Abstract
Despite the role of the estrogen receptor α (ERα) pathway as a key growth driver for breast cells, the phenotypic consequence of exogenous introduction of ERα into ERα-negative cells paradoxically has been growth inhibition. We mapped the binding profiles of ERα and its interacting transcription factors (TFs), FOXA1 and GATA3 in MCF-7 breast carcinoma cells, and observed that these three TFs form a functional enhanceosome that regulates the genes driving core ERα function and cooperatively modulate the transcriptional networks previously ascribed to ERα alone. We demonstrate that these enhanceosome occupied sites are associated with optimal enhancer characteristics with highest p300 co-activator recruitment, RNA Pol II occupancy, and chromatin opening. Most importantly, we show that the transfection of all three TFs was necessary to reprogramme the ERα-negative MDA-MB-231 and BT-549 cells to restore the estrogen-responsive growth resembling estrogen-treated ERα-positive MCF-7 cells. Cumulatively, these results suggest that all the enhanceosome components comprising ERα, FOXA1, and GATA3 are necessary for the full repertoire of cancer-associated effects of the ERα.
Keywords: enhanceosome, estrogen receptor α, FOXA1, GATA3, synthetic phenotypes
Introduction
Estrogen receptor α (ERα) is a member of the nuclear receptor superfamily that has broad impact on systems such as reproduction, cancer, bone, and cardiovascular biology. The basic regulatory function of ERα is to bind to its DNA recognition sequence also known as estrogen-response elements (EREs). The direct DNA binding, though necessary, is not sufficient to explain ERα-directed gene regulation. ERα recruits a variety of co-activators, corepressors, and chromatin remodeling enzymes to the promoter and enhancer regions but the range of possible combinations at each individual site and their distances from the transcription start sites (TSSs) of regulated genes suggest multiple regulatory mechanisms (Farnham, 2009). ERα is also known to commonly induce long-distance chromatin interactions with ERα binding sites and with TSS through chromatin looping (Fullwood et al, 2009), suggesting that higher dimensional structural order beyond the simple receptor–recognition motif interaction is essential in explaining ERα-directed transcriptional regulation. The finding that transcription factors (TFs) cluster at juxtaposed binding sites in the genome to form enhanceosomes further suggests that the totality of gene regulation by any TF will be dependent on a complex interaction between specific ERα binding, local configuration of co-occupying TFs, protein cofactors, chromatin conditions, and three dimensional interactions.
ERα is known as a ligand-activated TF that mediates the proliferative effects of estrogen (E2) in breast cancer cells. However, some physiologic and cellular contradictions have been previously noted in ERα biology. Garcia et al (1992) showed that the transfection of the ERα alone into ERα-negative cell lines has commonly no growth effect or even repress growth. This is also true for an important ERα-associated TF, FOXA1, where introduction of this gene represses cell growth (Wolf et al, 2007). We posited that these higher order regulatory mechanisms of ERα function such as the formation and composition of enhanceosomes may explain the establishment of transcriptional regulatory cassettes favoring either growth enhancement or growth repression.
In our previous (Lin et al, 2007) and recent (Joseph et al, 2010) studies, we have identified high confidence ERα binding sites in MCF-7 human mammary carcinoma cells. With known motif scanning and de novo motif finding methods, we identified that FOXA1 and GATA3 motifs were commonly enriched within ERα binding sites. FOXA1 has been extensively studied in the context of ERα biology and it is considered as a pioneering factor which prepares genomic sites for ERα binding (Cirillo et al, 2002). GATA3 has been found to be an essential TF for luminal development in mouse mammary models (Asselin-Labat et al, 2007). Moreover, numerous microarray studies have documented the co-expression of ERα, FOXA1, and GATA3 in primary breast tumors (Badve et al, 2007; Wilson and Giguere, 2008). Though this evidence suggests that the three TFs, ERα, FOXA1, and GATA3 may cluster on DNA binding sites and may be involved in the breast cancer phenotype, there is little understanding as to the nature of their coordinated interaction at the genome level or the biological consequences of their detailed interaction.
In the present study, we investigate the ERα-mediated transcriptional networks orchestrated with FOXA1 and GATA3 in breast cancer cells. We use chromatin immunoprecipitation-sequencing (ChIP-seq) to define the binding profiles of ERα, FOXA1, and GATA3 as to study the interplay among these TFs. We aim to dissect the roles of FOXA1 and GATA3 in regulating ERα action; to map the genomic effects of ERα, FOXA1, and GATA3 in altering the transcriptional activation in breast cancer cells; and to determine if FOXA1 and GATA3 are essential components of ERα-induced proliferation in breast cancer cells in response to estrogen stimulation.
Results
Mapping the binding profiles of ERα, FOXA1, and GATA3
We mapped the genome-wide in vivo binding sites of ERα, FOXA1, and GATA3 using the massively parallel ChIP-seq in MCF-7 cells before and after estradiol exposure. Using the peak calling algorithm MACS (Zhang et al, 2008), we found a total of 1990 high confidence ERα binding sites, 9337 FOXA1 binding sites, and 20 707 GATA3 binding sites in the vehicle-treated cells (i.e., without ligand). Upon E2 stimulation, we found a total of 19 412 high confidence ERα binding sites (an increase of ∼16.58-fold after normalization of library size, see details in Supplementary information and Supplementary Table I), 15 852 FOXA1 binding sites (an increase of ∼2.46-fold after normalization), and 38 530 GATA3 binding sites (an increase of ∼1.32-fold after normalization). Validation of randomly selected binding sites using ChIP-qPCR showed 100% concordance in calling bound sites (Supplementary Figure S1) and quantitatively, the ChIP-qPCR results for FOXA1 and GATA3 binding sites correlated well with the binding intensity measured by ChIP-seq (correlation coefficient R=0.63–0.81, see Supplementary Figure S1).
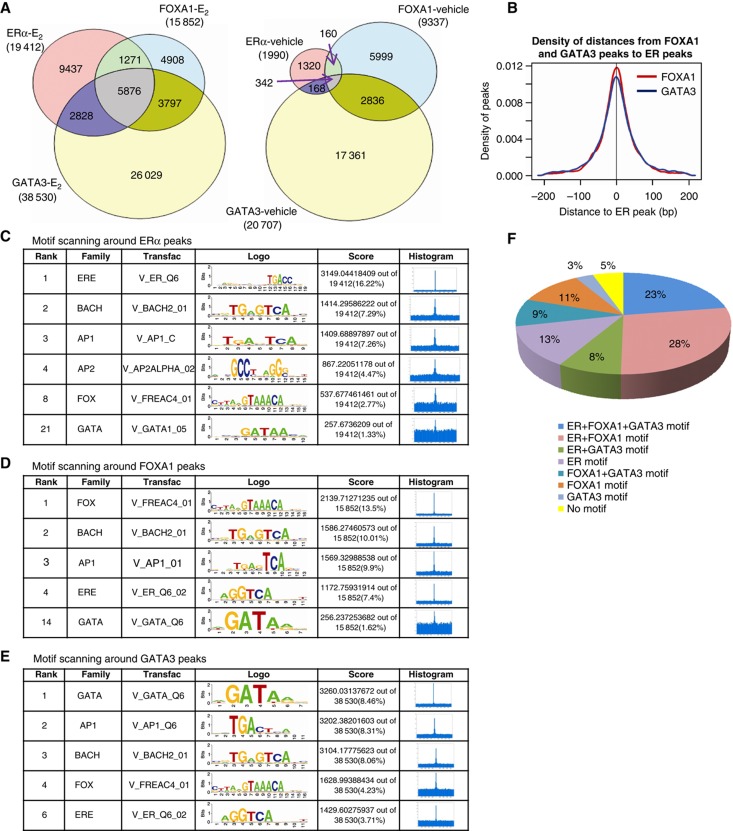
To assess how these TFs individually interact, we overlapped their binding profiles and found 37% of ∼19 k ERα binding sites showed FOXA1 colocalization, 45% of ERα binding sites overlapped with GATA3 binding sites, and as much as 30% of ERα binding sites were co-occupied by both FOXA1 and GATA3 (Figure 1A). Interestingly, the number of sites with occupancy of all three TFs increased from 342 (before estradiol exposure) to 5876 (after estradiol exposure). Figure 1B revealed that FOXA1 and GATA3 bindings are symmetrically distributed within 200 bp around the 5876 ERα, FOXA1, and GATA3 conjoint binding sites. The relative intensity of bindings as measured by TF occupancy at these conjoint sites highly correlated among the three TFs (R=0.48–0.63, Supplementary Figure S2). These results suggest that ERα, FOXA1, and GATA3 bind in a coordinated manner at ∼30% of all ERα binding sites after stimulation by ligand.
