Abstract
We have previously isolated mesenchymal stromal cells (MSCs) from the tracheal aspirates of premature neonates with respiratory distress. Although isolation of MSCs correlates with the development of bronchopulmonary dysplasia, the physiologic role of these cells remains unclear. To address this, we further characterized the cells, focusing on the issues of gene expression, origin, and cytokine expression. Microarray comparison of early passage neonatal lung MSC gene expression to cord blood MSCs and human fetal and neonatal lung fibroblast lines demonstrated that the neonatal lung MSCs differentially expressed 971 gene probes compared with cord blood MSCs, including the transcription factors Tbx2, Tbx3, Wnt5a, FoxF1, and Gli2, each of which has been associated with lung development. Compared with lung fibroblasts, 710 gene probe transcripts were differentially expressed by the lung MSCs, including IL-6 and IL-8/CXCL8. Differential chemokine expression was confirmed by protein analysis. Further, neonatal lung MSCs exhibited a pattern of Hox gene expression distinct from cord blood MSCs but similar to human fetal lung fibroblasts, consistent with a lung origin. On the other hand, limiting dilution analysis showed that fetal lung fibroblasts form colonies at a significantly lower rate than MSCs, and fibroblasts failed to undergo differentiation along adipogenic, osteogenic, and chondrogenic lineages. In conclusion, MSCs isolated from neonatal tracheal aspirates demonstrate a pattern of lung-specific gene expression, are distinct from lung fibroblasts, and secrete pro-inflammatory cytokines.
Introduction
With improvements in neonatal care, the survival of very premature infants has increased. However, enhanced survival has been accompanied by an increase in bronchopulmonary dysplasia (BPD), a fibrotic lung disease requiring supplemental oxygen for months or years [1]. Survivors of BPD have abnormal lung function even as young adults [2], making BPD one of the leading causes of lung disease in children. The lungs of infants dying from BPD show larger and fewer alveoli, as well as poorly formed secondary crests, indicating an interference with septation [3,4]. Alveolar septa are thickened with collagen and elongated cells resembling fibroblasts [5]. These cells are positive for both α-smooth muscle actin and transforming growth factor (TGF)-β, a stimulus for myofibroblastic differentiation [6]. These results indicate that BPD may result in part from the abnormal differentiation or proliferation of alveolar mesenchymal progenitor cells to myofibroblasts.
Our laboratory has previously isolated plastic-adherent, fibroblast-like cells from the tracheal aspirates of premature infants undergoing mechanical ventilation for respiratory distress [7–9]. These cells possess colony-forming potential, surface markers, and differentiation potential typically found in mesenchymal stem cells. Recent work suggests that there is a hierarchy of multipotent mesenchymal stromal cells (MSCs) ranging from true self-renewing stem cells with multilineage differentiation capacity to those with more restricted differentiation potential, until a state of complete restriction to the fibroblast is reached [10]. Since we have not thoroughly tested the clonogenicity or in vivo self-renewal of neonatal lung mesenchymal cells, we now refer to them as MSCs, which have a more restricted differentiation potential. Patients from whom MSCs are isolated require more days of mechanical ventilation and supplemental oxygen, and are more likely to develop BPD, than patients from whom these cells are not isolated [7,9]. MSCs differentiate to myofibroblasts in vitro [8]. Together, these results suggest that neonatal lung MSCs play a significant role in the pathogenesis of BPD.
At present, the fundamental nature, origin, and physiologic role of neonatal lung MSCs are unclear. For example, it has not been established whether neonatal lung MSCs originate from the lung or some other tissue. Adult bone marrow contains a minority population of cells with characteristics of MSCs [11]. More recently, it has been suggested that most organs apparently carry their own population of MSCs in a perivascular compartment that participate in tissue repair [12,13]. Consistent with this, MSCs of donor sex identity have been found in lung allografts years after transplantation [14]. It is also unclear whether MSCs function as precursor cells for lung lipofibroblasts or myofibroblasts, or whether they serve an immunomodulatory role. Finally, the extent to which lung MSCs differ from lung fibroblasts is not clear. We hypothesized that MSCs isolated from neonatal lungs express specific genes indicative of a lung origin. Further, based on the association of neonatal lung MSCs with adverse clinical outcomes [7,9], we hypothesized that, compared with structural lung mesenchymal cells, neonatal lung MSCs possess a pro-inflammatory phenotype. To address these hypotheses, we compared the gene expression pattern of neonatal lung MSCs to cord blood MSCs and diploid fetal and neonatal lung fibroblasts.
Materials and Methods
Patients
We examined tracheal aspirates from infants admitted to the University of Michigan C.S. Mott Children's Hospital Newborn Intensive Care Unit, as approved by the University of Michigan Investigational Review Board. Entry criteria included gestational age at birth less than or equal to 33 weeks, mechanical ventilation for respiratory distress, and age ≤7 days. Infants with acute sepsis were excluded. Infants were suctioned on an as-needed basis. Samples were collected after informed consent from 1 parent or legal guardian was obtained.
Cell culture
MSCs from tracheal aspirates of premature infants hospitalized at the University of Michigan were isolated as described previously [7], with some modifications. Specimens were centrifuged (1,200× g for 5 min at 15°C) and the cell pellet resuspended in 2 mL Dulbecco's modified essential medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine, and 2.5 μg/mL amphotericin B. Adherent cells were incubated at 37°C and in 5% CO2 and grown until colonies were established. Neonatal lung MSC colony forming units typically develop between 7 and 14 days after plating of tracheal aspirates [7]. Individual colonies were identified and isolated using cloning disks (PGC Scientifics, Frederick, MD). MSCs of passage 3 and 4 were studied.
Cord blood MSCs were obtained with Institutional Review Board approval from full-term normal deliveries at the University of Vermont, as described previously [15]. Briefly, cord blood monocytes were isolated by Ficoll gradient centrifugation (Fisher Bio-Reagents, Pittsburgh, PA), resuspended in 1:1 mixture of cord blood basal medium and human bone marrow MSC-conditioned medium, plated in standard culture dishes (Corning, Pittsburgh, PA), and maintained until colonies were established. Experiments were performed on passage 3 cells.
The primary fetal/neonatal diploid lung fibroblast lines MRC-5 (passage 16 cells isolated from 14 week fetus), MRC-9 (passage 7 cells isolated from 15 week fetus), and CCD34-Lu (passage 6 cells from a patient who died of Rh incompatibility) were purchased from ATCC (Rockville, MD). These lines were selected because they are closely related in origin and gestational age to neonatal lung MSCs. Cells were initially propagated in a T150 flask with ATCC-formulated Eagle's minimum essential medium with 10% fetal bovine serum. At approximately 70% confluence, cells were collected by trypsinization and stored in 10% DMSO at −80°F until use.
After initial cell collection, all cell types were grown under identical conditions in Dulbecco's modified essential medium supplemented with MSC grade 10% fetal bovine serum (Invitrogen, Carlsbad, CA), 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine, and 2.5 μg/mL amphotericin B. For all studies, the medium was changed every 2–3 days.
Differentiation
Passage 3 neonatal lung MSCs and fetal lung fibroblasts were subjected to differentiation assays along adipogenic, osteogenic and chrondrogenic lineages as previously described, with minor modifications [11]. For adipogenic differentiation, passage 3 neonatal lung MSCs were cultured in standard medium supplemented with dexamethasone (10 μM), isobutylmethylxanthine (100 μM), indomethacin (50 μM), and insulin (100 μg/mL). For osteogenic differentiation, cells were cultured in the medium supplemented with dexamethasone (0.1 μM), β-glycerophosphate (10 mM), and L-ascorbic acid (50 μg/mL). Cells were stained with oil red O and alizarin red S for identification of fat vacuoles and calcium deposits, respectively. Chondrogenic differentiation was performed by pelleting 1×105 cells by centrifugation at 1,200 rpm for 5 min. The pellet was placed in media supplemented with 10 ng/mL each of TGF-β1 and bone morphogenic protein-4 for 21 days, fixed in 10% formalin, and sectioned for Alcian blue staining.
