Abstract
Genes containing multiple pre-mRNA cleavage and polyadenylation sites, or polyA sites, express mRNA isoforms with variable 3′ untranslated regions (UTRs). By systematic analysis of human and mouse transcriptomes, we found that short 3′UTR isoforms are relatively more abundant when genes are highly expressed whereas long 3′UTR isoforms are relatively more abundant when genes are lowly expressed. Reporter assays indicated that polyA site choice can be modulated by transcriptional activity through the gene promoter. Using global and reporter-based nuclear run-on assays, we found that RNA polymerase II is more likely to pause at the polyA site of highly expressed genes than that of lowly expressed ones. Moreover, highly expressed genes tend to have a lower level of nucleosome but higher H3K4me3 and H3K36me3 levels at promoter-proximal polyA sites relative to distal ones. Taken together, our results indicate that polyA site usage is generally coupled to transcriptional activity, leading to regulation of alternative polyadenylation by transcription.
Keywords: 3′ end processing, 3′UTR, alternative polyadenylation, post-transcriptional control, transcription
Introduction
Expression of protein-coding genes in eukaryotes involves multiple transcriptional and post-transcriptional processes, which are increasingly found to be interconnected (Maniatis and Reed, 2002; Moore and Proudfoot, 2009). The 3′ end processing of pre-mRNAs, involving cleavage of nascent RNAs and synthesis of the poly(A) tail (Colgan and Manley, 1997), is critical for termination of transcription and interplays with pre-mRNA splicing (Buratowski, 2005; Millevoi and Vagner, 2010; Kuehner et al, 2011). A recent study also implicates its role in initiation of transcription (Mapendano et al, 2010). Pre-mRNA cleavage and polyadenylation, or mRNA polyadenylation, is carried out by the 3′ end processing complex which was recently found to comprise over 85 proteins in human cells (Shi et al, 2009). Interestingly, this complex includes not only the well-known core polyadenylation factors, or polyA factors for simplicity, such as CPSF, CstF, CF Im, and CF IIm proteins, but also proteins with roles in DNA damage repair, transcription, splicing, translation, etc., underscoring the diverse connections between mRNA polyadenylation and other cellular processes.
Over half of the human genes contain more than one polyA site (Tian et al, 2005; Yan and Marr, 2005), resulting in mRNA isoforms with different protein-coding regions and/or 3′ untranslated regions (3′UTRs). The pattern of alternative cleavage and polyadenylation, or alternative polyadenylation (APA), of genes is variable across tissues (Zhang et al, 2005; Wang et al, 2008), and is highly regulated during development and when cells change proliferation/differentiation states (Sandberg et al, 2008; Ji et al, 2009). In general, short 3′UTR isoforms resulting from usage of promoter-proximal polyA sites are relatively more abundant when cells are proliferative, transformed, or undifferentiated (Sandberg et al, 2008; Ji and Tian, 2009; Mayr and Bartel, 2009). Since 3′UTRs contain various cis elements involved in post-transcriptional gene regulation, such as mRNA localization, translation, and mRNA stability, APA can impact mRNA metabolism and protein expression level in the cytoplasm. Given that the predominant cis elements in 3′UTRs are those controlling mRNA stability, such as AU-rich elements, GU-rich elements, and microRNA target sites (Garneau et al, 2007; Vlasova et al, 2008; Bartel, 2009), it is conceivable that APA may have a widespread role in controlling mRNA half-life. Indeed, shortening of 3′UTRs has been shown to cause increased mRNA stability and higher protein expression for a number of oncogenes (Mayr and Bartel, 2009).
Several mechanisms that regulate APA have been reported. First, modulation of specific polyA factors has been shown to alter polyA site choice. For example, upregulation of CstF64 during B-cell maturation results in higher usage of a promoter-proximal polyA site in the IgM heavy chain gene (Takagaki et al, 1996), and knockdown of the 25-kDa subunit of CF Im was shown to alter APA for a number of genes in HeLa cells (Kubo et al, 2006). Consistently, a general inverse correlation between mRNA expression of polyA factors and global 3′UTR length was found in various tissues during development and in reprogrammed cells (Ji and Tian, 2009), indicating modulation of the general polyadenylation activity may be responsible for APA regulation in cell proliferation/differentiation. Second, various RNA binding proteins (RBPs) have been shown to modulate APA by interacting with cis elements adjacent to the polyA site (Millevoi and Vagner, 2010). An emerging theme is that some RBPs previously known to regulate pre-mRNA splicing may also have roles in mRNA polyadenylation (Licatalosi and Darnell, 2010).
Third, some proteins with apparent functions in gene transcription have been shown to regulate APA, such as ELL2, an RNA polymerase II (Pol II) elongation factor (Martincic et al, 2009), and Cdc73, a component of the PAF protein complex (PAFc) which associates with Pol II (Rozenblatt-Rosen et al, 2009). In addition, accumulating evidence suggests mRNA polyadenylation is extensively intertwined with transcription: Pol II itself is an essential polyA factor (Hirose and Manley, 1998) and its C-terminal domain (CTD) interacts with several other polyA factors and has been implicated in coupling pre-mRNA processing to transcription (McCracken et al, 1997; Ahn et al, 2004; Meinhart and Cramer, 2004; Adamson et al, 2005; Zhang and Gilmour, 2006); several polyA factors interact with the basal transcriptional machinery (Dantonel et al, 1997) and are present at the promoter region of genes (Venkataraman et al, 2005; Glover-Cutter et al, 2008); and several transcriptional factors have been shown to regulate 3′ end processing (Rosonina et al, 2003).
Here, we present several lines of evidence indicating that polyA site usage is generally coupled to transcriptional activity, contributing to a global correlation between the relative abundance of 3′UTR isoforms and gene expression level. Given the roles of 3′UTR in mRNA metabolism, this mechanism coordinates transcriptional regulation with post-transcriptional control via pre-mRNA processing.
Results
A general correlation between relative abundance of APA isoforms and gene expression level in human and mouse tissues and cells
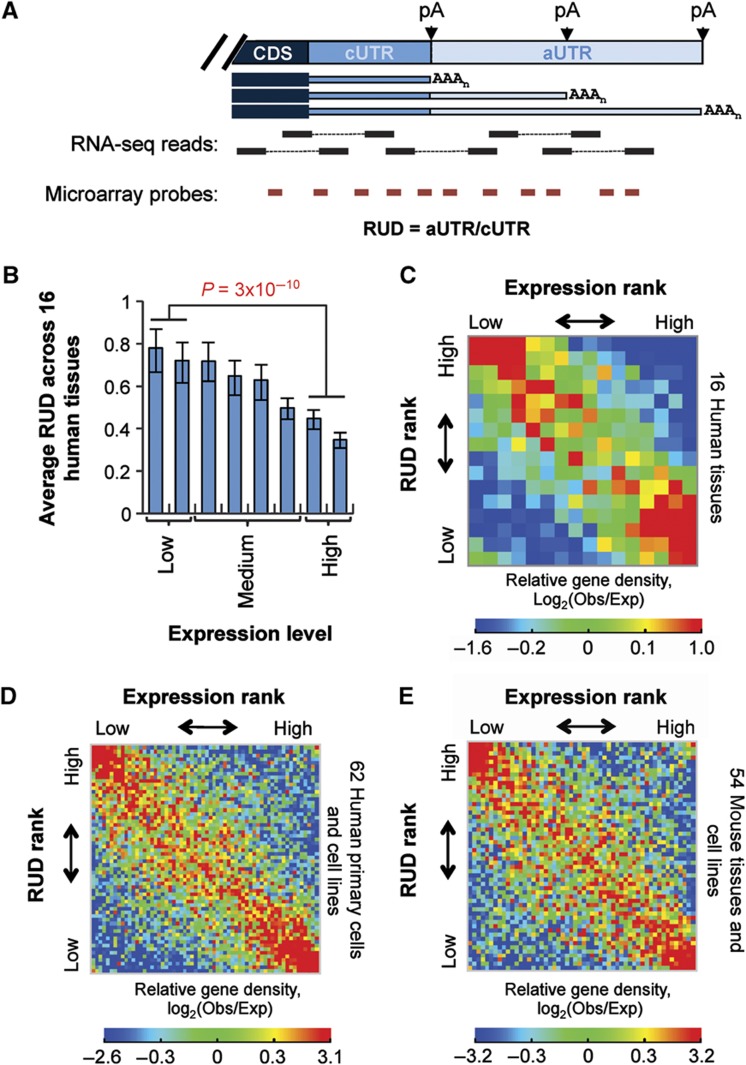
APA can lead to mRNA isoforms with short or long 3′UTRs (Figure 1A). Using a paired-end RNA-seq data set recently released from Illumina (Supplementary Table 1), we examined relative expression of APA isoforms across 16 human tissues. Our method detects 3′UTR length changes based on comparison of the RNA-seq reads mapped to constitutive and alternative portions of 3′UTR (named cUTR and aUTR, respectively), as defined by the 5′-most polyA site in the 3′UTR (Figure 1A). We developed a score named relative expression of isoforms using distal polyA sites (RUD) to represent the relative abundance of APA isoforms (see Materials and methods).