Figure 1.
The repertoire of ERα, FOXA1, and GATA3 in vivo bindings by massively parallel ChIP-seq approach. (A) Venn diagrams of in vivo bindings for ERα, FOXA1, and GATA3 upon and before E2 stimulation in MCF-7 cells. (B) The distance distribution of FOXA1 and GATA3 bindings around ERα sites, demonstrating that FOXA1 and GATA3 have similar distance distribution around ERα sites. (C–E) Motif scanning around ERα, FOXA1, and GATA3 peaks separately. (F) The distribution of ERα, FOXA1, and GATA3 motifs around ERα binding sites.
Motif analyses of the TFs binding
In order to determine the in vivo sequences enriched in the ERα, FOXA1, and GATA3 occupied sites, we used an in-house program CentDist (Zhang et al, 2011) for known motif scanning. This program not only allows for the identification of specific binding motifs, but also displays the position-distribution around the binding sites to indicate binding specificity. The motif position weight matrixes (PWM) from TRANSFAC (Matys et al, 2003) version 11.3 were used and the cutoff of PWM score was set to 1E−3. As expected, we found significant enrichment of individual ERE, FOXA1, and GATA3 motifs in the ERα, FOXA1, and GATA3 ChIP-seq libraries, respectively (Figure 1C–E). We also observed that the three ERE, FOXA1, and GATA3 binding motifs emerged together as the top enriched motifs in each set of the individual TF binding sites, suggesting a bias for recognition motifs for all three factors to be clustered together (Figure 1C–E). Besides FOXA1 and GATA3 motifs, AP1 and BACH motifs were also enriched in the ERα binding sites, which is in agreement with the previous finding reported by Bhat-Nakshatri et al (2008). Because of prior genetic data suggesting a role for FOXA1 and GATA3 in breast biology, we pursued the interaction of these three factors.
We specifically assessed the frequency of ERE, FOXA1, and GATA3 binding motifs around ERα binding sites. Figure 1F shows that 72% of the ERα sites have an ERE motif, 71% of ERα sites contain a FOXA1 motif, 43% of ERα sites contain a GATA3 motif, and 23% of the ERα sites contain all three motifs. Next, we asked whether these factors are physically colocalized. Using sequential ChIP followed by qPCR in randomly selected sites conjointly occupied by ERα+FOXA1 and ERα+GATA3, we found cooccupancy of these TFs at the overlap binding sites (Figure 2). These results suggest that the colocalization of the three factors at ERα occupied sites occur primarily through sequence recognition and not solely through a tethering mechanism involving only protein–protein interaction.
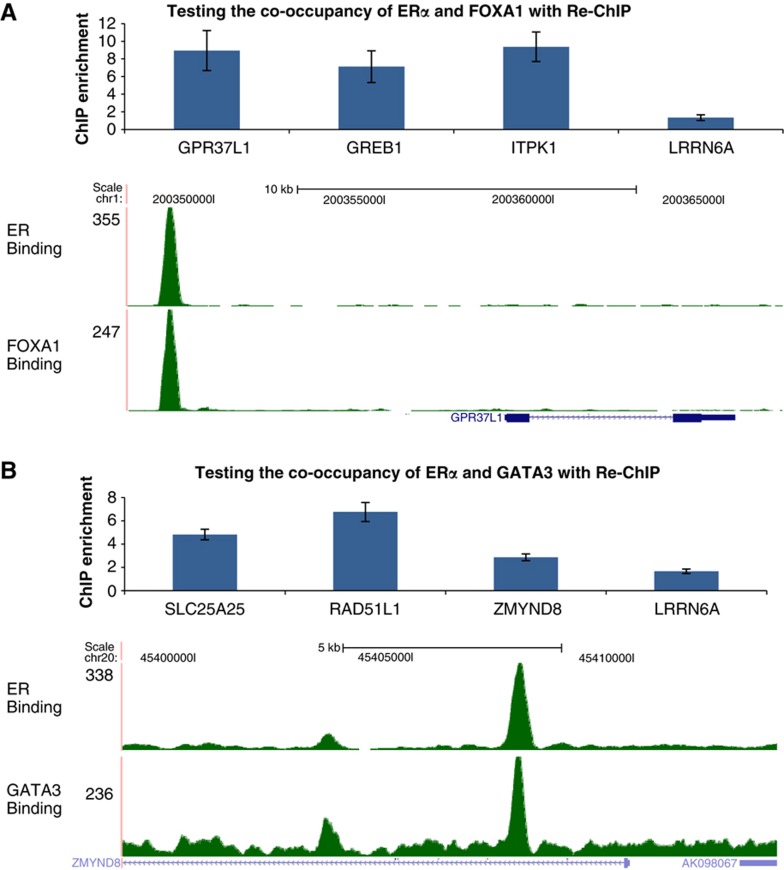
Figure 2.
The interaction between ERα, FOXA1, and GATA3 in breast cancer genome. The co-occupancy of ERα+FOXA1 (A) and ERα+GATA3 (B) to the target cis-regulatory regions as validated by sequential Re-ChIP assay. Genes nearby are used to label the peaks, and the tag densities around gene GPR37L1 with ERα+FOXA1 peaks and gene ZMYND8 with ERα+GATA3 peaks are shown as examples. A site near LRRN6A gene with only unique ERα binding is recruited as a negative control for the sequential Re-ChIP assay. The ChIP enrichment was computed by comparing with the IgG pull-down control. Mean values of at least two independent experiments are compared and standard errors are shown.
Progressive recruitment of ERα and FOXA1 to the cis-regulatory elements
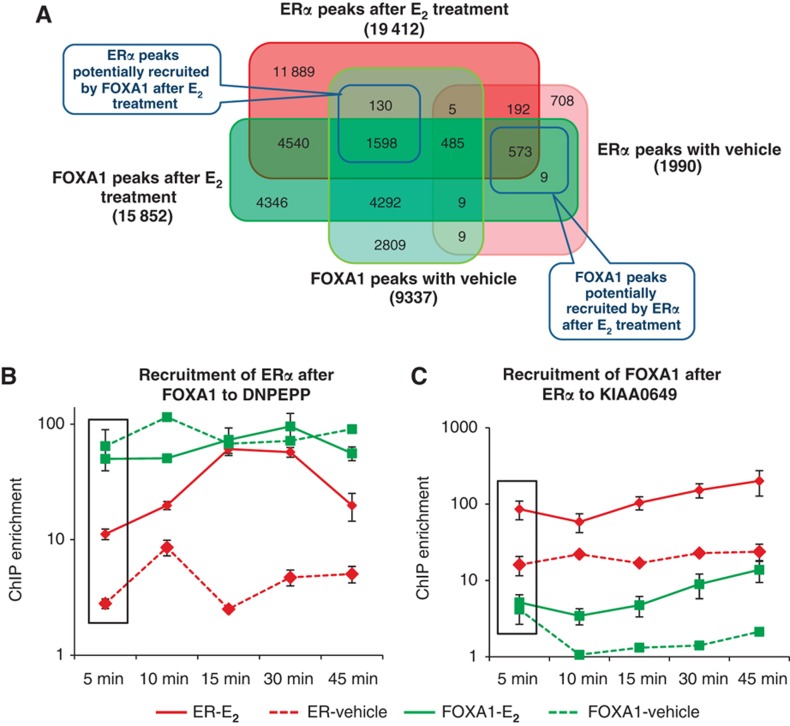
It was previously described that FOXA1 is a pioneering factor characterized by this sequence of events: FOXA1 binds to the condensed chromatin in the absence of E2 and opens the chromatin to facilitate the ERα binding upon E2 stimulation (Carroll et al, 2005; Hurtado et al, 2011). In addition, it was reported that FOXA1 bound at ERα sites before ligand stimulation followed by diminished FOXA1 occupancy after E2 exposure potentially through displacement by the activated ERα (Carroll et al, 2005). However, we observed both an increase in the number of FOXA1 binding sites (9337–15 852) and in the average level of occupancy at each site after ligand stimulation (on average, there were 2.58 reads per peak per million reads in the FOXA1 ChIP-seq library after E2 stimulation compared with 1.74 reads per peak per million reads before E2 stimulation). We found that 37% (7196/19 412) of the ERα binding sites after E2 stimulation were co-occupied by FOXA1 where 25% (508/1990) of the ERα binding sites in the absence of ligand were co-occupied by FOXA1 (Figure 3A). This is in accordance with the earlier observations that FOXA1 is preferentially associated with E2-bound ERα (Zhao et al, 2001).