Colony-forming assay
Three colonies arising from the initial plating of tracheal aspirates from 3 individual subjects were isolated as described above, and single cells were obtained by limiting dilution into 6-well plates. After incubation for 14 days, cells were washed with phosphate-buffered saline (PBS) and stained with 0.5% crystal violet (Sigma-Aldrich, St. Louis, MO) for 15 min at room temperature, and visible colonies counted. Diluted fetal lung fibroblasts were similarly incubated and stained for counting.
Flow cytometry
Passage 3 neonatal lung MSCs were harvested with Enzyme-Free Hanks-based Cell Dissociation Buffer (Gibco/Invitrogen, Carlsbad, CA), pelleted by centrifugation, and resuspended in PBS with 50% ethanol for permeabilization and storage. Before flow cytometry, cells were pelleted and resuspended in PBS. Isotype controls and primary antibodies against cell surface markers typically associated with MSCs (Stro-1, CD 34, CD45, CD73, CD90, and CD105; BD Pharmingen, San Diego, CA) were added to 100-μl cell suspensions for 1 h at 4°C. Cells were washed, and appropriate Alexa fluor 488–tagged secondary antibody (Molecular Probes, Portland, OR) was added at 1:1,000 dilution for 30 min at 4°C in the dark. After washing, cells were immediately analyzed in a flow cytometer (FACSCalibur; Becton-Dickinson, Franklin Lakes, NJ).
Gene array
We examined the gene expression profile of 4 individual neonatal lung MSC isolates, 2 individual cord blood MSC isolates, and 3 distinct fetal lung fibroblast lines using the Illumina HumanRefSeq-8 v3 expression BeadChip platform (San Diego, CA). This system covers >24,000 probes for >18,000 unique genes from the NCBI RefSeq database. After growth to approximately 70% confluence on a 100-mm plate, cells were serum-starved for 24 h to minimize differences in cell cycle-related gene expression, and total RNA was extracted using the RNeasy Plus Mini kit (Qiagen, Valencia, CA). Further preparation and analysis was carried out by the University of Michigan Sequencing Core, according to the chip manufacturer's recommended protocol. Hybridized biotinylated cRNA was detected with streptavidin-Cy3 and quantitated using the Illumina BeadArray Reader. Microarray results are deposited in the NCBI GEO database (accession number GSE24583). To validate the gene array, we performed quantitative 2-step real-time polymerase chain reaction (qPCR) using specific Syber green primers. All primers were designed and purchased from IDT (Coralville, IA).
Analysis of cytokine protein expression
Neonatal lung MSCs and fetal/neonatal lung fibroblast lines at 70% confluence were serum-deprived for 24 h and conditioned media were collected for measurement of IL-8, IL-6, CXCL1/GRO-α, CCL2/MCP-1, hepatocyte growth factor (HGF), and vascular endothelial growth factor (VEGF). Protein levels were measured by multiplex immunoassay (Bio-Rad, Hercules, CA). For these assays, cells were seeded at an initial concentration of 1×105 cells/mL.
Fluorescence microscopy
Neonatal lung MSCs, MRC-5 neonatal lung fibroblasts and cord blood MSCs were grown on collagen- or fibronectin-coated glass slides (BD Biosciences, San Jose, CA). Cells were fixed in 1% paraformaldehyde. Cells were permeabilized in 0.1% Triton X-100 in PBS. Alexa Fluor (AF) dye antibody conjugates were prepared as previously described [8]. Slides were probed with AF555-conjugated rabbit anti-human Wnt5a, AF488-conjugated rabbit anti-human FoxF1, AF633-conjugated rabbit anti-human Tbx3 (all from Abcam, Cambridge, MA), or similarly labeled rabbit IgG. Cells were imaged using a Zeiss LSM 150 confocal microscope (Thornwood, NY).
Statistical analysis
For microarray analysis, values for each probe signal on the BeadChip array were ranked relative to the signals of negative controls that were thermodynamically equivalent to the regular probes, but lack specific targets in the transcriptome. These data were extracted using GenomeStudio Data Analysis Software (Illumina). Detection P values were computed using a nonparametric method as 1−R/N, where R is the rank of the probe signal relative to the negative controls and N is number of negative controls. A stronger signal has less probability of being generated from nonspecific sources. Thus, the detection P value assigns the probability of specific probe binding to the target transcript. Differences in expression between lung-derived MSCs, cord blood MSCs, and fetal lung fibroblasts were analyzed using the Illumina Custom differential expression algorithm, and multiple tests were corrected by the Benjamini and Hochberg false discovery rate [16]. The differential P value reflects the probability of difference between groups for a given probe. Detection and differential P values of <0.05 were determined to be significant. Functional classifications of target genes was performed using the web-based program database for annotation, visualization, and integrated discovery (DAVID) [17,18]. In this analysis, each identified category is provided a P value against the likelihood of finding the identified category by random chance. An unpaired 2-way t-test was used to determine the significance of protein expression data.
Results
Patient data
Lung MSCs from 10 different neonates intubated for respiratory distress were used in this study. Characteristics of these patients are described in Table 1. The mean gestational age and birth weight were 29 weeks and 1,267 g, respectively. Six of 10 (60%) babies required supplemental oxygen at 36 weeks, a clinical definition of BPD [1,19].
Table 1.
Patient Characteristics
| Patient | Gender | Race | Gestational age (weeks) | Birthweight (g) | Lived | BPD | No. of (+) samples/total samples |
|---|---|---|---|---|---|---|---|
| 1 | Female | Caucacian | 28 5/7 | 1210 | Yes | Yes | 2/2 |
| 2 | Male | Caucacian | 28 | 800 | No | Yes | 1/2 |
| 3 | Female | African-American | 29 2/7 | 1080 | Yes | No | 1/1 |
| 4 | Female | White | 27 5/7 | 845 | Yes | Yes | 2/2 |
| 5 | Male | White | 27 | 1265 | Yes | Yes | 2/2 |
| 6 | Female | White | 27 6/7 | 1615 | Yes | Yes | 3/3 |
| 7 | Female | White | 28 5/7 | 1180 | Yes | Yes | 1/2 |
| 8 | Male | White | 31 3/7 | 1620 | Yes | No | 1/1 |
| 9 | Female | White | 29 6/7 | 1425 | Yes | No | 1/2 |
| 10 | Female | African-American | 32 3/7 | 1630 | Yes | No | 1/1 |
| Mean | 29 | 1267 | 9/10 | 6/10 | |||
| ±SD | ±1.5 | ±306 |
Mesenchymal cells from tracheal aspirates demonstrate cell surface markers and differentiation potential typical of MSCs
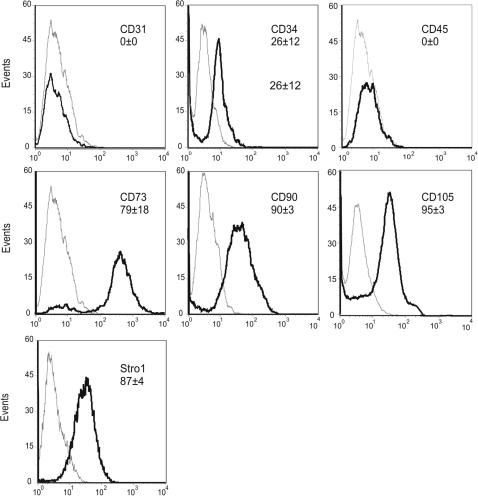
Plastic adherent cells from 3 neonatal tracheal aspirates were analyzed by flow cytometry (Fig. 1). As shown previously [7], MSCs were uniformly positive for Stro-1, CD73, CD90, and CD105, whereas negative for CD45. Cells showed weak staining for CD34, consistent with previous work demonstrating a population of CD34-positive in MSCs derived from second trimester fetal lung [20].
FIG. 1.
Immunophenotypic analysis of mesenchymal stromal cells (MSCs) by flow cytometry. Cells were stained for CD73, CD90, CD105, and Stro-1, each of which identifies MSCs. CD34 is mildly positive, and the endothelial and hematopoietic cell markers CD31 and CD45 are negative. Control reactions were performed with irrelevant isotype IgG controls (gray lines). Typical results from individual subjects are shown. The percentage of positive cells is shown for each marker (mean±SD, n=3).