Figure 1.
Gene expression level versus relative abundance of 3′UTR isoforms in human and mouse tissues and cells. (A) Schematic of APA. A gene with multiple polyA sites expresses isoforms with alternative 3′UTRs. Three polyA sites are shown. CDS, coding sequence; pA, polyA site; AAAn, poly(A) tail. The 3′UTR portion upstream of the first polyA site is called constitutive 3′UTR, or cUTR, and the downstream portion is called alternative 3′UTR, or aUTR. The ratio of RNA-seq read density or average microarray probe intensity of aUTR to that of cUTR is called the Relative expression of mRNA isoforms Using Distal polyA sites (RUD) score. (B) Average RUD scores for genes expressed at different levels across 16 human tissues (see Supplementary Figure 1 for plots of individual tissues). Genes were evenly divided into eight groups based on expression level. The average RUD score of genes in a group is plotted. Error bars represent 90% confidence intervals. P-value was based on T-test comparing highly expressed gene group (top 25%) with lowly expressed gene group (bottom 25%). (C) Gene density plot showing inverse correlation between RUD and gene expression level in 16 human tissues. Genes in all tissues were distributed in a 16 × 16 table, with columns corresponding to rank of gene expression level and rows to rank of RUD in 16 tissues. The number of genes in each cell of the table was normalized to an expected number derived from randomized data. Relative gene density, log2(Obs/Exp), where Obs is observed number of genes and Exp is expected number of genes, is represented in a heat map according to the color scheme shown in the figure. (D, E) As in (C), gene density plots showing inverse correlation between RUD and gene expression level in 62 human primary cells and cell lines (D) and 54 mouse tissues and cell lines (E). The data for (D) and (E) were based on Affymetrix GeneChip Exon Arrays.
Interestingly, we found that highly expressed genes in general tended to express short 3′UTR isoforms more frequently than lowly expressed genes in all examined tissues (Figure 1B; Supplementary Figure 1A). Overall, there were enrichments (∼2-fold over expected values) of genes with preferential expression of short 3′UTR isoforms when they were highly expressed (lower right corner in Figure 1C) and of genes with preferential expression of long 3′UTR isoforms when they were lowly expressed (upper left corner in Figure 1C). Conversely, there were depletions (∼3-fold below expected values) of genes with opposite trends (upper right and lower left corners in Figure 1C). A similar result was obtained by analyzing transcriptomes in 10 human tissues based on the single-end RNA-seq method (Pan et al, 2008; Wang et al, 2008; Supplementary Figure 1B and C).
We next analyzed two Affymetrix exon array data sets corresponding to 62 types of human primary cells and cell lines and 54 types of mouse tissues and cell lines (Supplementary Table 1), respectively. It is worth noting that data from Affymetrix exon arrays allow strand-specific analysis of gene expression, whereas the RNA-seq reads analyzed do not intrinsically reveal the strand information of transcription. This might be important for our analysis since previous reports have indicated that antisense transcripts are pervasive in human cells (He et al, 2008) and are involved in regulation of gene expression (Xu et al, 2011). Using RUD scores derived from microarray probe intensities (see Materials and methods and Figure 1A), we found that the connection between APA and gene expression is consistent with, but more obvious than, that observed using RNA-seq data, as indicated by gene enrichment and depletion values (Figure 1D and E). Therefore, we conclude that there is a general correlation between the relative abundance of 3′UTR isoforms and gene expression level.
Reporter assays confirmed regulation of polyA site choice by transcriptional activity
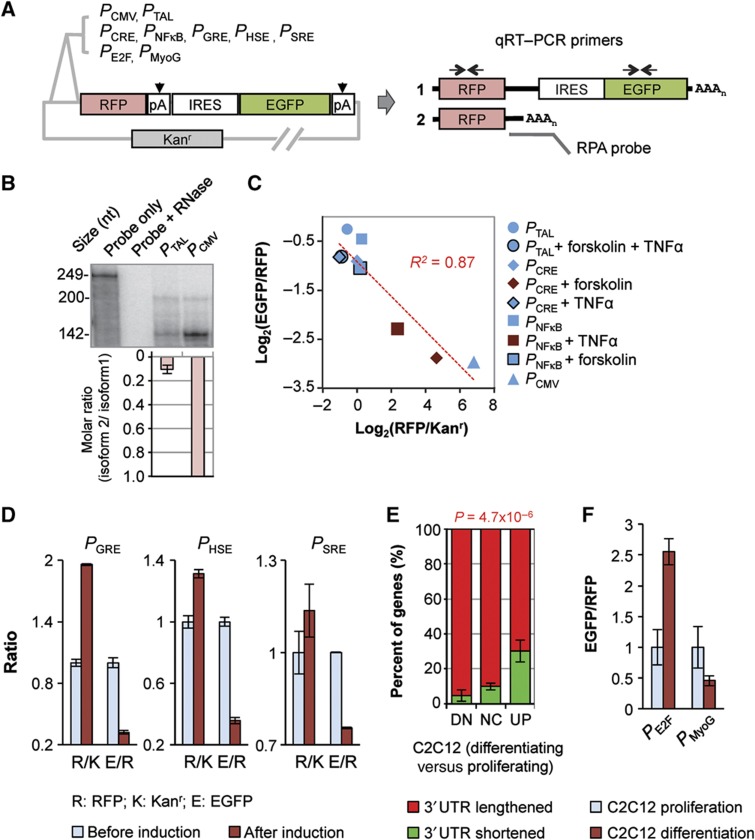
Difference in relative abundance between APA isoforms can be attributable to two potential mechanisms: (1) 3′UTR isoforms have different mRNA stabilities and/or (2) isoforms are differentially produced resulting from alternative 3′ end processing. The former mechanism has been shown for a number of genes (Edwalds-Gilbert et al, 1997; Mayr and Bartel, 2009). Thus, we wanted to know whether the latter could have a role in the correlation between relative abundance of APA isoforms and gene expression level. To this end, we made a set of constructs in which the same reporter gene, capable of expressing two APA isoforms, was under the control of different promoters (Figure 2A). This experimental design enables analysis of the effect of transcription on APA while minimizing the influence of mRNA stability. Using RNase protection assays (RPAs), we first confirmed the two expected APA isoforms (Figure 2B). In addition, we found that the CMV promoter (PCMV) resulted in much more (∼9-fold) expression of the short isoform relative to the long one than a basal TATA-like promoter (PTAL) (Figure 2B), indicating that polyA site choice can be controlled by promoter sequences.
Figure 2.
Reporter assays indicate regulation of polyA site choice by transcriptional activity. (A) Constructs used in this study. PCMV, PTAL, PCRE, PNFκB, PGRE, PHSE, PSRE, PE2F, and PMyoG are various promoters (see Materials and methods for details). RFP, IRES, EGFP, and Kanr are sequences encoding red fluorescent protein, internal ribosome entry site, enhanced green fluorescent protein, and kanamycin resistance gene, respectively. Kanr has its own promoter and polyA site. As shown in the graph, two polyA sites (pA) resulted in two transcript isoforms (1 and 2). Isoform 1 encodes RFP, IRES, and EGFP; and isoform 2 encodes RFP only. AAAn, poly(A) tail. qRT–PCR primers and RNase protection assay (RPA) probes are indicated in the graph, which were used to examine relative expression of the two isoforms. (B) RPA analysis of the isoforms expressed from the construct with PCMV or PTAL. Top, a representative autoradiograph of RPA results. The RPA probe is 249 nt in length. The 200-nt fragment corresponds to isoform 1 and the 142-nt fragment to isoform 2. The amount of sample loaded in each lane was adjusted to make the overall signal similar across samples. Bottom, normalized molar ratios of isoform 2 to isoform 1. The molar ratio of isoform 2 to isoform 1 was based on the amount of each RPA fragment quantified by PhosphorImager and the number of uracils in each fragment (UTP was used for probe labeling). The value for PCMV was set to 1, and error bars are standard deviation based on two experiments. (C) qRT–PCR analysis of expression level versus isoform ratio using cells transfected with constructs containing indicated promoters. Some of the transfected cells were treated with forskolin and/or TNFα as indicated in the graph. Expression level was measured by RFP/Kanr, and isoform ratio was measured by EGFP/RFP. R2 of linear regression is indicated. (D) qRT–PCR analysis of expression level and isoform ratio for constructs with PGRE, PHSE, and PSRE which were induced with dexamethasone, heat shock, and serum, respectively (see Materials and methods for details). R/K is RFP/Kanr and E/R is EGFP/RFP. Error bars are standard error of mean (s.e.m.) based on two experiments. (E) Percent of genes with 3′UTRs lengthened or shortened in three gene groups with different expression changes during differentiation of C2C12 cells. DN, downregulated; NC, no change; UP, upregulated. Error bars are standard deviation based on two samples; P-value is based on the Fisher's exact test comparing gene numbers in three groups. (F) qRT–PCR analysis of isoforms expressed from constructs with PE2F or PMyoG in proliferating and differentiating C2C12 cells. Isoform ratio was measured by EGFP/RFP. Error bars are standard error of mean (s.e.m.) based on two experiments. See Materials and methods for more technical details. Source data is available for this figure in the Supplementary Information.