Figure 3.
The progressive recruitment of ERα and FOXA1 to the cis-regulatory elements. (A) Venn diagram of ERα binding sites with vehicle, ERα binding sites after E2 treatment, FOXA1 binding sites with vehicle, and FOXA1 binding sites after E2 treatment. (B) The recruitment of ERα after FOXA1 binding in the synchronized MCF-7 cells upon E2 stimulation as validated by ChIP-qPCR in various time points. Two independent experiments have consistent results. Mean values and standard errors are shown. (C) The recruitment of FOXA1 after ERα binding in the synchronized MCF-7 cells upon E2 stimulation as validated by ChIP-qPCR in various time points. Two independent experiments have consistent results. Mean values and standard errors are shown.
If FOXA1 was a true pioneering factor, FOXA1 occupancy would be present in a significant percentage of ERα bound sites before estradiol exposure. However, our data revealed that only 11% (2218/19 412) of E2-induced ERα sites are occupied by FOXA1 before ligand exposure (Figure 3A). When we eliminate the number of basal ERα-bound sites before E2 stimulation, the percentage of FOXA1 sites that can recruit ERα is 19% (=(130+1598)/9337). This means that FOXA1 is a potential pioneering factor to recruit only a subset of ERα binding sites.
Interestingly, view from a different prospective, after excluding the number of basal FOXA1-bound sites before E2 stimulation, the percentage of ERα sites that can recruit FOXA1 after E2 exposure is 29% (=(573+9)/1990). Thus, ERα can also ‘pioneer’ a site for FOXA1 as efficiently as the converse even though, because of a much larger starting denominator (9337 sites versus 1990), it appears that FOXA1 is a better pioneering factor.
To confirm this ChIP-seq-based observation, we synchronized the cells with α-amanitin treatment followed by E2 stimulation and performed ChIP-qPCR over time on the nucleus lysates. In the sites where FOXA1 functions as the recruiting factor, we observed enrichment of FOXA1 occupancy as early as 5 min, followed by progressively increasing ERα occupancy at the later time points (e.g., after 10–15 min, example in Figure 3B). Conversely, in those sites where ERα functions as the recruiting factor, we show high ERα occupancy at early time points followed by increasing FOXA1 occupancy at the later time points (example in Figure 3C). Thus, we show that ERα and FOXA1 can equally function as recruiting factors for the other.
The formation of enhanceosome consisting of ERα, FOXA1, and GATA3 in breast cancer cells
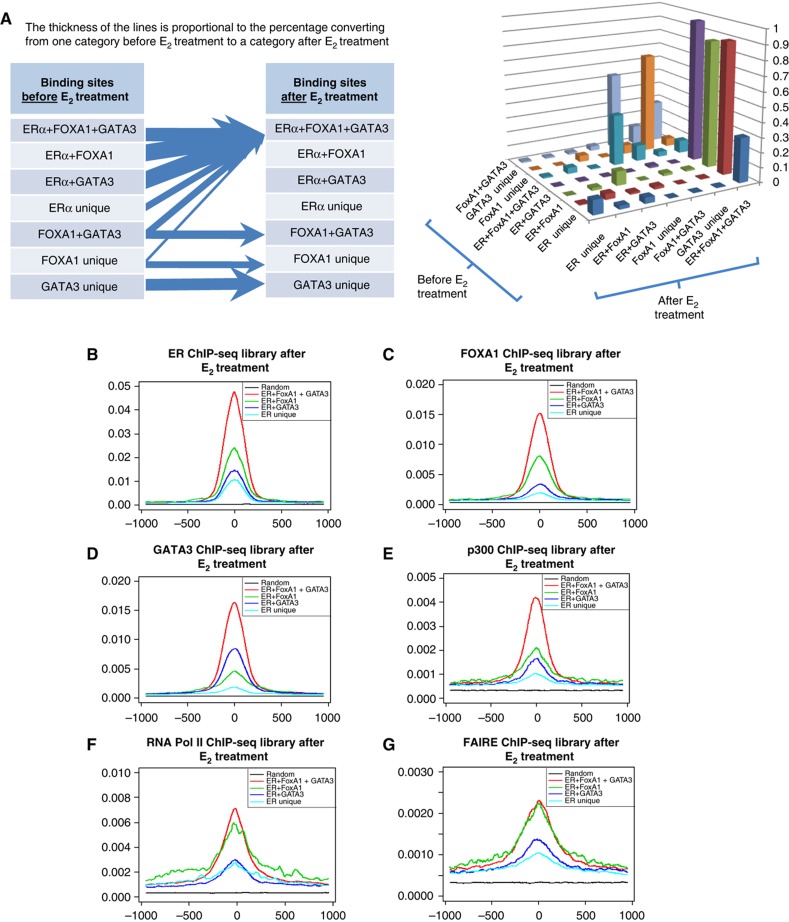
We have observed the colocalization of ERα, FOXA1, and GATA3 at genomic sites after ligand stimulation. We then wished to assess the dynamics of this recruitment by the three TFs in response to E2 stimulation. First, we grouped the different subsets of binding sites before E2 treatment as ERα unique, FOXA1 unique, GATA3 unique, ERα+FOXA1 overlap, ERα+GATA3 overlap, FOXA1+GATA3 overlap, and ERα+FOXA1+GATA3 overlap sites as shown in the Venn diagram of Figure 1A. We investigated how these different subsets of TF bindings clustered after E2 stimulation. We found (Figure 4A) a dramatic shift of single and double TF binding sites to sites occupied by the three TFs: >89% of vehicle-treated ERα+FOXA1 overlap sites, 86% of ERα+GATA3 overlap sites, 30% of ERα unique site, and 28% of the FOXA1+GATA3 overlap sites were shifted to the ERα+FOXA1+GATA3 overlap sites in response to E2 induction. By contrast, the FOXA1 unique and GATA3 unique sites (before ligand) showed little to no shift to the conjoint three factors occupancy state after E2 treatment. This suggests that estradiol activation induces the recruitment of FOXA1 and GATA3 with ERα at ERα binding sites.
Figure 4.
The formation of enhanceosome consists of ERα, FOXA1, and GATA3. (A) The dynamics of TFs binding before and after E2 stimulation. The different categories of ERα, FOXA1, and GATA3 binding sites in the Venn diagram before E2 stimulation will converge to ERα+FOXA1+GATA3 overlapped binding sites after E2 stimulation. (B–D) The enhanced ERα, FOXA1, and GATA3 occupancy at the ERα+FOXA1+GATA3 sites. The tag profiles around the binding sites are enriched after E2 stimulation (see Supplementary Figure S3 for tag profiles before E2 treatment). The tag profile is normalized by the number of peaks in the category and the sequencing depth from the corresponding ChIP-seq library. (E) The enhanced p300 co-activator recruitment to enhanceosome sites after E2 stimulation. (F) The highest association of RNA Pol II recruitment with enhanceosome sites after E2 stimulation. (G) The enhanceosome is correlated with chromatin opening as measured by FAIRE after E2 stimulation.
It has been previously shown that the functional utility of an ERα binding site is higher when these sites are marked by specific and quantitative chromatin signatures: functionally active sites have higher ERα occupancy, more open chromatin, and more likely to show p300 and RNA Pol II occupancy (Figure 4; Supplementary Figure S3). Figure 4B–D shows that the normalized tag profiles of ERα, FOXA1, and GATA3 at the binding sites are strongly enriched after E2 treatment for all the above marks with the ERα+FOXA1+GATA3 overlapping conjoint sites having the highest tag occupancy profile above all other colocalized categories. In addition, the triple TF overlap sites show the greatest occupancy of each individual TF. p300 co-activator possesses intrinsic histone acetyltransferase activity capable of modifying the chromatin organization and facilitating transcriptional initiation (Heintzman et al, 2007), and p300 enrichment is commonly found at the enhancer regions. We observed ERα+FOXA1+GATA3 overlap sites have the highest p300 co-activator occupancy (Figure 4E). Previously, we have determined that RNA Pol II co-binding at ERα binding sites is related to distant interactions linking the enhancer sites with the TSSs (Fullwood et al, 2009). In Figure 4F, we show that ERα+FOXA1+GATA3 overlap sites have the highest RNA Pol II occupancy with ERα+FOXA1 double overlap sites following closely. When the chromatin state was assessed by formaldehyde-assisted isolation of regulatory elements (FAIRE) (Joseph et al, 2010), we found that ERα+FOXA1+GATA3 and ERα+FOXA1 overlap sites have the highest association with chromatin opening (Figure 4G). This suggests that these triple overlap sites (ERα+FOXA1+GATA3) are potentially the most active enhancers affecting ERα transcriptional regulation and that there appears to be a hierarchy of associative effect: FOXA1 contributing the most to ERα enhancer function and GATA3 being less impactful (Figure 4F and G). This effect is independent of ERα occupancy intensity since the triple overlap/enhanceosome sites (ERα+FOXA1+GATA3) bear the marks for optimal enhancer with the highest p300 occupancy while the non-enhanceosome sites have less association with the enhancer marks though all these sites were from the top quartile of ERα sites of highest binding intensity (Supplementary Figure S4).
