Evolutionary conservation is widely used for assigning functions to proteins and genomic sequences. A complementary approach is discussed here whereby patterns of evolutionary divergence in regulatory programs provide novel insights into the mechanisms controlling gene expression.
Keywords: evolution, gene regulation, genomics
Abstract
The number of sequenced species is increasing at a staggering rate, calling for new approaches for incorporating evolutionary information in the study of biological mechanisms. Evolutionary conservation is widely used for assigning a function to new proteins and for predicting functional coding or non-coding sequences. Here, we argue for a complementary approach that focuses on the divergence of regulatory programs. Regulatory mechanisms can be learned from patterns of evolutionary divergence in regulatory properties such as gene expression, transcription factor binding or nucleosome positioning. We review examples of this concept using yeast as a model system, and highlight a hybrid-based approach that is highly instrumental in this analysis.
The basic approach: why comparing related species could help in identifying regulatory mechanisms
The ability to rapidly and cheaply sequence full genomes is revolutionizing biological research. However, the static genomic sequences conceal a highly dynamic program of DNA-associated processes such as transcription, binding of transcription factors or positioning of nucleosomes. This ‘regulatory program' is a defining property of an organism. Understanding this regulation and how it can be predicted from the genomic sequence are key challenges of genomic research. Comparative genome analysis of related species is highly instrumental in this regard, as it is used routinely for identifying conserved sequences and by that to predict cis-regulatory elements and other functional non-coding sequences (Kellis et al, 2003; Xie et al, 2005). Here, we will discuss the possibility of extending this comparative approach in two ways: first, by comparing also the regulatory programs of different species or strains (rather than just their genomic sequences) and second, by focusing on evolutionary divergence (rather than conservation).
The question of how gene expression evolves between closely related species has been studied extensively (King and Wilson, 1975; Carroll, 2005). Widespread inter-species differences were identified in high-throughput comparisons (Rifkin et al, 2003; Gilad et al, 2006; Khaitovich et al, 2006; Tirosh et al, 2006; Yanai and Hunter, 2009), and expression profiles vary greatly even between strains (or individuals) of the same species (Brem et al, 2002; Oleksiak et al, 2002; Denver et al, 2005; Kliebenstein et al, 2006; Landry et al, 2006). Evolutionary differences were also identified in other genomic properties, including transcription factor (TF) binding sites (Borneman et al, 2007; Kasowski et al, 2010; Schmidt et al, 2010; Zheng et al, 2010), nucleosome positioning (Field et al, 2009; Tsankov et al, 2010; Tirosh et al, 2010b), histone modifications (Nagarajan et al, 2010) and protein phosphorylation (Beltrao et al, 2009), leading to the conclusion that regulatory programs change quite rapidly.
Divergence of regulatory programs is encoded in the divergence of the genomic sequences. Inter-species comparison, therefore, provides a way to connect sequence and regulatory divergence. In this view, analyzing how the regulatory program diverged is analogous to studying how the regulatory program responds to multiple genetic perturbations. However, while genetic perturbations are normally studied one at a time (e.g., single-gene knockout), inter-species analysis uncovers the combined effects of numerous genetic perturbations. The disadvantage of this approach is that it becomes difficult to associate a regulatory change with specific genetic (sequence) perturbation. The advantage is that by combining many genetic perturbations, one can uncover general trends connecting the genetic perturbations with the regulatory programs.
In the sections below, we provide the following examples which employed this idea:
Patterns of expression divergence revealed a promoter architecture that governs gene expression variability.
Patterns of expression divergence in strains deleted of chromatin regulators revealed that chromatin regulators function as genetic capacitors of gene expression.
Patterns of divergence of antisense-containing genes suggested that antisense transcription induces a threshold-dependent transcriptional switch.
Patterns of divergence in nucleosome positioning describes both cis and trans influences on nucleosome positioning.
Patterns of divergence of mRNA degradation rate suggested a mechanistic coupling between transcription and mRNA degradation.
Patterns of allelic expression in a cross between diverged mice strains indicated instances of genomic imprinting.
The first five examples used related budding yeast strains (#3), or species (#1, 2, 4, 5) which share approximately the same set of genes, are almost completely syntenic and have high sequence similarity (∼80–90% identical nucleotide sequences; Kellis et al, 2003). The last example used different mice strains. Before describing these examples in detail, we discuss two general issues. First, we consider the relative contribution of natural selection (versus random drift) in shaping the observed patterns of divergence. Second, we discuss the general ‘hybrid approach' for dissecting regulatory changes into those that are generated in cis versus those generated in trans.
The contributions of positive selection versus neutral drift in shaping the regulatory divergence
As described above, we view the regulatory divergence between species as the result of a large collection of mutations. This enables us to deduce general trends connecting changes in genomic sequence to changes in the regulatory program. An underlying assumption here is that mutations accumulate largely at random. Yet, at least in some cases, recurrent patterns of regulatory divergence may reflect the signature of natural selection rather than the outcome of a random sampling of genetic mutations. For example, if certain aspects of regulation undergo frequent adaptive evolution (or conversely are kept constant by purifying selection) then these may appear as a recurrent pattern.
The contribution of positive selection to evolution of gene regulation received much attention (Khaitovich et al, 2004; Yanai et al, 2004; Jordan et al, 2005). The emerging conclusion is that adaptive regulatory changes are an exception rather than the rule, and that most of the observed regulatory changes are neutral. First, regulatory changes among closely related species are much more widespread than expected from the apparent differences in physiology between the species (often encompassing half of all genes) and second, most regulatory changes are small in magnitude and therefore are unlikely to carry phenotypic consequences: typical yeast inter-species differences in gene expression are in the order of 1.5-fold (Tirosh et al, 2009b), while a 2-fold reduction (heterozygote deletion) in most yeast genes has no effect on growth rates (Giaever et al, 1999). These and other considerations (Khaitovich et al, 2006) suggest that most of the changes observed in the regulatory program are neutral and therefore that the recurrent patterns may serve as good proxies for identifying regulatory mechanisms. Nevertheless, a ‘selective' explanation should still be considered when inferring a regulatory mechanism, as described in each of the sections below.