We then used several inducible promoters and examined their regulation of polyA site choice under induced or non-induced conditions. Using quantitative reverse transcription PCR (qRT–PCR) and constructs containing the cAMP response element (PCRE) or the NFκB binding site (PNFκB) in the promoter, which are inducible by forskolin or TNFα, respectively, we found that induction of PCRE and PNFκB led to conspicuously higher expression of the reporter gene, as expected, and more usage of the promoter-proximal polyA site (Figure 2C). In contrast, induction of PNFκB by forskolin or PCRE by TNFα had no effect on gene expression nor polyA site usage, indicating specificity of the regulation (Figure 2C). This result was also confirmed by fluorescence activated cell sorting analysis (Supplementary Figure 2). When the results for PTAL and PCMV were included, a good inverse correlation between gene expression level and usage of the distal polyA site was discerned (R2=0.87). Similar trends were also observed for constructs containing promoters with the glucocorticoid response element (PGRE), the heat-shock response element (PHSE), and the serum response element (PSRE) (Figure 2D). Thus, our reporter assays confirmed that short APA isoforms using promoter-proximal polyA sites are more likely to be produced when gene expression is induced, suggesting that polyA site usage is coupled to transcriptional activity.
We next wanted to know whether the transcription-coupled APA can be detected when cells change conditions in response to developmental and environmental signals, under which the global APA pattern can change (Ji and Tian, 2009). To this end, we first reanalyzed our previously published exon array data for mRNAs expressed in murine C2C12 myoblasts, in which genes tend to express long 3′UTR isoforms more frequently when cells switch from proliferation to differentiation (Ji et al, 2009). Consistent with the notion that polyA site usage is coupled to transcriptional activity, we found that upregulated genes were less likely to have 3′UTRs lengthened than downregulated ones (Figure 2E). This phenomenon was also observed when we analyzed data for breast cancer cell lines versus normal breast tissues and TNFα-treated lymphoblastoid cells versus non-treated ones (Supplementary Figure 3). In both cases, upregulated genes were more likely to express short 3′UTR isoforms than downregulated ones regardless of the overall trend of APA regulation in the cell. To validate this global analysis result, we made a construct containing a promoter with the E2F binding site (PE2F) and a construct containing the myogenin promoter (PMyoG). PE2F and PMyoG have been shown to be inhibited and activated, respectively, during C2C12 differentiation (Edmondson et al, 1992; Blais et al, 2005). In complete agreement with our global analysis result, more expression of the long 3′UTR isoform relative to the short one was observed for the construct containing PE2F in differentiating C2C12 cells as compared with proliferating ones; and the construct containing PMyoG showed the opposite trend (Figure 2F). Therefore, we conclude that modulation of polyA site choice by transcriptional activity can happen to genes with regulated expression when cells respond to developmental and environmental signals.
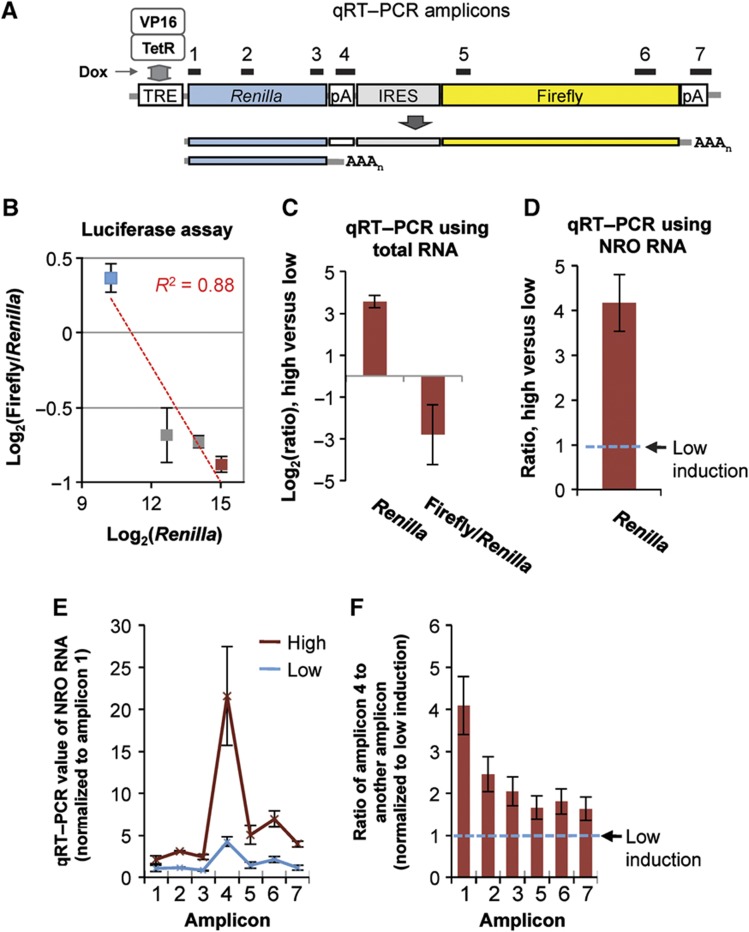
To address whether our reporter assay results could be generalized, we made another construct (pTRE-RIF) containing a different reporter gene and the tetracycline response element (TRE) in the promoter (Figure 3A), which can be activated by doxycycline (Dox) in HeLa Tet-On cells. Consistent with the results described above, activation of transcription resulted in higher gene expression and more usage of the proximal polyA site, as indicated both by luciferase assays (Figure 3B) and by qRT–PCR (Figure 3C). Since activation of transcription through TRE is mediated by the transcription activation domain from HSV VP16 (Figure 3A), this result also suggests that transcription factors may have an important role in the regulation of polyA site choice by transcriptional activity (see Discussion for more on this point).
Figure 3.
Pol II pausing at the polyA site correlates with polyA site usage and is regulated by transcriptional activity. (A) The pTRE-RIF vector used in this study. PTRE, promoter containing the tetracycline response element; Dox, doxycycline; TetR, tetracycline repressor; VP16, the transcription activation domain of HSV VP16. Renilla, IRES, and Firefly are sequences encoding Renilla luciferase, internal ribosome entry site, and firefly luciferase, respectively. There are two polyA sites (pA), resulting in two transcript isoforms. Isoform 1 encodes Renilla, IRES, and Firefly; and isoform 2 encodes Renilla only. qRT–PCR primers targeting different regions of the reporter gene are indicated in the graph (drawn to scale). (B) Dual-luciferase assay analysis of expression level versus isoform ratio. HeLa Tet-On cells transfected with pTRE-RIF were treated with 10 ng/ml, 100 ng/ml, 1 μg/ml, and 10 μg/ml Dox (corresponding to the data points shown from left to right) for induction of expression. Expression level was measured by Renilla and isoform ratio was measured by Firefly/Renilla. Error bars are standard error of mean (s.e.m.) based on two experiments. (C) qRT–PCR analysis of total RNA from cells treated with low (10 ng/ml) or high (10 μg/ml) doses of Dox. The Renilla region was used to indicate gene expression and Firefly/Renilla was used to indicate isoform ratio. Renilla value was the mean value base on amplicons 1, 2, and 3 as shown in (A), and Firefly value was the mean value based on amplicons 5 and 6 as shown in (A). Error bars are standard deviation based on multiple amplicons. (D) qRT–PCR analysis of nascent RNA using cells treated with low or high doses of Dox. The Renilla value based on amplicons 1, 2, and 3 was calculated, and its value for high induction was normalized to that for low induction (set to 1). (E) qRT–PCR analysis of NRO transcripts with different amplicons using cells treated with low or high doses of Dox. All amplicons were normalized to amplicon 1 (set to 1). (F) Data in (E) were reanalyzed to show that Pol II pausing at the proximal polyA site (amplicon 4) had the biggest difference between high induction of expression and low induction of expression. Each amplicon was first normalized to amplicon 4, and then the normalized value for high induction was normalized to that for low induction. Thus, the low induction value is 1 for all amplicons. Source data is available for this figure in the Supplementary Information.