It is known that TFs can interact through long-range chromatin interactions to regulate the transcriptional networks. Recently, a new method known as Chromatin Interaction Analysis with Paired-End-Tag sequencing (ChIA-PET) has been developed (Fullwood et al, 2009; Li et al, 2010) to characterize the long-range chromatin looping mediated by ERα in MCF-7 cell line. After re-analyzing the ERα ChIA-PET data, we observed that 81% of the 5067 interaction clusters have at least one anchor region (an ERα binding site associated with a distant chromatin interaction forming at least one loop) characterized by ERα+FOXA1+GATA3 colocalization. Furthermore, the interaction clusters from ChIA-PET can form complex clusters that organize the local genomic region into multiple loops. These complex interaction clusters also demarcate the most significant ERα-regulated genes (Fullwood et al, 2009). Of the 5067 ERα-mediated long-range interaction clusters, 4500 clusters are involved in complex interaction clusters and 567 clusters are involved in duplex interaction clusters. In all, 88% of the complex interaction clusters are associated with ERα+FOXA1+GATA3 overlapped binding sites, while 51% of the duplex interaction clusters have the support of ERα+FOXA1+GATA3 overlapped binding sites (Supplementary Figure S5). Since complex interaction clusters mark genes most responsive to E2 as compared with duplex clusters, these data support the notion that the presence of the ERα, FOXA1, and GATA3 putative enhanceosome is associated with genes that are most responsive to E2. A specific example of the ERα-mediated long-range interactions involving conjoint ERα, FOXA1, and GATA3 binding sites around the highly E2-responsive GREB1 gene is shown in Supplementary Figure S5. These triple TF conjoint binding sites are highly represented at the sites involved in frequent long-range chromatin interactions spanning 50 kb.
The impact of TFs binding on gene expression
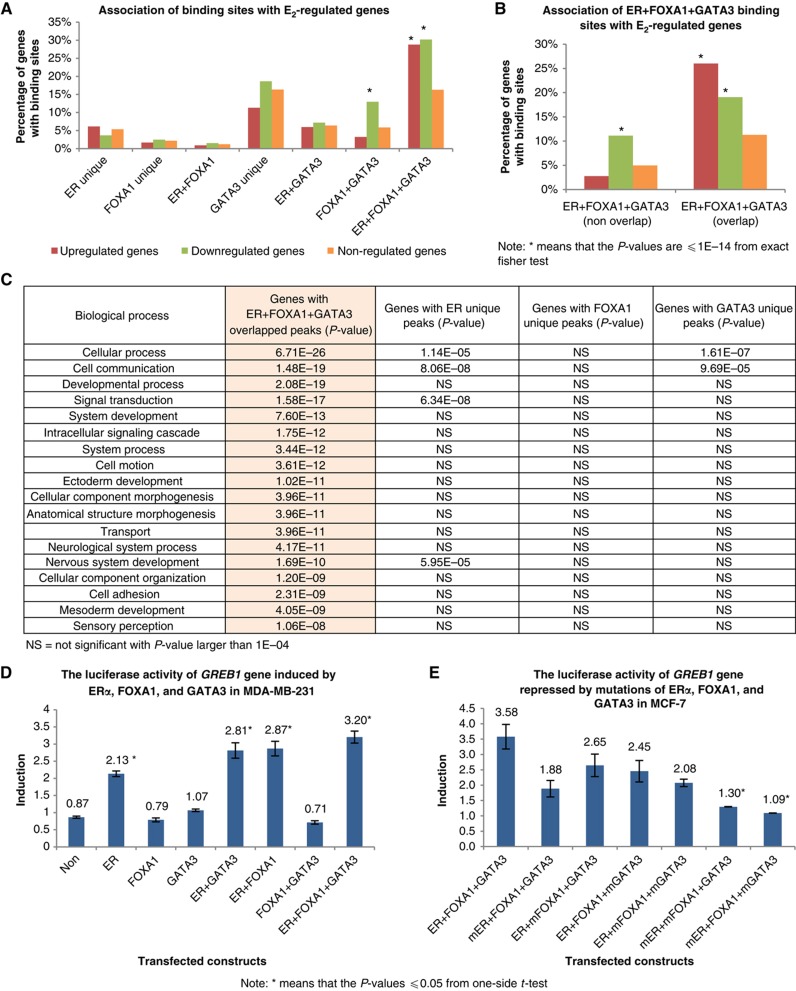
We have provided evidence that the clustering of ERα, FOXA1, and GATA3 at ERα binding sites is associated with chromatin characteristics of the most active ERα enhancers. To assess the effect of this enhanceosome presence on direct gene regulation, we performed a detailed microarray expression analysis to determine E2-responsive genes in MCF-7 cells. We found a total of 653 upregulated and 1249 downregulated genes in response to E2 stimulation (Supplementary Tables VIII and IX). We assigned a specific known gene to a binding site occupied by any combination of the three TFs if the peak of each TF category is the nearest and within 20 kb of TSS of that E2-responsive gene (see Supplementary information). Figure 5A shows that, for the genes with ERα, FOXA1, and GATA3 peaks or any combination, the biggest proportion of either upregulated or downregulated genes is from genes with adjacent ERα+FOXA1+GATA3 conjoint binding sites within 20 kb of their TSS (28 and 30%, respectively). Since the different binding sites are present in the genome at different frequencies, the ratio of regulated versus non-regulated genes for each binding site class can be used to normalize the differences. The proportion of upregulated genes with ERα, FOXA1, and GATA3 conjoint peaks is 2.3-fold of the non-regulated genes with the same configuration. This is contrasted by genes adjacent to other combinations of ERα, FOXA1, and GATA3 binding which do not show significant changes as compared with non-regulated genes. The only exception is the proportion of genes close to FOXA1+GATA3 co-bound sites, which are associated with greater downregulated genes. Despite this association, the percentage of downregulated genes putatively controlled jointly by FOXA1 and GATA3 is relatively small. Finally, the presence of the three TFs relative to a regulated gene may have two configurations: one where the ERα binding site has conjoint and therefore overlapping occupancy by all three TFs, and the other where the binding of the individual TFs is in proximity with each other and within 20 kb of a gene, but the binding sites are not overlapping (Figure 5B; see Supplementary information). When we analyzed the association of E2-regulated genes with these two categories (overlapping and non-overlapping), we found that the predominant association is between the conjoint binding sites and regulated genes (Figure 5B). Our results imply that ERα regulation of gene expression is closely linked to adjacency with sites that show conjoint binding with ERα, FOXA1, and GATA3 putatively forming an enhanceosome. Using Gene Ontology analysis (Thomas et al, 2003), we sought to further ascertain the importance of genes in proximity with enhanceosome binding as compared with binding of the individual TF components. We found that only genes associated with ERα+FOXA1+GATA3 binding have significant association (with P-values up to 6.7E−26) with specific biology processes known to be involved in ERα signaling (e.g., signal transduction and cell proliferation), molecular function (e.g., kinase and protein binding), and signaling pathways (e.g., PDGF signaling pathway, inflammation mediated by chemokine and cytokine signaling pathway) (Figure 5C; Supplementary Tables II and III). Thus, the identification of the ERα enhanceosome-associated genes allows for the identification of a ‘core’ set of ERα-regulated genes that are strongly associated with the cognate cellular functions previously known for ERα.
Figure 5.