Bead array results
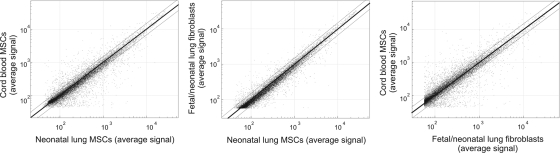
The concordance of gene expression between isolates, as measured by r2 values, was strong within the neonatal lung MSC (0.9372–0.9563), fetal lung fibroblast (0.9780–0.9854), and cord blood MSC (0.9521) groups. Intergroup r2 values were 0.9553 for the tracheal aspirate MSCs and fetal lung fibroblasts, 0.9016 between the tracheal aspirate MSCs and cord blood MSCs, and 0.8783 between cord blood MSCs and fetal lung fibroblasts (Fig. 2). Thus, with regard to the pattern of gene expression, tracheal aspirate MSCs resembled lung fibroblasts more than cord blood MSCs.
FIG. 2.
Concordance of gene probe detection P value; P≤0.05 between tracheal aspirate MSCs, cord blood MSCs, and fetal lung fibroblasts. Four MSC isolates, 3 fibroblast isolates, and 2 cord blood isolates were studied. Thin black lines signify 1.5-fold change based on average probe signal.
A detection P value of ≤0.05 identified 13,690 probes from neonatal lung MSCs, 12,939 probes from fetal lung fibroblasts, and 13,050 probes from cord blood MSCs. Comparing neonatal lung MSCs to cord blood MSCs, signals from 971 gene probes were statistically different (differential P value<0.05). As the Illumina Hum-Ref 8 bead array can detect statistical significance with as little as a 1.3-fold change based on average signal strength, we further narrowed our findings to include fold changes of ≥1.5. This resulted in 429 probes that were upregulated in neonatal MSCs compared with cord blood MSCs, and 282 that were downregulated. Among the upregulated genes were those involved in lung development, growth factor/cell signaling, inflammation, extracellular matrix, and lipid metabolism (Table 2, and Supplementary Table S1; Supplementary Data are available online at www.liebertonline.com/scd).
Table 2.
Selected Genes Differentially Expressed in Lung Mesenchymal Stromal Cells Relative to Cord Blood Mesenchymal Stromal Cells
| Symbol | Differential score P value | Fold increase | Definition |
|---|---|---|---|
| Development | |||
| HOXB5 | 0.04632 | 56.85 | Homeobox B5 |
| HOXA5 | 0.03145 | 36.98 | Homeobox A5 |
| FOXF1 | 0.00008 | 19.59 | Forkhead box F1 |
| LBH | 0.00035 | 10.40 | Limb bud and heart development homolog (mouse) |
| HOXB7 | 0.00522 | 9.77 | Homeobox B7 |
| WNT5A | 0.02439 | 6.51 | Wingless-type MMTV integration site family, member 5A |
| HOXB6 | 0.0352 | 5.84 | Homeobox B6 |
| HOXB3 | 0.00387 | 3.58 | Homeobox B3 |
| TBX3 | 0.03147 | 3.54 | T-box 3, transcript variant 1 |
| HOXB4 | 0.00313 | 3.22 | Homeobox B4 |
| HOXA2 | 0.0344 | 3.14 | Homeobox A2 |
| RUNX1 | 0 | 2.53 | Runt-related transcription factor 1, transcript variant 1 |
| HOXA4 | 0.00049 | 1.95 | Homeobox A4 |
| GLI2 | 0.01815 | 1.87 | GLI-Kruppel family member GLI2 |
| NOTCH1 | 0.00256 | 1.85 | Notch homolog 1, translocation-associated (Drosophila) |
| DKK3 | 0.00183 | 1.79 | Dickkopf homolog 3 (Xenopus laevis), transcript variant 1 |
| SMO | 0.00217 | 1.58 | Smoothened homolog (Drosophila) |
| SPRY2 | 0.00332 | 0.67 | Sprouty homolog 2 (Drosophila) |
| NOTCH2 | 0.00014 | 0.64 | Notch homolog 2 (Drosophila) |
| MESP1 | 0.01156 | 0.58 | Mesoderm posterior 1 homolog (mouse) |
| TWIST1 | 0.02902 | 0.53 | Twist homolog 1 |
| MEST | 0.00323 | 0.18 | Mesoderm specific transcript homolog (mouse), transcript variant 1 |
| Growth factor/cell signaling | |||
| IGFBP5 | 0.00856 | 5.23 | Insulin-like growth factor binding protein 5 |
| PDGFD | 0.01156 | 4.85 | Platelet derived growth factor D, transcript variant 1 |
| PDGFD | 0.02023 | 6.47 | Platelet derived growth factor D, transcript variant 2 |
| SOCS1 | 0.00539 | 2.47 | Suppressor of cytokine signaling 1 |
| BMP2K | 0.00001 | 2.25 | BMP2 inducible kinase, transcript variant 2 |
| 1EGFR | 0.00775 | 2.00 | Epidermal growth factor receptor, transcript variant 1 |
| JAK1 | 0.00496 | 1.83 | Janus kinase 1 |
| RARB | 0.00713 | 0.61 | Retinoic acid receptor, beta, transcript variant 1 |
| SMAD7 | 0.00003 | 0.49 | SMAD family member 7 |
| NGF | 0.00143 | 0.34 | Nerve growth factor (beta polypeptide) |
| STAT1 | 0.00658 | 0.33 | Signal transducer and activator of transcription 1, transcript variant α |
| STAT1 | 0.02745 | 0.31 | Signal transducer and activator of transcription 1, transcript variant β |
| Extracellular matrix | |||
| DCN | 0.01358 | 16.00 | Decorin, transcript variant C |
| LAMA2 | 0.03535 | 2.77 | Laminin, alpha 2, transcript variant 1 |
| COL6A3 | 0.00004 | 2.04 | Collagen, type VI, alpha 3 (COL6A3), transcript variant |
| COL5A1 | 0.01064 | 0.62 | Collagen, type V, alpha 1 |
| COL1A1 | 0.01543 | 0.53 | Collagen, type I, alpha 1 |
| LAMC2 | 0.00551 | 0.46 | Laminin, gamma 2, transcript variant 1 |
| FN1 | 0 | 0.22 | Fibronectin 1, transcript variant 6 |
| Inflammation | |||
| PDE5A | 0.02367 | 11.20 | Phosphodiesterase 5A, cGMP-specific, transcript variant 1 |
| PTGER2 | 0.04076 | 4.62 | Prostaglandin E receptor 2 (subtype EP2) |
| IRAK3 | 0.00775 | 3.80 | Interleukin-1 receptor-associated kinase 3 |
| NFKB1 | 0.00144 | 1.79 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| PLCG1 | 0.03371 | 1.51 | Phospholipase C, gamma 1, transcript variant 2 |
| IL1RAP | 0.01188 | 0.58 | Interleukin 1 receptor accessory protein, transcript variant 1 |
| TLR4 | 0.00001 | 0.28 | Toll-like receptor 4 |
| Lipid metabolism | |||
| OLR1 | 0.02776 | 7.27 | Oxidized low density lipoprotein receptor 1 |
| FASN | 0.01645 | 2.23 | Fatty acid synthase |
| ADFP | 0.03652 | 1.66 | Adipose differentiation-related protein |
| Others | |||
| CASP1 | 0.03147 | 7.71 | Caspase 1, apoptosis-related cysteine peptidase |
| CEBPD | 0.00747 | 6.19 | CCAAT/enhancer binding protein, delta |
| ABCC4 | 0 | 4.90 | ATP-binding cassette, sub-family C |
| ADM | 0.02776 | 3.42 | Adrenomedullin |
| HDAC1 | 0.00598 | 1.50 | Histone deacetylase 1 |
| THBS1 | 0.03937 | 0.46 | Thrombospondin 1 |
| ABCA3 | 0.02804 | 0.44 | ATP-binding cassette, sub-family A, member 3 |
| TAGLN | 0.04238 | 0.44 | Transgelin, transcript variant 1, mRNA |
| ODC1 | 0 | 0.42 | Ornithine decarboxylase 1 |
Diff Pval, differential score P value.