Nuclear run-on data support coupling of 3′ end processing to transcription
RNA polymerase II (Pol II) pauses at the polyA site for 3′ end processing (Nag et al, 2007; Glover-Cutter et al, 2008; West and Proudfoot, 2009), which can be detected by the nuclear run-on (NRO) method (Core et al, 2008; West and Proudfoot, 2009). We reasoned that if polyA site usage was coupled to transcriptional activity, Pol II pausing at the polyA site would be different for genes expressed at different levels. To this end, we carried out NRO with pTRE-RIF under low and high induction conditions using BrUTP to label nascent transcripts. Labeled nascent transcripts were immunoprecipitated and analyzed by qRT–PCR with primer sets targeting different regions of the reporter gene (Figure 3A). Consistent with the difference in total RNA expression, qRT–PCR of NRO RNA showed more nascent transcripts from the reporter gene when it was highly induced than when it was lowly induced (Figure 3D).
The qRT–PCR value for a given region also represents Pol II density in the region, reflecting its pausing kinetics. As shown in Figure 3E, we found that Pol II density was generally higher across the whole reporter gene when it is highly induced. A prominent peak can be discerned at the proximal polyA site under both induction conditions, suggesting significant pausing of Pol II in the region. Importantly, the difference in Pol II density at the proximal polyA site between high and low induction conditions is significantly greater than those at other regions (Figure 3F). Combined with the luciferase results and qRT–PCR data using total RNA, the NRO results indicate that Pol II pausing at the polyA site (1) correlates with polyA site usage and (2) can be modulated by transcriptional activity. Since NRO is not affected by mRNA stability, a common issue in analysis of steady-state mRNAs, this finding directly supports the notion that polyA site usage is coupled to transcriptional activity.
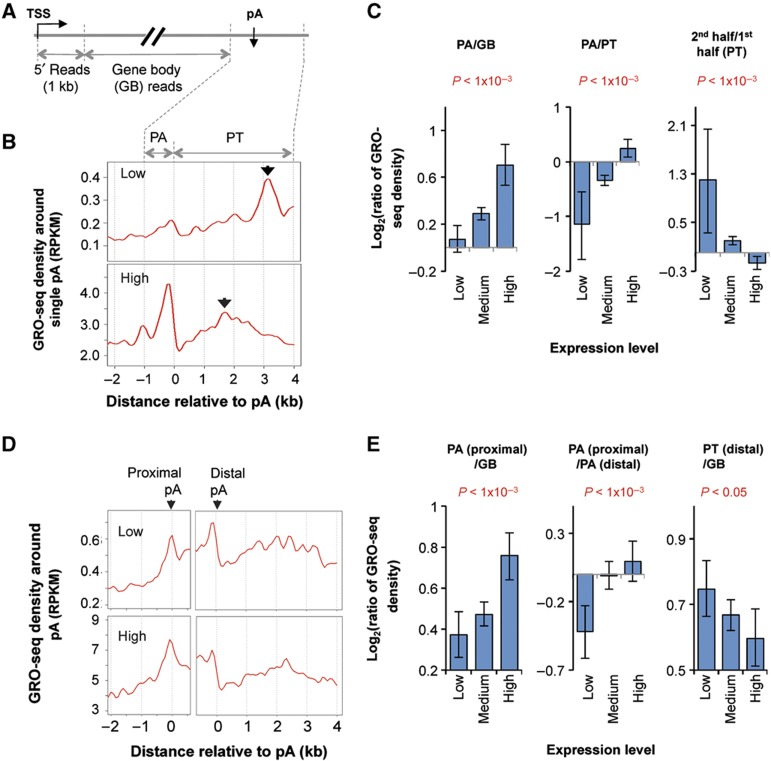
To examine regulation of 3′ end processing by transcription for endogenous genes, we carried out a global NRO experiment using deep sequencing (GRO-seq), similar to the method developed by the Lis group (Core et al, 2008). We obtained over 12.5 million uniquely mapped strand-specific GRO-seq reads for the nascent transcripts generated in NRO of C2C12 cells (see Materials and methods for details). We first examined genes with only one polyA site in the 3′-most exon (named single polyA site, Figure 4A). Consistent with the findings reported by the Lis group (Core et al, 2008), we observed two GRO-seq read peaks around the polyA site (Figure 4B): one spanned the 1-kilobase (kb) upstream region of the polyA site, peaking right before the polyA site; and the other spanned the 4-kb downstream region. As indicated previously (Core et al, 2008), the first peak corresponds to Pol II pausing at the polyA site, or polyA pausing, and the second one corresponds to Pol II pausing before termination, or pre-termination pausing. Using GRO-seq read density in the transcribed region to represent gene expression level (the first 1 kb region at the 5′ end was excluded to minimize influence of reads resulted from Pol II pausing at the promoter), we found that polyA pausing was more pronounced for highly expressed genes than for lowly expressed ones (Figure 4B and PA/GB in Figure 4C). This trend was also discernable for the ratio of polyA pausing to pre-termination pausing (PA/PT in Figure 4C). Interestingly, we found a shift of the pre-termination pausing peak toward the polyA site as gene expression level increased (see arrows in Figure 4B, and comparison of the first and second halves of the pre-termination pausing region in Figure 4C), suggesting that Pol II may terminate more rapidly on highly expressed genes than on lowly expressed ones.
Figure 4.
Global nuclear run-on deep sequencing (GRO-seq) indicates that Pol II pausing at the polyA site is coupled to transcriptional activity. (A) Schematic of using GRO-seq data in this study. GRO-seq reads mapped to the sense strand of genes were divided into four groups: 5′ reads, polyA site region (PA) reads, gene body (GB) reads, and pre-termination (PT) reads (see Materials and methods for details). (B) GRO-seq read density in the 3′ end region of genes with a single polyA site. Read density was presented as reads per kilobase per million mapped reads, or RPKM. Expressed genes were divided into three groups based on expression level, that is, low, medium, and high. Only data for low and high groups are shown. Arrows in the figures indicate peaks in the PT region. (C) Ratio of GRO-seq read density between different regions for genes expressed at different levels. The comparing regions are indicated in each graph. Error bars are 90% confidence intervals and the P-values indicate difference between highly and lowly expressed genes (see Materials and methods for details). (D) GRO-seq read density around 5′-most (proximal) and 3′-most (distal) polyA sites in the 3′-most exon of lowly expressed genes (top) and highly expressed genes (bottom). (E) As in (C), ratio of GRO-seq read density between different regions for genes expressed at different levels. The comparing regions are indicated in each graph. Only the 200-nt upstream region of the polyA site was used for the PA (proximal)/PA (distal) plot.
We next examined GRO-seq reads around alternative polyA sites. We focused on the 5′-most and 3′-most polyA sites in the 3′-most exons (Figure 4D). Consistent with the notion that the promoter-proximal polyA site is more likely to be used when gene expression is high, both Pol II pausing at the proximal polyA site (PA(proximal)/GB in Figure 4E) and the ratio of pausing between proximal and distal polyA sites (PA(proximal)/PA(distal) in Figure 4E) were much greater for highly expressed genes than for lowly expressed ones. Since measurement of Pol II pausing at the distal site may be affected by pre-termination pausing resulted from usage of upstream polyA sites, we calculated the ratio of pre-termination pausing after the distal polyA site to gene body. Interestingly, the ratio was significantly lower for highly expressed genes than for lowly expressed ones (PT(distal)/GB in Figure 4E), suggesting a greater loss of Pol II before reaching the distal polyA site for highly expressed genes. Taken together, our GRO-seq results based on endogenous genes further indicate that Pol II is more likely to pause at proximal polyA sites when genes are highly expressed. Notably, consistent results were also obtained using the data published by the Lis group, which was based on a human cell line (Supplementary Figure 4).
Regulation of APA by transcriptional activity correlates with differences in nucleosome positioning and histone methylations around alternative polyA sites
Transcriptional activity is known to impact epigenetic features such as nucleosome positioning and histone modifications (Barski et al, 2007; Campos and Reinberg, 2009; Schwartz et al, 2009). We reasoned that alteration of epigenetic features at alternative polyA sites would indicate change of transcriptional activity at these sites, supporting alternative polyA site usage. Since epigenetic features are not affected by RNA metabolism, such as stability, the result would address the connection between polyA site usage and transcriptional activity from a different perspective.
