Abstract
Oncogenes such as K-ras mediate cellular and metabolic transformation during tumorigenesis. To analyze K-Ras-dependent metabolic alterations, we employed 13C metabolic flux analysis (MFA), non-targeted tracer fate detection (NTFD) of 15N-labeled glutamine, and transcriptomic profiling in mouse fibroblast and human carcinoma cell lines. Stable isotope-labeled glucose and glutamine tracers and computational determination of intracellular fluxes indicated that cells expressing oncogenic K-Ras exhibited enhanced glycolytic activity, decreased oxidative flux through the tricarboxylic acid (TCA) cycle, and increased utilization of glutamine for anabolic synthesis. Surprisingly, a non-canonical labeling of TCA cycle-associated metabolites was detected in both transformed cell lines. Transcriptional profiling detected elevated expression of several genes associated with glycolysis, glutamine metabolism, and nucleotide biosynthesis upon transformation with oncogenic K-Ras. Chemical perturbation of enzymes along these pathways further supports the decoupling of glycolysis and TCA metabolism, with glutamine supplying increased carbon to drive the TCA cycle. These results provide evidence for a role of oncogenic K-Ras in the metabolic reprogramming of cancer cells.
Keywords: cancer, metabolic flux analysis, metabolism, Ras, transcriptional analysis
Introduction
Several decades ago Otto Warburg first described that tumors exhibit glycolytic metabolism with a reduced rate of oxidative phosphorylation, despite the availability of adequate oxygen. This phenomenon, known as the ‘Warburg effect,’ has been proposed to be a key driver of tumor progression (Warburg, 1956). Since then, several researchers have observed altered glucose metabolism associated with a high rate of lactate secretion in cancer cells and tumor tissues (Sauer et al, 1982; Steinberg et al, 1992). Furthermore, aerobic glycolysis represents a robust hallmark of cancer that is employed for tumor detection by positron emission tomography analysis with the 2-[18F]fluoro-2-deoxy-D-glucose tracer (Laing et al, 2009). However, we and others have recently provided evidence on the important anabolic role of glutamine in tumor cell proliferation (DeBerardinis et al, 2007; Yuneva et al, 2007; Gaglio et al, 2009). Indeed, by using different experimental approaches, it has been shown that glycolytic cancer cells consume more glutamine as compared with their normal counterparts to synthesize proteins, nucleotides and fatty acids and produce energy (Kovacevic and McGivan, 1983; Baggetto, 1992; DeBerardinis et al, 2007).
Several reports suggest that both nutrient uptake changes and metabolic alterations are under direct control of ras or myc oncogenes (Chiaradonna et al, 2006b; Wise et al, 2008; Vander Heiden et al, 2009). In particular, oncogenic Ras proteins, identified in 25% of human cancers (Bos, 1989), correlate with metabolic alterations, including increased glucose and glutamine consumption, lactic acid accumulation, altered expression of mitochondrial genes, increased reactive oxygen species (ROS) production and reduced mitochondrial activity (Bos, 1989; Vizan et al, 2005; Chiaradonna et al, 2006a; Yun et al, 2009; Weinberg et al, 2010). A result of this metabolic reprogramming is the dependence of K-Ras transformed cells on glucose and glutamine availability, since their withdrawal induces apoptosis and cell-cycle arrest, respectively (Ramanathan et al, 2005; Telang et al, 2006; Yun et al, 2009). However, the precise metabolic effects downstream of oncogenic Ras signaling in cancer cells have not been completely elucidated.
Due to the interconnected nature of metabolic pathways and the pleiotropic effects mediated by oncogenic signals, a systems approach is required to elucidate the mechanisms of such transforming events. To date, most large-scale analyses of tumor cells have employed microarray technology that provides a robust means of analyzing transcriptional modifications associated with physio/pathological changes (Ross et al, 2000; Scherf et al, 2000). However, various levels of post-transcriptional controls may not be captured by these analyses (Mata et al, 2005; Metallo and Vander Heiden, 2010), and new approaches are now emerging to increase our knowledge about cancer cell physiology (Liotta and Petricoin, 2000; Laubenbacher et al, 2009; Kreeger and Lauffenburger, 2010).
Metabolomic techniques offer a more direct means of studying metabolism at the systems level. Measuring metabolite concentrations, indeed, represents a sensitive approach that generates detailed ‘snapshots’ of biological processes (Hiller et al, 2009). Nevertheless, metabolic routes are best characterized by the measurement of fluxes, which describe the actual functionality of a given enzyme or pathway (Stephanopoulos and Vallino, 1991; Sauer, 2006). To this end, isotopic tracers and computational algorithms enable the quantitative estimation of intracellular fluxes and associated confidence intervals for a given system (Metallo et al, 2009; Hiller et al, 2010), and such methods are now effectively applied to mammalian cells (Vizan et al, 2005; Munger et al, 2008).
Therefore, to better understand the regulation of cancer cell metabolism and to identify key metabolic routes altered in K-Ras transformed cells, we have applied a systems-level approach based on the combined application of metabolic and transcriptional analyses. We have monitored the flux of 13C-labeled glucose and glutamine as well as [α15N]glutamine into downstream metabolites in normal and transformed cells and performed a detailed comparison with the transcriptional profiles obtained from the same cell lines. Herein, we show that K-ras oncogene expression enhances glucose uptake but decreases its utilization in the tricarboxylic acid (TCA) cycle and associated anabolic pathways. Furthermore, we show that while K-Ras transformation decreases overall flux through the TCA cycle, it increases utilization of the carbon backbone and amino-nitrogen moieties of glutamine either through TCA cycle or transamination activities in order to sustain biosynthetic reactions, including amino-acid, nucleotide and glutathione synthesis. Finally, we present evidence describing the dependence of K-Ras on glutamine metabolism, as inhibition of key enzymes along this pathway specifically compromises the proliferation of transformed cells.
Results
K-Ras transformation enhances glycolytic flux and decreases oxidative TCA cycle flux in mouse fibroblasts
We have previously shown that transformed NIH3T3 mouse fibroblasts expressing an oncogenic K-Ras protein (G12V) (Shih et al, 1981; Pulciani et al, 1982; Bossu et al, 2000) exhibit elevated sensitivity to glucose availability, reduced mitochondrial function linked to a decrease of Complex I activity and protein level, and reduced proliferation in response to glucose, glutamine or galactose shortage as compared with immortalized NIH3T3 mouse fibroblasts (Chiaradonna et al, 2005, 2006a; Gaglio et al, 2009; Baracca et al, 2010). NIH3T3 cells are a genetically well-characterized immortalized cell line that has long been established as a model of ‘normal’ cells for the study of cell transformation, as these cells undergo contact inhibition, exhibit no growth in soft agar, and do not form tumors in immunocompromised mice, in contrast to isogenic transformed lines (Bossu et al, 2000).
Therefore, we performed basic metabolic analyses to compare NIH3T3 cells (normal, N) and NIH3T3 K-Ras (transformed, T) 54 h (T54) after plating. This time point was chosen since it corresponds to the time in which the two cell lines begin to show a different proliferative ability (Chiaradonna et al, 2006b). As shown in Figure 1A, cell-specific glucose (Glc) and glutamine (Gln) consumption as well as lactate (Lac) and glutamate (Glu) secretion were measured. Significant increases in Glc uptake and Lac production in T cells were observed (Figure 1A), consistent with other reports linking ras mutations to aerobic glycolysis (Elstrom et al, 2004; Vizan et al, 2005). Although previous studies have shown an increase of Gln uptake in other kinds of cancer cell lines (Yuneva et al, 2007; Wise et al, 2008; Gao et al, 2009), we did not detect any significant changes in Gln uptake and Glu secretion between the two cell lines. To further define N- and T-cell metabolism, we have determined the relative abundance of various intracellular amino and organic acids by performing a targeted metabolomic analysis (Figure 1B). Notably, metabolites associated with glycolysis and biosynthetic processes were present in larger quantities in T cells, including pyruvate (Pyr), Lac, citrate (Cit), and aspartate (Asp). On the other hand, TCA cycle intermediates such as α-ketoglutarate (AKG), succinate (Suc), and fumarate (Fum), which arise primarily from oxidative metabolism, were less abundant in T cells as compared with N cells.
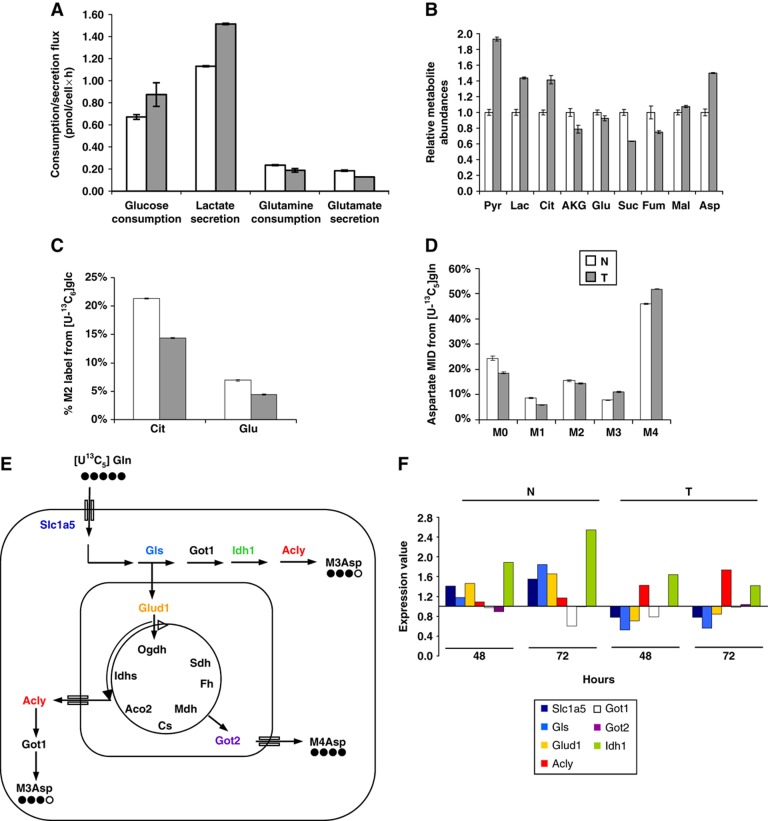
Figure 1.