The impact of enhanceosome on gene regulation. (A) The association of ERα, FOXA1, and GATA3 bindings with E2-regulated genes. The percentages of the upregulated and downregulated genes are significant from genes associated with ERα+FOXA1+GATA3 overlapped binding sites. (B) The association of TF bindings with E2-regulated genes in two different configurations where conjoint ERα+FOXA1+GATA3 bindings with overlapping occupancy by all three TFs and those non-overlapping individual ERα, FOXA1, and GATA3 bindings in close proximity within 20 kb of a TSS. (C) Gene ontology analysis of genes associated with different categories of ERα, FOXA1, and GATA3 bindings. The genes associated with ERα+FOXA1+GATA3 overlapped binding sites have significant functions, compared with genes only with individual unique ERα, FOXA1, and GATA3 bindings. (D) The presence of ERα, FOXA1, and GATA3 has induced the luciferase activity of GREB1 gene in MDA-MB-231 cells. The basal luciferase activity of GREB1 in MDA-MB-231 cells is used as the control reference. Mean values of three independent experiments are compared and standard errors are shown. (E) The loss of FOXA1 and/or GATA3 bindings has reduced the luciferase activity of GREB1 gene in MCF-7 cells. ‘mER’, ‘mFOXA1’, and ‘mGATA3’ denote mutated ERE, FOXA1, and GATA3 motif sequences around their respective binding sites near the GREB1 promoter. The basal luciferase activity of GREB1 in MCF-7 wild-type cells is served as the control reference. Mean values of three independent experiments are compared and standard errors are shown.
To further validate that ERα+FOXA1+GATA3 co-binding represents an optimal configuration for E2-mediated transcriptional activation, we have performed luciferase reporter assays on GREB1 locus that actively engages ERα enhanceosome sites in gene regulation. We cloned the promoter region of GREB1 that includes an ERα+FOXA1+GATA3 enhanceosome binding site into the pGL4-luciferase reporter construct and then transfected GREB1-luciferase promoter construct into ER-negative MDA-MB-231 cells, followed by transfection and overexpression of ERα, FOXA1, and/or GATA3. The individual presence of FOXA1 and GATA3 or combination of both only produced subtle changes to the GREB1 luciferase activity, demonstrating that the presence of FOXA1 and GATA3 alone or combination of both do not activate the transcription of GREB1 gene (Figure 5D). The presence of ERα induced the GREB1 luciferase activity to ∼246% (as compare with the control construct). The combination of ERα+FOXA1 and ERα+GATA3 has increased the luciferase activity to ∼330% (an increment of 26–32%). Interestingly, the assemblage of ERα+FOXA1+GATA3 provided the optimal ER responsiveness to 370% representing an additional 12–14% increment. This suggests that ERα provides the fundamental gene regulatory module but that FOXA1 and GATA3 incrementally improve ERα-regulated transcriptional induction.
Such artificial transfection reporter systems accentuate TF responses because of unnatural stoichiometries of the TFs. To further assess the interplay among ERα, FOXA1, and GATA3, we perturbed the binding of these TFs in MCF-7 cells through the site-directed mutagenesis assay and asked whether the loss of individual binding motifs would alter gene regulation under physiologic concentrations of the three TFs. Different GREB1-luciferase constructs with mutated ERE, FOXA1, or GATA3 motif at the specific ERα, FOXA1, and GATA3 binding sites were generated. The results revealed that individually mutated FOXA1 or GATA3 motif only imposed 25–30% loss of GREB1 luciferase activity (Figure 5E). Mutated ERE alone has repressed the luciferase measurement to ∼50%. Interestingly, combinatorial ERE+FOXA1 and ERE+GATA3 mutation further reduced the luciferase activity by ∼65–70%, suggesting that the effects of the individual TFs on this putative enhanceosome are additive. Here, we can build a hierarchy of TFs control, showing that ERα accounts for 50% of the transcriptional control while FOXA1 and GATA3 individually account for another 20% transcriptional control at the GREB1 gene regulatory locus.
FOXA1 and GATA3 are essential co-regulators in mediating the ERα-growth response
It is known that ERα is a ligand-activated TF that mediates the proliferative effects of E2 in breast cancer cells. Garcia et al (1992) showed inhibited growth in MDA-MB-231 cells with forced expression of ERα upon E2 treatment. The rationale for these different outcomes has remained elusive. We hypothesize that the absence of critical co-regulators such as FOXA1 and GATA3 is responsible for the ERα-response cassette.
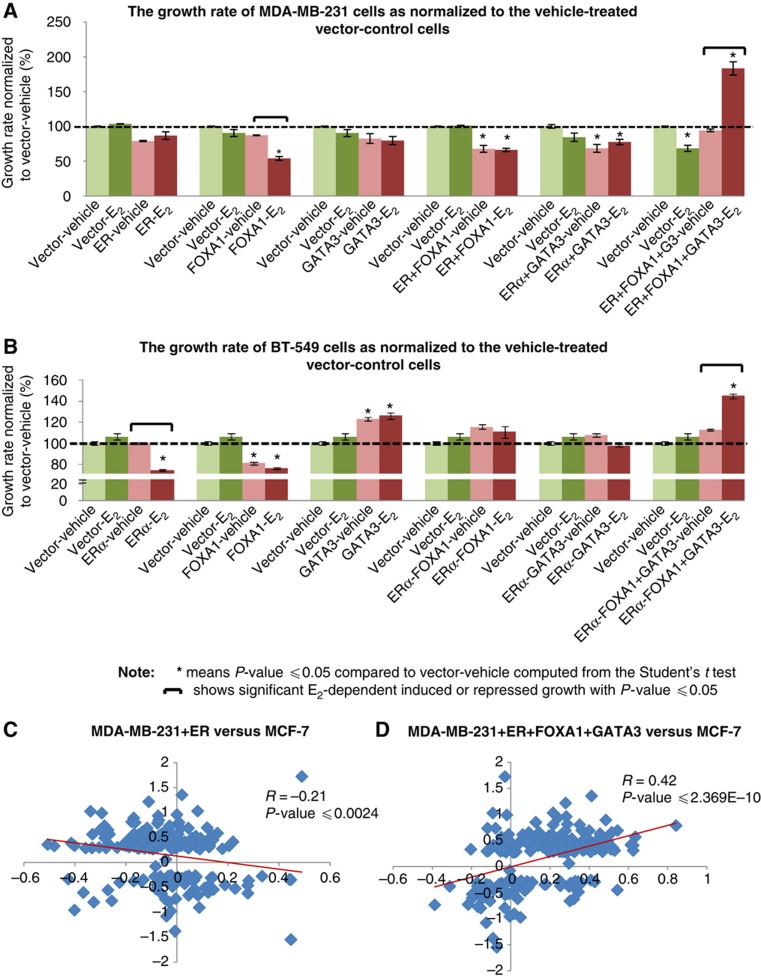
To test this hypothesis, we stably transfected the MDA-MB-231 cells with individual ERα, FOXA1, GATA3, or in combination. The induction of ERα, FOXA1, and GATA3 expressions following transfections was verified (Supplementary Figure S6). The cell proliferation in response to E2 stimulation was measured using two assays: WST-1 and cell count using Hoechst stain. In parallel to the report by Garcia et al (1992), we observed marginally inhibited growth in cells with forced expression of ERα and a greater inhibitory effect with forced expression of FOXA1. There was unaltered growth in cells with expression of GATA3. Co-expression of ERα and FOXA1; ERα and GATA3 exhibited inhibition of cell proliferation as compared with control cells. However, the co-expression of ERα together with FOXA1 and GATA3 resulted in marked induction of cell proliferation under E2 stimulation as assessed by either growth detection assays (Figure 6A; Supplementary Figures S7 and S8). We have recapitulated this cellular reprogramming in another ERα-negative breast cancer cell line, BT-549 and observed similar growth inhibition in BT-549 cells expressing ERα and FOXA1 individually (Supplementary Figures S9 and S10). We found minor induction of growth in GATA3-expressing BT-549 cells; however, this growth was independent of E2 stimulation. However, like MDA-MB-231 cells, we were able to induce E2-dependent growth in ERα+FOXA1+GATA3-expressing BT-549 cells (Figure 6B; Supplementary Figure S11). This suggests that only with the full activation of conjoint binding sites by the three TFs will the proliferative phenotype associated with ligand induced ERα be manifest. This further suggests that like induced pluripotent stem cells (iPS cells) only the combination of multiple factors (in this case, ERα, FOXA1, and GATA3) can transcriptionally reprogramme MDA-MB-231 and BT-549 cells to be estrogen responsive for growth. To assess the nature of this transcriptional reprogramming, we asked the question if the reprogrammed MDA-MB-231 cells display any similarity in the expression profile of the ERα-positive breast cancer cell line, MCF-7. We combined the E2-regulated genes from these differently transfected MDA-MB-231 cells, and compared their expression in these MDA-MB-231-transfected cells and MCF-7 cells. Strikingly, taking all differentially expressed genes in MDA-MB-231 sublines, we found that the expression profile of E2-induced ERα+FOXA1+GATA3-expressing MDA-MB-231 cells display a good positive correlation (R=0.42) with the E2-induced expression profile of MCF-7. By contrast, we observed a negative correlation between the expression profiles of MDA-MB-231 transfected with ERα only (R=−0.21) (Figure 6C and D; see Supplementary Table X for detailed analyses). In addition, when only cell-cycle, DNA replication, and proliferation genes were examined, again, there was positive correlation between MDA-MB-231 transfected with ERα+FOXA1+GATA3 and MCF-7 but no correlation between ERα-only MDA-MB-231 cells and MCF-7 (Supplementary Figure S12). Specific genes previously known to be E2 regulated in ERα-responsive cell lines (Frasor et al, 2003; Fullwood et al, 2009) such as CCND1, STC2, ADCY9, and BTG1 were also regulated in the same direction by ligand in the triple factor transfected MDA-MB-231 cells (Supplementary Table XI). Using the Ingenuity Pathway Analysis, we observed that the estrogen-responsive genes regulated in MDA-MB-231 transfectant cells were significantly associated with cell-cycle, cellular proliferation, and DNA replication functionalities (P-value=7.27E−12–1.28E−04, see Supplementary Figure S13A). Moreover, we found that there is upregulation of pro-proliferative cell-cycle genes in the ERα+FOXA1+GATA3-expressing MDA-MB-231 cells compared with ERα-only cells (Supplementary Figure S13B). Taken together, these results suggest that the presence of ERα, FOXA1, and GATA3 has transcriptionally reprogrammed the ERα-negative MDA-MB-231 cells to resemble the ERα-positive MCF-7 cells by recapitulating the estrogen-responsive cassette and manifesting the proliferative phenotype.