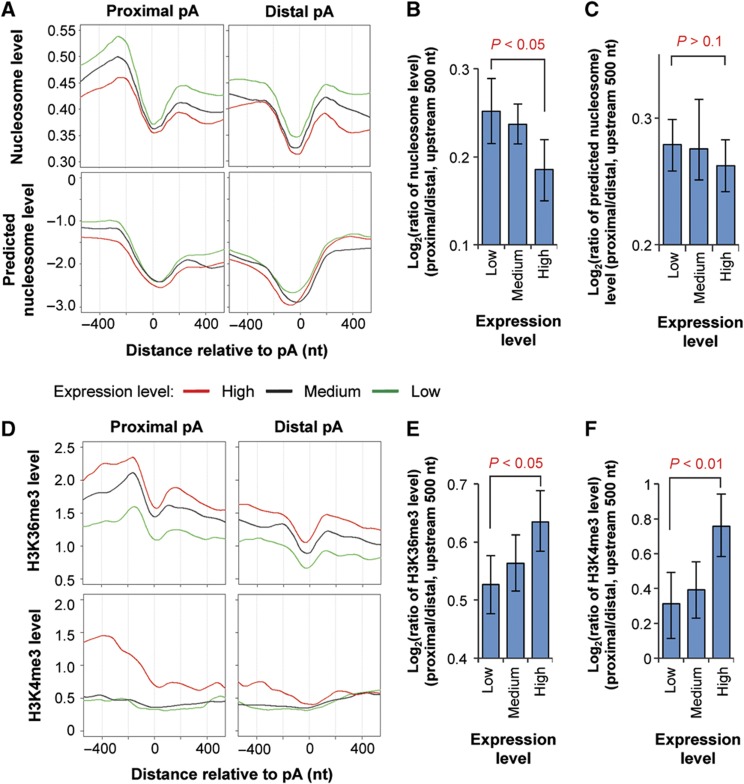
We first analyzed a data set generated by digestion of chromatin DNA from human resting T cells with micrococcal nuclease (Schones et al, 2008). Consistent with previous reports (Mavrich et al, 2008; Kaplan et al, 2009; Spies et al, 2009), depletion of nucleosome around the polyA site was detected for both proximal and distal polyA sites (Figure 5A). Interestingly, this depletion could be predicted using a computational model that was based solely on nucleotide content (Kaplan et al, 2009; Figure 5A, bottom), indicating important contribution of nucleotide composition to nucleosome positioning around the polyA site. However, highly expressed genes had a lower nucleosome level around the polyA site than lowly expressed genes, exceeding the difference predicted by the computational model, particularly in regions >200 nucleotides (nt) upstream or downstream of the polyA site. This result indicates transcriptional activity has an additional impact on nucleosome level around the polyA site. Importantly, highly expressed genes had a lower ratio of nucleosome level between proximal and distal polyA sites (upstream 500 nt used for analysis) than lowly expressed genes (Figure 5B). Since this difference could not be discerned using predicted nucleosome levels (Figure 5C), the nucleosome level difference between alternative polyA sites in genes expressed at different levels supports the notion that polyA site choice is connected to transcriptional activity.
Figure 5.
Nucleosome positioning and histone modifications around alternative polyA sites in genes expressed at different levels. (A) Top, nucleosome levels around the proximal (left) and distal (right) polyA sites in genes expressed at different levels. Proximal and distal polyA sites are the 5′-most and 3′-most sites in the 3′-most exon, respectively. The nucleosome level was based on data from Schones et al (2008) using human resting T cells. Nucleosome level is the average number of reads mapped to a position relative to the polyA site normalized to the total mappable read number in the sample. Genes were divided into three groups based on expression level, that is, low, medium, and high, as indicated by different colored lines. Bottom, predicted nucleosome levels around proximal (left) and distal (right) polyA sites using the computational model reported by Kaplan et al (2009). (B) Ratio of nucleosome level in the 500-nt upstream region of proximal polyA site to that of distal site for genes expressed at different levels. (C) As in (B) except that predicted nucleosome levels were analyzed. Error bars are 90% confidence intervals and P-values indicate difference between highly and lowly expressed genes (see Materials and methods for details). (D) As in (A), expect that H3K36me3 (top) and H3K4me3 (bottom) levels around the proximal (left) and distal (right) polyA sites were plotted. H3K36me3 and H3K4me3 levels were based on data from Barski et al (2007) using human resting T cells. (E) As in (B), except that H3K36me3 levels were analyzed. (F) As in (B), except that H3K4me3 levels were analyzed. See also Supplementary Figure 5 for H3K36me1, H3K4me1, and H3K4me2 in resting T cells, and H3K36me3 and H3K4me3 in mouse embryonic fibroblasts and neuronal progenitor cells.
To examine how regulation of APA by transcriptional activity correlates with changes in histone methylation levels, we analyzed a ChIP-seq data set for human resting T cells. As expected, H3K36me3 and H3K4me3 levels around the polyA site correlated with gene expression level (Figure 5D), although there are differences in their profiles. The H3K36me3 profiles showed a drop at the polyA site, presumably attributable to depletion of nucleosome. Significantly, the ratio of H3K36me3 level in the upstream region of proximal polyA site (within 500 nt) to that of distal site was significantly greater for highly expressed genes than that for lowly expressed ones (Figure 5E). This pattern was also corroborated by analysis of H3K36me3 levels in mouse embryonic fibroblasts (MEFs) and neuronal progenitor cells (NPCs), but was not discernable for H3K36me1 (Supplementary Figure 5).
Similar to the H3K36me3 result, the ratio of H3K4me3 level in the upstream region of proximal polyA site to that of distal site was significantly higher for highly expressed genes as compared with lowly expressed ones (Figure 5F). Interestingly, a marked drop of H3K4me3 level after the proximal polyA site can be discerned for highly expressed genes. The H3K4me3 result was also confirmed by using data for MEF and NPC (Supplementary Figure 5). A similar trend was found for H3K4me2, albeit less significant, but not for H3K4me1 (Supplementary Figure 5). Taken together, our results from analysis of epigenetic features indicate polyA site choice is connected to transcriptional activity and leaves epigenetic signatures.
Discussion
Here, we report a general correlation between the relative abundance of APA isoforms and gene expression level in human and mouse transcriptomes, and present several lines of evidence indicating that transcriptional activity regulates polyA site choice. The correspondence between APA and gene expression revealed in our study may be responsible for the coupled usage of alternative promoters and polyA sites previously reported for some genes (Costessi et al, 2006; Winter et al, 2007), and contribute to tissue-specific and condition-specific APA events involving transcription factors, such as neuronal activity-dependent polyA site selection mediated by MEF2 (Flavell et al, 2008).
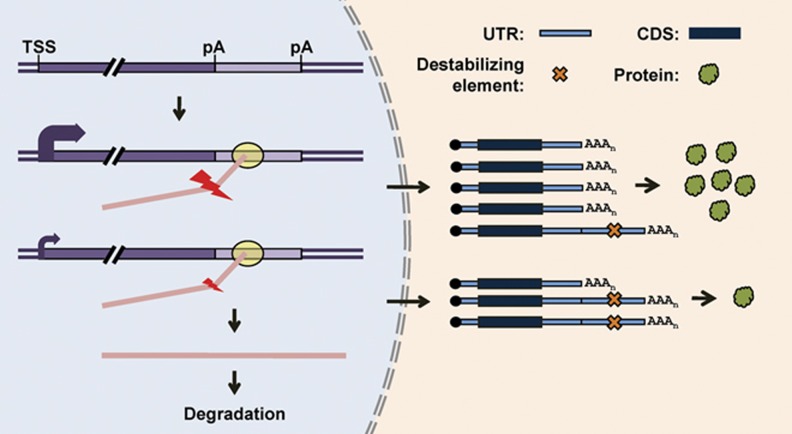
Regulation of polyA site choice by transcriptional activity results in preferential expression of long 3′UTR isoforms when gene expression is low and short 3′UTR isoforms when gene expression is high. Since short 3′UTRs are generally more stable due to avoidance of destabilizing elements in 3′UTRs (Mayr and Bartel, 2009) and escape from cellular mechanisms degrading long 3′UTRs (Hogg and Goff, 2010), more frequent production of short 3′UTR isoforms would make the overall expression level of a gene even higher and, conversely, more frequent production of long 3′UTR isoforms would have the opposite effect. Therefore, this mechanism can magnify the final outcome of transcriptional regulation (Figure 6): high transcriptional activity leads to more expression of mRNA isoforms with long half-lives and therefore high protein production capabilities; and low transcriptional activity leads to more expression of isoforms with short half-lives, and therefore low protein production capabilities. Conceivably, this mechanism can facilitate swift gene expression changes, which can be critical when cells respond to developmental and environmental signals.
Figure 6.