The ‘hybrid approach' for dissecting regulatory changes
Another key challenge is to associate regulatory changes with the causal genetic mutations. One way of doing that is using the genetical-genomics approach (Rockman and Kruglyak, 2006). The idea is to perform linkage analysis, comparing the segregation of regulatory changes with that of sequence polymorphisms in a panel of dozens of segregates from a cross between two strains. This approach is highly informative, but is also labor intensive. Furthermore, it can only be used for analyzing differences between strains of the same species but not for analyzing inter-species differences, as, by definition, inter-specific F1 hybrids are sterile and do not produce F2 segregants.
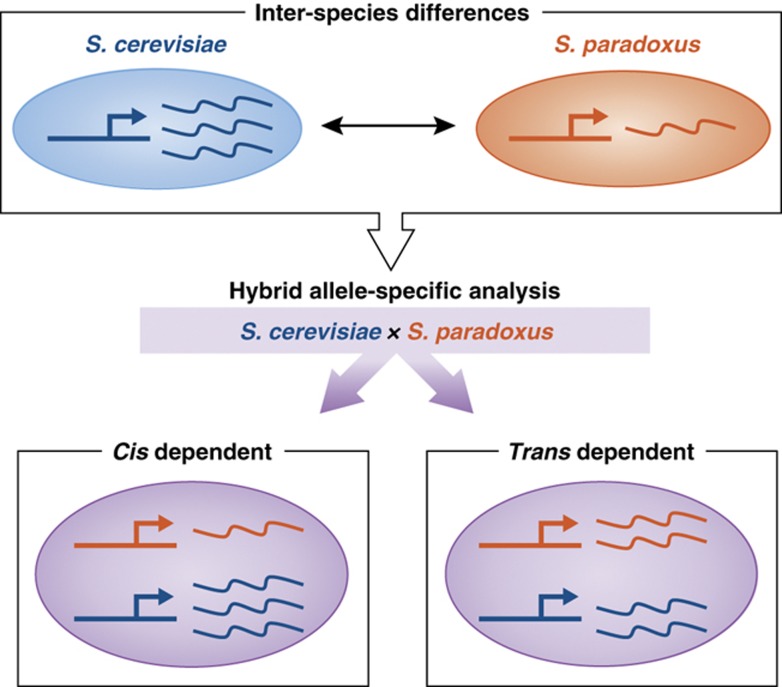
An alternative general approach which was used extensively in the studies we describe below is to ‘mix' the two genomes within one organism. This is done by generating a hybrid, which contains full copies of both genomes (Wittkopp et al, 2004; Figure 1). In the context of the hybrid, orthologous genes appear as different alleles of the same gene, and are subject to regulation by the same trans environment (e.g., same regulatory proteins). Thus, allele-specific differences in regulation must reflect mutations that are linked in cis to the gene itself and distinguish between its two orthologous alleles (i.e., mutations in the gene or its flanking regulatory sequences). The additional differences that are observed between orthologs in the two species, but not within the hybrid, correspond to the effects of trans mutations.
Figure 1.
Hybrid analysis distinguish between the effects of mutations in cis or trans. Comparison of gene expression between two species reveals differences due to the combined effects of cis and trans mutations (top). Analysis of a hybrid formed by mating the two species allows classification of the differences into those due to cis-acting and trans-acting mutations: two hybrid alleles (that correspond to orthologous genes of the parental species) reside in the same nucleus and are regulated by the same set of trans factors, thus avoiding any differential expression due to trans-acting mutations (bottom right). However, cis-acting mutations discriminate between the two alleles and thus maintain the inter-species differences (bottom left).
Genome-wide analysis of allele-specific expression was initially performed in humans (Yan et al, 2002; Verlaan et al, 2009), followed by extensive analysis of intra- and inter-specific F1 hybrids in flies (Wittkopp et al, 2004, 2008; Landry et al, 2005; McManus et al, 2010), yeasts (Sung et al, 2009; Tirosh et al, 2009b; Emerson et al, 2010a) and mice (Wang et al, 2010; Gregg et al, 2010a, 2010b). A related approach involves the substitution of individual chromosomes between flies (Lemos et al, 2008; Wang et al, 2008). Similarly, a mouse model of Down syndrome that contains an extra human chromosome 21 was used to study the role of cis- and trans-regulatory mutations in mammals (Wilson et al, 2008).
Collectively, these studies demonstrate that inter-species expression divergence is dominated by cis mutations, in contrast to expression divergence between strains, which is generated primarily by trans mutations (Wittkopp et al, 2008). At individual genes, the distinction between cis and trans effects is not sufficient to identify the causal mutations, but provides a useful classification of regulatory changes that hints where to look for the specific mutations. Importantly, cis mutations affecting different genes reflect independent evolutionary events and this can increase the significance of observed recurrent patterns (Bullard et al, 2010). A more complete discussion on the properties and roles of cis- and trans-acting mutations can be found in recent reviews (Thompson and Regev, 2009; Emerson and Li, 2010b).
‘Expression variability' and how it is linked to promoter architecture
Genes diverge at different rates. This was first observed in analysis of protein sequences: some proteins diverge quickly, whereas others remain largely the same between organisms. There are multiple reasons for that (Pal et al, 2006). For example, essential proteins are relatively conserved, likely due to the need to maintain their function. Highly expressed proteins are especially well conserved, possibly because they are subject to more stringent constraints for proper folding (Drummond and Wilke, 2008). One of the first questions that were asked when comparing the regulatory program is whether these differences in evolutionary rates generalize also to regulatory properties.
Not surprisingly, the rate by which gene expression diverges also differs between genes (Figure 2A). What was surprising, though, was the low correlation between this rate and evolutionary rate of the associated proteins. Although weak correlation was observed among mammalians (Khaitovich et al, 2005), no correlation was identified among yeasts (Tirosh and Barkai, 2008a). Thus, the fact that proteins are conserved in sequence did not imply their conservation in expression. Similarly, genes that diverged rapidly in expression often encoded for proteins with conserved sequence. The two modes of evolution, therefore, reflect distinct constraints. In yeast, only essentiality correlated with conservation both in expression and in sequence, while all other determinants were specific to one mode of evolution (Tirosh and Barkai, 2008a). Additional determinants that were either common or specific to the two modes of evolution were found in flies (Lemos et al, 2005) and in mammals (Liao and Zhang, 2006).
Figure 2.