Figure 6.
FOXA1 and GATA3 are essential components of E2-induced ERα-response casette. (A) The growth of MDA-MB-231 cells transfected with different combinations of TFs relative to the vehicle-treated MDA-MB-231 vector control cells at the final day of WST-1 measurement. (B) The recapitulation of reprogramming work in another ERα-negative BT-549 cells. The growth of BT-549 cells tranfected with different combinations of TFs relative to the vehicle-treated BT-549 vector control cells at the final day of WST-1 assay. (C) The comparison of gene profiles in the reprogrammed MDA-MB-231 and MCF-7 cells. The gene profiles of ERα-only MDA-MB-231 cells show weak correlation with the expression profiles of MCF-7 cells. (D) The ERα+FOXA1+GATA3-expressing MDA-MB-231 cells display good correlation with the expression profile of ERα-positive MCF-7 cells. The P-value for the differences between the two correlation coefficient stated in (C) and (D) is ⩽2.08E−11.
Finally, we asked if the reprogrammed MDA-MB-231 cells have acquired luminal cell characteristics. We investigated the expression of luminal and basal markers genes defined by Kao et al (2009) in the transfected MDA-MB-231 cells. The analysis revealed a modest but discernible induction of luminal markers genes and suppression of basal marker genes in the ERα+FOXA1+GATA3-expressing MDA-MB-231 cells as compared with the ERα only or vector control cells (Supplementary Figure S14; Supplementary Table XII). Moreover, we found that 63% of the luminal genes are associated with conjoint binding of the three TFs ERα+FOXA1+ GATA3 within 20 kb of the TSS, and only 13% of these luminal genes showing no proximity binding of any of these three TFs. On the other hand, 24% of the basal genes are associated with proximate conjoint ERα+FOXA1+GATA3 binding with 40% of these genes are not associated with any ERα, FOXA1, or GATA3 binding (see Supplementary Figure S15 and Supplementary Table XIII). This suggests that ERα+FOXA1+GATA3 binding exerts greater impact on regulating the transcription of luminal marker genes as compared with the basal marker genes. Taken together, we demonstrate that the co-expression of the ERα enhanceosome components, namely ERα, FOXA1, and GATA3, is required to approximate the appropriate luminal expression cassette.
Discussion
ER, as a prototype of a nuclear hormone receptor, mediates a broad range of cellular and physiologic functions with organ and context specificity. The most proximate form of regulatory control resides in the protein–DNA interaction of TF binding to their cognate recognition motifs and modified by cofactors. However, genome-wide studies of ERα binding show a dispersed occupancy pattern at binding sites bearing heterogenous recognition motifs that are, at the sequence level, also not well conserved in evolution (Kunarso et al, 2010). This binding site heterogeneity is normalized by chromatin looping to bring these distant and distributed enhancers in proximity to the regulated TSS (Fullwood et al, 2009). Herein, we show that FOXA1 and GATA3 are essential for optimal ERα binding to DNA, that FOXA1 and GATA3 are recruited as a complex to the most functional ERα binding sites after ligand activation, and that the binding of this tripartite enhanceosome complex of ERα, FOXA1, and GATA3 is necessary for optimal transcriptional activation in reporter gene assays. The enhanceosome assembly is recruited to sites bearing the three recognition motifs suggests that this complex formation is ‘hard-wired’ in the human genome and provides an evolutionary advantage. This notion is supported by the fact that the colocalization of the motifs for these TFs was found in 23 090 sites in the reference human genome, but in only 360 sites in a random nucleotide sequences for a 64-fold enrichment. This compares with an ∼18-fold enrichment for the ERE alone suggesting a strong evolutionary selection for the three TFs to be colocalized (see Supplementary Figure S16).
An enhanceosome has been defined as protein complex composed of a repertoire of TFs that binds to the ‘enhancer’ region of a gene and sequentially recruits components of the transcriptional machinery such as RNA polymerase to initiate the gene's transcription. Synergistic interplay among the members within the enhanceosome complex results in providing some functional specificity, and a multiple gene ‘fail-safe’ mechanism for controlling gene expression (Robert and Tom, 1994). It is suggested that an enhanceosome may provide functional redundancy that minimizes the chances that a gene may be switched off due to mutation, or permit activation of a gene by orchestrating multiple different signaling cascades (Farnham, 2009). The importance of enhanceosome formation is evidenced by the virus-inducible transcriptional activation of the human interferon-β (IFN-β) gene by the assembly of transcriptional activator (p50/p65), IRF-1, ATF-2, c-Jun, and high mobility group protein HMG I to the basal transcription complex (Thanos and Maniatis, 1995). Chen et al (2008) showed that TFs coordinately expressed in embryonic stem cell differentiation form specific enhanceosomes adjacent to cassettes of genes that demarcate different developmental functions. The present study provides evidence of how the ERα, FOXA1, and GATA3 enhanceosomes regulate this multifaceted transcriptional network operative in reproduction and cancer. Furthermore, we show that this ERα+FOXA1+GATA3 enhanceosome recruits distinct components of active transcription regulatory machinery, namely RNA Pol II and p300, an acetyltransferase associated with enhancer activity as well as chromatin opening. Interestingly, the ERα, FOXA1, and GATA3 binding were also coincided with retinoic acid receptor binding though the overlap is less frequent, suggesting that FOXA1 and GATA3 could have a broader ‘universal’ co-regulator function for nuclear hormone binding (Hua et al, 2009).
It is known that FOXA1 and GATA3 are important regulatory proteins in their own right. FOXA1 has winged helix domains that can structurally mimic histone H1 and H5, and thus permits its interaction with histone H3 and H4. This unique feature of FOXA1 allows it to bind to the specific DNA sequences on the nucleosome core and displace the linker histones, leading to de-compaction of chromatin and to facilitate the binding of other TFs (Clark et al, 1993; Cirillo et al, 1998; Kaestner, 2000). It is suggested that ERα+FOXA1-regulated network establishes an ‘one-step forward’ (through cyclin D1 induction) and ‘one-step backward’ (through p27KIP1 induction) manner to control cell-cycle progression in breast cancer cells (Nakshatri and Badve, 2009). Recent work by Lupien et al (2008) revealed that there was significant overlap of FOXA1 occupied sites on ERα cistrome, hence suggesting that FOXA1 contributed in the control of E2 signaling in breast cancer cells. In accordance with the report by Bernado et al (2010), we have found that siRNA-mediated knockdown of ERα reduces the levels of FOXA1 in MCF-7 cells, and similar attenuation of FOXA1 reduces the levels of ERα (data not shown).