K-Ras transformed fibroblasts decouple glycolysis and the TCA cycle. (A) Extracellular uptake and secretion of Glc, Lac, Gln, and Glu in N and T cells for 54 h of growth. (B) Relative metabolite abundances in N and T cells measured by GC/MS. (C) M2 labeling of Cit and Glu in N and T cells cultured with a 1:1 mixture of [1-13C]glucose and [U-13C6]glucose. (D) Mass isotopomer distribution (MID) of Asp in cells cultured in the presence of [U-13C5]glutamine. Error bars indicate s.e.m. (n=3). (E) Schematic representation of the metabolic routes involved in M3-M4 Asp labeling by using [U13C5]glutamine tracer. In the scheme are represented the key cytoplasmic and mitochondrial enzymes involved in these metabolic routes. (F) Absolute expression values of some genes, involved in the pathways described in the scheme (E), in N and T cell lines. The two cell lines, grown in 25 mM Glc+4 mM Gln (normal medium), were collected for transcriptional analysis at two different time points (48 and 72 h). The list of gene abbreviations is available in Supplementary information.
Isotopic tracers provide an effective means of determining the relative contribution of a substrate to a particular metabolic pathway, and mass spectrometry enables quantification of the relative abundance of molecules with different numbers of isotopic label in each metabolite pool (i.e., M0, M1, M2, … where M2 abundance includes all molecules with two 13C-labeled atoms). For instance, uniformly labeled Glc ([U-13C6]glucose) is converted to M3 Pyr (with three 13C atoms) and subsequently M2 Cit through the combined action of pyruvate dehydrogenase and citrate synthase. In the same way, M2 Cit, progressing along the TCA cycle, generates M2 AKG and Glu by TCA cycle-related reactions (i.e., transamination and reductive amination; Supplementary Figure S1). We cultured N and T cells in the presence of 13C-labeled glucose and quantified the mass isotopomer distribution (MID) of Cit and Glu via gas chromatography/mass spectrometry (GC/MS). These data indicated that T cells oxidize less Glc carbon to Cit and Glu, presumably diverting more Pyr to Lac, as evidenced by the significant decreases in M2 labeling observed in these metabolites (Figure 1C).
Consistent with the observation of decreased Glc oxidation, parallel [U-13C5]glutamine labeling experiments indicated that Gln carbon utilization for metabolic intermediate synthesis (e.g. Asp) was increased in T cells. We specifically observed a relative increase in M3 and M4 Asp abundance (Figure 1D), which are generated from M5 AKG downstream of reductive carboxylation by isocitrate dehydrogenase and oxidation by oxoglutarate dehydrogenase, respectively (Supplementary Figure S2). These results support the notion that Gln entering the TCA cycle of T cells may follow both the canonical forward oxidative reaction and a reductive carboxylation (Yoo et al, 2008). Moreover, given the significant role of Asp in amino-acid and nucleotide synthesis, these results suggest that T cells are strongly dependent on Gln carbon for biosynthesis (Weinberg et al, 2010).
Mass isotopomer data alone provides a somewhat limited view of cellular metabolism. To better characterize N- and T-cell metabolism at the systems level, we integrated our extracellular uptake and secretion results with MID data obtained by culturing the cells with [U-13C5]glutamine. Using these data, we estimated intracellular fluxes in each cell type using an elementary metabolite unit (EMU)-based 13C metabolic flux analysis (MFA) algorithm (Young et al, 2008). Upon obtaining acceptable fits, we performed a sensitivity analysis to generate 95% confidence intervals for each flux in the network (Antoniewicz et al, 2006; Antoniewicz et al, 2007). Flux results for N and T cells are listed in Table I, and the full dataset and model description are provided in Supplementary information and Supplementary Figures S3 and S4. These data indicate that transformation induces a significant reorganization of central carbon metabolism. T cells reduce flux through the PDH complex by ∼50% (i.e., flux values of Pyr → AcCoAmit+CO2). Furthermore, net fluxes throughout the TCA cycle were decreased by the same amount. Each of these changes was highly significant, as evidenced by the lack of overlap in confidence intervals between the two cell types. Although most exchange fluxes in reversible reactions of the TCA cycle could not be accurately determined, we observed no significant change in reductive flux through IDH or malic enzyme. Consistent changes in metabolic fluxes were observed when performing MFA using labeling data from cells cultured with 13C-glucose (see Supplementary Information for detailed results), though the observability of some fluxes (e.g. IDH exchange flux) was significantly decreased when using these tracers. These findings provide evidence that transformation causes cells to decouple glycolysis and the TCA cycle, with an overall decrease in activity of oxidative reactions within the TCA cycle.
Table 1. Experimentally determined fluxes and 95% confidence intervals estimated from MFA model using [U-13C5]glutamine as a tracer.
| Reaction | Normal |
Transformed |
T/N | ||
|---|---|---|---|---|---|
| Flux | Confidence interval | Flux | Confidence interval | ||
| Detailed description, assumptions, complete results, and data are available in Supplementary information. Net flux: (υF−υR) ( → ) and Exchange flux: min (υF,υR) (↔). Some scaling and undetermined exchange fluxes (i.e., confidence intervals of infinity, Inf) are omitted for brevity. Comparison of fluxes (Transformed versus Normal) is described as U=up, D=down, NC=no change, and ND=no detectable. Fluxes are listed × 1014 mol/(cell × h). | |||||
| Glycolysis | |||||
| Glucext → G6P | 65.5 | 61.2, 69.9 | 83.1 | 77.6, 88.6 | U |
| G6P → F6P | 62.5 | 58.2, 66.9 | 82.1 | 77.6, 87.6 | U |
| G6P↔F6P | 4.1 | 0.0, Inf | 18.6 | 0.0, Inf | ND |
| F6P → DHAP+GAP | 64.2 | 59.9, 68.5 | 82.4 | 76.9, 88.0 | U |
| DHAP → GAP | 64.2 | 59.9, 68.5 | 82.4 | 76.9, 88.0 | U |
| DHAP↔GAP | 36.7 | 0.0, Inf | 13.1 | 0.0, Inf | ND |
| GAP → 3PG | 129.2 | 120.6, 137.9 | 165 | 154.0, 176.1 | U |
| GAP↔3PG | 0 | 0.0, Inf | 0 | 0.0, Inf | ND |
| 3PG → Pyr | 129.2 | 120.6, 137.9 | 165 | 154.0, 176.1 | U |
| Pyr → Lac | 114.9 | 105.9, 123.4 | 158.1 | 146.9, 169.2 | U |
| Pyr↔Lac | 300.5 | 0.0, Inf | 9.2 | 0.0, Inf | ND |
| Lac → Lacext | 114.9 | 105.9, 123.4 | 158.1 | 146.9, 169.2 | U |
| Pentose phosphate pathway | |||||
| 6PG → P5P+CO2 | 3.00 | 2.71, 3.29 | 1.00 | 0.90, 1.10 | D |
| P5P+P5P → S7P+GAP | 0.83 | 0.73, 0.93 | 0.17 | 0.13, 0.20 | D |
| S7P+GAP → F6P+E4P | 0.83 | 0.73, 0.93 | 0.17 | 0.13, 0.20 | D |
| P5P+E4P → F6P+GAP | 0.83 | 0.73, 0.93 | 0.17 | 0.13, 0.20 | D |
| TCA cycle | |||||
| Pyrmit+CO2 → OAA | 0 | 0.0, 3.2 | 0 | 0.0, 1.9 | NC |
| Mal → Pyrmit+CO2 | 1.8 | 0.9, 5.9 | 1.8 | 0.7, 4.7 | NC |
| Mal↔Pyrmit+CO2 | 2.8 | 0.0, 3.2 | 1.6 | 0.0, 1.9 | NC |
| Pyrmit → AcCoAmit+CO2 | 16.1 | 13.5, 19.1 | 8.6 | 7.6, 9.6 | D |
| AcCoAmit+OAA → Cit | 16.1 | 13.5, 19.1 | 8.6 | 7.6, 9.6 | D |
| Cit → AKG+CO2 | 10.1 | 9.1, 11.2 | 5.4 | 4.9, 6.0 | D |
| Cit↔AKG+CO2 | 3.3 | 2.7, 3.9 | 3.7 | 3.2, 4.2 | NC |
| AKG → Suc+CO2 | 12.1 | 10.6, 13.7 | 7.4 | 6.1, 8.7 | D |
| Suc → Fum | 12.1 | 10.6, 13.7 | 7.4 | 6.1, 8.7 | D |
| Suc↔Fum | 37.2 | 0.0, Inf | 0 | 0.0, 1.5 | ND |
| Fum → Mal | 12.1 | 10.6, 13.7 | 7.4 | 6.1, 8.7 | D |
| Fum↔Mal | 0 | 0.0, Inf | 42 250 | 1.5, Inf | ND |
| Mal → OAA | 10.3 | 6.7, 11.3 | 5.6 | 3.5, 6.1 | D |
| Mal↔OAA | 36.2 | 0.0, Inf | 57 630 | 3.2, Inf | ND |
| Pyr → Pyrmit | 14 | 12.0, 17.0 | 7 | 6.0, 8.0 | D |
| Pyr↔Pyrmit | 74 | 39.0, 133 | 276 | 87.0, 4566 | U |
| Glutamine metabolism | |||||
| Glnext → Gln | 20 | 19.0, 22.0 | 16 | 15.0, 17.0 | D |
| Gln → Glu | 20 | 19.0, 21.0 | 16 | 15.0, 17.0 | D |
| Glu → Gluext | 18 | 16.0, 19.0 | 14 | 12.0, 15.0 | D |
| Glu → AKG | 2 | 1.0, 3.0 | 2 | 1.0, 3.0 | NC |
| Glu↔AKG | 168 | 115, 341 | 202 | 106, 1895 | U |
| Pyrmit+Glu → Ala+AKG | 0.15 | 0.13, 0.16 | 0.15 | 0.14, 0.17 | NC |
| OAA+Glu → Asp+AKG | 0.18 | 0.16, 0.20 | 0.18 | 0.16, 0.20 | NC |
Transcriptional profiling of N and T fibroblasts is largely coherent with metabolic flux analysis
Various reports have indicated that oncogenes alter the expression of genes encoding metabolic enzymes (Mathupala et al, 1997; Zhong et al, 1999). To identify oncogenic K-Ras-dependent transcriptional effects, we performed microarray transcriptional profiling of N and T cells grown as above for 48 and 72 h.