Expression divergence reflects an intrinsic tendency for expression variability that is encoded in promoter structure. (A) Comparison of genome-wide expression patterns among multiple species identifies genes whose expression patterns remain well conserved (low divergence) and genes whose expression diverged extensively (high divergence). (B) Expression divergence is correlated with responsiveness to environmental changes (data taken from Tirosh et al, 2006), and both measures are higher among OPN genes that contain a TATA-box (purple) than among DPN genes that lack a TATA-box (cyan) (Tirosh and Barkai, 2008b); empty black circles represent genes which do not fit with these two gene classes (e.g., intermediate nucleosome pattern between DON and OPN). (C) Schemes depicting the typical promoter structure of OPN genes with a TATA-box (top, purple) and DPN genes without a TATA-box (bottom, cyan). OPN promoters lack NFR, contain multiple TF binding sites (squares), a TATA-box and fuzzy nucleosomes (i.e., nucleosome positions vary across time and within a population; marked with a double-headed arrow). DPN promoters contain NFR, fewer TF binding sites and no TATA-box, and well-positioned nucleosomes.
One of the notable findings was that divergence of gene expression strongly correlates with other measures of expression variability on completely different time scales (Tirosh et al, 2006; Figure 2B). Genes that diverged rapidly in expression tended also to vary more when environmental conditions were modified. Moreover, these genes were more ‘noisy', displaying a wider variance in expression between identical cells subject to the same conditions (Newman et al, 2006). These observations suggested that evolutionary divergence is one facet of a more general property of expression variability: some genes are capable of broad changes in expression, whereas in others this capacity is limited (Tirosh et al, 2009a). Importantly, a ‘selective' explanation to the increased expression divergence of these genes was ruled out as these results were reproduced in analysis of mutation accumulation strains: strains that evolved in the laboratory while maintaining very low effective population sizes, thereby allowing non-lethal mutations to accumulate randomly with minimal effects of natural selection (Denver et al, 2005; Rifkin et al, 2005; Landry et al, 2007).
If expression variability is indeed a general property that differs between genes, it might be encoded in the promoter sequence. Indeed, bioinformatics analysis revealed a strong association between expression variability (on all time scales) and particular promoter structures (Figure 2B and C). First, promoters of variable genes have a TATA-box at much higher frequency than less-variable genes (Tirosh et al, 2006). Second, the organization of nucleosomes in variable genes typically lacks the Nucleosome Free Region (NFR) immediately upstream of the transcription start site, which is a characteristic of most genes. We, therefore, denoted this promoter structure as ‘OPN' for Occupied Proximal Nucleosome, in contrast to Depleted Proximal Nucleosome (DPN) genes (Tirosh and Barkai, 2008b). Notably, the association of this promoter organization with expression variability was not specific to yeast as similar observations were made in other organisms (Tirosh and Barkai, 2008b; Gilchrist et al, 2010).
A key challenge will be to understand how TATA and OPN promoters support higher expression variability. A hint to this question comes from the hybrid approach for distinguishing cis and trans effects. Although most of the expression divergence between S. cerevisiae and S. paradoxus reflect cis effects, the increased divergence associated with TATA and OPN was instead due to trans effects (Tirosh et al, 2009b). Coupled with the observation that these promoters are typically bound by (and affected by deletion of) a larger number of regulators compared with other promoters (Landry et al, 2007; Choi and Kim, 2008; Tirosh and Barkai, 2008b; Venters et al, 2011), these results suggest that there are simply more regulators (and thus possible trans-mutations) affecting these promoters. For example, in the OPN organization binding sites are more often covered by nucleosomes, an organization that may facilitate competition between the binding of TFs and nucleosomes and could thus increase the influence of various chromatin regulators.
Differences in expression variability between genes may be related to the general interplay between ‘robustness' and ‘evolvability'. An organism needs to be robust, namely maintain a reliable function under different conditions or when subjected to mutations. At the same time, it needs to maintain the ability to evolve in order to adapt to new environments, but this requires sensitivity to genetic mutations. The ability to control the plasticity of gene expression through its promoter structure may help in this interplay. Accordingly, genes that are required for the robustness of the organism will be maintained as lowly variable and their expression program will evolve slowly, whereas those that facilitate adaptation to new environments will be maintained as highly variable. In support of that, we observed that in yeast, genes of high expression variability and those with TATA and OPN promoters preferentially encoded proteins that interact with the environment, such as transporters, and as such may mediate the response and adaptation to environmental changes (Tirosh et al, 2006, 2009a; Zhang et al, 2009b).
Chromatin regulators as ‘capacitors' of gene expression variations
Another idea that arose from thinking about the interplay between robustness and evolvability is that of ‘genetic capacitors'. Robustness of the wild-type organism may be facilitated by proteins that act as ‘genetic capacitors' to reduce the effect of mutations. These capacitors enable the accumulation of mutations, or polymorphisms, that have no phenotypic consequences in normal conditions. The thought is that these mutations can support evolvability if capacitors are repressed under harsh conditions, so that the phenotypic effect of the accumulated mutations suddenly emerges. This model was proposed many years ago (Waddington, 1942), but only recently a striking example for a candidate ‘genetic capacitor', the heat-shock protein Hsp90, was identified (Rutherford and Lindquist, 1998; Queitsch et al, 2002).
A central question is whether chaperones, such as Hsp90, are unique in their capacity to buffer mutations or whether other protein capacitors can be identified. One line of thought suggested that, in fact, any regulatory protein with large-scale effects may serve in this capacity (Bergman and Siegal, 2003; Hermisson and Wagner, 2004; Levy and Siegal, 2008). The reason is that in the complex regulatory networks of cells, many epistatic effects are expected which means that mutations which do not have an effect in the wild-type organism, might still have a phenotypic consequence in the background of additional mutation. Large-scale regulators are particularly expected to be involved in such epistatic effects, and will therefore behave as effective capacitors: mutations that are neutral in the wild-type background will accumulate, but these mutations may have an effect when one of these regulators will be deleted or its function compromised.
