A comprehensive genome-scale metabolic network of Chlamydomonas reinhardtii, including a detailed account of light-driven metabolism, is reconstructed and validated. The model provides a new resource for research of C. reinhardtii metabolism and in algal biotechnology.
Keywords: Chlamydomonas reinhardtii , lipid metabolism, metabolic engineering, photobioreactor
Abstract
Metabolic network reconstruction encompasses existing knowledge about an organism's metabolism and genome annotation, providing a platform for omics data analysis and phenotype prediction. The model alga Chlamydomonas reinhardtii is employed to study diverse biological processes from photosynthesis to phototaxis. Recent heightened interest in this species results from an international movement to develop algal biofuels. Integrating biological and optical data, we reconstructed a genome-scale metabolic network for this alga and devised a novel light-modeling approach that enables quantitative growth prediction for a given light source, resolving wavelength and photon flux. We experimentally verified transcripts accounted for in the network and physiologically validated model function through simulation and generation of new experimental growth data, providing high confidence in network contents and predictive applications. The network offers insight into algal metabolism and potential for genetic engineering and efficient light source design, a pioneering resource for studying light-driven metabolism and quantitative systems biology.
Introduction
Algae have garnered significant interest in recent years for their potential commercial applications in biofuels (Hu et al, 2008; Hemschemeier et al, 2009) and nutritional supplements (Spolaore et al, 2006). Among eukaryotic microalgae, Chlamydomonas reinhardtii has arisen as the hallmark, model organism (Harris, 2001). C. reinhardtii has been widely used to study photosynthesis, cell motility and phototaxis, cell wall biogenesis, and other fundamental cellular processes (Harris, 2001).
Commercial use and basic scientific research of photosynthetic organisms could benefit from better understanding of how light is absorbed and affects cellular systems. The quality of light sources implemented in photobioreactors largely determines the efficiency of energy usage in industrial algal farming (Fernandes et al, 2010). Light spectral quality also affects how photon absorption induces various metabolic processes: photosynthesis, pigment and vitamin synthesis, and the retinol pathway required for phototaxis.
Metabolic network reconstruction provides a framework to integrate diverse experimental data for investigation of global properties of metabolism, and as such, can provide clear advantages as a mode of studying the effects of light upon a photosynthetic biological system if light input is accounted for explicitly. The standardized reconstruction process (Thiele and Palsson, 2010) yields a biochemically and genomically structured knowledgebase and, coupled with the standard simulation approach of flux balance analysis (FBA) (Orth et al, 2010), provides a basis for predictive phenotype modeling; both contexts have been used for a variety of applications (Durot et al, 2009; Oberhardt et al, 2009; Gianchandani et al, 2010), among them the design of genetic engineering strategies for production strains (Bro et al, 2006; Park et al, 2011). To date, however, photon flux, with associated spectral constraints, has not been integrated into a metabolic network reconstruction.
Characterizing algal metabolism is key to engineering production strains and framing the study of photosynthesis. Extensive literature on C. reinhardtii metabolism, reviewed in Stern et al (2008), and multiple metabolic mutants (Harris et al, 2008) provide a solid foundation for detailed characterization of its metabolic functions. The availability of complete genome sequence data for C. reinhardtii (Merchant et al, 2007) and its functional annotation have enabled bioinformatic approaches to inform the presence of genome-encoded enzymes (Grossman et al, 2007; Boyle and Morgan, 2009; Manichaikul et al, 2009). We have employed these resources to reconstruct and experimentally validate a genome-scale metabolic network of C. reinhardtii, the first network to account for detailed photon absorption permitting growth simulations under different light sources. This network accounts for the activity of substantially more genes with metabolic functions than existing reconstructions (Boyle and Morgan, 2009; Manichaikul et al, 2009).
Results
Reconstruction contents and advances
The genome-scale C. reinhardtii metabolic network (Figure 1A; Supplementary Figure S1; Supplementary Table S1; Supplementary Table S2; Supplementary Model S1) accounts for 1080 genes, associated with 2190 reactions and 1068 unique metabolites, and encompasses 83 subsystems distributed across 10 compartments. As per convention (Reed et al, 2003), we call this network iRC1080 based on the primary reconstructionist and the scope of genomic content. Of the putative protein-coding genes in the C. reinhardtii genome (http://augustus.gobics.de/predictions/chlamydomonas/augustus.u5.aa), an estimated 20% function in metabolism (Supplementary Table S3). iRC1080 accounts for the activity of >32% of the estimated genes with metabolic functions, a significant expansion over existing reconstructions (Boyle and Morgan, 2009; Manichaikul et al, 2009). iRC1080 is the most comprehensive metabolic network reconstruction of C. reinhardtii to date based on inclusion of pathways and a level of detail absent from previous reconstructions.
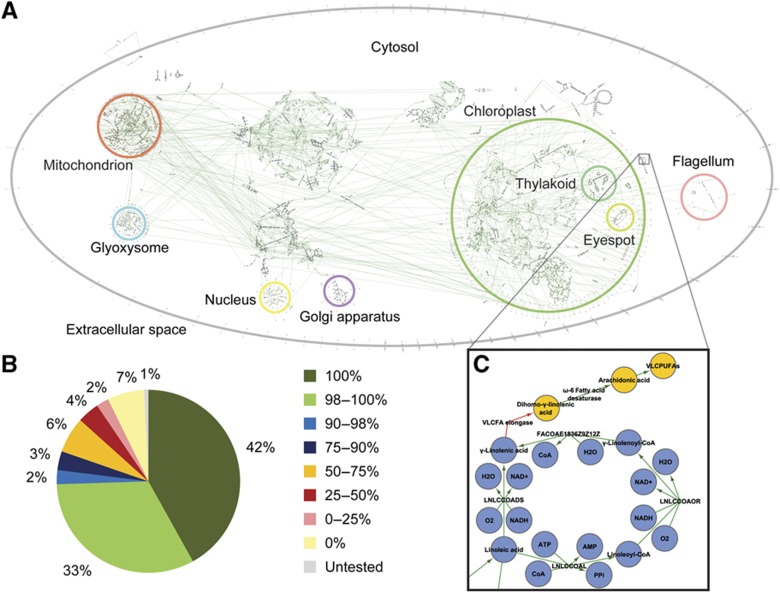
Figure 1.
Contents of the iRC1080 metabolic network reconstruction. (A) Compartmentalized network diagram. The full genome-scale metabolic network is depicted, denoting compartments. A high-resolution diagram without compartment labels is also available (Supplementary Figure S1). (B) Global transcript verification status. The graph shows the distribution of transcripts accounted for in the network categorized by their verification status. Color codes correspond to the noted percentage of transcript sequence verified experimentally. For example, 42% of transcripts in the network were verified experimentally by 100% sequence coverage. (C) Latent VLCPUFA pathway diagram. Blue nodes represent metabolites included in iRC1080, and orange nodes represent metabolites not included in iRC1080, hypothesized to be absent in C. reinhardtii. Green edges represent enzyme activities accounted for in our functional annotation, and the red edge represents the VLCFA elongase missing from our annotation and hypothesized to have been lost in C. reinhardtii's evolution. This pathway diagram also demonstrates the detail of the high-resolution network diagram (Supplementary Figure S1).
A major emergent feature of C. reinhardtii metabolism, apparent in Figure 1A, is the relative centrality of the chloroplast and its importance in light-driven metabolism. The chloroplast, including the thylakoid and eyespot subcompartments, accounts for >30% of the total reactions in the network and 9 of the 10 photon-utilizing reactions. The thylakoid contains essential pathways for photoautotrophic growth including photosynthesis, chlorophyll synthesis, and carotenoid synthesis, producing photoprotective pigments also valuable as fish feed additives and nutritional supplements for human consumption. The eyespot accounts for retinol metabolism, the mechanistic basis for phototaxis. Several pathways are partially duplicated across the chloroplast and other cellular compartments, in agreement with known biochemistry. A few crucial pathways are divided between the chloroplast and cytosol, including glycolysis and glycerolipid metabolism. Among the glycerolipids, triacylglycerides carrying high energy, long-chain fatty acids relevant for biofuel production accumulate substantially in microalgae. iRC1080 provides a thorough resource for studying these and other metabolic products and a basis for strain design for genetic engineering.
iRC1080 considerably expands lipid metabolic pathways over previous reconstructions. We compared the lipid pathways of iRC1080 with several previously published metabolic reconstructions (Duarte et al, 2007; Feist et al, 2007; Boyle and Morgan, 2009; Mo et al, 2009; Montagud et al, 2010) counting the number of genes, reactions, and chemically distinct lipid molecules included in pathways for each lipid class (Table I). The extent of gene, reaction, and metabolite content of lipid pathways in iR1080 is, in general, greater than previous reconstructions. The coverage of ketoacyl lipid chemical properties represented in each network was also analyzed for all metabolites in fatty acyl, glycerolipid, glycerophospholipid, and sphingolipid classes; the fraction of lipid metabolites in the networks that account for a given applicable property was determined (Table I). Lower coverage signifies incompletely specified molecular species and often lumped lipid reactions and metabolites. iRC1080 accounts explicitly for all metabolites in these pathways, providing sufficient detail to completely specify all individual molecular species: backbone molecule and its stereochemical numbering of acyl-chain positions; acyl-chain length; and number, position, and cis–trans stereoisomerism of carbon–carbon double bonds. This level of detail enables a significantly higher degree of precision in lipid studies and in metabolic engineering design involving these pathways.