These data, represented by heat maps in Supplementary Figure S5, indicate that several genes of the investment phase of glycolysis (Hk1 and Pfkp) as well as Pfkfb 2, 3 and 4, which through the regulation of fructose-2,6-biphosphate level allosterically control 6-phosphofructo-1-kinase, were consistently expressed at lower levels in T cells as compared with N cells (Supplementary Figure S5A) (green color). In addition, five genes encoding glycolytic enzymes (Gpi1, Pfkl, Pfkm, AldoA and Tpi1) were highly expressed, particularly at 48 h, in T cells as compared with N cells (red color) (Supplementary Figure S5A). Moreover, a high expression for genes involved in the pay-off phase of glycolysis, where ATP is generated, was observed. Indeed, almost all the genes of this phase, at both time points 48 and 72 h, were expressed at a higher level in T cells as compared with N ones (Gapdh, Pgk1, Eno1, Eno2, Pkm2, Ldha and Ldhb).
Consistent with the reduction of TCA flux that we observed, three genes encoding kinase that inhibit the PDH enzyme complex activity (Pdk1, Pdk3, and Pdk4) were significantly expressed in T cells (Supplementary Figure S5B). In addition, expression of Ogdh, which encodes one subunit of the oxoglutarate dehydrogenase complex, another key control point in the TCA cycle, was decreased in T cells (Supplementary Figure S5B). On the contrary, few genes encoding for positive regulators of TCA cycle, as Aco2, Sucla2 and Sucgl2, as well as Idh2, which is able to catalyze the reductive carboxylation of AKG, were expressed at higher levels in T cells throughout the time window of the analysis as compared with N ones (Supplementary Figure S5B). Taken together, these results indicate that numerous genes regulated by oncogenic K-Ras may drive the observed metabolic reprogramming, although we cannot exclude the possibility that other mechanisms (e.g., post-translational modifications, mass action, and allosteric enzyme regulation) also participate in the observed metabolic alterations of transformed cells.
Transcriptional analysis of glutamine entry into TCA cycle
Previous experiments (Figure 1) indicated that Gln contributes to precursors of cellular biomass (e.g., aspartate) to a greater extent in T cells as compared with N cells, despite the observed small decrease in uptake. These findings are consistent with other reports describing Myc-driven increases of Gln usage through glutaminolysis, mediated in part via expression of genes involved in Gln uptake and metabolism (Wise et al, 2008; Gao et al, 2009). To investigate this feature, we have analyzed the expression of genes encoding enzymes involved in Gln uptake and metabolism, with particular attention to those involved in metabolic pathways able to generate Asp. As shown in Figure 1E, Asp can be produced from Gln both in cytoplasm and in mitochondria through AKG by oxidative decarboxylation and reductive carboxylation. Gln enters into the cells by high-affinity glutamine importer Slc1a5 and is metabolized to Glu by glutaminase (Gls) (Figure 1E). Subsequently, Glu may be converted to AKG, either via transaminase activity of cytoplasmic and mitochondrial aspartate and alanine aminotransferases (Got1 and Got2 and Gpt1 and Gpt2, respectively) or via glutamate dehydrogenase-dependent oxidative deamination (Glud1) (Figure 1E). As shown in Figure 1F, Slc1a5, Gls1, and Glud1 were consistently expressed at lower levels in T cells (about two-fold) relative to N cells. On the contrary, Got1 and 2, showed an opposite behavior, with both genes slightly upregulated in T cells compared with N cells. These transcriptional changes may partially justify the slight reduction in Gln uptake (e.g., downregulation of Slc1a5 in transformed cells) shown in Figure 1A and suggest, by analysis of the metabolic routes depicted in Figure 1E, a relevant role of AKG for the generation of labeled M4 Asp through oxidative metabolism (see also transcriptional data in Figure 1F and Supplementary Figure S5). In addition, in T cells, higher expression of Idh2 (Supplementary Figure S5) and in particular of Acly, whose level was at least two-fold higher than N cells (Figure 1F), is coherent with the increased M3 Asp labeling previously shown. However, while transcriptional data are for the most part consistent with the reduction of glutamine uptake and its use for biosynthesis, not all metabolic fluxes are expected to be only regulated at the transcriptional level.
Transformed cells increase their use of glutamine nitrogen for biosynthetic processes
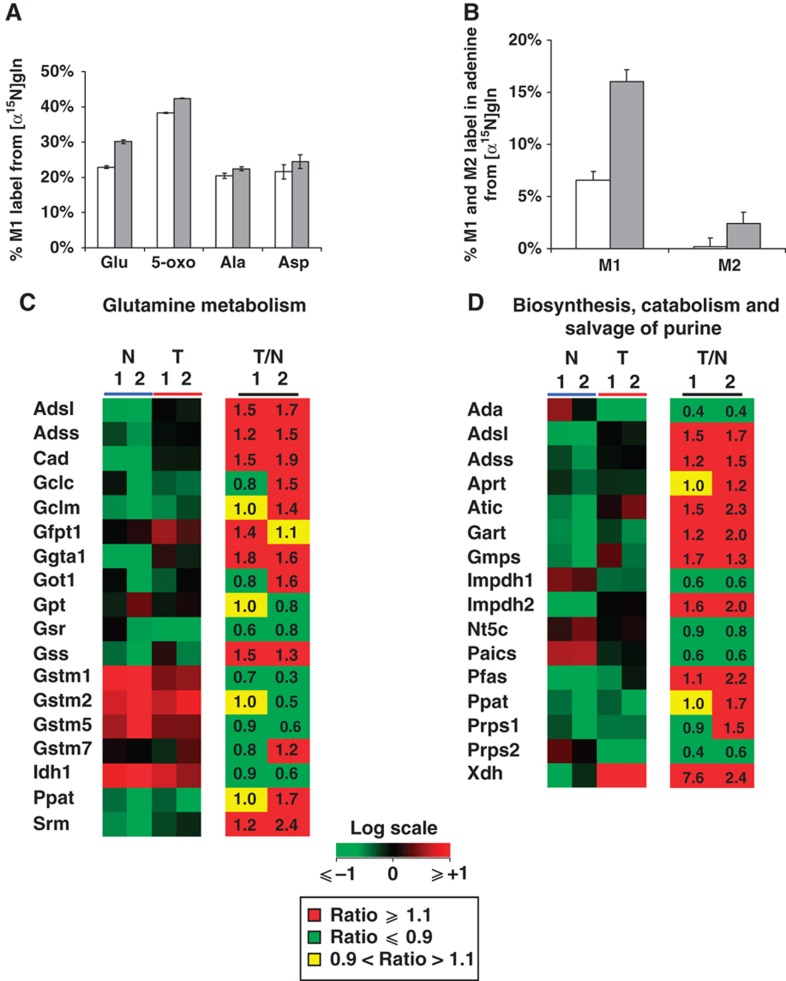
To further investigate the role of Gln in cellular biosynthesis, we performed a non-targeted tracer fate detection (NTFD) analysis using amino-labeled glutamine ([α-15N]glutamine) to identify and quantify metabolites containing glutamine-derived nitrogen. This method enables the detection and quantification of label in all observable metabolites downstream of the tracer substrate (Hiller et al, 2010). Cells require nitrogen atoms to build molecules such as nucleotides, amino acids, amino sugars with Gln and Glu representing the primary cellular reservoir of nitrogen. The amino nitrogen of Gln is retained on Glu and used as a nitrogen donor in transaminase reactions. As expected, the NTFD algorithm identified many amino acids and associated compounds that were highly labeled from [α-15N]glutamine tracer, including Glu, Ala, Asp, and 5-oxoproline (5-oxo) (Figure 2A). With the exception of Asp, each of these metabolites was more significantly labeled in T cells as compared with N cells. In addition, we observed that M1 and M2 labeling on the nucleobase adenine, generated from [α-15N]glutamine-derived nitrogen atoms from Asp, was over 2–3-fold higher in T cells compared with N control cells (Figure 2B).
Figure 2.
Labeled metabolites identified by NTFD analysis in cells cultured with [α-15N]glutamine for 54 h. (A) M1 labeling on amino acids and derivative metabolites in N (□) and T (▪) cells. (B) M1 and M2 labeling observed in free adenine in N (□) and T (▪) cells. (C) Heat maps of genes involved in Gln metabolism of N (blue line) and T (red line) cell lines, and corresponding ratios (T/N) between the two cell lines grown in normal medium and collected at two different time points (1=48 h and 2=72 h). (D) Heat maps of genes involved in purine metabolism of N (blue line) and T (red line) cell lines, and corresponding ratios (T/N) between the two cell lines grown in normal medium and collected at two different time points (1=48 h and 2=72 h). Color scale bar indicates the normalized expression intensity (log scale); in particular, low expression in green, high expression in red, and not changed expression in black. The ratio values are represented as Up—red color—when the ratio is ⩾1.1, down—green color—when the ratio is ⩽0.9 and no change—yellow color—when the ratio is between 0.9 and 1.1. The list of gene abbreviations is available in Supplementary information.
Transcriptional data support results from NTFD analysis with [α-15N]glutamine. Absolute gene expression values of N and T cells and their ratio (T/N ratio) as well, shown in Figure 2C, Supplementary Table S1, highlight the elevated expression of genes closely related to Gln utilization in T cells. In particular, the expression of genes involved in glutathione metabolism (e.g., Gss, Gclm), hexosamine metabolism (e.g., Gfpt1), and nucleotide biosynthesis (e.g., Cad, Adss, and Adsl) was increased in T cells as compared with N cells. Such transcriptional changes suggest increased activity of glutathione pathway and the enhancement of nucleotide synthesis, as indicated respectively by downstream metabolite 5-oxo labeling (Figure 2A) and M1 and M2 labeling on the nucleobase adenine (Figure 2B). In this regard, detailed analysis of genes specifically involved in purine metabolism (Figure 2D) have indicated a more sustained expression in T cells as compared with N cells. Altogether, these findings further suggest the main role of Gln in supporting anabolic processes of T cells as compared with N cells.