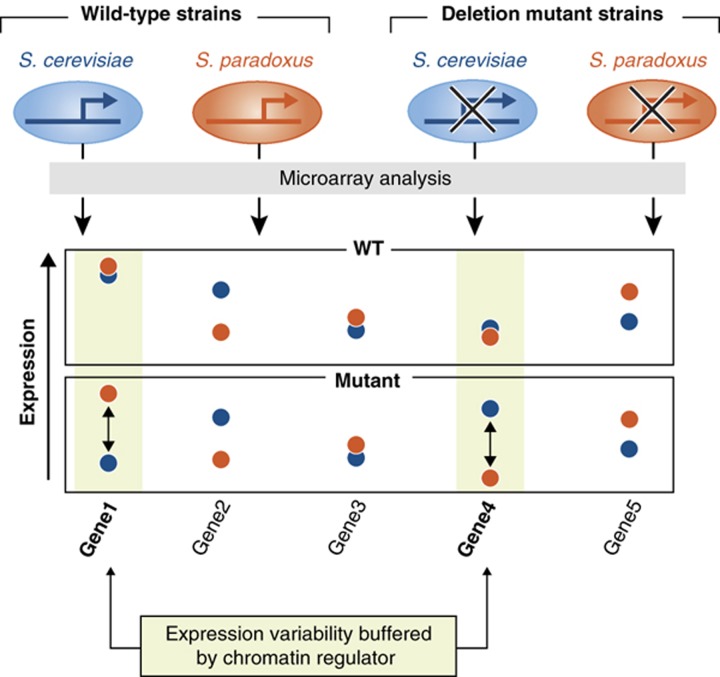
This idea was tested using an inter-species comparative approach (Tirosh et al, 2010a). The two yeast species, S. cerevisiae and S. paradoxus, differ in sequence and expression. The buffering hypothesis predicts, however, that many of the sequence differences are in fact buffered and therefore do not affect expression in wild-type cells. An expression effect will be observed when the capacitor protein is deleted, revealing the impact of hidden genetic changes. Comparing the expression profiles of S. cerevisiae and S. paradoxus, both for wild-type strains and for strains where specific chromatin regulators have been deleted, confirmed that this is the case (Figure 3). Deletions of each of the chromatin regulators that were examined had increased the amount of inter-species expression differences, consistent with the regulators acting as capacitors of gene expression variations. Furthermore, the hybrid analysis confirmed that these regulators buffer gene expression by acting in trans, as expected if they act primarily by influencing upstream regulatory signals.
Figure 3.
Chromatin regulators buffer gene expression variability. Comparison of inter-species expression differences between wild-type and (chromatin regulators) deletion strains shows increased expression differences among the deletion strains, indicating that chromatin regulators normally buffer the effects of hidden inter-species genetic variability.
Note that in this example we support a ‘selective' explanation whereby natural selection generated a bias toward mutations that affect gene expression in a mutant background, but not in the wild-type. Nonetheless, these results demonstrate that chromatin regulators effectively buffer gene expression and therefore that compromising their activity exposes hidden genetic variability.
Function of antisense transcription
Antisense transcription occurs frequently throughout the genomes of various organisms (He et al, 2008; Guell et al, 2009; Xu et al, 2009; Yassour et al, 2011). Several studies have shown that antisense transcription can, in certain cases, repress transcription of the sense genes by several mechanisms (Hongay et al, 2006; Camblong et al, 2007; Berretta et al, 2008), yet the frequency and mode of such repression remain poorly understood. Notably, the repressive effect of antisense on the sense transcript cannot be determined directly from steady-state expression levels but requires analysis of sense and antisense expression levels upon perturbations.
Steinmetz and colleagues compared sense and antisense expression among 2 diverged S. cerevisiae strains and 48 of their segregants, thus revealing the effects of numerous genetic changes (Xu et al, 2011). This analysis showed that genes associated with antisense transcription show an increased variability in expression among segregants, suggesting an effect similar to that of the TATA-box and occupied nucleosome patterns described above. Interestingly, increased variability of antisense-containing genes was due to similar induction but more efficient repression: antisense-containing genes were often completely ‘switched-off', while repression of other genes was more limited. In contrast, the induction (i.e. maximal levels) of antisense-containing genes did not differ from those of other genes, and thus the dynamic range and the variability of antisense-containing genes was typically larger than that of other genes. A ‘selective' explanation to this effect (i.e. that natural selection favors mutations that repress antisense-containing genes) seems unlikely, and instead these results suggest a model in which antisense transcription induces a threshold-dependent switch of sense transcription. According to this model, low sense transcription (e.g., in the absence of activation) is easily inhibited by the antisense transcription, but higher sense transcription (e.g., upon activation) ‘overcomes' this inhibition and eliminates the antisense effect. This model was further supported by direct experiments which demonstrated an inhibitory antisense effect that is abolished upon induction of the sense gene (Xu et al, 2011). In future work, it might also be useful to compare sense and antisense expression among different species and their hybrid.
Characterizing determinants of nucleosome positioning
Most studies on regulatory evolution focused on gene expression but recent work began to extend these studies to other properties such as TF binding, histone modifications and nucleosome occupancy. We will focus here on comparative studies of nucleosome occupancy, which provided insights into mechanisms that determine the positioning of nucleosomes along the genome.
Nucleosomes, the basic building block of chromatin, decrease DNA accessibility and thus the ability of regulatory proteins to bind specific DNA regions and exert their regulatory function (Li et al, 2007). A key issue is what determines the positioning of nucleosomes along the DNA. In particular, there is a contemporary debate about the relative importance of the local DNA sequence (the ‘genomic code'; Segal et al, 2006), compared with the contribution of trans factors such as chromatin remodelers, modifiers and TFs (Kaplan et al, 2009; Zhang et al, 2009a).