Comparing neonatal lung MSCs to fetal lung fibroblasts, there were 710 gene probes of statistically different strength. Using the 1.5-fold change cutoff, signals from 354 gene probes were increased and 110 probes were decreased. Among the upregulated genes were those involved in growth factor/cell signaling, transcription, extracellular matrix, inflammation, lipid metabolism, oxidative metabolism, and tight junctions (Table 3, and Supplementary Table S2). Complete results are provided in the online data supplement. We also analyzed these upregulated genes using the DAVID functional annotation tool [17,18]. Categories representing >5% of the differentially expressed genes and a P<0.01 were selected (Table 4). Unexpectedly, the top 3 gene ontology (GO) terms each involved lipid biosynthesis and metabolism.
Table 3.
Selected Genes Up-Regulated in Neonatal lung Mesenchymal Stromal Cells Relative to Fetal/Neonatal Lung Fibroblasts
| Symbol | Differential score P value | Fold increase | Definition |
|---|---|---|---|
| Growth factor/cell signaling | |||
| PTGS2 | 0.01108 | 3.37 | Homo sapiens prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| IRS2 | 0 | 3.24 | Insulin receptor substrate 2 |
| RHOB | 0 | 2.98 | ras homolog gene family, member B |
| POSTN | 0.00002 | 2.64 | Periostin, osteoblast specific factor |
| PDGFRB | 0.02029 | 2.02 | Platelet-derived growth factor receptor, beta polypeptide |
| FZD4 | 0.03599 | 1.99 | Frizzled homolog 4 (Drosophila) |
| BMP2K | 0.00011 | 1.96 | BMP2 inducible kinase, transcript variant 2 |
| ADCY3 | 0.02648 | 1.93 | Adenylate cyclase 3 |
| BMP2K | 0.00309 | 1.74 | BMP2 inducible kinase, transcript variant 1 |
| FYN | 0.00197 | 1.83 | FYN oncogene related to SRC, FGR, YES, transcript variant 3 |
| FYN | 0.0001 | 1.80 | FYN oncogene related to SRC, FGR, YES, transcript variant 2 |
| FYN | 0.01167 | 1.79 | FYN oncogene related to SRC, FGR, YES, transcript variant 1 |
| PIK3R1 | 0.0031 | 1.67 | Phosphoinositide-3-kinase, regulatory subunit 1 (α), transcript variant 1 |
| JAK1 | 0.03598 | 1.63 | Janus kinase 1 |
| TANK | 0.04767 | 1.58 | TRAF family member-associated NFKB activator, transcript variant 1 |
| PTEN | 0.00707 | 1.52 | Phosphatase and tensin homolog |
| ROCK1 | 0.00059 | 1.57 | Rho-associated, coiled-coil containing protein kinase 1 |
| BMP4 | 0.01126 | 0.56 | Bone morphogenetic protein 4, transcript variant 1 |
| SFRP1 | 0.00013 | 0.19 | Secreted frizzled-related protein 1 |
| Transcription | |||
| NKX3-1 | 0.04772 | 2.96 | NK3 homeobox 1 |
| TCF4 | 0.00002 | 2.19 | Transcription factor 4, transcript variant 2 |
| ATF4 | 0.0005 | 2.12 | Activating transcription factor 4, transcript variant 2 |
| GLI2 | 0.00344 | 2.09 | GLI-Kruppel family member GLI2 |
| KLF6 | 0.0292 | 1.91 | Kruppel-like factor 6, transcript variant 1 |
| KLF6 | 0.00381 | 1.88 | Kruppel-like factor 6, transcript variant 2 |
| CREBBP | 0.04601 | 1.64 | CREB binding protein, transcript variant 1 |
| CEBPG | 0.01062 | 1.60 | CCAAT/enhancer binding protein (C/EBP), gamma |
| GLI3 | 0.02884 | 1.63 | GLI-Kruppel family member GLI3 |
| ATF4 | 0.01673 | 1.57 | Activating transcription factor 4, transcript variant 1 |
| HDAC6 | 0.01317 | 1.53 | Histone deacetylase 6 |
| SP3 | 0.00093 | 1.52 | Sp3 transcription factor, transcript variant 2 |
| Extracellular matrix | |||
| COL8A1 | 0.0333 | 2.82 | Collagen, type VIII, alpha 1, transcript variant 2 |
| COL16A1 | 0 | 2.32 | Collagen, type XVI, alpha 1 |
| COL5A2 | 0.00101 | 2.12 | Collagen, type V, alpha 2 |
| COL5A1 | 0.01915 | 2.05 | Collagen, type V, alpha 1 |
| COL3A1 | 0.00002 | 1.97 | Collagen, type III, alpha 1 |
| COL8A1 | 0.04255 | 1.68 | Collagen, type VIII, alpha 1, transcript variant 1 |
| LAMB1 | 0.00984 | 1.56 | Laminin, beta 1 |
| VTN | 0.00739 | 0.56 | Vitronectin |
| Inflammation | |||
| IL8 | 0 | 24.50 | Interleukin 8 |
| IL6 | 0.00772 | 4.26 | Interleukin 6 |
| CXCL2 | 0.01386 | 3.75 | Chemokine (C-X-C motif) ligand 2 |
| NFKB1 | 0.0001 | 1.72 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| F10 | 0.03067 | 1.67 | Coagulation factor X |
| CCL20 | 0.04759 | 1.54 | Chemokine (C-C motif) ligand 20 |
| ADAM17 | 0.01387 | 1.62 | ADAM metallopeptidase domain 17 |
| MMP2 | 0.02185 | 1.53 | Matrix metallopeptidase 2 |
| IL17D | 0.00856 | 0.63 | Interleukin 17D |
| TIMP1 | 0.02736 | 0.57 | TIMP metallopeptidase inhibitor 1 |
| PLAU | 0.01751 | 0.40 | Plasminogen activator, urokinase |
| PTGER2 | 0 | 0.33 | Prostaglandin E receptor 2 (subtype EP2) |
| Lipid metabolism | |||
| FABP3 | 0.0445 | 5.95 | Fatty acid binding protein 3 |
| FADS1 | 0.00047 | 4.16 | Fatty acid desaturase 1 |
| FADS2 | 0.00736 | 3.60 | Fatty acid desaturase 2 (FADS2) |
| LDLR | 0.00001 | 2.61 | Low density lipoprotein receptor |
| FASN | 0.01282 | 2.48 | Fatty acid synthase |
| PPARG | 0 | 0.26 | Peroxisome proliferator-activated receptor gamma, transcript variant 2 |
| Oxidation | |||
| GPX7 | 0.00003 | 3.47 | Glutathione peroxidase 7 |
| NOX4 | 0.04887 | 3.09 | NADPH oxidase 4 |
| SOD2 | 0.03437 | 2.78 | Superoxide dismutase 2, mitochondrial, nuclear gene encoding mitochondrial protein, transcript variant 2 |
| Tight junctions | |||
| CGNL1 | 0.02648 | 3.06 | Cingulin-like 1 |
| PANX2 | 0.00001 | 2.32 | Pannexin 2 |
| TJP1 | 0.00096 | 1.65 | Tight junction protein 1 (zona occludens 1), transcript variant 2 |
| TJP1 | 0.02132 | 1.57 | Tight junction protein 1 (zona occludens 1), transcript variant 1 |
Diff Pval, differential score P value.
Table 4.
Functional Classifications of Target Genes Was Performed Using the Web-Based Database for Annotation, Visualization, and Integrated Discovery Program
| Term | Count | Percent | P value |
|---|---|---|---|
| Lipid biosynthetic process | 24 | 7.3 | 2.90E-10 |
| Cellular lipid metabolic process | 36 | 11 | 3.20E-10 |
| Lipid metabolic process | 38 | 11.6 | 5.40E-09 |
| Steroid metabolic process | 17 | 5.2 | 1.00E-07 |
| Alcohol metabolic process | 19 | 5.8 | 8.40E-06 |
| Biosynthetic process | 48 | 14.7 | 3.00E-05 |
| Response to stress | 35 | 10.7 | 2.80E-04 |
| Apoptosis | 27 | 8.3 | 7.00E-04 |
| Programmed cell death | 27 | 8.3 | 7.90E-04 |
| Cellular metabolic process | 164 | 50.2 | 1.10E-03 |
| Protein modification process | 48 | 14.7 | 1.30E-03 |
| Death | 27 | 8.3 | 1.70E-03 |
| Cell death | 27 | 8.3 | 1.70E-03 |
| Biopolymer modification | 48 | 14.7 | 2.90E-03 |
| Metabolic process | 176 | 53.8 | 3.40E-03 |
| Primary metabolic process | 161 | 49.2 | 4.20E-03 |
| Negative regulation of biological process | 33 | 10.1 | 4.60E-03 |
| Organ development | 35 | 10.7 | 4.90E-03 |
| Post-translational protein modification | 39 | 11.9 | 7.90E-03 |
| Negative regulation of cellular process | 31 | 9.5 | 8.20E-03 |
It has been demonstrated that MSCs and fibroblasts have characteristic Homeobox (Hox) expression signatures that are specific for their anatomical origin [21,22]. Comparison of the Hox gene profiles demonstrated similar expression patterns between neonatal lung MSCs and fetal lung fibroblasts (Table 5). The Hox expression profile for cord blood MSC was distinct, however. Together, these results are consistent with the notion that MSCs isolated from the tracheal aspirates of premature infants are derived from lung rather than hematopoietic tissue.