Table 1. Lipid pathway reconstruction properties in iRC1080 in comparison to other metabolic network reconstructions.
| Reconstructions |
||||||
|---|---|---|---|---|---|---|
| iRC1080 C. reinhardtii | [iNB305] C. reinhardtii | iSyn669 Synechocystis | iMM904 S. cerevisiae | iAF1260 E. coli | Recon 1 Homo sapiens | |
| aValues are the fraction of lipid metabolites in each network that account for each property, when applicable. | ||||||
| bGene transcripts (can be duplicated across lipid classes). | ||||||
| cLipid pathway reactions (non-transport). | ||||||
| dLipid metabolites (unique lipids). | ||||||
| Ketoacyl lipid chemical properties a | ||||||
| Backbone molecule | 1.00 | 0.94 | 1.00 | 1.00 | 1.00 | 1.00 |
| Stereochemical numbering | 1.00 | 0.00 | 0.60 | 0.85 | 1.00 | 0.00 |
| Acyl-chain length | 1.00 | 0.72 | 0.90 | 0.91 | 1.00 | 0.70 |
| C=C number | 1.00 | 0.72 | 0.75 | 0.91 | 1.00 | 0.70 |
| C=C positions | 1.00 | 0.00 | 0.80 | 0.42 | 0.91 | 0.60 |
| E–Z stereoisomerism | 1.00 | 0.00 | 0.80 | 0.50 | 0.42 | 0.53 |
| Fatty acyls | ||||||
| Gb | 64 | 7 | 13 | 32 | 26 | 91 |
| Rc | 167 | 41 | 71 | 108 | 139 | 233 |
| Md | 104 | 21 | 55 | 55 | 95 | 137 |
| Glycerolipids | ||||||
| Gb | 40 | 0 | 0 | 18 | 0 | 27 |
| Rc | 292 | 4 | 0 | 12 | 0 | 13 |
| Md | 135 | 4 | 2 | 4 | 7 | 4 |
| Glycerophospholipids | ||||||
| Gb | 47 | 0 | 8 | 46 | 22 | 87 |
| Rc | 126 | 5 | 7 | 52 | 227 | 51 |
| Md | 56 | 4 | 3 | 4 | 102 | 22 |
| Sphingolipids | ||||||
| Gb | 8 | 0 | 0 | 21 | 0 | 54 |
| Rc | 10 | 0 | 0 | 63 | 0 | 79 |
| Md | 6 | 0 | 0 | 31 | 0 | 59 |
| Sterol lipids | ||||||
| Gb | 22 | 0 | 1 | 32 | 0 | 87 |
| Rc | 34 | 0 | 3 | 49 | 0 | 156 |
| Md | 26 | 0 | 4 | 22 | 0 | 105 |
| Prenol lipids | ||||||
| Gb | 37 | 4 | 15 | 9 | 16 | 21 |
| Rc | 59 | 5 | 53 | 17 | 20 | 50 |
| Md | 43 | 4 | 42 | 15 | 17 | 41 |
| Total lipids | ||||||
| Gb | 218 | 11 | 37 | 158 | 64 | 367 |
| Rc | 688 | 55 | 134 | 301 | 386 | 582 |
| Md | 370 | 33 | 106 | 131 | 221 | 368 |
Experimental transcript verification
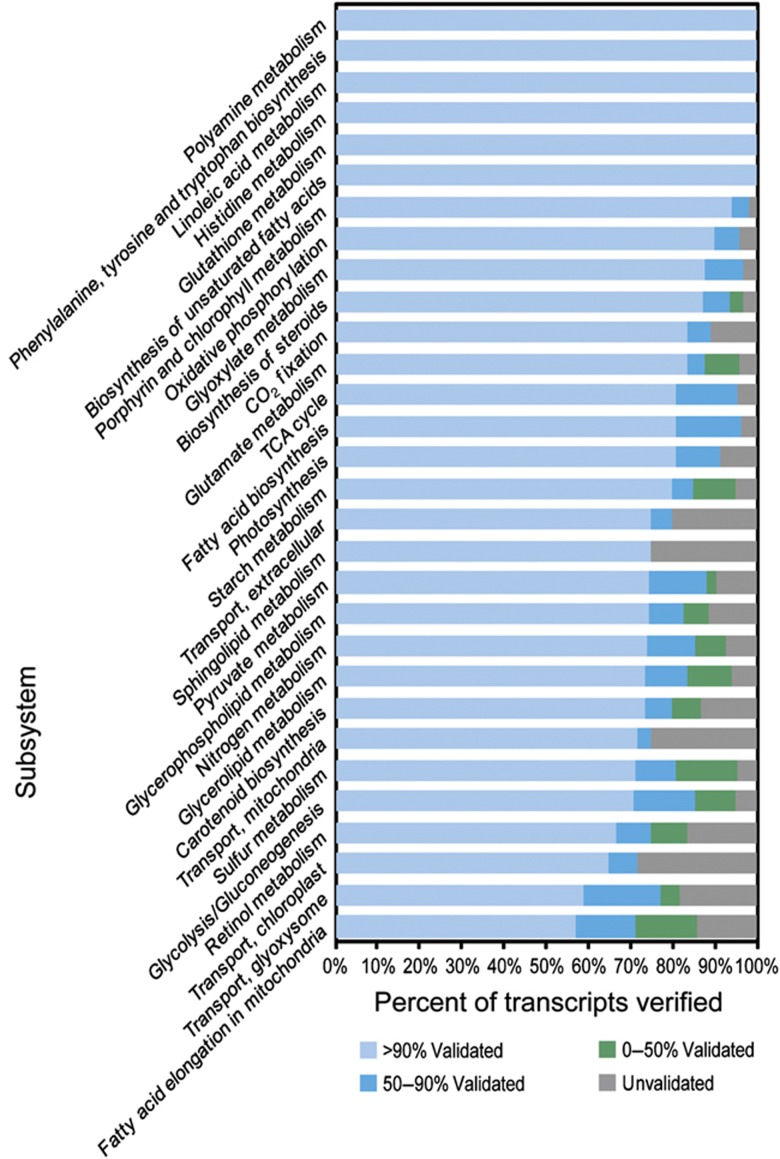
We have analyzed iRC1080 via experimental transcript verification under permissive growth conditions (Supplementary Table S4), representing the largest genome-scale transcript validation effort to date. More than 75% of included transcripts were verified at >90% sequence coverage, and 92% of tested transcripts were at least partially validated experimentally (i.e. a portion of the sequence was recovered in the sequenced transcripts) (Figure 1B). We also analyzed the strength of transcript verification by specific metabolic subsystems (Figure 2, a representative subset; Supplementary Figure S2, the full set). The full lengths of all transcripts associated with 10 subsystems were verified, notably including biosynthesis of unsaturated fatty acids, histidine metabolism, and phenylalanine, tyrosine and tryptophan biosynthesis, with 12, 12, and 24 transcripts, respectively. Many more subsystems were also well verified, 61 out of 76 gene-associated subsystems with >90% of associated transcripts at least partially validated. It should be noted that only sequencing reads that uniquely map to reference transcript sequences were counted toward the percentage of length validation; thus, sequencing reads unique enough to unambiguously specify the corresponding reference transcript were detected for every transcript with >0% validation. A few subsystems stood out as being more poorly verified, including chloroplast and mitochondrial transport systems and sphingolipid metabolism, all of which exhibited <80% of transcripts validated to any extent. This may reflect low expression level or imperfect structural annotation of these genes, particularly compartmental transporters. Low expression levels or complete deactivation of these genes is consistent with a hypothesized evolutionary trend (see below) in the case of sphingolipid metabolism.
Figure 2.
Experimental transcript verification by subsystem. The graph summarizes transcript verification status (see Materials and methods and Supplementary information for details) for 30 of the 76 gene-associated subsystems of iRC1080. Identical analysis for the full complement of 76 subsystems is also available (Supplementary Figure S2). The x axis corresponds to the percentage of subsystem-associated transcripts that were experimentally verified to the extent noted by the color code.
Evolution of latent lipid pathways
The comprehensive reconstruction of lipid metabolism in iRC1080 revealed hypothetical latent pathways, the functions of which have likely been lost through evolution. Previous studies established that C. reinhardtii lacks the practically ubiquitous membrane lipids phosphatidylcholine (Giroud et al, 1988) and phosphatidylserine (Riekhof et al, 2005). Similarly, our reconstruction suggests that C. reinhardtii also lacks very long-chain fatty acids (VLCFAs), their polyunsaturated analogs (VLCPUFAs) (Figure 1C), and ceramides.
Surveys of C. reinhardtii lipid species have not detected VLCFAs (Giroud et al, 1988; Giroud and Eichenberger, 1989; Tatsuzawa et al, 1996; Dubertret et al, 2002; Kajikawa et al, 2006; Lang, 2007), likely due to a lack of functional VLCFA elongase (Weers and Gulati, 1997; Guschina and Harwood, 2006; Kajikawa et al, 2006). No candidate VLCFA elongase was identified in our comprehensive functional annotation (Supplementary Table S3), and our annotation suggests several downstream gaps in arachidonic acid metabolism as well, corroborating this hypothesis. Arachidonic acid, the 20-carbon parent fatty acid of all VLCFAs and VLCPUFAs, is synthesized by a VLCFA elongase-catalyzed extension of γ-linolenic acid, which is present in C. reinhardtii (Griffiths et al, 2000). Notably, C. reinhardtii does encode a fatty acid desaturase that accepts arachidonic acid as substrate (Kajikawa et al, 2006) and, based on our functional annotation, encodes several other enzymes that act upon this substrate, indicating that algal ancestors likely had a functional VLCFA elongase.
