Abstract
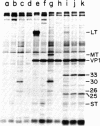
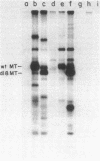
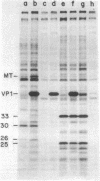
We have obtained antibodies specific for the polyoma virus middle-size tumor antigen (middle T antigen) by immunizing rabbits with a synthetic peptide, Lys-Arg-Ser-Arg-His-Phe, corresponding to the six carboxy-terminal amino acids of the middle T antigen predicted from the nucleotide sequence of polyoma DNA. The antipeptide serum precipitates the polyoma middle T antigen but not the small or large tumor antigens, and precipitation is inhibited in the presence of the peptide. Two cellular proteins, 30,000 and 26,000 daltons, are also precipitated specifically by the antipeptide serum and may have amino acid sequences related to the peptide. Two other cellular proteins, 33,000 and 25,000 daltons, are precipitated only in the presence of the peptide and may associate with it in cell extracts. Antisera directed against synthetic peptides are likely to be important in various ways, including the production of antibodies directed against particular determinants and the recognition of unknown proteins whose genes have been analyzed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Audibert F., Jolivet M., Chedid L., Alouf J. E., Boquet P., Rivaille P., Siffert O. Active antitoxic immunization by a diphtheria toxin synthetic oligopeptide. Nature. 1981 Feb 12;289(5798):593–594. doi: 10.1038/289593a0. [DOI] [PubMed] [Google Scholar]
- Benjamin T. L. Host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1970 Sep;67(1):394–399. doi: 10.1073/pnas.67.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart W., Hutchinson M. A., Hunter T. An activity phosphorylating tyrosine in polyoma T antigen immunoprecipitates. Cell. 1979 Dec;18(4):925–933. doi: 10.1016/0092-8674(79)90205-8. [DOI] [PubMed] [Google Scholar]
- FRIED M. CELL-TRANSFORMING ABILITY OF A TEMPERATURE-SENSITIVE MUTANT OF POLYOMA VIRUS. Proc Natl Acad Sci U S A. 1965 Mar;53:486–491. doi: 10.1073/pnas.53.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluck M. M., Benjamin T. L. Comparisons of two early gene functions essential for transformation in polyoma virus and SV-40. Virology. 1979 Jul 15;96(1):205–228. doi: 10.1016/0042-6822(79)90185-5. [DOI] [PubMed] [Google Scholar]
- Griffin B. E., Maddock C. New classes of viable deletion mutants in the early region of polyoma virus. J Virol. 1979 Sep;31(3):645–656. doi: 10.1128/jvi.31.3.645-656.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson M. A., Hunter T., Eckhart W. Characterization of T antigens in polyoma-infected and transformed cells. Cell. 1978 Sep;15(1):65–77. doi: 10.1016/0092-8674(78)90083-1. [DOI] [PubMed] [Google Scholar]
- Ito Y., Spurr N., Griffin B. E. Middle T antigen as primary inducer of full expression of the phenotype of transformation by polyoma virus. J Virol. 1980 Jul;35(1):219–232. doi: 10.1128/jvi.35.1.219-232.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. P., Crawford L. V. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979 Mar 15;278(5701):261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- Lania L., Gandini-Attardi D., Griffiths M., Cooke B., De Cicco D., Fried M. The polyoma virus 100K large T-antigen is not required for the maintenance of transformation. Virology. 1980 Feb;101(1):217–232. doi: 10.1016/0042-6822(80)90497-3. [DOI] [PubMed] [Google Scholar]
- Magnusson G., Berg P. Construction and analysis of viable deletion mutants of polyoma virus. J Virol. 1979 Nov;32(2):523–529. doi: 10.1128/jvi.32.2.523-529.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffhausen B. S., Benjamin T. L. Phosphorylation of polyoma T antigens. Cell. 1979 Dec;18(4):935–946. doi: 10.1016/0092-8674(79)90206-x. [DOI] [PubMed] [Google Scholar]
- Smith A. E., Smith R., Griffin B., Fried M. Protein kinase activity associated with polyoma virus middle T antigen in vitro. Cell. 1979 Dec;18(4):915–924. doi: 10.1016/0092-8674(79)90204-6. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Staneloni R. J., Fluck M. M., Benjamin T. L. Host range selection of transformation-defective hr-t mutants of polyoma virus. Virology. 1977 Apr;77(2):598–609. doi: 10.1016/0042-6822(77)90485-8. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G., Shinnick T. M., Green N., Liu F. T., Niman H. L., Lerner R. A. Chemical synthesis of a polypeptide predicted from nucleotide sequence allows detection of a new retroviral gene product. Nature. 1980 Oct 30;287(5785):801–805. doi: 10.1038/287801a0. [DOI] [PubMed] [Google Scholar]
- Türler H. The tumor antigens and the early functions of polyoma virus. Mol Cell Biochem. 1980 Sep 15;32(2):63–93. doi: 10.1007/BF00227801. [DOI] [PubMed] [Google Scholar]
- Walter G., Scheidtmann K. H., Carbone A., Laudano A. P., Doolittle R. F. Antibodies specific for the carboxy- and amino-terminal regions of simian virus 40 large tumor antigen. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5197–5200. doi: 10.1073/pnas.77.9.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. C., Hearing P., Rundell K. Cellular proteins associated with simian virus 40 early gene products in newly infected cells. J Virol. 1979 Oct;32(1):147–154. doi: 10.1128/jvi.32.1.147-154.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]