A model for regulation of APA by transcription and its impact on gene expression. A hypothetical gene is shown as a box with the transcription start site (TSS) and polyA sites (pA) indicated. High and low transcriptional activities are indicated by thick and thin curved arrows, respectively. Pol II is shown as a yellow oval. High and low polyA site usage is indicated by large and small lightning symbols. This model shows that high transcriptional activity leads to more polyA site usage, resulting in relatively higher expression of short 3′UTR isoforms whereas low transcriptional activity leads to lower polyA site usage, resulting in either relative higher expression of long 3′UTR isoforms or unprocessed pre-mRNAs that are subject to degradation in the nucleus. Since alternative 3′UTRs typically contain destabilizing elements, as indicated by various previous studies (Mayr and Bartel, 2009; Hogg and Goff, 2010), short 3′UTR isoforms are more stable and have higher protein expression capability than long isoforms.
Our result also suggests that downregulation of gene expression at the transcriptional level may lead to more unprocessed pre-mRNAs as a result of less usage of polyA site (Figure 6). This could lead to further inhibition of gene expression, because unprocessed pre-mRNAs are subject to degradation by the nuclear exosome (Houseley et al, 2006; Lykke-Andersen et al, 2009). On this note, a global analysis of RNA expression by tiling arrays indicated that most regions of the human genome can be transcribed (Johnson et al, 2005). However, intergenic transcripts outside the well-annotated genes generally have very low abundance (van Bakel et al, 2010). It is possible that transcription of some of these RNA species may result from the non-specific activity of Pol II, which, due to lack of coupling to 3′ end processing, leads to unprocessed transcripts that are rapidly degraded. Therefore, coupling 3′ end processing to transcription may have a role in augmenting the difference between specifically activated transcription and transcriptional noise in the cell.
A number of potential molecular mechanisms need to be considered to explain regulation of polyA site choice by transcriptional activity. First, transcription elongation, which can be controlled by gene promoters/enhancers, has been shown to regulate pre-mRNA splicing (Kornblihtt, 2007). However, unless the elongation rate constantly correlates with transcriptional activity, it is not likely to be a general mechanism responsible for global regulation of polyA site choice at different gene expression levels. Second, previous studies have shown that the CTD of Pol II is critical for coupling pre-mRNA processing to transcription (McCracken et al, 1997; Ahn et al, 2004; Meinhart and Cramer, 2004; Adamson et al, 2005). Phosphorylation of CTD is important for recruitment of polyA factors to the 3′ end of genes (Ahn et al, 2004). It remains to be seen, however, whether highly expressed genes have different CTD phosphorylation patterns than lowly expressed ones, leading to differential recruitment of polyA factors at the 3′ end.
Third, transcription factors can regulate mRNA polyadenylation. For example, some transcription factors were shown to stimulate PSF (Rosonina et al, 2005), which has a role in 3′ end processing and termination of transcription (Liang and Lutz, 2006; Kaneko et al, 2007). Notably, Nagaike et al (2011) have recently provided biochemical evidence in vitro that transcription activators stimulate transcription-coupled 3′ end processing through direct interaction with the PAFc, which was previously shown to be involved in 3′ end formation of yeast mRNAs and snRNAs (Penheiter et al, 2005; Sheldon et al, 2005), and to bind CPSF and CstF (Rozenblatt-Rosen et al, 2009). Interestingly, PAFc has also been shown to regulate histone methylation (Zhu et al, 2005; Jaehning, 2010), which appears consistent with our finding that the relative levels of H3K4me3 and H3K36me3 around alternative polyA sites are distinct for genes expressed at different levels. Therefore, it is plausible that the correspondence between APA and gene expression we observed here is due to recruitment of polyA factors to Pol II at the promoter region by transcription factors. This mechanism would make Pol II more ‘prepared’ to engage in 3′ end processing when a polyA site is encountered. Future studies need to further address whether different transcriptional factors involve different mechanisms in polyA factor recruitment.
The correlation between expression level and differences in the patterns of nucleosome positioning and histone methylation (H3K4me3 and H3K36me3) around alternative polyA sites not only supports the notion that polyA site choice is regulated by transcriptional activity but also raises the question as to whether epigenetic features can, in return, modulate APA, thereby forming a feedback regulatory mechanism. Histone methylation has been shown to facilitate pre-mRNA splicing by recruiting the spliceosome (Sims et al, 2007) and to regulate alternative splicing by affecting splicing regulators (Luco et al, 2010). DNA methylation, another type of epigenetic mark, has been shown to regulate APA in imprinted genes (Wood et al, 2008). It will be interesting in the future to examine how nucleosome remodeling and different types of histone methylation have roles in APA and its coupling to transcription.
Materials and methods
Data sets
Information about the data sets used in this study is listed in Supplementary Table 1. PolyA site information was obtained from PolyA_DB (Lee et al, 2007).
Analysis of RNA-seq data
Paired-end reads were mapped to hg18 genome using Tophat (Trapnell et al, 2009) with default parameters. Only uniquely mapped and properly paired reads were used for subsequent analyses. Properly paired reads are those with two pairing reads mapped to different strands of the same chromosome. See Supplementary Table 2 for the number of reads used. Gene expression levels were calculated using read density in the protein-coding region based on the reads per kilobase of mappable region per million mapped reads (RPKM) method (Mortazavi et al, 2008). A cutoff of RPKM=1 was used to select expressed genes, which resulted in similar numbers of expressed genes in different tissues to those reported by Ramskold et al (2009). The top 25%, middle 50%, and bottom 25% of expressed genes with respect to RPKM were considered as having high, medium, and low expression, respectively. The score for relative expression of isoforms using distal polyA sites (RUD) was based on the ratio of read density in aUTR to that in cUTR (Figure 1A). Therefore, a high RUD value indicates higher abundance of long 3′UTR isoform resulting from usage of promoter-distal polyA sites relative to short 3′UTR isoforms resulting from usage of promoter-proximal polyA sites. The transcription direction for each read pair was inferred based on overlap with RefSeq sequences and the 3′ end of the mapped read pair was used for RUD calculation. We required the 3′UTR of a surveyed gene do not overlap with any regions of other genes, regardless of transcription direction. This can minimize interference from antisense transcripts.
Analysis of exon array data
Exon array data were normalized by the Robust Multichip Average (RMA) method and were corrected for hybridization bias using the COSIE program (Gaidatzis et al, 2009). Expressed genes were selected by the Detection Above Background (DABG) method. Gene expression level was calculated using probesets in constitutive exons, based on NCBI cDNAs/ESTs (Lee et al, 2008). The RUD score was based on the ratio of average probeset intensity in aUTR to that in cUTR, as previously described (Ji et al, 2009). For both human and mouse data sets, we first calculated mean for each probeset across replicates and then combined all samples for analysis. For gene expression regulation in differentiation of C2C12 cells, 1 × standard deviation of log2(ratio of expression) was used to group differentially expressed genes. APA regulation was based on analysis of RUD values using the Significance Analysis of Microarray (SAM) method with FDR<0.05 as cutoff for selection of significant events, as previously described (Ji et al, 2009).
Analysis of GRO-seq data
GRO-seq reads for C2C12 cells were mapped to the mouse genome (mm9) using Bowtie (Langmead et al, 2009) allowing up to three mismatches. Unmapped reads were trimmed to the first 38 nt and mapped to the genome again by Bowtie allowing up to three mismatches. This approach resulted in 12 511 052 uniquely mapped reads (69% of total). The 5′ end position of each read was used to indicate its location. GRO-seq reads were examined in four regions of a gene: (1) 5′ region, 1 kb downstream of the transcription start site (TSS); (2) polyA region (PA), 1 kb upstream of the polyA site; (3) gene body region (GB), whole gene region excluding 5′ and PA regions; (4) pre-termination region (PT), 4 kb downstream of the polyA site. When there were multiple polyA sites in the 3′-most exon, the 5′-most site was used to define GB. Gene expression level was based on read density (RPKM) in GB and PA. RPKM>0.04 was used as cutoff to select expressed genes. The top 25%, middle 50%, and bottom 25% of expressed genes with respect to RPKM were considered as having high, medium, and low expression, respectively. To minimize interference from downstream genes, we selected only genes that were not followed by any RefSeq-supported genes with the same transcriptional direction in the 6-kb downstream region. For analysis of alternative polyA sites, we selected only the distal polyA sites that were not preceded by any polyA sites in the upstream 400 nt region and used only the upstream 200 nt region of proximal or distal polyA sites. The same method was used to analyze the GRO-seq data generated by the Lis group for IMR90 cells, except that the PT region was 3 kb.