Multiple lines of evidence uncovered during the reconstruction also support the absence of ceramides in C. reinhardtii. Our functional annotation did not uncover a convincing candidate for ceramide synthetase (EC:2.3.1.24), a required enzyme for ceramide synthesis, nor, to our knowledge, has one been discovered by previous efforts, including C. reinhardtii enzyme annotations of the Kyoto Encyclopedia of Genes and Genomes. Similarly, our functional annotation suggests substantial gaps downstream in the sphingolipid metabolic pathway. As aforementioned, C. reinhardtii also lacks VLCFAs, and VLCFA-CoA is a required substrate for the ceramide synthetase reaction (Hills and Roscoe, 2006). Finally, our experimental transcript analysis failed to verify 2 out of 8 transcripts associated with sphingolipid metabolism (Figure 2) that were included in iRC1080, 1 of 2 serine C-palmitoyltransferases and a putative sphingosine 1-phosphate aldolase. This result may reflect still further gene function loss in this pathway, perhaps occurring more recently in evolutionary time given that our functional annotation actually detected candidate sequences for these enzymes. Considering this evidence, we suggest that the evolutionary history of C. reinhardtii includes the loss of ceramide metabolism, although this hypothesis remains to be verified. Annotated enzymes in this pathway separated from the broader network by gaps may represent multifunctional proteins or proteins that have evolved to function in a pathway distinct from ceramide synthesis. These gaps in C. reinhardtii metabolism not only increase understanding of the evolution of algal lipid pathways but also represent potential targets for genetic engineering in an effort to expand the diversity of lipids this alga can synthesize. Such engineering efforts serve as valuable test cases for engineering industrial strains and could improve C. reinhardtii as a model alga for biofuel development.
Modeling metabolic light usage
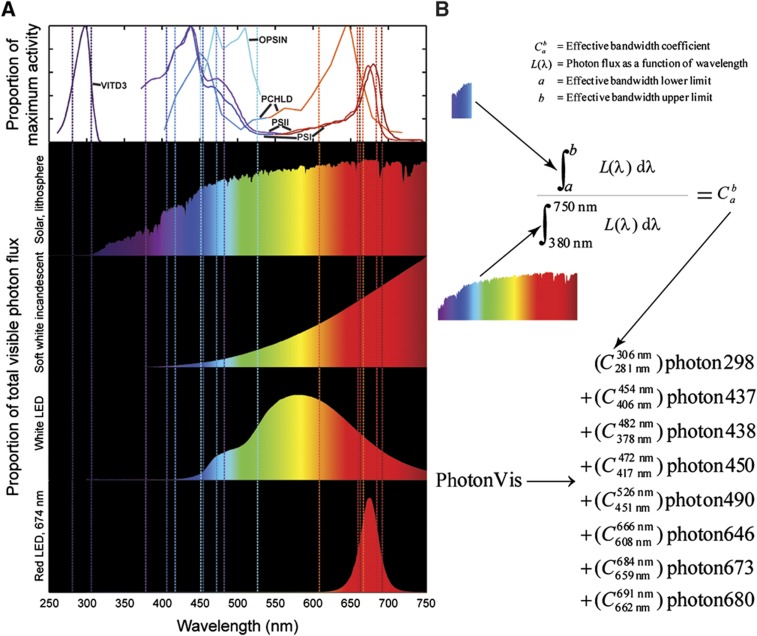
Our reconstruction accounted for effective light spectral ranges by analyzing biochemical activity spectra (Figure 3A), either reaction activity or absorbance at varying light wavelengths. Defining effective spectral bandwidths associated with each photon-utilizing reaction enabled our network to model growth under different light sources via stoichiometric representation of the spectral composition of emitted light, which we term prism reactions. The coefficients for different photon wavelengths in prism reactions correspond to the ratios of photon flux in the defined effective spectral ranges to the total photon flux in the visible spectrum emitted by a given light source (Figure 3A and B). In this manner, it is possible to distinguish the amount of emitted photons that drive different metabolic reactions. We created prism reactions for 11 distinct light sources (Supplementary Figure S3), covering most sources that have been used in published studies for algal and plant growth including solar light, various light bulbs, and LEDs.
Figure 3.
Analysis of light spectra. (A) Activity and irradiance spectra. The top graph displays activity spectra for photon-utilizing reactions included in iRC1080. The abbreviated reactions are defined as follows: VITD3, vitamin D3 synthesis; OPSIN, rhodopsin photoisomerase; PCHLD, both protochlorophyllide photoreductase and divinylprotochlorophyllide photoreductase; PSI, photosystem I; PSII, photosystem II. The y axis for the activity spectra is the fraction of maximum-measured activity with respect to each noted reaction. Four of the eleven sample irradiance spectra (Supplementary Figure S3) are depicted with y axes set as the percentage of total visible photon flux at each wavelength (x axis). Effective spectral bandwidths are denoted by vertical dashed lines color coded to match the activity spectra for each reaction. (B) Prism reaction derivation. The photon flux from wavelengths a to b is normalized by the total visible photon flux from 380 to 750 nm to yield the effective spectral bandwidth coefficient C. The coefficients for each range are compiled into a single prism reaction for a given light source, representing the composition of emitted light as defined by photon-utilizing metabolic reactions. Equation variables are defined at top.
The network reconstruction provides a detailed account of metabolic photon absorption by light-driven reactions. Photosystems I and II in iRC1080 stoichiometrically absorb photons according to the Z-scheme (Berg et al, 2007). The light-dependent protochlorophyllide oxidoreductases require a single photon per catalysis as demonstrated in wheat (Griffiths et al, 1996). Extrapolation of UVB energy requirements for spontaneous provitamin D3 conversion to vitamin D3 (Bjorn, 2007) based on the average photon energy in the UVB range suggests a stoichiometric ratio of approximately one. Two phototactic rhodopsins, reactants of the rhodopsin photoisomerase reaction, are encoded by C. reinhardtii, one requiring a single photon and one requiring two photons for activation; the average effective stoichiometric photon count was measured to be 1.6 (Hegemann and Marwan, 1988).
A prism reaction is the intermediate step between light input and the specific photon-utilizing metabolic reactions mentioned above. Flux through the photon exchange reaction ‘EX_photonVis(e)’ represents the total metabolically active photon flux incident upon the cell. Flux passing through this exchange reaction then passes through a single user-specified prism reaction, for example ‘PRISM_solar_litho,’ and is distributed across specific spectral ranges. These ranges are specified explicitly in the photon-dependent metabolic reaction formulas (Supplementary Table S2), thereby making these reactions wavelength specific. Flux through the photon-dependent metabolic reactions is then propagated through the network. Excess wavelength-specific photon fluxes that are not absorbed metabolically leave the system via demand reactions, for example ‘DM_photon298(c),’ completing the pathway of light through the network.
To accurately model metabolic activity of a photosynthetic organism, it is also important to consider regulatory effects resulting from lighting conditions. Indeed, light and dark conditions have been shown to affect metabolic enzyme activity in C. reinhardtii at multiple levels: transcriptional regulation (Bohne and Linden, 2002), chloroplast RNA degradation (Salvador et al, 1993), translational regulation (Cahoon and Timko, 2000), and thioredoxin-mediated enzyme regulation (Lemaire et al, 2004). As a preliminary attempt to incorporate light and dark regulatory effects, literature was reviewed to identify such regulation upon enzymes in iRC1080 (Supplementary Table S5), focusing mainly on thioredoxin regulation of chloroplast enzymes since most published data relate to this mode. In the absence of activity spectra for these effects, it is not yet possible to represent these effects via prism reactions. Therefore, we modeled regulation with Boolean reaction flux constraints following published approaches (Covert et al, 2001).
Environmental and genetic validation of iRC1080
Implementing light-regulated constraints and basic environmental exchange constraints (Supplementary Table S6) yielded photoautotrophic, heterotrophic, and mixotrophic models from iRC1080. We simulated various growth conditions (Supplementary Table S7) and all gene knockouts for which phenotypes have been published and are assessable in our network (Supplementary Table S8) to validate the predictive ability of the models. All 30 validations involving environmental parameters displayed very close agreement with experimental results (Supplementary Table S7). Of particular note is the ability of our photosynthetic model in sunlight to accurately recapitulate O2-PAR (photosynthetically active radiation) energy conversion efficiency, predicting an efficiency of 2% compared with the experimental result (Greenbaum, 1988) of 1.3–4.5%. Of the 14 gene knockouts simulated, 7 were partially or completely validated relative to experimental results (Supplementary Table S8). The unconfirmed gene knockout phenotypes may result from network errors or an incomplete set of constraints in the model (e.g. enzyme capacity, regulatory, thermodynamic, or other constraints). No internal model reactions were constrained in the models except indirectly via constraints on the input exchanges and the few explicitly noted Boolean regulatory constraints imposed (Supplementary Table S5). The unconfirmed knockout phenotypes were investigated through model analysis and literature search, although in most cases, current literature evidence could not completely explain these discrepancies, leaving them to be fully accounted for by future studies.