Table 5.
Expression of Homeobox Genes
| |
Neonatal lung mesenchymal stromal cells |
Fetal lung fibroblasts |
Cord blood mesenchymal stromal cells |
|||
|---|---|---|---|---|---|---|
| Symbol | Average probe signal | Detection P value | Average probe signal | Detection P value | Average probe signal | Detection P value |
| HOXA2 | 149.4 | 0 | 132.7 | 0 | 47.6 | 0.57078 |
| HOXA4 | 88.1 | 0.00452 | 79.9 | 0.01205 | 45.2 | 0.85542 |
| HOXA5 | 1575.5 | 0 | 2462.3 | 0 | 42.6 | 0.95633 |
| HOXA6 | 65.5 | 0.02108 | 97.1 | 0.00301 | 59.6 | 0.06928 |
| HOXA9 | 41 | 0.59639 | 58.5 | 0.03614 | 52.7 | 0.18223 |
| HOXB1 | 94.7 | 0.00301 | 93.7 | 0.00301 | 96.7 | 0.00602 |
| HOXB2 | 610.3 | 0 | 397.9 | 0 | 288.9 | 0 |
| HOXB3 | 163.6 | 0 | 98.6 | 0.00301 | 45.7 | 0.76506 |
| HOXB4 | 162 | 0 | 132.7 | 0 | 50.3 | 0.33434 |
| HOXB5 | 3331.2 | 0 | 3890.6 | 0 | 58.6 | 0.07681 |
| HOXB6 | 65.9 | 0.02108 | 65.6 | 0.01657 | 72.8 | 0.01657 |
| HOXB6 | 397.2 | 0 | 355.2 | 0 | 68 | 0.02259 |
| HOXB7 | 439.6 | 0 | 516 | 0 | 45 | 0.80723 |
| HOXB8 | 48.2 | 0.14006 | 258.1 | 0 | 44.6 | 0.9006 |
| HOXC11 | 72.1 | 0.01657 | 68.5 | 0.01657 | 76.2 | 0.01355 |
| HOXC4 | 41.6 | 0.51355 | 59.2 | 0.03614 | 47.1 | 0.68524 |
| HOXC6 | 47.1 | 0.16566 | 172.6 | 0 | 52.5 | 0.2259 |
| HOXC8 | 45.3 | 0.24849 | 95 | 0.00301 | 51.5 | 0.28313 |
| HOXD12 | 152 | 0 | 142.9 | 0 | 133.4 | 0 |
| HOXD13 | 109.4 | 0.00301 | 104.9 | 0.00301 | 97.3 | 0.00602 |
| HOXD3 | 73.6 | 0.01657 | 69.4 | 0.01657 | 83.3 | 0.01054 |
Bold font indicates that the given probe achieved statistical significance for detection. Only Hox genes expressed in at least 1 cell line were shown.
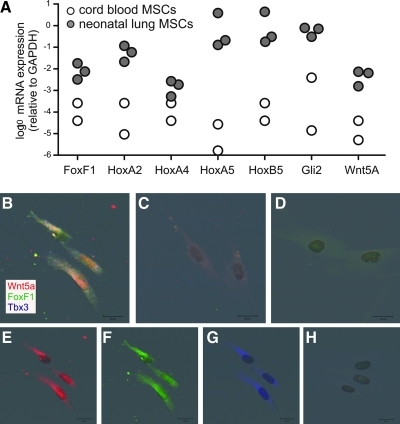
To validate the gene array, we performed qPCR for 7 lung-specific transcription factors that, according to the gene array, were significantly overexpressed in lung MSCs compared with cord blood MSCs. Gene expression of FoxF1, HoxA2, HoxA4, HoxA5, HoxB5, Gli2, and Wnt5A was 1–5 logs higher in neonatal lung MSCs than cord blood MSCs (Fig. 3A). Cells were also examined by fluorescent microscopy. Neonatal lung MSCs demonstrated increased expression of FoxF1, Wnt5a, and Tbx3 compared with MRC-5 fetal lung fibroblasts or cord blood MSCs (Fig. 3B).
FIG. 3.
Neonatal lung MSCs express higher levels of lung-specific transcription factors than cord blood MSCs. (A) mRNA expression of FoxF1, HoxA2, HoxA4, HoxA5, HoxB5, Gli2, and Wnt5A was assessed by qPCR. Data were normalized to GAPDH. (B–H). Immunocytochemistry of neonatal lung MSCs, fetal lung fibroblasts, and cord blood MSCs. Cells were probed for Wnt5a (red), FoxF1 (green), and Tbx3 (blue). Colocalization is white. (B) Neonatal lung MSCs. (C) MRC-5 fetal lung fibroblasts. (D) Cord blood MSCs. (E–G) Individual channels for Wnt5a, FoxF1, and Tbx3. (H) MRC-5 fetal lung fibroblasts stained with labeled control IgGs.
Lung MSCs secrete greater levels of cytokines than fetal lung fibroblasts
As shown in Table 4, the mRNA expression of a number of cytokines, including IL8 and IL6, was significantly increased in neonatal lung MSCs compared with fetal lung fibroblasts. In addition, there were other cytokines, including CXCL1 and CXCL5, expressed in neonatal lung MSCs but not in fetal lung fibroblasts (data not shown). Differences in CXCL1/GRO-α, CXCL8/IL-8, and IL-6 were also present at the protein level (Fig. 4). Compared with fetal/neonatal lung fibroblasts, MSCs showed no increase in the secretion of CCL2/MCP-1, HGF, or VEGF.
FIG. 4.
Cytokine secretion by neonatal lung MSCs and fetal lung fibroblasts. Passage 3 neonatal lung MSCs secreted greater amounts of CXCL1/GRO-α, CXCL-8/IL-8, and IL-6 (n=9) compared with the 3 isolates of fetal/neonatal lung fibroblasts tested (*P<0.05, unpaired t-test). There was no statistical difference in the secretion of CCL2/MCP-1, hepatocyte growth factor, or vascular endothelial growth factor.
Differences between MSCs from patients developing BPD and MSCs from patients who do not develop chronic lung disease
Of the 9 MSC isolates tested for cytokine/growth factor protein secretion, 5 were isolated from patients who later developed BPD and 4 were isolated from patients who did not. Cells isolated from patients with BPD showed significantly decreased secretion of CXCL1/GRO-α and HGF (Fig. 4, P<0.05, unpaired t test), an epithelial repair factor that has been shown to suppress bleomycin-induced lung fibrosis [23–25]. On the other hand, cells from BPD patients showed greater levels of VEGF (Fig. 3, P<0.05, unpaired t test), a growth factor that has been associated with pulmonary fibrosis [26]. Together, these results suggest that neonatal lung MSCs from patients with BPD maintain a stable anti-inflammatory, pro-fibrotic phenotype.