K-Ras transformed human cells show metabolic alterations similar to those observed in mouse transformed cells
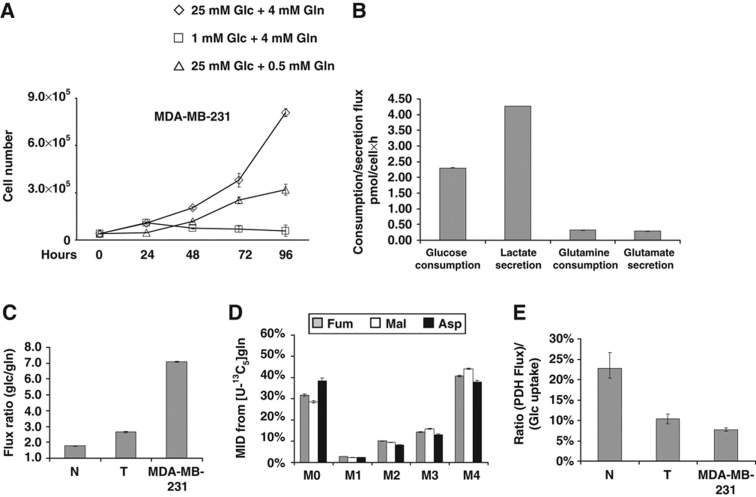
To test whether human cancer cells exhibit similar metabolic alterations to the previously described mouse model of transformation, we next sought to expand our metabolic and transcriptional analyses to the MDA-MB-231 human breast carcinoma cell line, which express an activated K-Ras protein (G13D). To assess the glycolytic phenotype of MDA-MB-231 cells, asynchronous cells grown in high and low Glc (25 and 1 mM, respectively), were followed in a time course of 96 h. As shown in Figure 3A, MDA-MB-231 cells grown in 25 mM Glc continued to proliferate and reached a much higher cell density compared with those in 1 mM Glc. In fact, in low Glc cells completely lost their proliferation ability and showed evident signs of cell death at 48 h, as revealed by proliferation curves (Figure 3A) and by morphological analysis (data not shown). In addition, MDA-MB-231 cells are dependent upon Gln for proliferation; even low levels of Gln (0.5 mM) significantly limited growth compared with normal culture (Figure 3A). Since the effects of Glc and Gln shortage on cell viability and proliferation of MDA-MB-231 cells were very similar to those observed in the mouse model of K-Ras-dependent transformation, we analyzed the metabolic phenotype of these human cancer cells in greater detail. Extracellular flux quantifications indicated that MDA-MB-231 cells convert Glc to Lac at approximately stoichiometric levels (i.e., 2:1; Figure 3B). Indeed GC/MS analysis of cells cultured with a 1:1 mixture of [1-13C]glucose and [U-13C6]glucose have demonstrated that at least 90% of Lac was derived from Glc (i.e., M3 Lac abundance was measured at 44.6±0.1%). Compared with murine N and T cells, MDA-MB-231 cells consume much higher amounts of Glc relative to Gln (Figure 3C). However, direct oxidative conversion of Gln still accounted for as much as 40% of the Fum, Mal, and Asp pools in cells cultured with [U-13C5]glutamine (see M4 mass isotopomer abundances; Figure 3D). Taken together, these data indicate that, much like transformed mouse cells, MDA-MB-231 cells divert most Glc to Lac and rely on Gln carbon for anaplerosis.
Figure 3.
MDA-MB-231 human cancer cell line exhibits enhanced glycolysis and reduced TCA cycle activity. (A) MDA-MB-231 cells were plated at 3000 cells/cm2 in 6-well plates in normal medium. Culture medium was replaced after 18 h with normal medium (◊), or a medium containing 1 mM Glc+4 mM Gln (□) or 25 mM Glc+0.5 mM Gln (▵). Then, the cells were collected and counted at indicate time points. (B) Extracellular uptake of Glc and Gln and secretion of Lac and Glu after 54 h. (C) Ratio of Glc uptake to Gln uptake. (D) MID of Fum, Mal, and Asp in cells cultured in the presence of [U-13C5]glutamine. (E) Ratio of PDH flux to Glc uptake. Error bars indicate s.e.m. (n=3) using propagation of error.
Using extracellular flux data and MIDs obtained from MDA-MB-231 cells cultured for 54 h with [U-13C5]glutamine, we performed MFA to determine intracellular fluxes throughout central carbon metabolism. The results of this analysis (Supplementary Figure S6; Supplementary Table S4) indicated that, in a similar way to mouse T cells, MDA-MB-231 cells exhibit high glycolytic flux and significantly lower TCA cycle activity. This comparison is best demonstrated by plotting the ratio of PDH flux to Glc consumption (Figure 3E). While Pyr oxidation flux is ∼25% of Glc uptake in control N cells, cells with oncogenic K-Ras exhibit a PDH flux ⩽10% of Glc consumption. Therefore, oncogenic K-Ras consistently drives the decoupling of central carbon metabolism, increasing the glycolytic flux to Lac and decreasing oxidative TCA flux in cells with different genetic backgrounds (Figures 1 and 3). Importantly, MDA-MB-231 cells carry a mutant p53 (Olivier et al, 2002), whereas K-Ras-NIH3T3 cells have wild-type p53 (Wadhwa et al, 1999). Given the fact that recent literature results have implicated the p53 tumor suppressor protein as a regulator of mitochondrial respiration and the Warburg effect (Matoba et al, 2006), we tested the effect of p53 status on these metabolic changes to ensure our results were specific to oncogenic K-Ras. Comparing p53+/+ wild-type (HCT116 p53+/+) or a p53−/− KO (HCT116 p53−/−) colon carcinoma cells, which express oncogenic K-Ras (Bunz et al, 1998), we observed no significant differences in proliferation under nutrient deprivation (Supplementary Figure S7A and B) or intracellular metabolite abundances (Supplementary Figure S7C–F). These data indicate that oncogenic K-Ras, at least in these cellular models, contributes to cancer-specific metabolic changes in a way that is not dependent on the presence of wild-type p53.
In order to analyze the relationship between our metabolic analysis and transcriptional profiling, microarray datasets of MDA-MB-231 cells and normal breast tissues were recovered from publicly accessible gene expression profile dataset (GEO and CellMiner). Data were analyzed as previously described (Balestrieri et al, 2009), and results indicated that several glycolytic genes (i.e., Hk1, Pfkp, Eno1, and Ldha) were expressed at a higher level in cancer cells as compared with normal breast tissue (Supplementary Figure S8A), in agreement with the more active glycolysis shown in Figure 3B. Differences in gene expression were also observed for several TCA cycle genes (Supplementary Figure S8B).
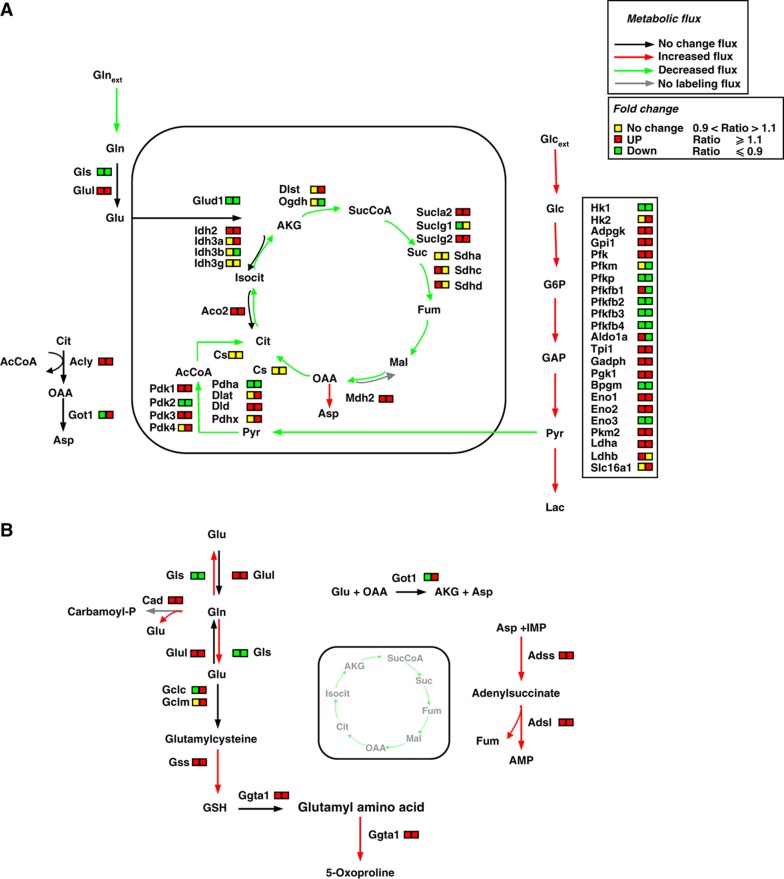
These findings, collectively, suggest that oncogenic K-Ras transformation induces cell-specific transcriptional programs that have a role in the metabolic alterations we observed (i.e., increased glycolytic activity) and highlight the utility of systems-level approaches. Figure 4 depicts a schematic view of metabolism, indicating the changes we observed upon K-Ras transformation using 13C MFA and transcriptional profiling. In particular, it can be observed that both K-Ras transformed mouse and human cells, as shown by previous metabolic analyses, divert most part of consumed Glc to Lac (Figure 4A, red arrows). Such a preferential metabolic flux is supported by the observed high level of expression of significant glycolytic genes in both cell lines (i.e., Adpgk, Gadph, Pgk, Eno 1 and 2, Pkm2 and Ldha; Figure 4A, right box). In addition, the reduced flux of pyruvate entering into mitochondria (Figure 4A, green arrow) and the deregulation of several genes encoding TCA cycle enzymes (Figure 4A) in part provide an explanation of the decreased TCA cycle flux observed in both transformed cells lines (Figure 4A, green arrows). At the same time, our metabolic data indicate that such a reduced TCA cycle flux in transformed cells has an important anabolic role and is essentially fueled by Gln, as suggested by the identification of labeled Asp, a product of Gln-derived TCA cycle intermediates (Figure 4A). Furthermore, the important role of Gln in supporting anabolic processes of cancer cells (Figure 4B, red arrows) is suggested by the identification of labeled metabolites containing Gln-derived nitrogen and associated increases in the expression of genes involved in glutathione metabolism (Gclm, Gss, and Ggta1), hexosamine metabolism (Gfpt1), and nucleotide biosynthesis (Cad, Adss, and Adsl) (Figure 4B). Taken together, these findings indicate a major alteration in cancer cell metabolism with a decoupling between Glc and Gln fates.