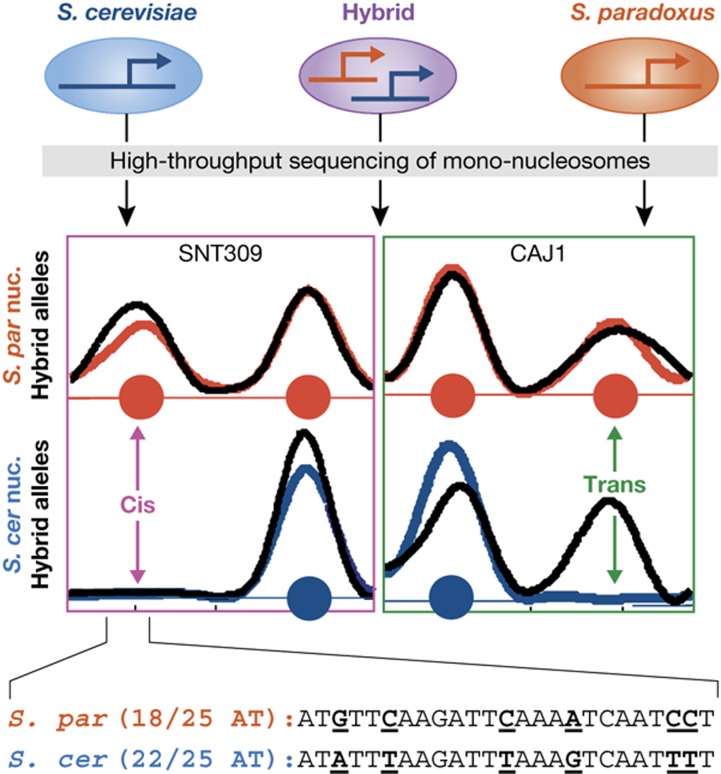
Comparative analysis provided insights into these questions (Tirosh et al, 2010b; Figure 4). Nucleosome patterns of S. cerevisiae differ in many sites from the orthologous S. paradoxus patterns. Notably, the hybrid analysis mapped ∼70% of the differences as cis (i.e., resulting from changes in the local DNA sequence) and ∼30% as trans (i.e., resulting from mutations that effect regulatory proteins). These results provide a general estimation for the relative roles of local DNA sequence versus regulatory proteins in controlling nucleosome positioning, although this analysis might be biased by natural selection. The ability to compare genomic sequences at positions of cis-dependent differences was further informative for evaluating the role of sequence patterns in controlling nucleosome positions: among the various patterns that were proposed to control nucleosome positioning, only the presence of AT-rich elements had a significant effect, suggesting that AT-rich elements are the dominant feature of local DNA sequence with respect to nucleosome positioning. A ‘selective' explanation is highly unlikely in this case, as variation in other sequence patterns was observed but was not associated with changes in nucleosome positioning. Recent analysis of sequence-derived models of nucleosome positioning has independently reached a similar conclusion (Tillo and Hughes, 2009).
Figure 4.
Inter-species and hybrid analysis uncovers determinants of nucleosome positioning. Inter-species comparison (blue and red correspond to S. cerevisiae and S. paradoxus, respectively) and hybrid analysis (black curves) of nucleosome positioning characterizes changes due to cis and trans mutations (Tirosh et al, 2010b). Nucleosomes found in only one of the species and in the corresponding hybrid allele reflect the effect of local (cis) mutations (right). Nucleosome found in only one of the species but in both hybrid alleles reflect the trans effect of distal mutations through the activity of a chromatin-related protein or RNA (left). Sequence analysis at positions of cis changes can suggest which sequence patterns influence nucleosome positioning. For example, the inset shows sequences of the two species within a region that is bound by a nucleosome only in S. paradoxus, demonstrating that inter-species substitutions changed the frequency of AT bases between 22/25 (S. cerevisiae) and 18/25 (S. paradoxus). This is consistent with a nucleosome-disfavoring effect of AT-rich sequences, as observed systematically for inter-species nucleosome differences, and is also observed in other studies (Tillo and Hughes, 2009).
Interestingly, local divergence of AT-rich sequences not only affected the positioning of the closest nucleosomes but also influenced the positioning of multiple surrounding nucleosomes (Tirosh et al, 2010b). A large (∼50 bp) shift in the position of a single nucleosome was typically associated with gradually smaller shifts of several (∼3–6) adjacent nucleosomes at both directions. Furthermore, the large shifts were normally consistent with the predicted effect of local sequence changes, while the smaller shifts of adjacent nucleosomes were not, suggesting that sequence-dependent changes in the positioning of a single nucleosome (e.g., by nucleosome-disfavoring sequences) propagated to adjacent nucleosomes. Such propagation is consistent both with the statistical positioning model (Kornberg and Stryer, 1988; Mavrich et al, 2008) and with active nucleosome packing by chromatin remodeling (Zhang et al, 2011). Regardless of the specific mechanism involved, the widespread propagation of nucleosome shifts indicates that apart from direct regulation by cis (sequence) and trans (regulators) elements, nucleosome positioning depends on the state of the surrounding chromatin.
Coupling of transcription and mRNA degradation
Gene expression reflects the combined influence of a multitude of regulatory processes, and in recent years it has become clear that many of these apparently discrete processes are in fact coordinated (Maniatis and Reed, 2002; Proudfoot et al, 2002; Hagiwara and Nojima, 2007; Nagaike et al, 2011). Interestingly, in addition to coordination between consecutive steps, several observations have suggested that transcription in the nucleus may be coupled to the degradation of mRNAs in the cytoplasm. Such coupling was observed in the response of yeast to stresses (Shalem et al, 2008), and is supported by the finding that individual protein complexes regulate both transcription and mRNA degradation (Rpb4/7 and Ccr4-Not) (Collart, 2003; Goler-Baron et al, 2008). However, the scope of this coordination and its underlying mechanisms are poorly understood.
Comparative analysis of mRNA degradation rates and mRNA levels between S. cerevisiae and S. paradoxus supported this notion (Dori-Bachash et al, 2011). This analysis demonstrated that most genes that diverged in mRNA degradation rates also diverged in transcription. The mode of this coordination was surprising: increased mRNA degradation was typically associated with increased (rather than decreased) mRNA levels, indicating that transcriptional changes generated opposite effects to those of mRNA degradation (Figure 5). The more intuitive mode of coordination, whereby transcription and mRNA degradation act together to either increase or decrease mRNA levels, was observed only for few genes.
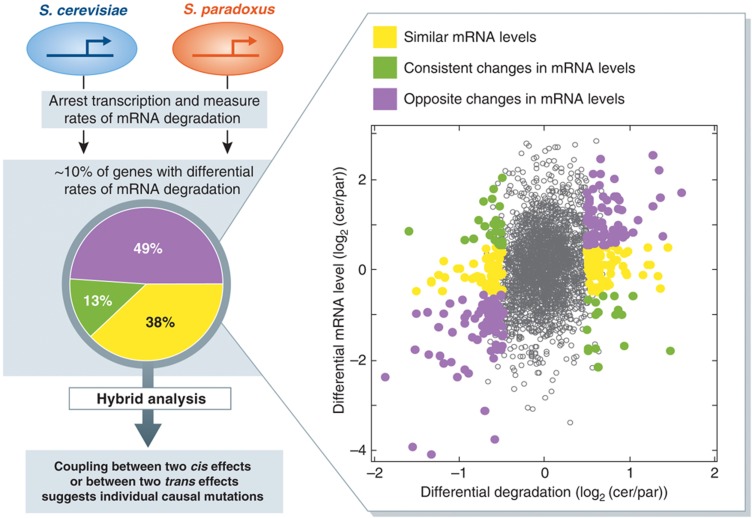
Figure 5.