Analysis of nucleosome positioning and histone methylation patterns
The data for nucleosome positioning and histone methylation were from Schones et al (2008) and Barski et al (2007), respectively. For the nucleosome positioning data, we extended reads to 147 bp, the length of DNA bound by a full nucleosome. Gene expression levels were based on microarray data of human T cells using the Affymetrix MAS5 method. The bottom and top 20% of genes with respect to probe intensity values were considered as lowly and highly expressed genes, respectively. The ratio of read density in the 500-nt upstream region of proximal polyA site to that of distal site was calculated to indicate relative level at the proximal versus distal polyA sites. Nucleosome level prediction using nucleotide content was based on a program from http://genie.weizmann.ac.il/software/nucleo_genomes.html (Kaplan et al, 2009). Histone modification patterns in MEFs and NPCs were analyzed by the same method.
Statistical analysis
To compare various features between highly and lowly expressed genes, we used a data resampling method based on bootstrapping (Venables and Ripley, 2002). This was carried out by resampling genes in two comparing groups 1000 times to derive a P-value based on how many times one group has a higher or lower mean value than the other. This method was also used to derive 90% confidence intervals for various data.
Gene density plot
We used the gene density plot to examine the correlation between the relative abundance of 3′UTR isoforms and gene expression level. As described above, the RUD score was used to represent the relative abundance of APA isoforms. In the gene density plot, genes expressed in each sample were summarized in an N × N table according to the RUD rank (row) and gene expression rank (column) across a sample set, where N is the number of samples in the set. The number of genes in each cell of the table is an observed value. The expected number of genes for each cell was calculated using randomized data with shuffled RUD and gene expression ranks. The ratio of observed number of genes (Obs) to expected number of genes (Exp) of a cell indicates the extent of enrichment (when ratio >1), or of depletion (when ratio <1), of genes in the cell; and the log2(ratio) values of the table are represented in a heat map using R.
Constructs used in this study
Constructs containing PTAL, PCRE, PNFκB, PGRE, PHSE, and PSRE were constructed by replacing the CMV promoter (PCMV) in pRiG-77S.AD (Ji et al, 2009) with fragments containing promoter sequences from corresponding constructs included in the Mercury™ Pathway Profiling System (Clontech) by PCR (primers: 5′-GCGCATTAATGAGCTCTTACGCGTTCTAGC and 5′-CGATGCTAGCCGATTCGAAGCTTCTGCTTC) and restriction enzymes AseI and NheI. Constructs containing PE2F and PMyoG were constructed by replacing the CMV promoter in pRiG-77S.AE (Ji et al, 2009) with respective DNA fragments and restriction enzymes AseI and NheI. PMyoG was derived from the myogenin promoter (−395 to +39 nt surrounding the TSS; Edmondson et al, 1992) using PCR (primers: 5′-CGATATTAATGGATTTTCAAGACCCCTTCC and 5′-GGCCGCTAGCAAGGCTTGTTCCTGCCACT) and C2C12 genomic DNA, and PE2F was derived from the E2F luciferase reporter vector (Panomics) using PCR (primers: 5′-CGATATTAATCTAGCCTTGGCGGGAGATA and 5′-GGCCGCTAGCTTACCAACAGTACCGGAATGC). The pTRE-RIF vector was constructed by cloning a fragment encoding the Renilla luciferase from pRL-CMV (Promega), a fragment encoding the firefly luciferase from pGL3-Basic (Promega), and a fragment containing a polyA site and an IRES sequence from pRiG-77S.AD into the pTRE-Tight vector (Clontech).
Cell culture and reporter assays
Experiments using constructs containing PCMV, PTAL, PCRE, PNFκB, PGRE, PHSE, or PSRE were carried out in Human Embryonic Kidney (HEK) 293 cells, which were maintained in Dulbecco's Modified Eagles Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Transfection was carried out using Lipofectamine 2000 (Invitrogen). The following conditions were used to induce PCRE, PNFκB, and PGRE: 5 μM forskolin (Fisher) for PCRE, 0.1 μg/ml human TNFα (Sigma) for PNFκB, and 5 μM Dexamethasone (Sigma) for PGRE. Cells were treated with inducing agents 16 h after transfection. Six hours after treatment, total cellular RNA was extracted using TRIzol (Invitrogen). To induce PHSE, cells were incubated at 45°C for 15 min. To induce PSRE, cells were first grown in the reduced serum media Opti-MEM (Invitrogen) and then in DMEM with 10% FBS.
Constructs containing PE2F or PMyoG were studied in proliferating and differentiating C2C12 cells. Briefly, C2C12 cells were maintained at 30–70% confluency in DMEM supplemented with 10% FBS. Transfection was carried out using Lipofectamine 2000 (Invitrogen) when the confluency of cells was 30%. After 16 h, cells were split into a proliferation group and a differentiation group. For the proliferation group, cells were still maintained in DMEM with 10% FBS. For the differentiation group, cells were switched to DMEM with 2% horse serum when they reached 90% confluency. qRT–PCR was carried out 24 h after transfection.
pTRE-RIF was studied in HeLa Tet-On cells (gift from Andrew Harris, UMDNJ). Briefly, cells were maintained in DMEM supplemented with 10% FBS and 200 μg/ml G418. Transfection of pTRE-RIF was carried out using jetPEI (Polyplus-transfection). Cells were treated with different doses of Dox (Sigma) right after transfection. After 24 h, cell lysis and luciferase assay were carried out using the Dual-Luciferase Reporter Assay System (Promega).
RNase protection assay
The DNA template for RPA probe was produced by putting a fragment surrounding the proximal polyA site of pRiG-77S.AD into the pcDNA3.1 vector (Invitrogen) using restriction enzymes (XhoI and BamHI). The 32P-labeled antisense RNA probe was generated by in vitro transcription using the MAXIscript kit (Ambion). RPA assays were carried out using the RPA III kit (Ambion).
Quantitative real-time reverse transcription PCR
For qRT–PCR, total cellular RNA was treated with DNase I and reverse transcribed using the oligo-dT primer. qRT–PCR was carried out using the Maxima SYBR Green/Rox qPCR Master Mix (Fermentas) with primers targeting RFP (5′-GCCCCGTAATGCAGAAGAAG and 5′-CTTCAGGGCCTTGTGGATCT), EGFP (5′-GGGCACAAGCTGGAGTACAACT and 5′-ATGTTGTGGCGGATCTTGAAG), or the kanamycin resistance gene (5′-GCCGAATATCATGGTGGAAA and 5′-AATATCACGGGTAGCCAACG).
NRO assay
NRO assays using BrUTP to label newly synthesized RNA were based on the methods developed by the Lis group (Core et al, 2008) and the Fu group (Lin et al, 2008). Briefly, ∼1 × 107 cells were washed three times in cold PBS on ice, followed by incubation in 10 ml ice-cold swelling buffer (10 mM Tris–HCl pH 7.5, 3 mM CaCl2, 2 mM MgCl2) for 5 min. Cells were collected with a scraper and pelleted with 500 g at 4°C for 10 min. Nuclei were isolated by pipetting cells up and down 20 times using a cut P1000 tip in 1 ml lysis buffer (swelling buffer with 0.5% NP-40, 10% glycerol, 20 U/ml RNasin). Nuclei were washed and pelleted in the lysis buffer, resuspended in 1 ml freezing buffer (50 mM Tris–HCl pH 8.3, 40% glycerol, 5 mM MgCl2, 0.1 mM EDTA). Nuclei were pelleted again with 1000 g at 4°C for 5 min, and resuspended in 100 μl of freezing buffer. An equal volume (100 μl) of reaction buffer (10 mM Tris–HCl pH 8.0, 5 mM MgCl2, 1 mM DTT, 300 mM KCl, 20 U of RNase inhibitor, 1% sarkosyl, 0.5 mM of BrUTP, ATP, and GTP, and 5 μM CTP) was added to carry out NRO at 30°C for 5 min.
NRO reporter assay
For NRO of reporter constructs, transfected HeLa Tet-On cells were treated with 10 ng/ml or 10 μg/ml Dox. After 16 h, cells were harvested for NRO as described above. The NRO reaction was stopped by adding Trizol into the reaction mix. RNA was then extracted, and treated with DNase I to remove DNA. Newly synthesized labeled RNA was pulled down by anti-BrdU antibody conjugated to the protein G Dynabeads (Invitrogen) in the binding buffer (10 mM Tris–HCl pH 7.4, 500 mM NaCl, 2.5 mM MgCl2, 0.5% Triton X-100, 0.5 μg/μl yeast RNA, 0.1 μg/μl yeast tRNA) at 4°C for 2 h. After immunoprecipitation (IP), beads were washed with the washing buffer (10 mM Tris–HCl pH 7.4, 500 mM NaCl, 2.5 mM MgCl2, 0.5% Triton X-100) six times. RNA was eluted from the beads by the RLT buffer (Qiagen) supplemented with yeast RNA (0.5 μg/μl). RNA was then purified by the RNeasy kit (Qiagen) and used for qRT–PCR with random hexamers as primer for the RT step. Primers used for qRT–PCR of NRO transcripts are shown in Supplementary Table 3.