Figure 4.
Schematic integrative representation of metabolic fluxes and transcriptional data in normal and mouse K-Ras transformed cells. (A, B) Integration maps of metabolic routes identified in T cells as compared with N cells by using flux analysis and transcriptional data. The ratio between T and N cells of both metabolic fluxes and gene expression values are represented by a color code (as indicated in the upper right legends) as follow: black (no change value), red (increased value), and green (decreased value) for flux analysis and Up—red color—T/N ratio ⩾1.1, down—green color—T/N ratio ⩽0.9 and no change—yellow color—T/N ratio between 0.9 and 1.1 for transcriptional data. The list of gene abbreviations used in the figure can be found in ‘Supplementary information’. Metabolic flux analysis of central carbon metabolism was obtained by using the data derived from [U-13C5] glutamine, a mixture of [U-13C6] glucose and 1-13C] glucose (A) and [α-15N]glutamine (B) as specific isotopic tracers. The net and exchange fluxes were calculated as described under ‘Materials and methods’. Transcriptional data of N and T regulated genes were obtained as described in ‘Materials and methods’.
Metabolic alterations are dependent upon oncogenic K-Ras activation and glutamine utilization
To directly assess the role of oncogenic K-Ras in the previously described metabolic alterations of transformed cells, we performed several metabolic analyses by taking advantage of a cell line, NIH3T3-GEF-DN (reverted cells, R), stably expressing a dominant-negative Guanine Exchange Factor CDC25 (GEF-DN) that specifically attenuates oncogenic K-Ras activation (Vanoni et al, 1999; Bossu et al, 2000). Notably, R cells exhibit changes consistent with a regression of cellular transformation, including Ras-GTP levels, morphology, anchorage-independent growth, reduction of Ras-dependent tumor formation in nude mice, glucose and glutamine dependence and mitochondrial dysfunction to a phenotype similar to that observed in N cells (Chiaradonna et al, 2006b; Gaglio et al, 2009).
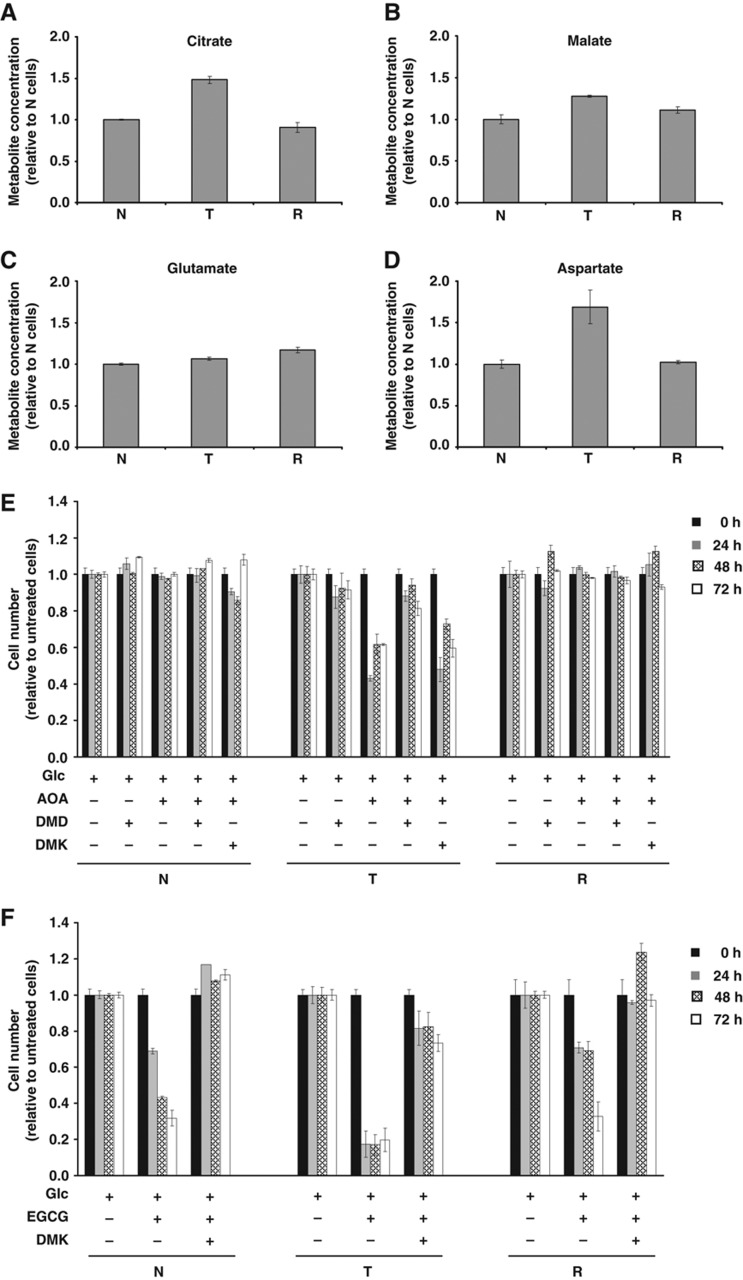
To compare the metabolic phenotype of R cells with N and T cells, the intracellular concentration of Cit and Mal (Figure 5A and B) or Glu and Asp (Figure 5C and D) was measured by enzymatic assays in cells cultured for 54 h in optimal growth conditions (25 mM Glc+4 mM Gln). Results indicated that N and R cells contained similar abundances of these metabolites, while T-cell metabolic abundances were similar to those previously observed using GC/MS. To further detail the metabolic characteristics of R cells as compared with N and T cell lines and evaluate whether Gln utilization is a general requirement for the proliferation of K-Ras transformed cells, we performed cell proliferation analyses in medium containing low Gln+ each of two cell permeable glutamine products, dimethyl aspartate (DMD) and dimethyl α-ketoglutarate (DMK). As shown in Supplementary Figure S9, Gln shortage caused a proliferation reduction only in T cells, which was partially rescued by addition of DMD or DMK (Supplementary Figure S9).
Figure 5.
Relative metabolites concentrations in N, T, and R cell lines and effect of aminooxyacetate (AOA) and epigallocatechin gallate (EGCG) treatments on their proliferation. Evaluation of Cit (A), Mal (B), Glu (C), and Asp (D) intracellular concentrations was carried out after 54 h of grown in normal medium by using enzymatic assays. Analysis of AOA (E) and EGCG (F) treatments on N, T, and R cell lines proliferation. Cells, seeded at density of 3000 cells/cm2, after 18 h and following a complete medium replacement, were treated with 100 μM AOA, or 1 mM dimethyl aspartate (DMD), or 2 mM dimethyl α-ketoglutarate (DMK) or 20 μM EGCG and counted at indicated time points. Error bars indicate s.e.m. (n=3).
To further test whether Gln metabolism induced in T cells was dependent on the K-ras oncogene, we treated N, T, and R cell lines with aminooxyacetate (AOA), an inhibitor of aminotransferase activity (Figure 5E) and epigallocatechin gallate (EGCG), an inhibitor of glutamate dehydrogenase (GDH) activity (Figure 5F). As shown in Figure 5E, only T-cell proliferation was inhibited by AOA treatment (Figure 5E, compare middle panel versus left panel:N cells and right panel: R cells) and rescued by DMD addition and not by DMK addition (Figure 5E). These data suggest that aminotransferase activity is necessary to sustain cancer cell growth and Gln-derived Asp is an important anabolic precursor for cancer cell growth.
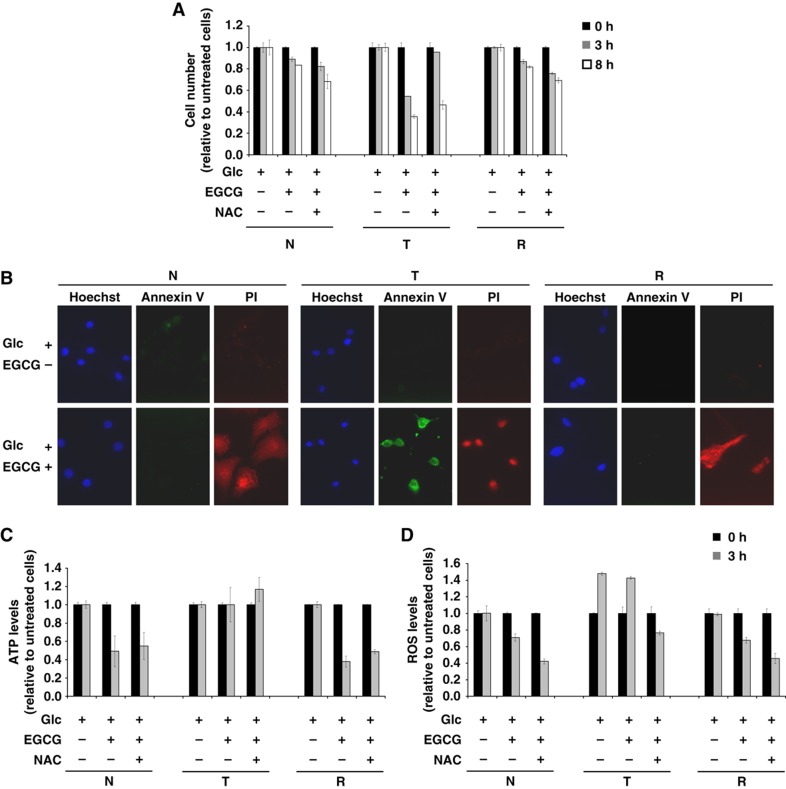
Analogous proliferation experiments were performed in N, T, and R cell lines upon treatment with EGCG. As shown in Figure 5F, the treatment induced cell death in all cell lines. Nevertheless, the reduction of cell number was markedly greater in T cells compared with that observed in N and R cells. Importantly, addition of DMK completely rescued viability in all the three cell lines (Figure 5F). Given the significant level of cell death observed in all cells at 24 h, we investigated the effects of EGCG on cell survival at earlier time points (between 0 and 8 h) to better understand the mechanism of cytotoxicity. EGCG reduced viability in all three cell lines in a time-dependent manner, with T cells being the most sensitive to EGCG treatment (Figure 6A, light gray bars). Interestingly, the antioxidant N-acetyl-L-cysteine (NAC) protected only T cells from EGCG-mediated cytotoxicity (Figure 6A), suggesting a specific role of ROS accumulation for their cell death. To ascertain whether EGCG induced a different type of cell death in T cells as compared with N and R cells, cells were treated for 8 h with EGCG and stained with Annexin V (AV) and propidium iodide (PI), indicators of late apoptosis and of plasma membrane permeability, respectively. Untreated cells exhibited an AV-negative/PI-negative pattern showing viable cells (Figure 6B, upper panels), whereas N and R cells treated with EGCG for 8 h exhibited an AV-negative/PI-positive pattern, showing a dead population with loss of plasma membrane integrity, typical of necrosis (Figure 6B, bottom panels). T cells treated with EGCG contained a population of AV-positive/PI-positive pattern, showing a typically apoptotic population with an increased AV signal. These results were confirmed also by cell morphology analysis after EGCG treatment. While EGCG induced flattened and attached cell remnants in N and R cells (almost all PI-positive cells), EGCG-treated T cells were detached and rounded cells (data not shown).