Inter-species comparison of mRNA degradation rates suggests a mechanistic coupling between transcription and mRNA degradation. Transcription of S. cerevisiae and S. paradoxus was chemically arrested and mRNA levels were measured at several time points after transcription arrest, enabling the estimation of mRNA degradation rates (Dori-Bachash et al, 2011). Inter-species differential degradation was identified for ∼10% of the genes, and these were further divided into three classes: (i) genes for which mRNA levels did not differ significantly between the two species (yellow); (ii) genes for which mRNA levels differed significantly between the two species in the direction that is consistent with the changes in mRNA degradation, that is, the species with higher degradation rate for a given gene had lower mRNA level for that gene (green); (iii) genes for which mRNA levels differed significantly between the two species but in the opposite direction to that which is expected by the changes in mRNA degradation, that is, the species with higher degradation rate for a given gene had higher mRNA level for that gene (purple). Notably, approximately half of the differentially degraded genes belonged to the purple class, indicating a widespread coupling between opposite effects in transcription and mRNA degradation. Similar analysis of mRNA degradation within the hybrid further indicated that such coupled effects (transcription and mRNA degradation) are almost exclusively due to the same type of mutation (cis or trans), suggesting that the same individual mutations have influenced both processed through a mechanistic coupling (Dori-Bachash et al, 2011).
Coordinated evolutionary changes may reflect co-evolution, that is, independent mutation events whose coordination may be facilitated by natural selection. Alternatively, it could reflect a mechanistic coupling whereby individual mutations generate the two opposing effects of transcription and mRNA degradation. It is this latter possibility that is particularly intriguing, as it may indicate that this unappreciated mechanism of regulation is in fact quite common. To distinguish between these possibilities, we turned to the hybrid approach and examined whether coordinated changes in transcription and mRNA degradation are generated by the same or by different types of mutations (cis or trans) (Dori-Bachash et al, 2011). Strikingly, we found that cis changes in mRNA degradation are coordinated only with cis changes (but not with trans changes) in transcription, and similarly, that trans changes in mRNA degradation are coordinated only with trans changes in transcription (Figure 5). Since co-evolution through natural selection would occur between independent sets of mutations, this result rules out a ‘selective' explanation and suggests that coordinated changes in transcription and mRNA degradation are generated by the same individual mutations.
Analysis of the sets of genes with coupled changes in cis or trans further suggested possible coupling mechanisms (Dori-Bachash et al, 2011). First, trans-coupled genes were associated with the two complexes known to regulate both transcription and mRNA degradation (Rpb4/7 and Ccr4-Not). Second, two additional RNA-binding factors (Npl3 and Pab1) were enriched with trans-coupled genes, consistent with previous findings that these factors shuttle between the nucleus and the cytoplasm (Lei et al, 2001; Brune et al, 2005). Third, cis-coupled promoters were enriched with diverged TF binding sites, suggesting that modulation of transcription regulation in the nucleus (TF binding) may in some cases signal to the mRNA degradation apparatus in the cytoplasm, perhaps through shuttling of transcription-associated molecules. Accordingly, promoters might encode not only for transcription but also for degradation of the associated mRNA.
Characterizing genomic imprinting in the mouse brain
Although we have so far discussed only studies in yeast, the basic concept of analyzing regulatory divergence to elucidate mechanisms of regulation is a general one and can be applied in other species. An important issue that arises in mammalian hybrids is that allele-specific expression differences reflect both the effects of cis-regulatory mutations but also the phenomenon of genomic imprinting, namely the preferential expression of the parental or maternal alleles of certain genes through epigenetic mechanisms (Wood and Oakey, 2006). These effects can be distinguished by using reciprocal crosses, where the parental strain of one cross serves as the maternal strain for the second cross and vice versa. An effect of cis-regulatory mutations should be consistent among the reciprocal crosses, with higher expression of the same allele regardless of its parental origin, while an effect of genomic imprinting should switch between the reciprocal crosses, consistently favoring either the parental or the maternal allele. Allele-specific expression analysis of reciprocal crosses from divergent strains therefore simultaneously characterizes the effects of cis-regulatory mutations and those of genomic imprinting.
Recent studies used this approach to gain insight into the scope and properties of genomic imprinting in the mouse brain (Babak et al, 2008; Gregg et al, 2010a, 2010b; Wang et al, 2010). Note that in this case divergence between mouse strains was not studied directly but was required in order to discriminate between alleles and identify instances of genomic imprinting. Imprinting was observed at >1300 loci, indicating a widespread influence on gene regulation (Gregg et al, 2010b). Most imprinted loci show moderate preferential expression of one allele, rather than strict monoallelic expression, and reside within clusters of imprinted genes. Interestingly, notable differences were observed between brain regions and developmental stages. For example, expression was biased toward the maternal allele in the developing brain, but toward the paternal allele in the adult brain. In addition to biasing gene expression according to the sex of the parent, genomic imprinting also depended on the sex of the offspring: imprinting in the hypothalamus was more common in females than in males, and the female-specific imprinting was preferentially biased toward the parental allele, perhaps indicating increased parental influence on certain brain functions in females (Gregg et al, 2010a).
Concluding remarks
Most studies comparing gene expression in related species were motivated by two challenges: to uncover conserved patterns inferred to be functionally important and to discover adaptive evolutionary changes that drive new phenotypes. Here, we presented a complementary approach that uses evolutionary comparisons as means for obtaining basic insights on gene regulation. This approach builds on the fact that such comparisons expose the regulatory effects of thousands of genetic changes at a single experiment. We described five studies that used this approach in yeast and a related approach that was employed in mice. Crucial to these studies was the analysis of allele-specific expression in hybrids to classify the regulatory changes into those due to mutations in cis or trans.
We anticipate that future studies will continue to explore the evolution of diverse regulatory mechanisms and further expand to additional regulatory processes such as translational control and protein–protein interactions. Recent studies began to examine the evolutionary divergence of protein abundance, demonstrating significant differences from divergence of mRNA levels (Foss et al, 2007; Fu et al, 2007; Laurent et al, 2010). Integration of various data sets of regulatory divergence would be needed to bridge this gap and understand how different mechanisms of regulation contribute to divergence of the final outcome, namely protein levels and activity. Such integrative analysis would uncover interplay and coordination among regulatory processes.