Two discrepancies may result from incomplete genome functional annotation or missing constraints. Knockout of mitochondrial NADH:ubiquinone oxidoreductase complex I (EC:1.6.5.3) in the model fails to recapitulate a reduced heterotrophic growth phenotype (Remacle et al, 2001a). The NDA2 and NDA3 genes can substitute completely for this activity in the current model. Sequence-based localization analysis places both proteins in the mitochondria, but this may be incorrect as a recent study suggests that both may be plastid localized (Desplats et al, 2009). Two other network reactions can also substitute for the reduction of ubiquinone, succinate dehydrogenase (ubiquinone) (EC:1.3.5.1) and electron transfer flavoprotein-ubiquinone oxidoreductase (EC:1.5.5.1). The cytochrome c oxidase complex IV (EC:1.9.3.1) knockout does not result in an obligate photoautotrophic phenotype (Remacle et al, 2001b) in the model because the cytochrome c peroxidase (EC:1.11.1.5) reaction is capable of compensating. The C. reinhardtii CCPR1 protein is homologous to mitochondrial cytochrome c peroxidases from a number of species, but no focused studies have been carried out to provide further evidence for this enzyme. In the model, the complex IV and CCPR1 double knockout is an obligate photoautotroph. These discrepancies point out important genes that should be the focus of subsequent experimentation in order to more clearly understand these metabolic phenotypes.
Another discrepancy may result from missing thermodynamic constraints. The zeaxanthin epoxidase (EC:1.14.13.90) knockout does not preclude antheraxanthin, violaxanthin, or neoxanthin production (Baroli et al, 2003) in the model because violaxanthin de-epoxidase (EC:1.10.99.3) reactions compensate. This substitution depends on the reversibility of these de-epoxidase reactions and may point to missing thermodynamic constraints or to undiscovered regulation under this condition.
Two discrepancies result from the lack of accounting for kinetics of the reactions of ribulose-1,5-bisphosphate carboxylase oxygenase (RuBisCO) from the model. Both phosphoglycolate phosphatase (EC:3.1.3.18) (Suzuki et al, 1990) and glycolate dehydrogenase (EC:1.1.99.14) (Nakamura et al, 2005) deficient mutants require high CO2 for photoautotrophic growth in vivo, not recapitulated in simulations. This phenotype results from dominance of the oxygenase over carboxylase activity of RuBisCO under lower CO2 conditions, both reactions sharing the same catalytic site. In vivo, these two mutants are deficient in the salvage of carbon from 2-phosphoglycolate, a product of the oxygenase activity of RuBisCO. Although these two reactions are carried out by the same enzyme in the model, their fluxes are treated as independent and not competitive; due to an absence of kinetic parameters in the model, the effect of relative CO2 and O2 concentrations upon RuBisCO activity cannot be explicitly expressed. Because the carboxylase activity more efficiently promotes growth, both high and low CO2 conditions drive only this reaction and not the oxygenase reaction in the model; therefore, the salvage pathway is unnecessary in the model to achieve wild-type growth rates.
Finally, two mutant phenotype discrepancies in the model result from complex compensatory pathways that convert an input carbon source to the mutant-required carbon source. The high CO2 requirement for photoautotrophic growth due to knockout of the chloroplast carbonic anhydrase (EC:4.2.1.1) (Spalding et al, 1983; Funke et al, 1997) can be compensated for in the model by activity of a six-reaction pathway of pyrimidine metabolism leading from bicarbonate incorporation via carbamoyl-phosphate synthase (EC:6.3.5.5) to conversion to CO2 via orotidine-5′-phosphate decarboxylase (EC:4.1.1.23). The chloroplast ATP synthase (EC:3.6.3.14) deficient mutant (Smart and Selman, 1991; Dent et al, 2005; Drapier et al, 2007) with an acetate-requiring phenotype can be compensated for in the model by a complex pathway consisting of >15 reactions by which CO2 is converted to acetate, which is then used in pathways similar to those supporting heterotrophic growth. Although this complex pathway has many branch points, it is notable that chloroplast malate dehydrogenase (EC:1.1.1.40) and the diffusion of pyruvate between the cytosol and chloroplast are essential to coupling the CO2 fixation reactions to pyruvate metabolism and ultimate conversion to acetate but are not essential to the wild-type photoautotrophic or heterotrophic models. Loss of either of these conditionally essential reactions prevents the CO2-to-acetate conversion and recapitulates the acetate-requiring phenotype. Given the complexity of these compensatory pathways, a number of possible missing constraints could explain their inactivity in vivo under photosynthetic conditions, and the model offers a starting point to explore possible targets of regulation under these conditions.
Gene essentiality analysis
To demonstrate the prospective use of iRC1080 in predicting phenotypic outcomes of genetic manipulations of C. reinhardtii, comprehensive essentiality analysis of all simulated single-gene knockouts was performed in models under four basic environmental conditions: growth in sunlight with and without acetate, aerobic growth in dark on acetate, and anaerobic subsistence in dark on starch. Phenotypes were defined as growth equivalent to wild-type, reduced growth relative to wild-type, or lethal based on the comparative objective fluxes of the mutant and wild-type models (Supplementary Table S9). A lethal phenotype was defined as no flux through the biomass reaction (defined as the objective function) in the mutant. Simulation results exhibited distinct metabolic system dependencies under each condition. There were 201 and 144 lethal knockouts in the model with sunlight and with and without acetate, respectively. There were 147 and only 3 lethal knockouts in the aerobic and anaerobic dark model, respectively. The metabolic processes associated with essential genes were ranked, and the three subsystems associated with the essential genes were compared under each condition. Photosynthesis, porphyrin and chlorophyll metabolism, and phenylalanine, tyrosine, and tryptophan biosynthesis were the most essential subsystems in light without acetate. Phenylalanine, tyrosine, and tryptophan biosynthesis, porphyrin and chlorophyll metabolism, and purine metabolism were the most essential subsystems in light with acetate. Expectedly, photosynthesis is most crucial for photoautotrophic growth and not required in the presence of acetate. The dark, aerobic condition had the same top ranked essential subsystems as in the mixotrophic condition, which is also expected as amino acids, chlorophyll, and nucleotides make up a high proportion of the required biomass components under both conditions. For subsistence in dark on starch, glycolysis/gluconeogenesis, starch metabolism, and starch and sucrose metabolism were the most essential subsystems, paralleling the expected core pathways for ATP maintenance with starch breakdown. While these predicted genotype–phenotype relationships demonstrate a compelling prospective use of the network, the majority of the mutant phenotypes remain to be validated experimentally; however, these predictions could be used to help define the scope and expected outcomes of such future studies.
Light-source-specific model validations
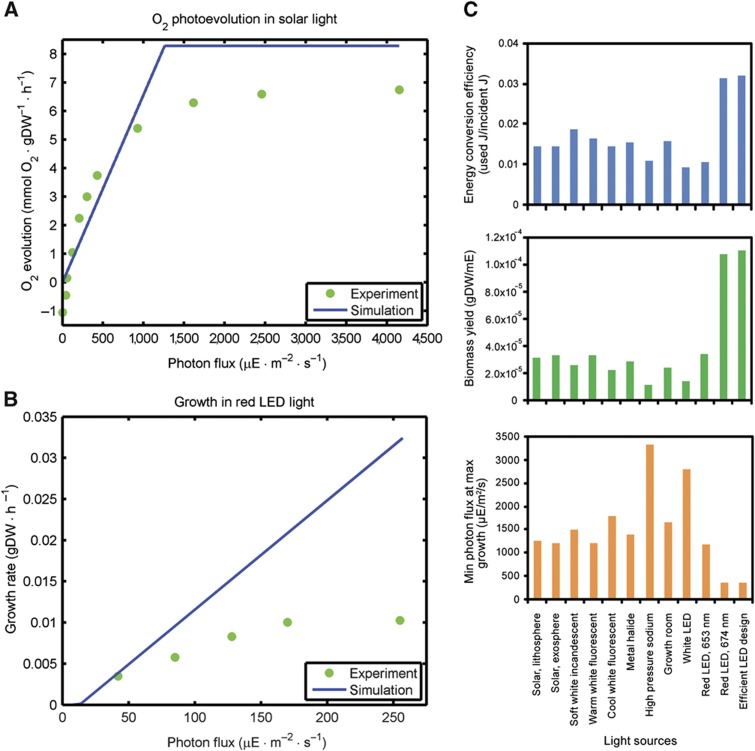
Next, we performed more extensive validations of light models grown under specific light sources at varying intensities. Varying sunlight intensity in our model and evaluating photosynthetic O2 evolution, we observed that the model reached photosynthetic saturation at light intensity consistent with experimental measurement (Polle et al, 2003) (Figure 4A). Our model under red LED light (653 nm) also showed fair agreement with our experimentally measured maximum growth rate across the range of unsaturated photon flux (Figure 4B), despite divergence above the experimental saturation point. The principal explanation for this divergence lies in the relative CO2 supplies of the experimental setup and the model. All reported photoautotrophic model simulations utilize the same maximum CO2 exchange constraint corresponding to the maximum-measured cellular uptake rate under non-CO2-limiting conditions (Supplementary Table S6), while the CO2 supply in our bioreactor setup was clearly growth-limiting given that the light-saturated maximum growth rate was 0.01 gDW/h, much lower than the maximum growth rate of 0.14 gDW/h under non-CO2-limiting conditions (Janssen et al, 2000). It should also be noted that the linearity of the simulation trends is a property of steady-state system modeling, which is incapable of kinetic representation of growth shifts observable in the in vivo experiments. For further validation, we present that the maximum biomass yield under incandescent white light is 5.7 × 10−5 gDW/mE (Janssen et al, 2000), in close agreement with our analogous prediction of 2.6 × 10−5 gDW/mE (Figure 4C). Similarly, our predicted biomass yield on 674 nm peak LED light of 1.1 × 10−4 gDW/mE is on the same order of magnitude as our experimental results for C. reinhardtii under 660 nm peak LED light near growth-saturating photon flux, 4.3 × 10−4 gDW/mE. This agreement is striking given that the network explicitly accounts for the spectral photon flux of these light sources and the subsequent processing of this energy to generate all of the constituents of biomass without any parameter fitting to the experimental data. Together, these results constitute an important validation of our models using three different light sources.