GRO-seq
C2C12 cells grown to ∼80% confluency in DMEM+10% FBS were used for NRO, as described above. After NRO, the reaction mix was treated with 50 U DNase I at 37°C for 1 h followed by incubation at 55°C for 1 h with an equal volume of buffer S containing 20 mM Tris–HCl pH 7.4, 2% SDS, 10 mM EDTA, 200 mg/ml Proteinase K. RNA was extracted by phenol/chloroform twice, followed by ethanol precipitation. Purified RNA was fragmented to ∼100 nt by the RNA Fragmentation Reagents kit (Ambion) at 70°C for 15 min, and was subjected to IP with the anti-BrdU antibody (2 mg/reaction, Sigma) conjugated on the protein G Sepharose (GE Healthcare). Immunoprecipitated RNA was extracted by phenol/chloroform and ethanol precipitation. The IP step was repeated twice to obtain pure nascent RNA. Purified nascent RNA was treated with shrimp alkaline phosphatase (Roche) at 1 U/reaction at 37°C for 30 min to remove the 3′ phosphate group from RNA, followed by addition of the 5′ phosphate group with T4 kinase (NEB) at 10 U/reaction at 37°C for 1 h. Treated RNA was purified and used to prepare a sequencing library using the Illumina Small RNA Sample Prep Kit (v1.5). Deep sequencing was carried out on an Illumina Genome Analyzer IIx, which generated over 12.5 million uniquely mapped single reads (76 nt).
Supplementary Material
Supplementary Figures S1–5, Supplementary Tables S1–3
Acknowledgments
We thank Gary P Schroth at Illumina for providing RNA-seq data and James L Manley, Takashi Nagaike, Dinghai Zheng, Michael B Mathews, and Carol S Lutz for helpful discussions. This work was funded by a grant from NIH (R01 GM084089) to BT.
Author contributions: ZJ, WL (Luo), and BT conceived and designed research and wrote the paper; ZJ, WL (Luo), WL (Li), MH, ZP, and YZ performed research; ZJ, WL (Luo), WL (Li), and BT analyzed data.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adamson TE, Shutt DC, Price DH (2005) Functional coupling of cleavage and polyadenylation with transcription of mRNA. J Biol Chem 280: 32262–32271 [DOI] [PubMed] [Google Scholar]
- Ahn SH, Kim M, Buratowski S (2004) Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD (2005) An initial blueprint for myogenic differentiation. Genes Dev 19: 553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S (2005) Connections between mRNA 3′ end processing and transcription termination. Curr Opin Cell Biol 17: 257–261 [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D (2009) Histones: annotating chromatin. Annu Rev Genet 43: 559–599 [DOI] [PubMed] [Google Scholar]
- Colgan DF, Manley JL (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev 11: 2755–2766 [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costessi L, Devescovi G, Baralle FE, Muro AF (2006) Brain-specific promoter and polyadenylation sites of the beta-adducin pre-mRNA generate an unusually long 3′-UTR. Nucleic Acids Res 34: 243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L (1997) Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389: 399–402 [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Cheng TC, Cserjesi P, Chakraborty T, Olson EN (1992) Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol Cell Biol 12: 3665–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C (1997) Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res 25: 2547–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME (2008) Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60: 1022–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidatzis D, Jacobeit K, Oakeley EJ, Stadler MB (2009) Overestimation of alternative splicing caused by variable probe characteristics in exon arrays. Nucleic Acids Res 37: e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ (2007) The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126 [DOI] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL (2008) RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol 15: 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW (2008) The antisense transcriptomes of human cells. Science 322: 1855–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Manley JL (1998) RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395: 93–96 [DOI] [PubMed] [Google Scholar]
- Hogg JR, Goff SP (2010) Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D (2006) RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539 [DOI] [PubMed] [Google Scholar]
- Jaehning JA (2010) The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta 1799: 379–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B (2009) Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA 106: 7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Tian B (2009) Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One 4: e8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Edwards S, Shoemaker D, Schadt EE (2005) Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet 21: 93–102 [DOI] [PubMed] [Google Scholar]
- Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL (2007) The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev 21: 1779–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E (2009) The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR (2007) Coupling transcription and alternative splicing. Adv Exp Med Biol 623: 175–189 [DOI] [PubMed] [Google Scholar]
- Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H (2006) Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3′-UTRs. Nucleic Acids Res 34: 6264–6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner JN, Pearson EL, Moore C (2011) Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol 12: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Park JY, Tian B (2008) Identification of mRNA polyadenylation sites in genomes using cDNA sequences, expressed sequence tags, and Trace. Methods Mol Biol 419: 23–37 [DOI] [PubMed] [Google Scholar]
- Lee JY, Yeh I, Park JY, Tian B (2007) PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res 35: D165–D168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Lutz CS (2006) p54nrb is a component of the snRNP-free U1A (SF-A) complex that promotes pre-mRNA cleavage during polyadenylation. RNA 12: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB (2010) RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 11: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD (2008) The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol 15: 819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T (2010) Regulation of alternative splicing by histone modifications. Science 327: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Brodersen DE, Jensen TH (2009) Origins and activities of the eukaryotic exosome. J Cell Sci 122: 1487–1494 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Reed R (2002) An extensive network of coupling among gene expression machines. Nature 416: 499–506 [DOI] [PubMed] [Google Scholar]
- Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH (2010) Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell 40: 410–422 [DOI] [PubMed] [Google Scholar]
- Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C (2009) Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol 10: 1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, Gilmour DS, Albert I, Pugh BF (2008) Nucleosome organization in the Drosophila genome. Nature 453: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP (2009) Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL (1997) The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361 [DOI] [PubMed] [Google Scholar]
- Meinhart A, Cramer P (2004) Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430: 223–226 [DOI] [PubMed] [Google Scholar]
- Millevoi S, Vagner S (2010) Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res 38: 2757–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ (2009) Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nag A, Narsinh K, Martinson HG (2007) The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase. Nat Struct Mol Biol 14: 662–669 [DOI] [PubMed] [Google Scholar]
- Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL (2011) Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell 41: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA (2005) A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell 20: 213–223 [DOI] [PubMed] [Google Scholar]
- Ramskold D, Wang ET, Burge CB, Sandberg R (2009) An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Bakowski MA, McCracken S, Blencowe BJ (2003) Transcriptional activators control splicing and 3′-end cleavage levels. J Biol Chem 278: 43034–43040 [DOI] [PubMed] [Google Scholar]
- Rosonina E, Ip JY, Calarco JA, Bakowski MA, Emili A, McCracken S, Tucker P, Ingles CJ, Blencowe BJ (2005) Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol 25: 6734–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M (2009) The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci USA 106: 755–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB (2008) Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K (2008) Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G (2009) Chromatin organization marks exon-intron structure. Nat Struct Mol Biol 16: 990–995 [DOI] [PubMed] [Google Scholar]
- Sheldon KE, Mauger DM, Arndt KM (2005) A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol Cell 20: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR III, Frank J, Manley JL (2009) Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ III, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D (2007) Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 28: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB (2009) Biased chromatin signatures around polyadenylation sites and exons. Mol Cell 36: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL (1996) The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941–952 [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS (2005) A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR (2010) Most ‘dark matter’ transcripts are associated with known genes. PLoS Biol 8: e1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, Ripley BD (eds) (2002) In Modern Applied Statistics with S, 4th edn. New York: Springer [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM (2005) Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev 19: 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, Bohjanen PR (2008) Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell 29: 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Proudfoot NJ (2009) Transcriptional termination enhances protein expression in human cells. Mol Cell 33: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Kunath M, Roepcke S, Krause S, Schneider R, Schweiger S (2007) Alternative polyadenylation signals and promoters act in concert to control tissue-specific expression of the Opitz Syndrome gene MID1. BMC Mol Biol 8: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, Bourc'his D, Oakey RJ (2008) Regulation of alternative polyadenylation by genomic imprinting. Genes Dev 22: 1141–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Clauder-Munster S, Smolik M, Huber W, Steinmetz LM (2011) Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol 7: 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Marr TG (2005) Computational analysis of 3′-ends of ESTs shows four classes of alternative polyadenylation in human, mouse, and rat. Genome Res 15: 369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lee JY, Tian B (2005) Biased alternative polyadenylation in human tissues. Genome Biol 6: R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gilmour DS (2006) Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol Cell 21: 65–74 [DOI] [PubMed] [Google Scholar]
- Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D (2005) The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev 19: 1668–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–5, Supplementary Tables S1–3