Figure 6.
(A) Effects of EGCG inhibitor at short term in N, T, and R cell lines. Cells, seeded at density of 3000 cells/cm2, after 18 h and following a complete medium replacement, were treated with 20 μM EGCG and/or 5 mM NAC and counted at indicated time points. (B) Analysis of cell death by fluorescence microscopy using PI (red color), Annexin V (green color), and Hoechst for nuclei staining (Blue color). The cells were plated at 3000 cells/cm2 on microglasses, treated as above described and analyzed after 8 h. The images were acquired by × 60 objective using Metamorph 7 software. (C) Analysis of intracellular ATP level by enzymatic assays. The cells were treated as above described and collected at indicated time points. (D) Intracellular ROS levels were measured by using 5 mM DCFH2-DA staining. The cells were treated as above described and collected at indicated time points. Error bars indicate s.e.m. (n=3).
Given the fact that ATP depletion is more likely to induce necrosis, while ROS accumulation is more typically associated with apoptosis, we measured intracellular ATP and ROS levels upon 3 h of EGCG treatment in each cell line. As shown in Figure 6C, EGCG caused a significant decrease of intracellular ATP levels in N and R cells, which could not be rescued by NAC treatment. On the contrary, ROS levels were high and almost constant in T cells regardless of EGCG treatment, whereas N and R cells experienced a decrease in ROS levels upon EGCG treatment (Figure 6D). Absolute levels of intracellular ROS in untreated T cells at 3 h were about two-fold higher as compared with N and R cells (e.g., N=193.5; T=317.2; R=141.7), and addition of NAC reduced ROS levels in all cells (Figure 6D). Altogether, these findings suggest that GDH activity, through different mechanisms is necessary for the survival of all three cell lines, and its inhibition may cause in N and R cells an energy derangement followed by necrosis, due to ATP depletion, and in T cells an antioxidant depletion that ultimately may bring to apoptosis. In conclusion, these results suggest a role of glutamine in endogenous antioxidant production, for instance glutathione synthesis, further confirming the anabolic role of glutamine in T cells.
Discussion
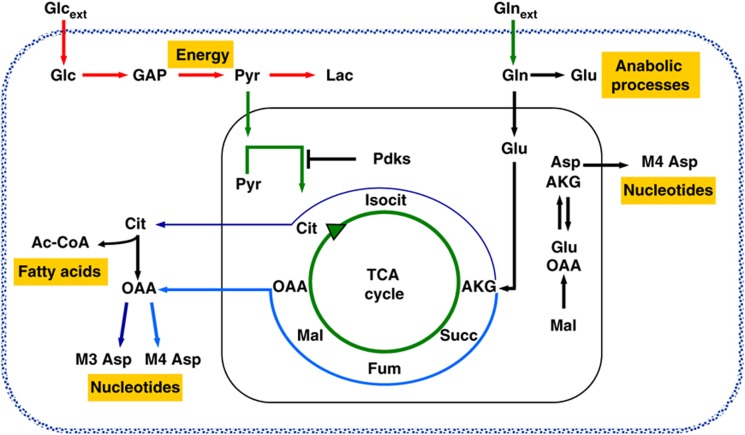
Cancer is a complex disease that arises from numerous molecular changes on a various cellular pathways (Mazurek et al, 1998; Hanahan and Weinberg, 2000). Among these alterations is the reprogramming of metabolic pathways associated with bioenergetics and cellular biosyntheses, and metabolic interventions are now emerging as potential therapeutic targets (Michelakis et al, 2010; Vander Heiden et al, 2010; Wise and Thompson, 2010). In this paper, we used 13C MFA, NTFD analysis, and transcriptional profiling to provide a comprehensive analysis of cancer cell metabolism and of its regulation by the K-ras oncogene. The results, summarized in Figure 7, highlight the reprogramming of metabolic pathways that occurs upon introduction of mutant forms of K-Ras. In cancer cells, most consumed Glc is diverted to Lac (Figure 7, red arrows), while the oxidation of Pyr to AcCoA via PDH is consistently decreased (Figure 7, green arrows). This decrease results in a marked reduction of TCA cycle flux (Figure 7, green circle arrow), presumably caused by expression of Pdk genes. It is intriguing to note that both mouse and human transformed cells herein analyzed, present a substantially lower mitochondrial Complex I activity (Ishikawa et al, 2008; Baracca et al, 2010). Consistent with such a decrease in glucose utilization through the TCA cycle, in cancer cells Gln becomes the more relevant source of both carbon and nitrogen for cellular biosynthesis (Figure 7, black, blue, and sky-blue arrows), through aminotransferase (Asp labeling) and glutamate dehydrogenase activities (Figure 7, black, blue, and sky-blue arrows).
Figure 7.
Schematic representation of Glc and Gln pathways decoupling in K-Ras cancer cells. Glc provides the main energy source for cancer cells with oncogenic K-Ras. Instead, Gln provides the carbon and nitrogen for the synthesis of biomass building blocks (amino acids, nucleotides, and fatty acids). Red arrows represent enhanced flux, green arrows represent reduced flux, black arrows represent no change flux, blue arrow represents forward flux of TCA cycle, and sky-blue represents reductive carboxylation flux.
In conclusion, in K-Ras transformed cells the decoupling between Glc and Gln metabolism leads to an efficient utilization of both carbon and nitrogen from Gln into biomass building blocks (amino acids and nucleotides) and glutathione. Both, sustaining growth and the ability to quench ROS production, may promote cancer cell proliferation and/or survival. The strong dependence of transformed cell growth on Gln availability is, therefore, not surprising (Gaglio et al, 2009; Wang et al, 2010; Weinberg et al, 2010). Our analysis also suggests that transcriptional changes contribute to the observed metabolic reprogramming in mouse and human transformed cells. In the future, detailed analyses of the various enzymatic steps involved in the growth-sustaining metabolism as well as genes regulating these reactions may shed light on the molecular basis of transformation. In addition, these results may enable the identification of fragile points in the metabolic network and more effective, targeted cancer therapeutics.
Materials and methods
Cell culture
Mouse embryonic fibroblast NIH3T3 cells (CRL-1658; American Type Culture Collection) and a K-Ras transformed NIH3T3-derived cell line, 226.4.1 (Pulciani et al, 1985), were routinely grown in Dulbecco's modified Eagle's medium containing 10% newborn calf serum, 4 mM L-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin (normal growth medium) (all from Invitrogen, Carlsbad, CA, USA) at 37°C in a humidified atmosphere of 5% CO2. The NIH-GEF-DN (reverted cell line), stably and constitutively expressing the dominant-negative mutant Cdc25MmW1056E (GEF-DN; Bossu et al, 2000), was maintained in normal growth medium supplemented with 0.7 mg/ml geneticin (G418; Sigma-Aldrich Inc., St Louis, MO, USA). MDA-MB-231 human breast carcinoma cell line, obtained from the European Institute of Oncology (Milano, Italy), was grown in Dulbecco's modified Eagle's medium containing 5% fetal bovine serum, 4 mM L-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin (normal growth medium) (all from Invitrogen, Carlsbad, CA, USA) at 37°C in a humidified atmosphere of 5% CO2. HCT116 p53+/+ and HCT116 p53−/− cells, obtained from the European Institute of Oncology (Milano, Italy), were cultured with complete DMEM plus 10% FBS. Cells were passed using trypsin-EDTA (Invitrogen) and maintained in culture for 48 h before manipulation.
Cell proliferation analysis and cell treatments
For long-term studies, cells were plated at the density of 3000 cells/cm2 in complete growth medium. After 18 h, the cells were washed twice with phosphate-buffered saline (PBS) and incubated in a media with different glucose concentrations (25 and 1 mM glucose) or different glutamine concentrations (4 and 0.5 mM glutamine) (time 0). When indicated, cells were treated with 100 μM AOA (O-(Carboxymethyl)hydroxylamine hemihydrochloride), or 20 μM (-)-EGCG, or 2 mM dimethyl 2-Oxoglutarate -DMK- or 1 mM L-Aspartic acid β-methyl ester hydrochloride -DMD- (Sigma-Aldrich Inc.). For short-term studies, cells were plated and grown as above and then treated with 20 μM EGCG or 5 mM NAC (Sigma-Aldrich Inc.). To measure cell proliferation, harvested cells were counted by Burker chamber.
Microarray data collection
Microarray transcriptional profiles were extracted from two different datasets. The mouse time course data were obtained from a transcription profiles collection of NIH3T3 and NIH3T3 K-Ras mouse fibroblasts, generated in our laboratory, grown in 25 mM glucose+4 mM glutamine, along a time course of 72 h. In particular, the cells used for the time course transcription analysis, were collected at 18 h from the initial seeding, time corresponding to the change of medium (indicated as T0) and at 24, 48 and 72 h (NCBI GEO database accession from GSM741354 to GSM741361 and from GSM741368 to GSM741375 of NIH3T3 and NIH3T3 K-Ras, respectively). For each time points, labeled cRNA was generated by using the Affymetrix One-Cycle Target Labeling and Control Reagent kit (Affymetrix Inc., Santa Clara, CA, USA), following the manufacturer's protocol and was hybridized using Affymetrix Genechips (Mouse Genome 430 2.0 Array) in order to determine the global gene expression patterns. Arrays were washed and scanned on the Affymetrix Complete GeneChip® Instrument System and processed into CEL files.