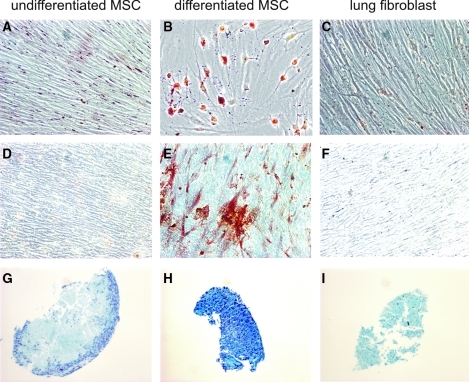
Lung MSCs, but not fetal lung fibroblasts, undergo differentiation along mesenchymal lineages and form colonies at a high rate
Given the similarities in Hox gene expression between neonatal lung MSCs and fetal/neonatal lung fibroblasts, we performed additional studies comparing the differentiation and colony forming potential of these cells. As shown previously [16], MSC colonies from neonatal tracheal aspirate underwent adipogenic, osteogenic, and chondrogenic differentiation (Fig. 5). Cord blood MSCs were also capable of differentiation along the 3 mesenchymal lineages [15]. Fetal lung fibroblasts treated under the same differentiating conditions failed to undergo differentiation. Next, to determine colony-forming potential, limiting dilution was performed to generate single cells. On average, 73% of MSCs formed colonies when followed over a 14-day period. These results demonstrate that neonatal lung MSCs are not dependent on the presence of other cells for survival. However, when cells from 3 fetal lung fibroblast lines were tested, only 10% of fetal/neonatal lung fibroblasts formed colonies (P=0.006). Together with the cytokine expression data, these results provide strong evidence that neonatal lung MSCs represent a distinct subset of lung mesenchymal cells and are qualitatively different from lung fibroblasts.
FIG. 5.
Differentiation capability of neonatal lung MSCs and fetal lung fibroblasts. Under adipogenic conditions, lipid droplets are revealed with staining by with oil red O (A–C). Osteogenic growth conditions create calcium deposits seen after staining with alizarin red (D–F). Chondrogenic differentiation promotes production of glycosaminoglycans, which are detected by staining with Alcian blue (G–I). Neonatal lung MSCs, but not fetal lung fibroblasts, differentiate along these lineages. We analyzed 3 MSC isolates, 3 fibroblast isolates, and 2 cord blood MSC isolates, and all yielded the same results.
Discussion
Our laboratory has isolated plastic-adherent, fibroblast-like cells from the tracheal aspirates of premature infants undergoing mechanical ventilation for respiratory distress. These cells have the cell surface markers and in vitro differentiation potential of MSCs [27]. Patients from whom MSCs are isolated require significantly more days of mechanical ventilation and supplemental oxygen, and develop BPD at a significantly higher rate than patients from whom these cells were not isolated [9]. In the present study, study subjects again demonstrated a high prevalence of BPD; 60% developed BPD, compared with the reported 25% of very low birth weight premature infants [1]. These data suggest that MSCs play a role in lung injury, and are not simply the result of endotracheal intubation. In this report, we further describe these cells to better understand the role of MSCs in lung injury, focusing on the issues of gene expression, origin, and cytokine expression
Until now, we had not examined the origin of tracheal aspirate neonatal MSCs. Although MSCs were first identified in bone marrow, MSCs of donor sex identity have been found in lung allografts years after transplantation, suggesting that MSCs may originate from the lung tissue itself [14], perhaps from the perivascular compartment [12,13]. It has also been shown that bone-marrow-derived progenitor cells may contribute to the population of lung cells found in the murine lung after either bleomycin [28] or naphthalene-induced [29] lung injury. To examine this question, we took advantage of the fact that MSCs and fibroblasts have characteristic Hox expression signatures that are highly specific for their organ of origin [21,22]. Patterns of MSC Hox gene expression are not altered by the presence of hematopoietic cells or by differentiation [22]. These data are consistent with the notion that MSCs primarily colonize tissues as constituents of the wall of ingrowing blood vessels [12,13]. Although cells from neonatal tracheal aspirates and cord blood each met criteria for MSCs [27], they showed distinct patterns of Hox gene expression. Several Hox genes implicated in lung development, including Hoxa2, a4, a5, and b5, were expressed in neonatal lung MSCs but absent in cord blood MSCs [27,30–33]. On the other hand, there was a notable similarity of Hox gene expression between neonatal tracheal aspirate MSCs and fetal lung fibroblasts, consistent with a common origin.
Additionally, in contrast to cord blood MSCs, neonatal lung MSCs expressed several transcription factors important in pulmonary development. Wnt5a, Tbx2, Tbx3, FoxF1, and Gli2 were each significantly upregulated compared with cord blood MSCs. Wnt5a transcripts are detected in mesenchymal and epithelial compartments of the developing lung. Wnt5a knockout mice demonstrate truncation of the trachea and overexpansion of the distal airways [34,35], highlighting the importance of Wnt5a in distal lung morphogenesis. Forkhead Box F1 (FoxF1) is involved in the regulation of mesenchymal–epithelial interactions critical for lung morphogenesis. FoxF1 heterozygous null mice show defects in alveolarization and vasculogenesis [36]. FoxF1 heterozygotes also exhibit decreased expression of members of the Brachyury T-Box gene family Tbx 2–5. Tbx 2–5 encode DNA-binding transcription factors that are expressed in the developing lung mesenchyme and regulate branching morphogenesis [37,38]. Together with the Hox gene expression profile, these data strongly suggest that MSCs from neonatal tracheal aspirates originate in the lung itself, rather than a peripheral site such as blood or bone marrow.
Although neonatal lung MSCs and fetal/neonatal lung fibroblasts demonstrated a similar pattern of Hox gene expression, comparison of gene expression patterns also revealed important differences, most notably in the areas of inflammation and lipid metabolism. Increases in cytokine gene expression were confirmed by analysis of protein levels. The strong induction of pro-inflammatory cytokines is consistent with the notion that MSCs modulate the inflammatory response [39]. Based on the highly significant induction of genes involved in lipid biosynthesis and metabolism, as assessed using the DAVID functional annotation tool [17,18], as well as the capacity of these cells to undergo adipogenic differentiation, we speculate that neonatal lung MSCs may also serve as precursors for alveolar lipofibroblasts. These cells provide type II cells with neutral lipid for surfactant synthesis [40] and may play a role in alveolarization [41].
Based the capacity of MSCs to accumulate contractile proteins under the influence of TGF-β [8], they may also differentiate into alveolar myofibroblasts. In the mouse, α-actin- and elastin-expressing myofibroblasts are required for the formation of pulmonary alveolar secondary septi [42,43]. However, excessive proliferation or differentiation of myofibroblasts, as found in infants with BPD [6], can impair alveolarization. Interestingly, neonatal lung MSCs also appear to express numerous genes that encode products associated with either alveolar development (adipose differentiation-related protein, adrenomedullin) [44,45] or fibrosis (Nox4, coagulation factor X) [46,47]. Thus, the physiologic role of neonatal lung MSCs may depend on the differential translation of such genes, as regulated by such clinical factors as mechanical ventilation, supplemental oxygen, and infection. Consistent with this, we found MSCs isolated from patients who ultimately developed BPD elaborated decreased levels of CXCL1/GRO-α and HGF, and increased levels of VEGF, compared with isolates from patients without chronic lung disease. Together, these results suggest that neonatal lung MSCs from patients with BPD maintain a stable anti-inflammatory, pro-fibrotic phenotype that leads to disorganized lung repair.
Finally, we compared the differentiation and colony-forming potential of neonatal lung MSCs and fetal/neonatal lung fibroblasts. We confirmed that the plastic-adherent, colony-forming cells isolated from the tracheal aspirates of premature infants with respiratory distress meet the criteria for MSCs on the basis of cell surface markers, ability to differentiate along adipogenic, osteogenic, and chondrogenic lineages, and ability to generate colonies [27]. These data also support the notion that MSCs do not require support from other cells to expand in an in vitro environment. In contrast, fetal lung fibroblasts generated colonies to a much lower extent than MSCs, and failed to undergo significant morphological changes in differentiation media.