GATA3 has essential roles in the mammary gland morphogenesis and lactogenesis. Inactivation of GATA3 resulted in diminished mammary epithelial structure, severely impaired lactogenesis and disrupted differentiation of luminal progenitor cells into ductal and alveolar cells (Asselin-Labat et al, 2007). Moreover, GATA3 is also involved in the positive crossregulatory loop with ERα in breast cancer cells in mediating the E2 signaling (Eeckhoute et al, 2007). Clinically, both FOXA1 and GATA3 are known to be co-expressed in ER-positive breast cancers. In addition, Mehra et al (2005) reported that low levels of GATA3 were strongly associated with larger tumor size, positive lymph node status, higher histology grade, ERα-negative status, Her2-neu overexpression as well as increased risk for recurrence and metastasis. Taken together, we posit that such a complex regulatory and functional interaction of three TFs each subserving important functions is another evolutionary strategy to ensure the balanced co-regulation of gene networks important in mammalian reproduction.
Here, we have shown that the effects of the ERα+FOXA1+GATA3 enhanceosome expression are the regulation of the major important E2-responsive genes associated with various signaling pathways, biology processes, and molecular functions previously ascribed to ERα alone. Though the presence of an ERα is necessary for E2-induced growth in responsive cells, its presence is not sufficient for cellular proliferation, and in fact, the introduction of ERα or FOXA1 into ERα-negative cell lines such as MDA-MB-231 leads to cell-cycle arrest. Importantly, we show that transfection of the three TFs into the ERα-negative cell line, MDA-MB-231, could reprogram the cell to be estrogen responsive for cell proliferation, counteracting the growth inhibitory action of unaided FOXA1 or ERα. This cellular reprogramming is correlated with reconstruction of the approximate transcriptional cassette of the modified MDA-MB-231 to partially resemble that of E2-stimulated MCF-7 cells. Thus, it appears that the primary role of FOXA1 and ERα alone in breast cancer cells is as a growth or tumor inhibitor, but that the conditional expression of ERα, FOXA1, and GATA3 reverses this state to that of growth induction.
Intriguingly, enforced expression of the triple factors, ERα, FOXA1, and GATA3, also induced a modest basal to luminal expression cassette change by reducing the basal signature and increasing the luminal signature in MDA-MB-231 cells not seen in the ERα alone-transfected clone. Our results suggest that the conjoint effects by the three TFs could formulate a luminal cassette and then manifest the proliferative phenotype in response to estrogen stimulation.
Our work also sheds some light on the functional role of FOXA1 which is thought to be a pioneering factor for nuclear hormone receptors such as the ER and androgen receptor (Carroll et al, 2005). As a pioneering factor, FOXA1 may function to open chromatin structures so as to facilitate ERα binding to its cognate response elements. Indeed, our chromatin model predictive of ERα binding includes FOXA1 occupancy in the preligand (before E2 exposure) state (Joseph et al, 2010). However, these studies did not examine the dynamic relationship of ERα and FOXA1 occupancy before and after ligand exposure. Our results suggest that ERα is as likely to be a pioneering factor to recruit FOXA1 as the converse.
Recently, Eeckhoute et al (2009) reported that a significant fraction of FOXA1-bound sites have a relatively closed chromatin conformation that is unrelated to gene expression, suggesting that FOXA1 may require a repertoire of collaborating TFs to promote chromatin opening. Our findings suggest that ERα is one such collaborating TF with GATA3 playing a more minor role.
Taken together, we have uncovered the functional importance of an enhanceosome comprising ERα, FOXA1, and GATA3 in the estrogen responsiveness of ERα-positive breast cancer cells. This enhanceosome exerts significant combinatorial control of the transcriptional network regulating growth and proliferation of ERα-positive breast cancer cells.
Materials and methods
ChIP assay
The ChIP assays were carried out as described previously (Lin et al, 2007). Briefly, the serum-depleted MCF-7 cells were treated with 10 nM E2 or vehicle control for 45 min. Cells were crosslinked with 1% formaldehyde for 10 min at room temperature, followed by 125 mM glycine treatment to inactivate the crosslinking. Chromatin extracts were fragmentized to an average size of 500 bp with sonication, followed by overnight immunoprecipitation at 4°C. The protein–DNA complex was de-crosslinked with overnight incubation at 65°C. DNA extraction was performed and qPCR validation was carried out using SYBR Green chemistry. The ChIP samples were then subjected to ChIP-seq on Solexa platform. Antibodies used in these ChIP experiments are listed in Supplementary Table IV.
Binding sites analysis
The short reads from ChIP-seq libraries were aligned to the human genome hg18 using Batman with at most two mismatches, and only the uniquely mapped reads were extracted for further analysis. The ChIP-seq for ERα, FOXA1, RNA pol II, and FAIRE libraries have been described in Joseph et al (2010). Here, we used the Model-based Analysis for ChIP-Seq (MACS) to call the peaks for all the three TFs (Zhang et al, 2008) with default parameters. The peaks were reported as the summit of the enriched regions. The number of binding sites for each ChIP-seq library is shown in Supplementary Table I. The binding sites were validated with qPCR using the specific primer sets. The primers are designed around the binding sites and qPCR was performed using SYBR Green Chemistry and 5 μM of primers (listed in Supplementary Table VII) in ABI7500 Read-time PCR System (Applied Biosystems).
The overlap of peaks from two libraries is defined as the peaks within genomic distance 200 bp. The motif scanning was done with the program CentDist (Zhang et al, 2011) with motif PWM from TRANSFAC version 11.3 (Matys et al, 2003) with FDR<1E−3.
Sequential ChIP
The ChIP assay was carried out as described previously. The elute from the first round of immunoprecipitation with ERα antibody was subjected to second round of immunoprecipitation using FOXA1 and GATA3, respectively, followed by qPCR to validate co-occupancy of ERα+FOXA1 and ERα+GATA3 to the target sites. The primers used for Re-ChIP are listed in Supplementary Table V.
Cells synchronization
The MCF-7 cells were grown in phenol red-free DMEM with 5% charcoal dextran-treated FBS (CDFBS) for 3 days before subjecting to 2.5 μM α-amanitin treatment for 2 h. The cells were washed with PBS twice, followed by E2 or vehicle control treatment for 45 min. Cells were harvested at the 5-min interval and subjected to ChIP-qPCR assays. The primers used to study the progressive recruitment of ERα and FOXA1 are listed in Supplementary Table VI.
Microarray gene expression study on MCF-7 cells
The MCF-7 cells were grown in phenol red-free DMEM with 5% CDFBS for 3 days before E2 stimulation. Total RNA was harvested at 3, 6, 9, 12, 24, and 48 h after E2 treatment using RNeasy Kit (Qiagen). The quality of RNA samples was verified with Bioanalyzer before proceeding to Affymetrix microarray experiments. The microarray data from E2 stimulation were normalized against the data with vehicle treatment, log-transformed, and median normalized. The upregulated and downregulated genes were called with 1.2-fold change (which corresponds to 0.263 after log2 transformation).
Microarray gene expression study on the transfected MDA-MB-231 cells
The transfected MDA-MB-231 cells were grown in phenol red-free RPMI with 5% CDFBS before subjecting to E2 stimulation. Total RNA was harvested at days 2 and 10 using RNeasy Kit (Qiagen) and labeled using the TargetAmp™ Nano-g™ Biotin-aRNA Labeling Kit (Epicentre) before proceeding to Illumina microarray experiment. The microarray data from E2 stimulation were normalized against the data with vehicle treatment, log-transformed, and median normalized. The upregulated and downregulated genes were called with 1.2-fold change.
Association of TF binding with gene expression
The binding sites were associated with the nearest TSSs within 20 kb (see Supplementary information for detailed description).
Site-mutagenesis experiments
The promoter region of GREB1 gene was cloned into pGL4-luciferase construct using the In-fusion kit (Clontech). Several primers with the mutated ERα, FOXA1, and GATA3 binding motifs were designed and mutagenesis experiment was carried out with Stratagene Site-directed Mutagenesis Kit. The sequences of the GREB1-luc construct as well as the mutated-luc constructs were verified with sequencing.
Expression clones
The expression clones encoding ESR1, FOXA1, and GATA3 were purchased from Genecopoeia and verified by sequencing.
Luciferase reporter assays
The MDA-MB-231 cells were transfected with the reporter construct together with different combination of TFs. The MCF-7 cells were transfected with the reporter construct together with the mutated ERα, FOXA1, and GATA3 constructs. A Renilla luciferase plasmid was co-transfected as an internal control. Dual-luciferase reporter kit (Promega) was employed to measure the luciferase activity relative renilla activity using GloMax 96 microplate luminometer (Promega).
Transfection experiments
The MDA-MB-231 and BT-549 cells were stably transfected with ERα, FOXA1, and GATA3 using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. An empty vector transfection was included as a negative control. The G418-selected clonal cells were verified for their ERα, FOXA1, and GATA3 expression with western blot using ERα (sc-543, Santa-Cruz), FOXA1 (ab-5089, Abcam), and GATA3 (sc-269, Santa-Cruz) antibodies at the dilution of 1:500.