Figure 4.
Photosynthetic model simulation results. (A) O2 photoevolution under solar light. Simulated (blue line) and experimentally measured (green dots) O2 evolution are compared. (B) Photosynthetic growth under red LED light. Simulations were performed using the 653-nm prism reaction, and experimentally grown culture was exposed to 660 nm LED light. Simulated (blue line) and experimentally measured (green dots) growth are compared. (C) Efficiency of light utilization. The minimum photon flux required for maximum-simulated growth (bottom), biomass yield (middle), and energy conversion efficiency (top) are presented for 11 light sources derived from measured spectra and for the designed growth-efficient LED.
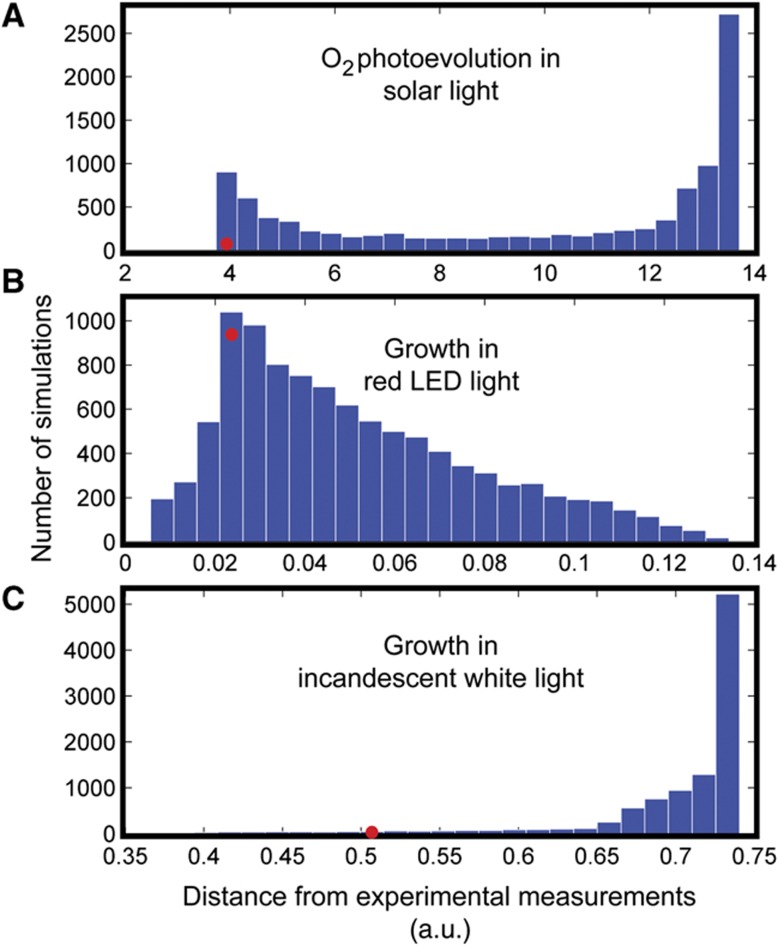
To quantitatively evaluate the significance of the agreement between our reported model simulations using prism reactions derived through analysis of irradiance spectra and experimental measurements under the three light sources reported above, we compared the reported simulation results for each of these light sources with an unbiased sample of results representative of potential solutions achievable using our network. We sampled the space of possible light models by generating random prism reactions with the same total metabolically active photon flux. To obtain stoichiometric coefficients for a random prism reaction, a set of random fractions of the sum of stoichiometric coefficients of the prism reaction representing the evaluated light source was generated, contingent upon resulting in the same sum of coefficients. The simulations as reported above for sunlight, red LED, and white incandescent light were repeated using such random prism reactions. The Euclidean distance between the simulated and experimental results was compared with the distribution of distances for 10 000 randomly sampled results (Figure 5). The probability of randomly achieving experimental agreement closer than seen in our simulations was determined empirically based on these distributions of distances. Only 77 of 10 000 randomized simulations (Figure 5A) had experimental agreement better than the simulated oxygen photoevolution under sunlight (Figure 4A), yielding an empirical P-value of 0.0077, and indicating our model had experimental agreement statistically significantly better than a random model constrained to have the same total metabolically active photon flux. Simulated growth under 665 nm peak LED (Figure 4B) had a suggestive P-value of 0.1947 (Figure 5B), although the reported simulation was still closer to experiment than the mean of randomized simulations. Our simulated growth under white incandescent light was statistically significantly closer to experiment (Janssen et al, 2000) than random (Figure 5C) with a P-value of 0.0285. This analysis shows that the reported model for each of these light sources is exceptionally close to recapitulating experimental results and thus serves as an excellent validation. These results indicate that the network has the capacity to broadly differentiate light-dependent growth based on spectral properties and that the formulation of a prism reaction serves to accurately narrow the space of possible flux distributions relevant to a specific light source.
Figure 5.
Distributions of randomly sampled distances from experimental measurements. (A) O2 photoevolution under solar light. (B) Photosynthetic growth under red LED light. (C) Photosynthetic growth under white incandescent light. All three distance distributions result from 10 000 unbiased sampling results in which random prism reactions were generated with the same total metabolically active photon flux as the given light source. Each distribution is depicted in 25 equal-sized bins. The red dot in each plot is placed over the bin in which the distance of the reported simulation result for the given light source falls; the vertical placement of each red dot indicates the number of randomly sampled distances within the same bin that are less than that of the reported result.
Application of iRC1080 to evaluate light source efficiency and design
Our photosynthetic model was applied prospectively to evaluate the efficiency of light utilization under different light sources. The photon energy conversion efficiency (Supplementary Equation 1) and biomass yield on light (Supplementary Equation 2) were computed for each light source given the minimum incident photon flux required to achieve maximum growth rate (Figure 4C); the minimum photon flux for maximum growth rate is the growth-saturating photon flux value for a given light source. One clear result is that red LEDs provide the greatest efficiency in terms of both absorbed photon energy and biomass yield, about two and three times as efficient as can be optimally achieved in sunlight by these respective measures. Although experimental growth data for validation is only presented for three light sources, simulation results are presented for all 11 light sources for which irradiance spectra were obtainable (Figure 4C). This analysis demonstrates the prospective extensibility of the network and modeling approach to any possible lighting condition, natural or manmade, for which an irradiance spectrum can be measured.
Given the capability of our photosynthetic model to evaluate light source efficiency, we applied it to design an LED spectrum providing maximum photon utilization efficiency for growth (Supplementary Figure S3). The result was a 677-nm peak LED spectrum with a total incident photon flux of 360 μE/m2/s (Figure 4C; Supplementary Figure S3), which is quite close to the 674-nm LED with a minimum incident photon flux of 362 μE/m2/s for maximum growth. This result suggests that for the simple objective of maximizing growth efficiency, LED technology has already reached an effective theoretical optimum, which is further supported by experimental measurements of the spectral peak of light absorption for green algae (Akkerman et al, 2002) and the quantum action spectrum of land plants (Barta et al, 1992) (Supplementary Table S7).
Discussion
We have presented a genome-scale network reconstruction of C. reinhardtii metabolism, well validated in content and function, and its application for detailed modeling of diverse light sources. Initial model validations also highlight the need for more experimental studies to uncover regulatory mechanisms in order to expand understanding of the complexity of light regulation of algal metabolism. This open research topic presents important challenges and opportunities in enumerating such effects on a genome scale.
Given the importance of lipid metabolism in biofuel production, iRC1080 was reconstructed enumerating all lipids supported by evidence in the literature and genome functional annotation. The capacity of iRC1080 as a knowledgebase was demonstrated through analysis of lipid metabolism to generate novel hypotheses about latent metabolic pathways resulting from algal evolution. In particular, the exclusion of certain enzymatic reactions in VLCFA and sphingolipid pathways from iRC1080 suggests evolutionary recession of these pathways in C. reinhardtii, a hypothesis supported by undetected lipids in experimental measurements, gaps in genome functional annotation for these enzymes, and incomplete transcript verification for other enzymes included in these pathways. Not only do these network gaps reflect the relatively simple lipid biosynthetic capabilities of C. reinhardtii among microalgae, but their identification suggests gene insertions that could expand its lipid metabolic repertoire, relevant for industrial and scientific purposes. Of particular interest may be the potential for enabling algal synthesis of essential fatty acids for human health such as docosahexaenoic acid (Yashodhara et al, 2009). Candidate enzymes for the conversion of arachidonic acid to essential fatty acids downstream of the apparently absent VLCFA elongase reaction are present in our functional annotation.