We acknowledge some drawbacks of our study. First, several factors may affect gene expression and cytokine production from the cells, making comparisons difficult. For example, we studied commercially available, higher passage fetal and neonatal lung fibroblasts, rather than primary neonatal lung fibroblasts. Two of these lines were obtained from extremely early lungs, raising the possibility of maturational differences in gene expression. Differences in methods of cell isolation and health status may also have affected our results. Due to ethical concerns, the neonatal primary cells are not readily available. However, one could argue that use of higher passage fetal lung fibroblast cell lines might select for more rapidly dividing primitive cells, or favor the acquisition of mutations enhancing colony forming potential. Nevertheless, fetal lung fibroblasts showed minimal ability to form clones after limiting dilution, or to differentiate among mesenchymal lineages. Second, whereas Hox gene and transcription factor profiles strongly suggest that neonatal tracheal aspirate MSCs from tracheal aspirates are indeed lung-derived, we acknowledge the possibility that they could originate in the bone-marrow and undergo change to a lung phenotype upon exposure to the pulmonary milieu. Answering this question conclusively would require more sophisticated methods such as lineage tagging, which cannot be accomplished in newborn infants. However, as we noted above, it has been shown that patterns of MSC Hox gene expression are not altered by the presence of hematopoietic cells or by differentiation [22]. Third, our study does not address the potential role of lung- or bone marrow-derived MSCs in the pathogenesis or treatment of neonatal lung injury. In contrast to our study, in which MSCs are associated with an adverse pulmonary outcome, administration of bone-marrow-derived MSCs, or their conditioned media, has been shown to ameliorate hyperoxia-induced murine neonatal lung injury [48,49]. The reason for these discrepant outcomes is unclear, but the most obvious explanation likely relates to the different microenvironments to which lung- and bone marrow-derived MSCs are exposed. Finally, as touched on above, the study of cells derived from newborn human infants makes it impractical to perform the experimental interventions necessary to definitively prove MSC origin or function. Nevertheless, we believe that our careful characterization of these cells, combined with outcomes data and the development of appropriate animal models, will ultimately link lung MSCs to lung injury and repair.
In conclusion, we have isolated and characterized MSCs from tracheal aspirates of premature neonates with respiratory distress. This present study strengthens the concept that these cells are indeed progenitor cells, as evidenced by their ability to form clones from single cells and differentiate along mesenchymal lineages, in contrast to fetal lung fibroblasts. Further, based on their Hox gene profile and expression of lung transcription factors, our data support the notion that these cells are lung-derived. Finally, compared with fetal/neonatal lung fibroblasts, MSCs demonstrate increased expression of pro-inflammatory cytokines. Further studies will be required to understand the precise physiological role of these cells in lung development and repair.
Supplementary Material
Acknowledgments
This work was supported by NIH HL90134 (to M.B.H.). The authors thank Dr. Jeffrey L. Spees, University of Vermont College of Medicine, for his assistance in obtaining cord blood-derived mesenchymal stem cells.
Author Disclosure Statement
The authors declare no commercial associations that might create a conflict of interest in connection with submitted article.
References
- 1.Jobe AH. Bancalari E. Bronchopulmonary dysplasia. Am J Respir Crit Care Med. 2001;163:1723–1729. doi: 10.1164/ajrccm.163.7.2011060. [DOI] [PubMed] [Google Scholar]
- 2.Eber E. Zach MS. Paediatric origins of adult lung disease {bullet} 8: long term sequelae of bronchopulmonary dysplasia (chronic lung disease of infancy) Thorax. 2001;56:317–323. doi: 10.1136/thorax.56.4.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hussain NA. Siddiqui NH. Stocker JR. Pathology of arrested acinar development in postsurfactant bronchopulmonary dysplasia. Hum Pathol. 1998;29:710–717. doi: 10.1016/s0046-8177(98)90280-5. [DOI] [PubMed] [Google Scholar]
- 4.Coalson JJ. Pathology of new bronchopulmonary dysplasia. Semin Neonatol. 2003;8:73–81. doi: 10.1016/s1084-2756(02)00193-8. [DOI] [PubMed] [Google Scholar]
- 5.Bhatt AJ. Pryhuber GS. Huyck H. Watkins RH. Metlay LA. Maniscalco WM. Disrupted pulmonary vasculature and decreased vascular endothelial growth factor, Flt-1, and TIE-2 in human infants dying with bronchopulmonary dysplasia. Am J Respir Crit Care Med. 2001;164:1971–1980. doi: 10.1164/ajrccm.164.10.2101140. [DOI] [PubMed] [Google Scholar]
- 6.Toti P. Buonocore G. Tanganelli P. Catella AM. Palmeri ML. Vatti R. Seemayer TA. Bronchopulmonary dysplasia of the premature baby: an immunohistochemical study. Pediatr Pulmonol. 1997;24:22–28. doi: 10.1002/(sici)1099-0496(199707)24:1<22::aid-ppul4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 7.Hennrick KT. Keeton AG. Nanua S. Kijek TG. Goldsmith AM. Sajjan US. Bentley JK. Lama VN. Moore BB. Schumacher RE. Thannickal VJ. Hershenson MB. Lung cells from neonates show a mesenchymal stem cell phenotype. Am J Respir Crit Care Med. 2007;175:1158–1164. doi: 10.1164/rccm.200607-941OC. [DOI] [PubMed] [Google Scholar]
- 8.Popova AP. Bozyk PD. Goldsmith AM. Linn MJ. Lei J. Bentley JK. Hershenson MB. Autocrine production of TGF-b1 promotes myofibroblastic differentiation of neonatal lung mesenchymal stem cells. Am J Physiol Lung Cell Mol Physiol. 2010;298:L735–L743. doi: 10.1152/ajplung.00347.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popova AP. Bozyk PD. Bentley JK. Linn MJ. Goldsmith AM. Schumacher RE. Filbrun AG. Weiner GM. Hershenson MB. Isolation of mesenchymal stromal cells from tracheal aspirates of premature infants predicts bronchopulmonary dysplasia. Pediatrics. 2010;126:e1127–e1133. doi: 10.1542/peds.2009-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarugaser R. Hanoun L. Keating A. Stanford WL. Davies JE. Human mesenchymal stem cells self-renew and differentiate according to a deterministic hierarchy. PLoS One. 2009;4:e6498. doi: 10.1371/journal.pone.0006498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pittenger MF. Mackay AM. Beck SC. Jaiswal RK. Douglas R. Mosca JD. Moorman MA. Simonetti DW. Craig S. Marshak D. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 12.da Silva Meirelles L. Chagastelles PC. Nardi NB. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci. 2006;119:2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 13.Crisan M. Yap S. Casteilla L. Chen C-W. Corselli M. Park TS. Andriolo G. Sun B. Zheng B. Zhang L. Norotte C. Teng P-N. Traas J. Schugar R. Deasy BM. Badylak S. Buhring H-J. Giacobino J-P. Lazzari L. Huard J. Péault B. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Lama VN. Smith L. Badri L. Flint AJ. Andrei A-C. Murray S. Wang Z. Liao H. Toews GB. Krebsbach PH. Peters-Golden M. Pinsky DJ. Martinez FJ. Thannickal VJ. Evidence for tissue-resident mesenchymal stem cells in human adult lung from studies of transplanted allografts. J Clin Invest. 2007;117:989–996. doi: 10.1172/JCI29713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sueblinvong V. Loi R. Eisenhauer PL. Bernstein IM. Suratt BT. Spees JL. Weiss DJ. Derivation of lung epithelium from human cord blood-derived mesenchymal stem cells. Am J Respir Crit Care Med. 2008;177:701–711. doi: 10.1164/rccm.200706-859OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benjamini Y. Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 17.Dennis G. Sherman BT. Hosack DA. Yang J. Gao W. Lane HC. Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 18.Huang DW. Sherman BT. Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Ehrenkranz RA. Walsh MC. Vohr BR. Jobe AH. Wright LL. Fanaroff AA. Wrage LA. Poole K for the National Institutes of Child Health Human Development Neonatal Research Network. Validation of the National Institutes of Health Consensus Definition of Bronchopulmonary Dysplasia. Pediatrics. 2005;116:1353–1360. doi: 10.1542/peds.2005-0249. [DOI] [PubMed] [Google Scholar]