Cell proliferation assays
The transfected MDA-MB-231 and BT-549 cells were seeded in 96-well plate and subjected to 10 nM E2 or vehicle treatment. The culture media were changed every 3 days and the cell proliferation was assayed with WST-1 (Roche) using Sunrise microplate absorbance reader system (Tecan). Another cell growth assay assessed by cell number count was performed. The cells are fixed with 4% paraformaldehyde followed by permeabilization with 0.1% Triton-X at room temperature. The cells were then stained with 2.5 μg/ml of Hoechst for 10 min before proceeding to cell counting scan on Cellomics ArrayScan VTi machine (Thermo Scientific).
Data release
The raw ChIP-seq sequences and processed data can be accessed from NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE23701, GSE23893, GSE26831, and GSE29073. The gene expression data can be accessed from NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE30574.
Supplementary Material
Supplemental Tables SI-VI, Supplemental Figures S1-16
List of primers used in ChIP-qPCR study
List of E2 up-regulated genes in MCF-7 cells
List of E2 down-regulated genes in MCF-7 cells
List of all the E2-regulated genes in the transfected MDA-MB-231 cells compare to MCF-7 cells
List of previously reported E2-regulated genes that regulate in our MCF-7 and transfected MDA-MB-231-ER+FOXA1+GATA3 cells
The expression of luminal and basal marker genes in MCF-7 and transfected MDA-MB-231 cells
List of luminal and basal marker genes with ER, FOXA1 and GATA3 binding
Acknowledgments
We thank Jane S Thomsen, Roy Joseph, Sabry M Hamza, Edwin WE Cheung, Yuriy L Orlov, and Kartiki V Desai for advice and participation in discussion. We acknowledge members of the sequencing team from Genome Institute of Singapore for supporting the Solexa ChIP-sequencing efforts. This research work was funded by Agency of Science, Technology and Research (A*STAR) of Singapore and the 6th European Community Framework Program grant CRESCENDO (FP6-018652) to ETL. SLK is a graduate student from National University of Singapore, supported by the Genome Institute of Singapore Scientific Staff Development Award.
Author contributions: SLK and ETL conceptualized and designed the experimental strategy. SLK coordinated the study and performed all the experiments. GLL performed all the computational and statistical analyses. SLL provided the technical assistance. WK provided advice for the computational analyses. SLK, GLL, and ETL interpreted the results and wrote the manuscript. ETL provided supervision for SLK graduate study.
Footnotes
The authors declare that they have no conflict of interest.
References
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, Lindeman GJ, Visvader JE (2007) Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol 9: 201–209 [DOI] [PubMed] [Google Scholar]
- Badve S, Turbin D, Thorat MA, Morimiya A, Nielsen TO, Perou CM, Dunn S, Huntsman DG, Nakshatri H (2007) FOXA1 expression in breast cancer—correlation with luminal subtype A and survival. Clin Cancer Res 13: 4415–4421 [DOI] [PubMed] [Google Scholar]
- Bernado GM, Lozada KL, Miedler JD, Harburg G, Hewitt SC, Mosley JD, Godwin AK, Korach KS, Visvader JE, Kaestner KH, Abdul-Karim FW, Montana MM, Keri RA (2010) FOXA1 is an essential determinant of ERa expression and mammary ductal morphogenesis. Development 137: 2045–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat-Nakshatri P, Wang G, Appaiah H, Luktuke N, Carroll JS, Geistlinger TR, Brown M, Badve S, Liu Y, Nakshatri H (2008) AKT alters genome-wide estrogen receptor a binding and impacts estrogen signaling in breast cancer. Mol Cell Biol 28: 7487–7503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK et al. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS (2002) Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9: 279–289 [DOI] [PubMed] [Google Scholar]
- Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim EY, Clark KL, Burley SK, Zaret KS (1998) Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J 17: 244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK (1993) Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364: 412–420 [DOI] [PubMed] [Google Scholar]
- Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M (2007) Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res 67: 6477–6483 [DOI] [PubMed] [Google Scholar]
- Eeckhoute J, Lupien M, Meyer CA, Verzi MP, Shivdasani RA, Liu XS, Brown M (2009) Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res 19: 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham PJ (2009) Insights from genomic profiling of transcription factors. Nat Rev Genet 10: 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasor J, Danes JM, Komm B, Chang KCN, Lyttle R, Katzenellenbogen BS (2003) Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144: 4562–4574 [DOI] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY et al. (2009) An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462: 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M, Derocq D, Freiss G, Rochefort H (1992) Activation of estrogen receptor transfected into a receptor-negative breast cancer cell line decreases the metastatic and invasive potential of the cells. Proc Natl Acad Sci USA 89: 11538–11542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Hua S, Kittler R, White KP (2009) Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell 137: 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS (2011) FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43: 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph R, Orlov YL, Huss M, Sun W, Kong SL, Ukil L, Pan YF, Li G, Lim M, Thomsen JS, Ruan Y, Clarke ND, Prabhakar S, Cheung E, Liu ET (2010) Integrative model of genomic factors for determining binding site selection by estrogen receptor α. Mol System Biol 6: 456; DOI: 10.1038/msb2010109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner KH (2000) The hepatocyte nuclear factor 3 (HNF3 or FOXA) family in metabolism. Trends Endocrinol Metab 11: 281–285 [DOI] [PubMed] [Google Scholar]
- Kao J, Salari K, Bocanegra M, Choi Y, Girard L, Gandhi J, Kwei KA, Hernandez-Boussard T, Wang P, Gazdar AF, Minna JD, Pollack JR (2009) Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. Plos One 4: e6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia N-Y, Jeyakani J, Hwang C, Lu X, Chan Y-S, Ng H-H, Bourque G (2010) Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 42: 631–634 [DOI] [PubMed] [Google Scholar]
- Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi H-S, Tennakoon C, Wei C-L, Ruan Y, Sung W-K (2010) ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol 11: R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, Yeo A, George J, Kuznetsov VA, Lee YK, Charn TH, Palanisamy N, Miller LD, Cheung E, Katzenellenbogen BS, Ruan Y et al. (2007) Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet 3: e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M (2008) FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S et al. (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra R, Varambally S, Ding L, Shen R, Sabel MS, Ghosh D, Chinnaiyan AM, Kleer CG (2005) Identification of GATA3 as a breast cancer prognostic marker by global gene expression meta-analysis. Cancer Res 65: 11259–11264 [DOI] [PubMed] [Google Scholar]
- Nakshatri H, Badve S (2009) FOXA1 in breast cancer. Expert Rev Mol Med 11: e8. [DOI] [PubMed] [Google Scholar]
- Robert T, Tom M (1994) Transcriptional activation: a complex puzzle with few easy pieces. Cell 77: 5–8 [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T (1995) Virus induction of human IFN-β gene expression requires the assembly of an enhanceosome. Cell 83: 1091–1100 [DOI] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A (2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 13: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BJ, Giguere V (2008) Meta-analysis of human cancer microarrays reveals GATA3 is integral to the estrogen receptor alpha pathway. Mol Cancer 7: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf I, Bose S, Williamson EA, Miller CW, Karlan BY, Koeffler HP (2007) FOXA1: growth inhibitor and a favorable prognostic factor in human breast cancer. Int J Cancer 120: 1013–1022 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chang CW, Goh WL, Sung WK, Cheung E (2011) CENTDIST: discovery of co-associated factors by motif distribution. Nucleic Acids Res 39(Suppl 2): W391–W399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao HH, Herrera RE, Coronado-Heinsohn E, Yang MC, Ludes-Meyers JH, Seybold-Tilson KJ, Nawaz Z, Yee D, Barr FG, Diab SG, Brown PH, Fuqua SA, Osborne CK (2001) Forkhead homologue in rhabdomyosarcoma functions as a bifunctional nuclear receptor-interacting protein with both coactivator and corepressor functions. J Biol Chem 276: 27907–27912 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Tables SI-VI, Supplemental Figures S1-16
List of primers used in ChIP-qPCR study
List of E2 up-regulated genes in MCF-7 cells
List of E2 down-regulated genes in MCF-7 cells
List of all the E2-regulated genes in the transfected MDA-MB-231 cells compare to MCF-7 cells
List of previously reported E2-regulated genes that regulate in our MCF-7 and transfected MDA-MB-231-ER+FOXA1+GATA3 cells
The expression of luminal and basal marker genes in MCF-7 and transfected MDA-MB-231 cells
List of luminal and basal marker genes with ER, FOXA1 and GATA3 binding