit H, et al. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Naas T, et al. 2008. Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene. Antimicrob. Agents Chemother. 52:1257–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kitchel B, et al. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cuzon G, et al. 2010. Worldwide diversity of Klebsiella pneumoniae that produce beta-lactamase blaKPC-2 gene. Emerging Infect. Dis. 16:1349–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marchaim D, Navon-Venezia S, Schwaber MJ, Carmeli Y. 2008. Isolation of imipenem-resistant Enterobacter species: emergence of KPC-2 carbapenemase, molecular characterization, epidemiology, and outcomes. Antimicrob. Agents Chemother. 52:1413–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cai JC, Zhou HW, Zhang R, Chen GX. 2008. Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob. Agents Chemother. 52:2014–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sidjabat HE, et al. 2009. Interspecies spread of Klebsiella pneumoniae carbapenemase gene in a single patient. Clin. Infect. Dis. 49:1736–1738 [DOI] [PubMed] [Google Scholar]
- 9. Goren MG, et al. 2010. Transfer of carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patient. Emerging Infect. Dis. 16:1014–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gootz TD, et al. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City Hospital. Antimicrob. Agents Chemother. 53:1998–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen P, et al. 2009. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53:4333–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mathers AJ, et al. 2009. Fatal cross infection by carbapenem-resistant Klebsiella in two liver transplant recipients. Transpl. Infect. Dis. 11:257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Begun J, Sifri CD, Goldman S, Calderwood SB, Ausubel FM. 2005. Staphylococcus aureus virulence factors identified by using a high-throughput Caenorhabditis elegans-killing model. Infect. Immun. 73:872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O’Toole GA, et al. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91–109 [DOI] [PubMed] [Google Scholar]
- 15. John JF, Jr, Twitty JA. 1986. Plasmids as epidemiologic markers in nosocomial gram-negative bacilli: experience at a university and review of the literature. Rev. Infect. Dis. 8:693–704 [DOI] [PubMed] [Google Scholar]
- 16. O’Brien TF, et al. 1985. Intercontinental spread of a new antibiotic resistance gene on an epidemic plasmid. Science 230(4721):87–88 [DOI] [PubMed] [Google Scholar]
- 17. Tompkins LS, Plorde JJ, Falkow S. 1980. Molecular analysis of R-factors from multiresistant nosocomial isolates. J. Infect. Dis. 141:625–636 [DOI] [PubMed] [Google Scholar]
- 18. Bebora LC, Oundo JO, Yamamoto H. 1994. Resistance of E. coli strains, recovered from chickens to antibiotics with particular reference to trimethoprim-sulfamethoxazole (septrin). East Afr. Med. J. 71:624–627 [PubMed] [Google Scholar]
- 19. Wiener J, et al. 1999. Multiple antibiotic-resistant Klebsiella and Escherichia coli in nursing homes. JAMA 281:517–523 [DOI] [PubMed] [Google Scholar]
- 20. Bingen EH, et al. 1993. Molecular epidemiology of plasmid spread among extended broad-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates in a pediatric hospital. J. Clin. Microbiol. 31:179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Corkill JE, Anson JJ, Hart CA. 2005. High prevalence of the plasmid-mediated quinolone resistance determinant qnrA in multidrug-resistant Enterobacteriaceae from blood cultures in Liverpool, UK. J. Antimicrob. Chemother. 56:1115–1117 [DOI] [PubMed] [Google Scholar]
- 22. Tato M, et al. 2007. Complex clonal and plasmid epidemiology in the first outbreak of Enterobacteriaceae infection involving VIM-1 metallo-beta-lactamase in Spain: toward endemicity? Clin. Infect. Dis. 45:1171–1178 [DOI] [PubMed] [Google Scholar]
- 23. Rasheed JK, et al. 2008. Detection of the Klebsiella pneumoniae carbapenemase type 2 carbapenem-hydrolyzing enzyme in clinical isolates of Citrobacter freundii and K. oxytoca carrying a common plasmid. J. Clin. Microbiol. 46:2066–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goren MG, et al. 2010. Transfer of carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patient. Emerging Infect. Dis. 16:1014–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pitout JD, Laupland KB. 2008. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8:159–166 [DOI] [PubMed] [Google Scholar]
- 26. Rodríguez-Baño J, et al. 2006. Bacteremia due to extended-spectrum beta-lactamase-producing Escherichia coli in the CTX-M era: a new clinical challenge. Clin. Infect. Dis. 43:1407–1414 [DOI] [PubMed] [Google Scholar]
- 27. Pitout JD, Hanson ND, Church DL, Laupland KB. 2004. Population-based laboratory surveillance for Escherichia coli-producing extended-spectrum beta-lactamases: importance of community isolates with blaCTX-M genes. Clin. Infect. Dis. 38:1736–1741 [DOI] [PubMed] [Google Scholar]
- 28. Arpin C, et al. 2009. Nationwide survey of extended-spectrum β-lactamase-producing Enterobacteriaceae in the French community setting. J. Antimicrob. Chemother. 63:1205–1214 [DOI] [PubMed] [Google Scholar]
- 29. Lee K, et al. 2001. Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 7:88–91 [DOI] [PubMed] [Google Scholar]
- 30. Moland ES, Kim SY, Hong SG, Thompson KS. 2008. Newer β-lactamases: clinical and laboratory implications, part II. Clin. Microbiol. Newslett. 30:79–85 [Google Scholar]
- 31. Clinical and Laboratory Standards Institute 2008. Performance standards for antimicrobial susceptibility testing; 17th informational supplement, 2008; CLSI document M100-S18. Clinical and Laboratory Standards Institue, Wayne, PA [Google Scholar]
- 32. Ribot EM, et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Organization of the Tn4401 transposon. (A) Schematic representation of the genetic organization of transposon Tn4401 (~10 kb) with designated open reading frames, including blaKPC. Gray triangles with horizontal lines depict primer sets and PCR products of elements spanning all open reading frames; sizes of products are listed below. Adapted from the work of Naas et al. (3). (B) Representative electrophoresis gel with 100-bp ladder from a representative CRE isolate (Cfr1047) demonstrating the presence of all predicted products numbered in panel A: L, ladder; 1, tnpR-tnpA; 2, tnpA-istA; 3, istA-istB; 4, istB-blaKPC; 5, blaKPC-tnpA*. Asterisks indicate the tnpA gene of ISKpn6. Download Figure S1, PDF file, 0.1 MB.
Schematic of the ARB-PCR method, used to determine the Tn4401 insertion site. Round 1 PCR is performed using the intertransposon outward-facing primer AM14 and arbitrary primer Arb1.1 with a fixed “tail” (black square), at 10 times the concentration. Round 2 PCR enriches for the region flanking the transposon by using the nested intertransposon outward-facing primer AM11 and primer Arb2.1 at 10 times the concentration, which anneals to the fixed tail of Arb1.1. Unidirectional sequencing is performed using a third nested intertransposon primer. Download Figure S2, PDF file, 0.1 MB.
Representations of outbreak isolates or plasmid carriage during the investigation. (A) Representation of the outbreak by species and strain of the CRE isolate through the outbreak. (B) Representation of the outbreak by the blaKPC-containing plasmid of the CRE isolate through the outbreak. Download Figure S3, PDF file, 0.6 MB.
Primers used in the study.