Acknowledgments
We thank members of the Barkai laboratory for helpful discussions. This work was supported by the NIH (P50GM068763), the Binational Science Foundation and the European Research Council (Ideas) (to NB). IT is supported by the Clore center at the Weizmann Institute of Science.
Footnotes
The authors declare that they have no conflict of interest.
References
- Babak T, Deveale B, Armour C, Raymond C, Cleary MA, van der Kooy D, Johnson JM, Lim LP (2008) Global survey of genomic imprinting by transcriptome sequencing. Curr Biol 18: 1735–1741 [DOI] [PubMed] [Google Scholar]
- Beltrao P, Trinidad JC, Fiedler D, Roguev A, Lim WA, Shokat KM, Burlingame AL, Krogan NJ (2009) Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol 7: e1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman A, Siegal ML (2003) Evolutionary capacitance as a general feature of complex gene networks. Nature 424: 549–552 [DOI] [PubMed] [Google Scholar]
- Berretta J, Pinskaya M, Morillon A (2008) A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev 22: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, Wang LY, Gerstein M, Snyder M (2007) Divergence of transcription factor binding sites across related yeast species. Science 317: 815–819 [DOI] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L (2002) Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755 [DOI] [PubMed] [Google Scholar]
- Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K (2005) Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA 11: 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard JH, Mostovoy Y, Dudoit S, Brem RB (2010) Polygenic and directional regulatory evolution across pathways in Saccharomyces. Proc Natl Acad Sci USA 107: 5058–5063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F (2007) Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 131: 706–717 [DOI] [PubMed] [Google Scholar]
- Carroll SB (2005) Evolution at two levels: on genes and form. PLoS Biol 3: e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Kim YJ (2008) Epigenetic regulation and the variability of gene expression. Nat Genet 40: 141–147 [DOI] [PubMed] [Google Scholar]
- Collart MA (2003) Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313: 1–16 [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Streelman JT, Kim SK, Lynch M, Thomas WK (2005) The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet 37: 544–548 [DOI] [PubMed] [Google Scholar]
- Dori-Bachash M, Shema E, Tirosh I (2011) Coupled evolution of transcription and mRNA degradation. PLoS Biol 9: e1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO (2008) Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134: 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Hsieh LC, Sung HM, Wang TY, Huang CJ, Lu HH, Lu MY, Wu SH, Li WH (2010a) Natural selection on cis and trans regulation in yeasts. Genome Res 20: 826–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Li WH (2010b) The genetic basis of evolutionary change in gene expression levels. Philos Trans R Soc Lond B Biol Sci 365: 2581–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Fondufe-Mittendorf Y, Moore IK, Mieczkowski P, Kaplan N, Lubling Y, Lieb JD, Widom J, Segal E (2009) Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat Genet 41: 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss EJ, Radulovic D, Shaffer SA, Ruderfer DM, Bedalov A, Goodlett DR, Kruglyak L (2007) Genetic basis of proteome variation in yeast. Nat Genet 39: 1369–1375 [DOI] [PubMed] [Google Scholar]
- Fu N, Drinnenberg I, Kelso J, Wu JR, Paabo S, Zeng R, Khaitovich P (2007) Comparison of protein and mRNA expression evolution in humans and chimpanzees. PLoS One 2: e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, Davis RW (1999) Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet 21: 278–283 [DOI] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP (2006) Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature 440: 242–245 [DOI] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K (2010) Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143: 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goler-Baron V, Selitrennik M, Barkai O, Haimovich G, Lotan R, Choder M (2008) Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev 22: 2022–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C (2010a) Sex-specific parent-of-origin allelic expression in the mouse brain. Science 329: 682–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C (2010b) High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science 329: 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guell M, van Noort V, Yus E, Chen WH, Leigh-Bell J, Michalodimitrakis K, Yamada T, Arumugam M, Doerks T, Kuhner S, Rode M, Suyama M, Schmidt S, Gavin AC, Bork P, Serrano L (2009) Transcriptome complexity in a genome-reduced bacterium. Science 326: 1268–1271 [DOI] [PubMed] [Google Scholar]
- Hagiwara M, Nojima T (2007) Cross-talks between transcription and post-transcriptional events within a ‘mRNA factory'. J Biochem 142: 11–15 [DOI] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW (2008) The antisense transcriptomes of human cells. Science 322: 1855–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J, Wagner GP (2004) The population genetic theory of hidden variation and genetic robustness. Genetics 168: 2271–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay CF, Grisafi PL, Galitski T, Fink GR (2006) Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127: 735–745 [DOI] [PubMed] [Google Scholar]
- Jordan IK, Marino-Ramirez L, Koonin EV (2005) Evolutionary significance of gene expression divergence. Gene 345: 119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E (2009) The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowski M, Grubert F, Heffelfinger C, Hariharan M, Asabere A, Waszak SM, Habegger L, Rozowsky J, Shi M, Urban AE, Hong MY, Karczewski KJ, Huber W, Weissman SM, Gerstein MB, Korbel JO, Snyder M (2010) Variation in transcription factor binding among humans. Science 328: 232–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254 [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Enard W, Lachmann M, Paabo S (2006) Evolution of primate gene expression. Nat Rev Genet 7: 693–702 [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Paabo S (2005) Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309: 1850–1854 [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Weiss G, Lachmann M, Hellmann I, Enard W, Muetzel B, Wirkner U, Ansorge W, Paabo S (2004) A neutral model of transcriptome evolution. PLoS Biol 2: E132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC (1975) Evolution at two levels in humans and chimpanzees. Science 188: 107–116 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, West MA, van Leeuwen H, Kim K, Doerge RW, Michelmore RW, St Clair DA (2006) Genomic survey of gene expression diversity in Arabidopsis thaliana. Genetics 172: 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, Stryer L (1988) Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res 16: 6677–6690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL (2007) Genetic properties influencing the evolvability of gene expression. Science 317: 118–121 [DOI] [PubMed] [Google Scholar]
- Landry CR, Oh J, Hartl DL, Cavalieri D (2006) Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene 366: 343–351 [DOI] [PubMed] [Google Scholar]
- Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL (2005) Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics 171: 1813–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent JM, Vogel C, Kwon T, Craig SA, Boutz DR, Huse HK, Nozue K, Walia H, Whiteley M, Ronald PC, Marcotte EM (2010) Protein abundances are more conserved than mRNA abundances across diverse taxa. Proteomics 10: 4209–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Krebber H, Silver PA (2001) Messenger RNAs are recruited for nuclear export during transcription. Genes Dev 15: 1771–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Araripe LO, Fontanillas P, Hartl DL (2008) Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proc Natl Acad Sci USA 105: 14471–14476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Bettencourt BR, Meiklejohn CD, Hartl DL (2005) Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Mol Biol Evol 22: 1345–1354 [DOI] [PubMed] [Google Scholar]