The models developed from iRC1080 provide a platform for prediction of phenotypic outcomes of system perturbations, light source evaluation and design, and genetic engineering design for production of biofuels and other commodity chemicals. We demonstrated an approach applying iRC1080 to the design of an energetically efficient light source for growth, a novel application of metabolic networks. Other light sources may be more efficient for other metabolic objectives or under other environmental conditions or genetic backgrounds. This result could be of significant interest to the metabolic engineering and bioreactor-design communities because it demonstrates that our network and light-modeling approach are capable of accurately predicting light source efficiencies in terms of a metabolic objective.
The prism reactions developed and applied in this study to quantitatively integrate spectral quality with biological activity represent a significant integration of diverse data types for biological system modeling, which hopefully will encourage a new paradigm for systems biology. This modeling approach could be used for applications beyond light source design, including as a metabolic basis for studying and simulating phototaxis. Given the acquisition of appropriate biological spectral activity data, this approach could be extended to other biological light-response phenomena and other organisms. The importance of understanding how light parameters affect biological systems may also extend beyond natural phenomena with recent progress in protein engineering leading to chimeric light-inducible proteins (Shimizu-Sato et al, 2002; Levskaya et al, 2005).
The iRC1080 network and presented metabolic modeling represent a milestone in systems biology. Our network provides a broad knowledgebase of the biochemistry and genomics underlying global metabolism of a photoautotroph, and our modeling of light-driven metabolism exemplifies how integration of largely unvisited data types, such as physicochemical environmental parameters, can expand the diversity of applications of metabolic networks.
Materials and methods
Metabolic network reconstruction
Building from our previously published reconstruction of C. reinhardtii central metabolism (Manichaikul et al, 2009), iAM303, the iRC1080 network was reconstructed in a bottom–up manner according to current standards (Thiele and Palsson, 2010) on a pathway-by-pathway basis, drawing biochemical, genomic, and physiological evidence from >250 publications (Supplementary Table S2). The genomic evidence was derived from our own functional annotation (Supplementary Table S3) of metabolic enzymes, coenzymes, and transport proteins. Network gap-filling was performed to make pathways functional and account for dead-end metabolites. Global quality control checks were then performed, including elemental balancing and elimination of as many internal thermodynamically infeasible loops and new photon-driven, input-only pathways as possible (Supplementary Figure S4; Supplementary information). We also accounted for subcellular compartment pH in the protonation states of metabolites as much as possible.
iRC1080 is publicly available at http://www.ebi.ac.uk/biomodels (Accession: MODEL1106200000) and as Supplementary Model S1.
Functional annotation of transcripts
Functional annotation for iRC1080 was performed using a consensus of two separate approaches. In the first approach, gene models (http://augustus.gobics.de/predictions/chlamydomonas/augustus.u5.aa) from the Augustus update 5 (Au5) of C. reinhardtii genome assembly version JGI v4.0 were functionally annotated by assigning enzyme classification (EC) terms using BLASTP results against UniProt (http://www.uniprot.org/) and AraCyc (http://www.arabidopsis.org/biocyc/) enzyme protein sequences and their EC annotations as the basis. The second approach followed from mapping of Au5 gene models to annotated JGI v3.1 gene models, for which EC terms and Gene Ontology annotation were assigned using a combination of BLASTP, AutoFACT, InterProScan, and PRIAM. The comprehensive annotation is presented in Supplementary Table S3. See Supplementary information for full details.
Growth simulations
Simulation procedures consisted of FBA (Orth et al, 2010) and flux variability analysis (FVA) (Mahadevan and Schilling, 2003) as implemented in the COBRA toolbox (Becker et al, 2007), testing model capabilities while optimizing biomass functions to simulate growth (Supplementary Table S10) or subsistence on starch by optimizing ATP maintenance. FBA is a widely used simulation approach for large-scale, constraint-based metabolic models and has become a standard method in the systems biology field with a long history of success (Gianchandani et al, 2010). Different environmental conditions were modeled by appropriately setting reaction flux constraints in iRC1080 (Supplementary Table S6) including environmental exchanges, non-growth associated ATP maintenance, O2 photoevolution, starch degradation, and light- or dark-regulated enzymatic reactions (Supplementary Table S5).
C. reinhardtii strains and growth conditions
For transcript verification experiments, C. reinhardtii strain CC-503 was grown in tris-acetate-phosphate medium containing 100 mg/l carbamicillin without agitation, at room temperature (22–25°C) and under continuous illumination with cool white light at a photosynthetic photon flux of 60 μE/m2/s.
For growth experiments under 660 nm peak LED light (Supplementary Figure S5), C. reinhardtii strain UTEX2243 was grown in a bubble column photobioreactor at 23–27°C with P49 medium. The total volume of algal culture was 300 ml, and the gas supply was 180 ml/min air with 2.5% CO2. The 660-nm peak LED light supply was set at 10 kHz frequency and different duty cycles to get varied average photon fluxes.
Transcript verification by sequencing
ORF amplicons were generated from C. reinhardtii cells by RT–PCR from RNA or PCR from Gateway clones. The Roche 454FLX Titanium sequencing system was used for sequencing of the generated ORF amplicons according to the manufacturer's instructions. The generated data were processed using the GS FLX data analysis software v2.3. Minimum overlap length of 40 nucleotides and minimum overlap identity of 90% were used to align the sequencing reads against the Au5 reference sequences. ORFs encoding transporter proteins were verified by capillary Sanger sequencing.
Prism reaction derivation
Spectral bandwidths that effectively drive each photon-utilizing reaction in iRC1080 were determined from published experimental activity spectral data or absorbance data. Effective spectral bandwidths were defined as the full width half maximum of activity, denoted by color-paired dashed lines in Figure 3A. The effective spectral bandwidths were used to derive stoichiometric coefficients of the prism reactions used to quantitatively represent different light sources from the composition of their published irradiance spectra, converted to photon flux units according to Supplementary Equations 3 and 4. Coefficients for each of the effective spectral bandwidths were computed based on Equation 1.
 |
Each coefficient represents the ratio of photon flux in the defined effective bandwidth to total visible photon flux. Definite integrals in Equation 1 were approximated using the trapezoidal rule. For each light source, all effective bandwidth coefficients were compiled into a single reaction in the form of Equation 2.
 |
Constraints on prism reaction fluxes (Supplementary Table S6) were derived from the total visible photon flux, the definite integral of the spectrum from 380 to 750 nm. The total experimentally measured emitted visible photon flux was converted to model units of incident photon flux using the values in Supplementary Table S11 and Supplementary Equations 5 and 6. Prism reactions for 11 different light sources (Supplementary Figure S3) were generated.
Random sampling of prism reaction space and significance test
For a given prism reaction, first the sum of the stoichiometric coefficients was calculated, representing the total quantity of metabolically active photons per incident photon from the specified light source. Next, to sample the space of prism reactions, 10 000 random prism reactions with the same sum of stoichiometric coefficients were generated and used in growth simulations. In these simulations, input photon flux was constrained to the reported experimental values, generating a set of simulated results (biomass or photosynthetically evolved O2 flux, depending on the experimental parameter) with one value corresponding to each experimental data point. The Euclidean distance between the sampled and experimental results was calculated for each of the 10 000 randomized prism reactions (Figure 5). The significance of the experimental agreement with simulations reported for a given prism reaction derived directly from analysis of irradiance spectra was established by comparison between the corresponding Euclidean distance and the distribution of distances from the randomly sampled prism reactions. Probability of achieving equal or closer results to experiments by chance was computed as the proportion of smaller values in the randomly sampled distribution of 10 000 distances.
Procedure for efficient LED design
Multiple iterations of FVA were used to maximize growth while minimizing the energy of the sum of individual wavelengths of model photon flux. The ratios of these individual wavelength photon fluxes to total photon flux were set as stoichiometric coefficients for a theoretical maximum-efficiency prism reaction. The Euclidean vector distance was computed (Supplementary Figure S6) between this set of coefficients and prism reaction coefficients calculated for an LED spectrum of the same shape as the experimentally measured 674 nm peak LED but centered at varying wavelengths across the visible spectrum, with a total photon flux equal to the total theoretical maximum-efficiency photon flux. The spectrum corresponding to the minimum distance was taken as the solution and subsequently tested through growth simulation.
Supplementary Material
Supplementary Methods, Supplementary Figures S4–6, Supplementary Table S1
High-resolution network diagram. This network diagram displays all metabolites (nodes) and reactions (complex edges) included in iRC1080. Metabolites are color-coded based on compartment localization. Reversibility and irreversibility are represented by the placement of arrows at the ends of edges connecting metabolites (i.e. substrates of irreversible reactions do not have arrows on the edges connecting to them). Labels follow the abbreviation conventions used in iRC1080 (Supplementary Table S2). Visually resolving node and edge labels requires zooming to at least 6400% on most displays.
Complete transcript verification by subsystem. The transcript verification status for all gene-associated subsystems of iRC1080 is displayed. The graph follows the same format as Figure 2.
Complete activity and irradiance spectra. The irradiance spectra are shown for all light sources used in this study. The graphs follow the same format as Figure 3A.