Human raw expression data for U133A arrays were obtained from publicly accessible gene expression profile database GEO (http://www.ncbi.nlm.nih.gov/geo/) or CellMiner (http://discover.nci.nih.gov/cellminer/home.do). In particular, we used the transcription profiles data of the human breast cancer cell line MDA-MB-231 (CellMiner accession 13409hg133a21) and of the human normal breast tissue (NCBI GEO accession GSM44683). Both mouse and human transcriptional data were separately submitted on the same procedure of normalization and filtering of noise, in order to obtain a good degree of comparison across the different experiments. Briefly, the data files (CEL files) were imported into GeneSpring GX 11.0.2 software (Agilent Technologies Inc.), by using RMA method, were normalized and summarized as probe-level intensity measurements (Irizarry et al, 2003). In particular, the algorithm implemented in GeneSpring, starting from the perfect-match probe-level data of a set of arrays, performs the background correction, the normalization and finally summarize the results as a set of expression measures for each probe set. Subsequently, also ‘per-gene normalization’ was performed as described in GeneSpring's manual. Altogether, these procedures permitted to obtain the relative expression of each probe set in a log2 scale with the value 0 as center. This scale of measurement was used for the heat maps showed in the paper. From these two datasets (mouse and human) have been identified and gathered the expression levels for genes encoding proteins involved in glycolysis, TCA cycle and glutamine metabolism, at 48 and 72 h for mouse datasets and, at time indicated by the authors for both human tissue and cell line samples. Total selected mouse and human genes and their intensity expression levels have been reported in Supplementary Dataset.
Metabolite extraction
For labeling experiments and GC/MS-based metabolomics, asynchronous NIH3T3, NIH3T3 K-Ras, and MDA-MB-231 were plated at 3000 cells/cm2 in 6-well plates with normal growth medium. In all, 18 h after seeding, cells were washed with PBS and incubated in different DMEM (Sigma) plus 10% dialyzed serum supplemented with unlabeled glucose or glutamine and either 4 mM [U-13C5]glutamine or a 25 mM 1:1 mixture of [U-13C6]glucose and [1-13C]glucose. After 54 h, spent medium was collected and analyzed for glucose, lactate, and glutamine consumption on a YSI7100 analyzer. Cells were quickly rinsed with PBS to minimize spent medium carryover and quenched with 0.4 ml ice-cold methanol. An equal volume of water containing 1 μg norvaline internal standard was added, and cells were collected by scraping with a pipette tip. Two volumes of chloroform were added, and the cells were vortexed at 4°C for 30 min. Samples were centrifuged at 3000 g for 10 min, and the aqueous phase was collected in a new tube and evaporated under airflow at room temperature.
Derivatization and GC/MS measurements
Dried polar metabolites were dissolved in 60 μl of 2% methoxyamine hydrochloride in pyridine (Pierce), sonicated for 30 min, and held at 37°C for 2 h. After dissolution and reaction, 90 μl MBTSTFA+1% TBDMCS (Pierce) was added and samples were incubated at 55°C for 60 min. GC/MS analysis was performed using an Agilent 6890 GC equipped with a 30-m DB-35MS capillary column connected to an Agilent 5975BMS operating under electron impact (EI) ionization at 70 eV, 1 μl of sample was injected in splitless mode at 270°C, using helium as the carrier gas at a flow rate of 1 ml/min. The GC oven temperature was held at 100°C for 3 min and increased to 300°C at 3.5°C/min for a total run time of ∼60 min. The MS source and quadrupole were held at 230 and 150°C, respectively, and the detector was operated in selected ion monitoring mode. MIDs were obtained for each measured metabolite and incorporated with extracellular flux measurements for flux determination. For determination of relative metabolite abundances, the integrated signal of all potentially labeled ions for each metabolite fragment was normalized by the signal from norvaline and the per well cell number (obtained by counting surrogate plates).
Flux estimation
Intracellular fluxes were estimated for a model reaction network by minimizing the lack of fit between actual and simulated flux and GC/MS measurements. The network contained simplified versions of glycolysis, the PPP, anaplerotic reactions, the TCA cycle, and amino-acid biosynthesis. Specific assumptions regarding the network and metabolism that were made for the sake of the flux analysis are listed in Supplementary information. We additionally calculated 95% confidence intervals for each flux using parameter continuation (Antoniewicz et al, 2006). All flux simulation, estimation, and continuation in this study were conducted using Metran, a flux analysis tool built upon an EMU framework (Antoniewicz et al, 2007; Yoo et al, 2008; Young et al, 2008). When required, MIDs were corrected for natural isotope abundance using in house algorithms adapted from Fernandez et al (1996). Simulated and measured MIDs for MFA of N and T cells are presented in Supplementary Figures S2 and S3 ([U-13C5]glutamine) or S10 and S11 ([U-13C6]glucose+[1-13C]glucose). Data for MDA-MB-231 cell MFA ([U-13C5]glutamine) are presented in Supplementary Figure S6.
Non-targeted tracer fate detection
The NTFD analysis was performed to determine the metabolic fate of the α-amine nitrogen atom of glutamine (Hiller et al, 2010). Cells were incubated for 54 h as described above, but instead of unlabeled glutamine, a 1:1 mixture of unlabeled and [α-15N]glutamine (Cambridge Isotope Laboratories) was employed for each growth condition. Since it is a requirement of NTFD, additional cells were cultivated with completely unlabeled glutamine as reference. Both cultivations were performed in triplicate and intracellular metabolites were extracted and measured as described above. The raw GC/MS data were used as input for the NTFD software and all detected labeled compounds for all growth conditions were matched using algorithms of the Metabolite Detector software package (Hiller et al, 2009). Finally, the NIST reference library was employed for compound identification (Babushok et al, 2007).
Metabolite assay
For enzymatic assay quantitation of aspartate, citrate, malate, and glutamate, asynchronous NIH3T3, NIH3T3 K-Ras, and NIH-GEF-DN were plated at 3000 cells/cm2 with normal growth medium. In all, 18 h after seeding, cells were washed with PBS and incubated in medium with 25 mM glucose and 4 mM glutamine. After 54 h, the cells were washed twice with PBS cold and collected using trypsin-EDTA (Invitrogen). The cells were counted and 500 000 cell aliquots were rapidly homogenized with appropriate assay buffer on ice. The samples were centrifuged at 15 000 g for 10 min to remove cell debris and deproteinized using a perchloric acid/KOH protocol (BioVision). The samples were added into triplicate wells of a 96-well plate and the volume was brought to 50 μl with assay buffer. The samples were incubated for 30 min at room temperature or at 37°C, protected from light. Then, OD was measured in a microplate reader.
Intracellular ATP quantification
Intracellular ATP levels were measured using CellTiter Glo® luciferin-luciferase assay (Promega) according to the manufacturer's protocol. In particular, 100 μl of cell suspension containing 104 cells was added to an equal volume of CellTiter Glo Reagent in a single well of 96-well plate. The emitted luminescence was collected after incubation at the wavelength of 560 nm using Cary Eclipse spectrofluorimeter (Varian) and the luminescence values were converted in ATP quantities after the setting of a calibration curve.
ROS levels measurement
Detection of ROS levels was carried out through flow cytometric analyses using a FACScan flow cytometer (Becton-Dickinson) with CellQuest software (BD Biosciences). ROS levels were measured through staining plated cells with 5 μM dichloro-dihydro-fluoresceine-diacetate (DCFH2-DA; Molecular Probes, Invitrogen) for 30 min at 37°C. Then, the cells were trypsinized, collected in ice and acquired by FACScan. Analysis of flow cytometric data was carried out using WinMDI software.
AV/PI cell death assay
Cell death was estimated using the Apoptosis Detection Kit (Immunological Sciences). Cells were plated on coverslips previously treated with 2% gelatin. Cells on coverslips were stained by adding 50 μl of binding buffer, 5 μl of Annexin V-FITC, and 5 μl of PI. After 15 min of incubation at 37°C in the dark, cells were treated with 5 μg/ml Hoechst (Invitrogen) for 5 min at RT. The cover glasses, mounted in DABCO, were analyzed under a Nikon ECLIPSE 90i fluorescence microscope equipped with a b/w CCD camera (Hamamatsu-CoolSNAP, Hamamatsu Corporation Japan), using Plan Apo objective ( × 60 oil; numerical aperture 0.75 and 1.4, respectively). The images were acquired using the imaging software Metamorph 7, and then processed in Adobe Photoshop CS3 with adjustments of brightness and contrast.
Supplementary Material
Supplementary figures S1–11, Supplementary Tables S1–4
Gene expression data
Acknowledgments
We acknowledge support from NIH grant 1R01 DK075850-01. C Metallo is supported by a postdoctoral fellowship from the American Cancer Society. D Gaglio is supported by a postdoctoral fellowship from Tecnomed Foundation. L Alberghina is supported by MIUR, FIRB-Italbionet. F Chiaradonna is supported by Italian Government Grant (PRIN and FAR). We thank Pier Giuseppe Pelicci for human breast cancer cells MDA-MB-231 and human colon cancer cell HCT116 p53+/+ and HCT116 p53−/−; and Roberta Palorini for technical assistance.