- Levy SF, Siegal ML (2008) Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biol 6: e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Liao BY, Zhang J (2006) Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during mammalian evolution. Mol Biol Evol 23: 1119–1128 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Reed R (2002) An extensive network of coupling among gene expression machines. Nature 416: 499–506 [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF (2008) A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Coolon JD, Duff MO, Eipper-Mains J, Graveley BR, Wittkopp PJ (2010) Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res 20: 816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL (2011) Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell 41: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan M, Veyrieras JB, de Dieuleveult M, Bottin H, Fehrmann S, Abraham AL, Croze S, Steinmetz LM, Gidrol X, Yvert G (2010) Natural single-nucleosome epi-polymorphisms in yeast. PLoS Genet 6: e1000913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846 [DOI] [PubMed] [Google Scholar]
- Oleksiak MF, Churchill GA, Crawford DL (2002) Variation in gene expression within and among natural populations. Nat Genet 32: 261–266 [DOI] [PubMed] [Google Scholar]
- Pal C, Papp B, Lercher MJ (2006) An integrated view of protein evolution. Nat Rev Genet 7: 337–348 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ (2002) Integrating mRNA processing with transcription. Cell 108: 501–512 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S (2002) Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624 [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Houle D, Kim J, White KP (2005) A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature 438: 220–223 [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP (2003) Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet 33: 138–144 [DOI] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L (2006) Genetics of global gene expression. Nat Rev Genet 7: 862–872 [DOI] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S (1998) Hsp90 as a capacitor for morphological evolution. Nature 396: 336–342 [DOI] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, Talianidis I, Flicek P, Odom DT (2010) Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J (2006) A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Dahan O, Levo M, Martinez MR, Furman I, Segal E, Pilpel Y (2008) Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol Syst Biol 4: 223 10.1038/msb.2008.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung HM, Wang TY, Wang D, Huang YS, Wu JP, Tsai HK, Tzeng J, Huang CJ, Lee YC, Yang P, Hsu J, Chang T, Cho CY, Weng LC, Lee TC, Chang TH, Li WH, Shih MC (2009) Roles of trans and cis variation in yeast intraspecies evolution of gene expression. Mol Biol Evol 26: 2533–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DA, Regev A (2009) Fungal regulatory evolution: cis and trans in the balance. FEBS Lett 583: 3959–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Hughes TR (2009) G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics 10: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N (2008a) Evolution of gene sequence and gene expression are not correlated in yeast. Trends Genet 24: 109–113 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Barkai N (2008b) Two strategies for gene regulation by promoter nucleosomes. Genome Res 18: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N, Verstrepen KJ (2009a) Promoter architecture and the evolvability of gene expression. J Biol 8: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N (2009b) A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324: 659–662 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Sigal N, Assia Y, Barkai N (2010a) Chromatin regulators as capacitors of interspecies variations in gene expression. Mol Syst Biol 6: 43510.1038/msb.2010.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Sigal N, Barkai N (2010b) Divergence of nucleosome positioning between two closely related yeast species: genetic basis and functional consequences. Mol Syst Biol 6: 36510.1038/msb.2010.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Weinberger A, Carmi M, Barkai N (2006) A genetic signature of interspecies variations in gene expression. Nat Genet 38: 830–834 [DOI] [PubMed] [Google Scholar]
- Tsankov AM, Thompson DA, Socha A, Regev A, Rando OJ (2010) The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol 8: e1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, Pugh BF (2011) A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell 41: 480–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlaan DJ, Ge B, Grundberg E, Hoberman R, Lam KC, Koka V, Dias J, Gurd S, Martin NW, Mallmin H, Nilsson O, Harmsen E, Dewar K, Kwan T, Pastinen T (2009) Targeted screening of cis-regulatory variation in human haplotypes. Genome Res 19: 118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH (1942) Canalization of development and the inheritance of acquired characters. Nature 150: 563–565 [DOI] [PubMed] [Google Scholar]
- Wang HY, Fu Y, McPeek MS, Lu X, Nuzhdin S, Xu A, Lu J, Wu ML, Wu CI (2008) Complex genetic interactions underlying expression differences between Drosophila races: analysis of chromosome substitutions. Proc Natl Acad Sci USA 105: 6362–6367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Soloway PD, Clark AG (2010) Paternally biased X inactivation in mouse neonatal brain. Genome Biol 11: R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VL, Fisher EM, Tavare S, Odom DT (2008) Species-specific transcription in mice carrying human chromosome 21. Science 322: 434–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG (2004) Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG (2008) Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet 40: 346–350 [DOI] [PubMed] [Google Scholar]
- Wood AJ, Oakey RJ (2006) Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet 2: e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M (2005) Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Clauder-Munster S, Smolik M, Huber W, Steinmetz LM (2011) Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol 7: 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM (2009) Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW (2002) Allelic variation in human gene expression. Science 297: 1143. [DOI] [PubMed] [Google Scholar]
- Yanai I, Graur D, Ophir R (2004) Incongruent expression profiles between human and mouse orthologous genes suggest widespread neutral evolution of transcription control. OMICS 8: 15–24 [DOI] [PubMed] [Google Scholar]
- Yanai I, Hunter CP (2009) Comparison of diverse developmental transcriptomes reveals that coexpression of gene neighbors is not evolutionarily conserved. Genome Res 19: 2214–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour M, Pfiffner J, Levin JZ, Adiconis X, Gnirke A, Nusbaum C, Thompson DA, Friedman N, Regev A (2011) Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol 11: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K (2009a) Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol 16: 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Qian W, Zhang J (2009b) Positive selection for elevated gene expression noise in yeast. Mol Syst Biol 5: 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF (2011) A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332: 977–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Zhao H, Mancera E, Steinmetz LM, Snyder M (2010) Genetic analysis of variation in transcription factor binding in yeast. Nature 464: 1187–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]