Complete iRC1080 network data. All data included in the iRC1080 network are presented here including but not limited to lower and upper flux bounds (LB and UB, respectively) and all literature references used during the reconstruction. The flux bounds highlighted in yellow represent model input and output parameters that were varied when performing the various simulations presented in this study, and the values in these columns only represent defaults and not the actual values used in any specific simulation.
Transcript functional annotation. The full set of metabolic functional annotation of transcripts is presented, including but not limited to those functions and transcripts represented in iRC1080. The rightmost column denotes the method by which the annotation was obtained: 1 corresponds to the first method we implemented for annotation, 2 corresponds to the second method we implemented for annotation, 3 corresponds to both methods, TCDB signifies annotation resulting from a bidirectional BLAST search comparing TCDB to the C. reinhardtii genome (JGI v4.0), and literature references signify those annotations taken directly from published literature.
Transcript verification status. The percentage of sequence length verification is given for all experimentally tested transcripts included in iRC1080 (see Transcript verification experiments in the Supplementary Methods for more details). Those transcripts verified by capillary Sanger sequencing are denoted by an asterisk.
Light and dark-regulated reaction constraints. Reaction regulation under light and dark conditions, used in this study for constraint-based modeling, are summarized.
Basic modeling constraints. Precise values of parameters used to constrain models in simulations are presented.
Environmental validations. Outcomes of simulations are compared to experimentally measured results to validate model function with respect to environmental parameters.
Genetic validations. Outcomes of knock-out simulations are compared to the analogous experimentally measured mutant phenotypes to validate model function with respect to genetic parameters.
Gene-knockout lethality. Growth or subsistence, in the case of anaerobic dark growth, phenotypes of all single-gene knockout simulations are presented. Phenotypes are classified in relation to the objective flux achieved relative to the wild type simulation: wild type phenotype (WT), reduced relative to wild type phenotype (R), and lethal (L).
Biomass functions. Stoichiometric coefficients for all biomass components accounted for to simulate growth in this study are presented under three growth conditions: photoautotrophic, heterotrophic, and mixotrophic.
Constants for calculations. The basic cellular parameters assumed for calculations in this study and the derived conversion factors (see Dimensional and effective photon flux conversion factor derivation in the Supplementary Methods) are presented.
Acknowledgments
We thank Harish Nagarajan for his critical assessment of this work. This research was supported by the Office of Science (Biological and Environmental Research), US Department of Energy, Grant DE-FG02-07ER64496 (to JP and KS-A), New York University Abu Dhabi Research funds (to KS-A), and Institute Sponsored Research funds from the Dana-Farber Cancer Institute Strategic Initiative (to CCSB). RLC was supported by the National Science Foundation IGERT training Grant DGE0504645, and EFYH was supported by the Jane Coffin Childs Memorial Fund for Medical Research.
Author contributions: RLC wrote the manuscript and developed the light-modeling approach. RLC and AM reconstructed the metabolic network and performed model simulations and analysis. LG, YS and TH performed transcript verification experiments and associated computational analyses. EFYH and SB performed genome functional annotation. WF performed bioreactor growth experiments. B∅P, JAP, and KS-A designed the study.
References
- Akkerman I, Janssen M, Rocha J, Wijffels RH (2002) Photobiological hydrogen production: photochemical efficiency and bioreactor design. Int J Hydrogen Energy 27: 1195–1208 [Google Scholar]
- Baroli I, Do AD, Yamane T, Niyogi KK (2003) Zeaxanthin accumulation in the absence of a functional xanthophyll cycle protects Chlamydomonas reinhardtii from photooxidative stress. Plant Cell 15: 992–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barta DJ, Tibbitts TW, Bula RJ, Morrow RC (1992) Evaluation of light emitting diode characteristics for a space-based plant irradiation source. Adv Space Res 12: 141–149 [DOI] [PubMed] [Google Scholar]
- Becker SA, Feist AM, Mo ML, Hannum G, Palsson BO, Herrgard MJ (2007) Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc 2: 727–738 [DOI] [PubMed] [Google Scholar]
- Berg J, Tymoczko J, Stryer L (2007) Biochemistry. New York, USA: W.H. Freeman [Google Scholar]
- Bjorn L (2007) Photobiology: The Science of Life and Light. Dordrecht, Netherlands: Springer [Google Scholar]
- Bohne F, Linden H (2002) Regulation of carotenoid biosynthesis genes in response to light in Chlamydomonas reinhardtii. Biochim Biophys Acta 1579: 26–34 [DOI] [PubMed] [Google Scholar]
- Boyle NR, Morgan JA (2009) Flux balance analysis of primary metabolism in Chlamydomonas reinhardtii. BMC Syst Biol 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bro C, Regenberg B, Forster J, Nielsen J (2006) In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab Eng 8: 102–111 [DOI] [PubMed] [Google Scholar]
- Cahoon AB, Timko MP (2000) yellow-in-the-dark mutants of Chlamydomonas lack the CHLL subunit of light-independent protochlorophyllide reductase. Plant Cell 12: 559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert MW, Schilling CH, Palsson B (2001) Regulation of gene expression in flux balance models of metabolism. J Theor Biol 213: 73–88 [DOI] [PubMed] [Google Scholar]
- Dent RM, Haglund CM, Chin BL, Kobayashi MC, Niyogi KK (2005) Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol 137: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats C, Mus F, Cuine S, Billon E, Cournac L, Peltier G (2009) Characterization of Nda2, a plastoquinone-reducing type II NAD(P)H dehydrogenase in chlamydomonas chloroplasts. J Biol Chem 284: 4148–4157 [DOI] [PubMed] [Google Scholar]
- Drapier D, Rimbault B, Vallon O, Wollman FA, Choquet Y (2007) Intertwined translational regulations set uneven stoichiometry of chloroplast ATP synthase subunits. EMBO J 26: 3581–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BO (2007) Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA 104: 1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubertret G, Gerard-Hirne C, Trémolières A (2002) Importance of trans-Δ3-hexadecenoic acid containing phosphatidylglycerol in the formation of the trimeric light-harvesting complex in Chlamydomonas. Plant Physiol Biochem 40: 829–836 [Google Scholar]
- Durot M, Bourguignon PY, Schachter V (2009) Genome-scale models of bacterial metabolism: reconstruction and applications. FEMS Microbiol Rev 33: 164–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BO (2007) A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol 3: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes BD, Dragone GM, Teixeira JA, Vicente AA (2010) Light regime characterization in an airlift photobioreactor for production of microalgae with high starch content. Appl Biochem Biotechnol 161: 218–226 [DOI] [PubMed] [Google Scholar]
- Funke RP, Kovar JL, Weeks DP (1997) Intracellular carbonic anhydrase is essential to photosynthesis in Chlamydomonas reinhardtii at atmospheric levels of CO2. Demonstration via genomic complementation of the high-CO2-requiring mutant ca-1. Plant Physiol 114: 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianchandani EP, Chavali AK, Papin JA (2010) The application of flux balance analysis in systems biology. Wiley Interdiscip Rev Syst Biol Med 2: 372–382 [DOI] [PubMed] [Google Scholar]
- Giroud C, Eichenberger W (1989) Lipids of Chlamydomonas reinhardtii. Incorporation of [14C]acetate, [14C]palmitate and [14C]oleate into different lipids and evidence for lipid-linked desaturation of fatty acids. Plant Cell Physiol 30: 121–128 [Google Scholar]
- Giroud C, Gerber A, Eichenberger W (1988) Lipids of Chlamydomonas reinhardtii. Analysis of molecular species and intracellular site(s) of biosynthesis. Plant Cell Physiol 29: 587–595 [Google Scholar]
- Greenbaum E (1988) Energetic efficiency of hydrogen photoevolution by algal water splitting. Biophys J 54: 365–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G, Leverentz M, Silkowski H, Gill N, Sanchez-Serrano JJ (2000) Lipid hydroperoxide levels in plant tissues. J Exp Bot 51: 1363–1370 [PubMed] [Google Scholar]
- Griffiths WT, McHugh T, Blankenship RE (1996) The light intensity dependence of protochlorophyllide photoconversion and its significance to the catalytic mechanism of protochlorophyllide reductase. FEBS Lett 398: 235–238 [DOI] [PubMed] [Google Scholar]
- Grossman AR, Croft M, Gladyshev VN, Merchant SS, Posewitz MC, Prochnik S, Spalding MH (2007) Novel metabolism in Chlamydomonas through the lens of genomics. Curr Opin Plant Biol 10: 190–198 [DOI] [PubMed] [Google Scholar]
- Guschina IA, Harwood JL (2006) Lipids and lipid metabolism in eukaryotic algae. Prog Lipid Res 45: 160–186 [DOI] [PubMed] [Google Scholar]
- Harris E, Stern D, Witman G (2008) The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and Its Laboratory Use. Amsterdam, Netherlands: Academic Press [Google Scholar]
- Harris EH (2001) Chlamydomonas as a model organism. Annu Rev Plant Physiol Plant Mol Biol 52: 363–406 [DOI] [PubMed] [Google Scholar]
- Hegemann P, Marwan W (1988) Single photons are sufficient to trigger movement responses in Chlamydomonas reinhardtii. Photochem Photobiol 48: 99–106 [Google Scholar]
- Hemschemeier A, Melis A, Happe T (2009) Analytical approaches to photobiological hydrogen production in unicellular green algae. Photosynth Res 102: 523–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills MJ, Roscoe TJ (2006) Synthesis of Structural and Storage Lipids by the ER. Berlin/Heidelberg: Springer [Google Scholar]
- Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A (2008) Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J 54: 621–639 [DOI] [PubMed] [Google Scholar]
- Janssen M, de Winter M, Tramper J, Mur LR, Snel J, Wijffels RH (2000) Efficiency of light utilization of Chlamydomonas reinhardtii under medium-duration light/dark cycles. J Biotechnol 78: 123–137 [DOI] [PubMed] [Google Scholar]
- Kajikawa M, Yamato KT, Kohzu Y, Shoji S, Matsui K, Tanaka Y, Sakai Y, Fukuzawa H (2006) A front-end desaturase from Chlamydomonas reinhardtii produces pinolenic and coniferonic acids by omega13 desaturation in methylotrophic yeast and tobacco. Plant Cell Physiol 47: 64–73 [DOI] [PubMed] [Google Scholar]
- Lang ID (2007) New Fatty Acids, Oxylipins and Volatiles in Microalgae, PhD Thesis, Göttingen: Mathematisch-naturwissenschaftliche Fakultäten, Georg-August-Universität Göttingen [Google Scholar]
- Lemaire SD, Guillon B, Le Marechal P, Keryer E, Miginiac-Maslow M, Decottignies P (2004) New thioredoxin targets in the unicellular photosynthetic eukaryote Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 101: 7475–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, Voigt CA (2005) Synthetic biology: engineering Escherichia coli to see light. Nature 438: 441–442 [DOI] [PubMed] [Google Scholar]
- Mahadevan R, Schilling CH (2003) The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng 5: 264–276 [DOI] [PubMed] [Google Scholar]
- Manichaikul A, Ghamsari L, Hom EF, Lin C, Murray RR, Chang RL, Balaji S, Hao T, Shen Y, Chavali AK, Thiele I, Yang X, Fan C, Mello E, Hill DE, Vidal M, Salehi-Ashtiani K, Papin JA (2009) Metabolic network analysis integrated with transcript verification for sequenced genomes. Nat Methods 6: 589–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo ML, Palsson BO, Herrgard MJ (2009) Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst Biol 3: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagud A, Navarro E, Fernandez de Cordoba P, Urchueguia JF, Patil KR (2010) Reconstruction and analysis of genome-scale metabolic model of a photosynthetic bacterium. BMC Syst Biol 4: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Tanaka S, Teko Y, Mitsui K, Kanazawa H (2005) Four Na+/H+ exchanger isoforms are distributed to Golgi and post-Golgi compartments and are involved in organelle pH regulation. J Biol Chem 280: 1561–1572 [DOI] [PubMed] [Google Scholar]
- Oberhardt MA, Palsson BO, Papin JA (2009) Applications of genome-scale metabolic reconstructions. Mol Syst Biol 5: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Thiele I, Palsson BO (2010) What is flux balance analysis? Nat Biotechnol 28: 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Kim TY, Lee KH, Lee SY (2011) Fed-batch culture of Escherichia coli for L-valine production based on in silico flux response analysis. Biotechnol Bioeng 108: 934–946 [DOI] [PubMed] [Google Scholar]
- Polle JE, Kanakagiri SD, Melis A (2003) tla1, a DNA insertional transformant of the green alga Chlamydomonas reinhardtii with a truncated light-harvesting chlorophyll antenna size. Planta 217: 49–59 [DOI] [PubMed] [Google Scholar]
- Reed JL, Vo TD, Schilling CH, Palsson BO (2003) An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol 4: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle C, Baurain D, Cardol P, Matagne RF (2001a) Mutants of Chlamydomonas reinhardtii deficient in mitochondrial complex I: characterization of two mutations affecting the nd1 coding sequence. Genetics 158: 1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle C, Duby F, Cardol P, Matagne RF (2001b) Mutations inactivating mitochondrial genes in Chlamydomonas reinhardtii. Biochem Soc Trans 29: 442–446 [DOI] [PubMed] [Google Scholar]
- Riekhof WR, Sears BB, Benning C (2005) Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: discovery of the betaine lipid synthase BTA1Cr. Eukaryot Cell 4: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador ML, Klein U, Bogorad L (1993) Light-regulated and endogenous fluctuations of chloroplast transcript levels in Chlamydomonas. Regulation by transcription and RNA degradation. Plant J 3: 213–219 [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato S, Huq E, Tepperman JM, Quail PH (2002) A light-switchable gene promoter system. Nat Biotechnol 20: 1041–1044 [DOI] [PubMed] [Google Scholar]
- Smart EJ, Selman BR (1991) Isolation and characterization of a Chlamydomonas reinhardtii mutant lacking the gamma-subunit of chloroplast coupling factor 1 (CF1). Mol Cell Biol 11: 5053–5058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MH, Spreitzer RJ, Ogren WL (1983) Carbonic anhydrase-deficient mutant of Chlamydomonas reinhardii requires elevated carbon dioxide concentration for photoautotrophic growth. Plant Physiol 73: 268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolaore P, Joannis-Cassan C, Duran E, Isambert A (2006) Commercial applications of microalgae. J Biosci Bioeng 101: 87–96 [DOI] [PubMed] [Google Scholar]
- Stern D, Harris E, Witman G (2008) The Chlamydomonas Sourcebook: Organellar and Metabolic Processes. Amsterdam, Netherlands: Academic Press [Google Scholar]
- Suzuki K, Marek LF, Spalding MH (1990) A photorespiratory mutant of Chlamydomonas reinhardtii. Plant Physiol 93: 231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuzawa H, Takizawa E, Wada M, Yamamoto Y (1996) Fatty acid and lipid composition of the acidophilic green alga Chlamydomonas sp. J Phycol 32: 598–601 [Google Scholar]
- Thiele I, Palsson BO (2010) A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc 5: 93–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weers PMM, Gulati RD (1997) Growth and reproduction of Daphnia galeata in response to changes in fatty acids, phosphorus, and nitrogen in Chlamydomonas reinhardtii. Limnol Oceanogr 42: 1584–1589 [Google Scholar]
- Yashodhara BM, Umakanth S, Pappachan JM, Bhat SK, Kamath R, Choo BH (2009) Omega-3 fatty acids: a comprehensive review of their role in health and disease. Postgrad Med J 85: 84–90 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods, Supplementary Figures S4–6, Supplementary Table S1
High-resolution network diagram. This network diagram displays all metabolites (nodes) and reactions (complex edges) included in iRC1080. Metabolites are color-coded based on compartment localization. Reversibility and irreversibility are represented by the placement of arrows at the ends of edges connecting metabolites (i.e. substrates of irreversible reactions do not have arrows on the edges connecting to them). Labels follow the abbreviation conventions used in iRC1080 (Supplementary Table S2). Visually resolving node and edge labels requires zooming to at least 6400% on most displays.
Complete transcript verification by subsystem. The transcript verification status for all gene-associated subsystems of iRC1080 is displayed. The graph follows the same format as Figure 2.
Complete activity and irradiance spectra. The irradiance spectra are shown for all light sources used in this study. The graphs follow the same format as Figure 3A.
Complete iRC1080 network data. All data included in the iRC1080 network are presented here including but not limited to lower and upper flux bounds (LB and UB, respectively) and all literature references used during the reconstruction. The flux bounds highlighted in yellow represent model input and output parameters that were varied when performing the various simulations presented in this study, and the values in these columns only represent defaults and not the actual values used in any specific simulation.
Transcript functional annotation. The full set of metabolic functional annotation of transcripts is presented, including but not limited to those functions and transcripts represented in iRC1080. The rightmost column denotes the method by which the annotation was obtained: 1 corresponds to the first method we implemented for annotation, 2 corresponds to the second method we implemented for annotation, 3 corresponds to both methods, TCDB signifies annotation resulting from a bidirectional BLAST search comparing TCDB to the C. reinhardtii genome (JGI v4.0), and literature references signify those annotations taken directly from published literature.
Transcript verification status. The percentage of sequence length verification is given for all experimentally tested transcripts included in iRC1080 (see Transcript verification experiments in the Supplementary Methods for more details). Those transcripts verified by capillary Sanger sequencing are denoted by an asterisk.
Light and dark-regulated reaction constraints. Reaction regulation under light and dark conditions, used in this study for constraint-based modeling, are summarized.
Basic modeling constraints. Precise values of parameters used to constrain models in simulations are presented.
Environmental validations. Outcomes of simulations are compared to experimentally measured results to validate model function with respect to environmental parameters.
Genetic validations. Outcomes of knock-out simulations are compared to the analogous experimentally measured mutant phenotypes to validate model function with respect to genetic parameters.
Gene-knockout lethality. Growth or subsistence, in the case of anaerobic dark growth, phenotypes of all single-gene knockout simulations are presented. Phenotypes are classified in relation to the objective flux achieved relative to the wild type simulation: wild type phenotype (WT), reduced relative to wild type phenotype (R), and lethal (L).
Biomass functions. Stoichiometric coefficients for all biomass components accounted for to simulate growth in this study are presented under three growth conditions: photoautotrophic, heterotrophic, and mixotrophic.
Constants for calculations. The basic cellular parameters assumed for calculations in this study and the derived conversion factors (see Dimensional and effective photon flux conversion factor derivation in the Supplementary Methods) are presented.