Author contributions: LA and GS planned the project. DG, CMM, LA, GS, and FC conceived and designed the research. DG, CMM, LA, GS, and FC wrote the manuscript. FC and DG performed transcriptome experiments. CB performed transcriptional data analysis. DG, CMM, and PAG performed metabolic analysis. CMM and KH performed statistical computations of metabolic analysis. DG and LSD performed cell biology experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Antoniewicz MR, Kelleher JK, Stephanopoulos G (2006) Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng 8: 324–337 [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G (2007) Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab Eng 9: 68–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babushok VI, Linstrom PJ, Reed JJ, Zenkevich IG, Brown RL, Mallard WG, Stein SE (2007) Development of a database of gas chromatographic retention properties of organic compounds. J Chromatogr 1157: 414–421 [DOI] [PubMed] [Google Scholar]
- Baggetto LG (1992) Deviant energetic metabolism of glycolytic cancer cells. Biochimie 74: 959–974 [DOI] [PubMed] [Google Scholar]
- Balestrieri C, Alberghina L, Vanoni M, Chiaradonna F (2009) Data recovery and integration from public databases uncovers transformation-specific transcriptional downregulation of cAMP-PKA pathway-encoding genes. BMC Bioinformatics 10(Suppl 12): S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baracca A, Chiaradonna F, Sgarbi G, Solaini G, Alberghina L, Lenaz G (2010) Mitochondrial Complex I decrease is responsible for bioenergetic dysfunction in K-ras transformed cells. Biochim Biophys Acta 1797: 314–323 [DOI] [PubMed] [Google Scholar]
- Bos JL (1989) Ras oncogenes in human cancer: a review. Cancer Res 49: 4682–4689 [PubMed] [Google Scholar]
- Bossu P, Vanoni M, Wanke V, Cesaroni MP, Tropea F, Melillo G, Asti C, Porzio S, Ruggiero P, Di Cioccio V, Maurizi G, Ciabini A, Alberghina L (2000) A dominant negative RAS-specific guanine nucleotide exchange factor reverses neoplastic phenotype in K-ras transformed mouse fibroblasts. Oncogene 19: 2147–2154 [DOI] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science (New York, NY) 282: 1497–1501 [DOI] [PubMed] [Google Scholar]
- Chiaradonna F, Gaglio D, Vanoni M, Alberghina L (2006a) Expression of transforming K-Ras oncogene affects mitochondrial function and morphology in mouse fibroblasts. Biochim Biophys Acta 1757: 1338–1356 [DOI] [PubMed] [Google Scholar]
- Chiaradonna F, Magnani C, Sacco E, Manzoni R, Alberghina L, Vanoni M (2005) Acquired glucose sensitivity of K-ras transformed fibroblasts. Biochem Soc Trans 33: 297–299 [DOI] [PubMed] [Google Scholar]
- Chiaradonna F, Sacco E, Manzoni R, Giorgio M, Vanoni M, Alberghina L (2006b) Ras-dependent carbon metabolism and transformation in mouse fibroblasts. Oncogene 25: 5391–5404 [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB (2007) Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA 104: 19345–19350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J (2003) Targeting RAS signalling pathways in cancer therapy. Nature Rev 3: 11–22 [DOI] [PubMed] [Google Scholar]
- Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, Thompson CB (2004) Akt stimulates aerobic glycolysis in cancer cells. Cancer Res 64: 3892–3899 [DOI] [PubMed] [Google Scholar]
- Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H (1996) Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom 31: 255–262 [DOI] [PubMed] [Google Scholar]
- Gaglio D, Soldati C, Vanoni M, Alberghina L, Chiaradonna F (2009) Glutamine deprivation induces abortive s-phase rescued by deoxyribonucleotides in K-ras transformed fibroblasts. PLoS One 4: e4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV (2009) c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458: 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- Hiller K, Hangebrauk J, Jager C, Spura J, Schreiber K, Schomburg D (2009) MetaboliteDetector: comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis. Anal Chem 81: 3429–3439 [DOI] [PubMed] [Google Scholar]
- Hiller K, Metallo CM, Kelleher JK, Stephanopoulos G (2010) Nontargeted elucidation of metabolic pathways using stable-isotope tracers and mass spectrometry. Anal Chem 82: 6621–6628 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics (Oxford, England) 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J (2008) ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science (New York, NY) 320: 661–664 [DOI] [PubMed] [Google Scholar]
- Kovacevic Z, McGivan JD (1983) Mitochondrial metabolism of glutamine and glutamate and its physiological significance. Physiol Rev 63: 547–605 [DOI] [PubMed] [Google Scholar]
- Kreeger PK, Lauffenburger DA (2010) Cancer systems biology: a network modeling perspective. Carcinogenesis 31: 2–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing RE, Nair-Gill E, Witte ON, Radu CG (2009) Visualizing cancer and immune cell function with metabolic positron emission tomography. Curr Opini Genet Dev 20: 100–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubenbacher R, Hower V, Jarrah A, Torti SV, Shulaev V, Mendes P, Torti FM, Akman S (2009) A systems biology view of cancer. Biochim Biophys Acta 1796: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liotta L, Petricoin E (2000) Molecular profiling of human cancer. Nature Rev 1: 48–56 [DOI] [PubMed] [Google Scholar]
- Mata J, Marguerat S, Bahler J (2005) Post-transcriptional control of gene expression: a genome-wide perspective. Trends Biochem Sci 30: 506–514 [DOI] [PubMed] [Google Scholar]
- Mathupala SP, Rempel A, Pedersen PL (1997) Aberrant glycolytic metabolism of cancer cells: a remarkable coordination of genetic, transcriptional, post-translational, and mutational events that lead to a critical role for type II hexokinase. J Bioenerg Biomembr 29: 339–343 [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM (2006) p53 regulates mitochondrial respiration. Science (New York, NY) 312: 1650–1653 [DOI] [PubMed] [Google Scholar]
- Mazurek S, Grimm H, Wilker S, Leib S, Eigenbrodt E (1998) Metabolic characteristics of different malignant cancer cell lines. Anticancer Res 18: 3275–3282 [PubMed] [Google Scholar]
- Metallo CM, Vander Heiden MG (2010) Metabolism strikes back: metabolic flux regulates cell signaling. Genes Dev 24: 2717–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Walther JL, Stephanopoulos G (2009) Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. J Biotechnol 144: 167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelakis ED, Sutendra G, Dromparis P, Webster L, Haromy A, Niven E, Maguire C, Gammer TL, Mackey JR, Fulton D, Abdulkarim B, McMurtry MS, Petruk KC (2010) Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med 2: 31ra34. [DOI] [PubMed] [Google Scholar]
- Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD (2008) Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol 26: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P (2002) The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mut 19: 607–614 [DOI] [PubMed] [Google Scholar]
- Pulciani S, Santos E, Lauver AV, Long LK, Robbins KC, Barbacid M (1982) Oncogenes in human tumor cell lines: molecular cloning of a transforming gene from human bladder carcinoma cells. Proc Natl Acad Sci USA 79: 2845–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulciani S, Santos E, Long LK, Sorrentino V, Barbacid M (1985) Ras gene amplification and malignant transformation. Mol Cell Biol 5: 2836–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A, Wang C, Schreiber SL (2005) Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc Natl Acad Sci USA 102: 5992–5997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO (2000) Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet 24: 227–235 [DOI] [PubMed] [Google Scholar]
- Sauer LA, Stayman JW III, Dauchy RT (1982) Amino acid, glucose, and lactic acid utilization in vivo by rat tumors. Cancer Res 42: 4090–4097 [PubMed] [Google Scholar]
- Sauer U (2006) Metabolic networks in motion: 13C-based flux analysis. Mol Syst Biol 2: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, Kohn KW, Reinhold WC, Myers TG, Andrews DT, Scudiero DA, Eisen MB, Sausville EA, Pommier Y, Botstein D, Brown PO, Weinstein JN (2000) A gene expression database for the molecular pharmacology of cancer. Nat Genet 24: 236–244 [DOI] [PubMed] [Google Scholar]
- Shih C, Padhy LC, Murray M, Weinberg RA (1981) Transforming genes of carcinomas and neuroblastomas introduced into mouse fibroblasts. Nature 290: 261–264 [DOI] [PubMed] [Google Scholar]
- Steinberg P, Storkel S, Oesch F, Thoenes W (1992) Carbohydrate metabolism in human renal clear cell carcinomas. Lab Invest 67: 506–511 [PubMed] [Google Scholar]
- Stephanopoulos G, Vallino JJ (1991) Network rigidity and metabolic engineering in metabolite overproduction. Science (New York, NY) 252: 1675–1681 [DOI] [PubMed] [Google Scholar]
- Telang S, Yalcin A, Clem AL, Bucala R, Lane AN, Eaton JW, Chesney J (2006) Ras transformation requires metabolic control by 6-phosphofructo-2-kinase. Oncogene 25: 7225–7234 [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science (New York, NY) 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Christofk HR, Schuman E, Subtelny AO, Sharfi H, Harlow EE, Xian J, Cantley LC (2010) Identification of small molecule inhibitors of pyruvate kinase M2. Biochem Pharmacol 79: 1118–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoni M, Bertini R, Sacco E, Fontanella L, Rieppi M, Colombo S, Martegani E, Carrera V, Moroni A, Bizzarri C, Sabbatini V, Cattozzo M, Colagrande A, Alberghina L (1999) Characterization and properties of dominant-negative mutants of the ras-specific guanine nucleotide exchange factor CDC25(Mm). J Biol Chem 274: 36656–36662 [DOI] [PubMed] [Google Scholar]
- Vizan P, Boros LG, Figueras A, Capella G, Mangues R, Bassilian S, Lim S, Lee WN, Cascante M (2005) K-ras codon-specific mutations produce distinctive metabolic phenotypes in NIH3T3 mice [corrected] fibroblasts. Cancer Res 65: 5512–5515 [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Takano S, Mitsui Y, Kaul SC (1999) NIH 3T3 cells malignantly transformed by mot-2 show inactivation and cytoplasmic sequestration of the p53 protein. Cell Res 9: 261–269 [DOI] [PubMed] [Google Scholar]
- Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, Cerione RA (2010) Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 18: 207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O (1956) On the origin of cancer cells. Science (New York, NY) 123: 309–314 [DOI] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS (2010) Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A 107: 8788–8793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB (2008) Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A 105: 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Thompson CB (2010) Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci 35: 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK (2008) Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem 283: 20621–20627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, Walther JL, Antoniewicz MR, Yoo H, Stephanopoulos G (2008) An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnol Bioeng 99: 686–699 [DOI] [PubMed] [Google Scholar]
- Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, Schmidt K, Willson JK, Markowitz S, Zhou S, Diaz LA Jr, Velculescu VE, Lengauer C, Kinzler KW, Vogelstein B, Papadopoulos N (2009) Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science (New York, NY) 325: 1555–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y (2007) Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol 178: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW (1999) Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res 59: 5830–5835 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures S1–11, Supplementary Tables S1–4
Gene expression data