- 20.in't Anker PS. Noort WA. Scherjon SA. Kleijburg-van der Keur C. Kruisselbrink AB. van Bezooijen RL. Beekhuizen W. Willemze R. Kanhai HH. Fibbe WE. Mesenchymal stem cells in human second-trimester bone marrow, liver, lung, and spleen exhibit a similar immunophenotype but a heterogeneous multilineage differentiation potential. Haematologica. 2003;88:845–852. [PubMed] [Google Scholar]
- 21.Chang HY. Chi J-T. Dudoit S. Bondre C. van de Rijn M. Botstein D. Brown PO. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci USA. 2002;99:12877–12882. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ackema KB. Charite J. Mesenchymal stem cells from different organs are characterized by distinct topographic Hox codes. Stem Cells and Development. 2008;17:979–992. doi: 10.1089/scd.2007.0220. [DOI] [PubMed] [Google Scholar]
- 23.Yaekashiwa M. Nakayama S. Ohnuma K. Sakai T. Abe T. Satoh KEN. Matsumoto K. Nakamura T. Takahashi T. Nukiwa T. Simultaneous or Delayed Administration of Hepatocyte Growth Factor Equally Represses the Fibrotic Changes in Murine Lung Injury Induced by Bleomycin. A Morphologic Study. Am J Respir Crit Care Med. 1997;156:1937–1944. doi: 10.1164/ajrccm.156.6.9611057. [DOI] [PubMed] [Google Scholar]
- 24.Dohi M. Hasegawa T. Yamamoto K. Marshall BC. Hepatocyte Growth Factor Attenuates Collagen Accumulation in a Murine Model of Pulmonary Fibrosis. Am J Respir Crit Care Med. 2000;162:2302–2307. doi: 10.1164/ajrccm.162.6.9908097. [DOI] [PubMed] [Google Scholar]
- 25.Gazdhar A. Fachinger P. van Leer C. Pierog J. Gugger M. Friis R. Schmid RA. Geiser T. Gene transfer of hepatocyte growth factor by electroporation reduces bleomycin-induced lung fibrosis. Am J Physiol Lung Cell Mol Physiol. 2007;292:L529–L536. doi: 10.1152/ajplung.00082.2006. [DOI] [PubMed] [Google Scholar]
- 26.Hamada N. Kuwano K. Yamada M. Hagimoto N. Hiasa K. Egashira K. Nakashima N. Maeyama T. Yoshimi M. Nakanishi Y. Anti-vascular endothelial growth factor gene therapy attenuates lung injury and fibrosis in mice. J Immunol. 2005;175:1224–1231. doi: 10.4049/jimmunol.175.2.1224. [DOI] [PubMed] [Google Scholar]
- 27.Dominici M. Le Blanc K. Mueller I. Slaper-Cortenbach I. Marini F. Krause D. Deans R. Keating A. Prockop D. Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 28.Hashimoto N. Jin H. Liu T. Chensue SW. Phan SH. Bone marrow-derived progenitor cells in pulmonary fibrosis. J Clin Invest. 2004;113:243–252. doi: 10.1172/JCI18847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong AP. Keating A. Lu W-Y. Duchesneau P. Wang X. Sacher A. Hu J. Waddell TK. Identification of a bone marrow-derived epithelial-like population capable of repopulating injured mouse airway epithelium. J Clin Invest. 2009;119:336–348. doi: 10.1172/JCI36882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boucherat O. Franco-Montoya ML. Thibault C. Incitti R. Chailley-Heu B. Delacourt C. Bourbon JR. Gene expression profiling in lung fibroblasts reveals new players in alveolarization. Physiol Genomics. 2007;32:128–141. doi: 10.1152/physiolgenomics.00108.2007. [DOI] [PubMed] [Google Scholar]
- 31.Mandeville I. Aubin J. LeBlanc M. Lalancette-Hebert M. Janelle MF. Tremblay GM. Jeannotte L. Impact of the loss of Hoxa5 function on lung alveogenesis. Am J Pathol. 2006;169:1312–1327. doi: 10.2353/ajpath.2006.051333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volpe MV. Vosatka RJ. Nielsen HC. Hoxb-5 control of early airway formation during branching morphogenesis in the developing mouse lung. Biochim Biophys Acta. 2000;1475:337–345. doi: 10.1016/s0304-4165(00)00087-8. [DOI] [PubMed] [Google Scholar]
- 33.Volpe MV. Pham L. Lessin M. Ralston SJ. Bhan I. Cutz E. Nielsen HC. Expression of Hoxb-5 during human lung development and in congenital lung malformations. Birth Defects Res A Clin Mol Teratol. 2003;67:550–556. doi: 10.1002/bdra.10086. [DOI] [PubMed] [Google Scholar]
- 34.Yamaguchi TP. Bradley A. McMahon AP. Jones S. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development. 1999;126:1211–1223. doi: 10.1242/dev.126.6.1211. [DOI] [PubMed] [Google Scholar]
- 35.Li C. Xiao J. Hormi K. Borok Z. Minoo P. Wnt5a participates in distal lung morphogenesis. Dev Biol. 2002;248:68–81. doi: 10.1006/dbio.2002.0729. [DOI] [PubMed] [Google Scholar]
- 36.Kalinichenko VV. Lim L. Stolz DB. Shin B. Rausa FM. Clark J. Whitsett JA. Watkins SC. Costa RH. Defects in pulmonary vasculature and perinatal lung hemorrhage in mice heterozygous null for the Forkhead Box f1 transcription factor. Dev Biol. 2001;235:489–506. doi: 10.1006/dbio.2001.0322. [DOI] [PubMed] [Google Scholar]
- 37.Chapman DL. Garvey N. Hancock S. Alexiou M. Agulnik SI. Gibson-Brown JJ. Cebra-Thomas J. Bollag RJ. Silver LM. Papaioannou VE. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn. 1996;206:379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 38.Cebra-Thomas JA. Bromer J. Gardner R. Lam GK. Sheipe H. Gilbert SF. T-box gene products are required for mesenchymal induction of epithelial branching in the embryonic mouse lung. Dev Dyn. 2003;226:82–90. doi: 10.1002/dvdy.10208. [DOI] [PubMed] [Google Scholar]
- 39.Jarvinen L. Badri L. Wettlaufer S. Ohtsuka T. Standiford TJ. Toews GB. Pinsky DJ. Peters-Golden M. Lama VN. Lung resident mesenchymal stem cells isolated from human lung allografts inhibit T cell proliferation via a soluble mediator. J Immunol. 2008;181:4389–4396. doi: 10.4049/jimmunol.181.6.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Besnard V. Wert SE. Stahlman MT. Postle AD. Xu Y. Ikegami M. Whitsett JA. Deletion of Scap in alveolar type II cells influences lung lipid homeostasis and identifies a compensatory role for pulmonary lipofibroblasts. J Biol Chem. 2009;284:4018–4030. doi: 10.1074/jbc.M805388200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGowan SE. Torday JS. The pulmonary lipofibroblast (lipid interstitial cell) and its contributions to alveolar development-1. Ann Rev Physiol. 1997;59:43–62. doi: 10.1146/annurev.physiol.59.1.43. [DOI] [PubMed] [Google Scholar]
- 42.Boström H. Willetts K. Pekny M. Levéen P. Lindahl P. Hedstrand H. Pekna M. Hellström M. Gebre-Medhin S. Schalling M. Nilsson M. Kurland S. Törnell J. Heath JK. Betsholtz C. PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell. 1996;85:863–873. doi: 10.1016/s0092-8674(00)81270-2. [DOI] [PubMed] [Google Scholar]
- 43.Lindahl P. Karlsson L. Hellstrom M. Gebre-Medhin S. Willetts K. Heath J. Betsholtz C. Alveogenesis failure in PDGF-A-deficient mice is coupled to lack of distal spreading of alveolar smooth muscle cell progenitors during lung development. Development. 1997;124:3943–3953. doi: 10.1242/dev.124.20.3943. [DOI] [PubMed] [Google Scholar]
- 44.Rehan VK. Sugano S. Wang Y. Santos J. Romero S. Dasgupta C. Evidence for the presence of lipofibroblasts in human lung. Exp Lung Res. 2006;32:379–393. doi: 10.1080/01902140600880257. [DOI] [PubMed] [Google Scholar]
- 45.Vadivel A. Abozaid S. van Haaften T. Sawicka M. Eaton F. Chen M. Thebaud B. Adrenomedullin promotes lung angiogenesis, alveolar development and repair. Am J Respir Cell Mol Biol. 2010;43:152–160. doi: 10.1165/rcmb.2009-0004OC. [DOI] [PubMed] [Google Scholar]
- 46.Hecker L. Vittal R. Jones T. Jagirdar R. Luckhardt TR. Horowitz JC. Pennathur S. Martinez FJ. Thannickal VJ. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nat Med. 2009;15:1077–1081. doi: 10.1038/nm.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scotton CJ. Krupiczojc MA. Konigshoff M. Mercer PF. Lee YCG. Kaminski N. Morser J. Post JM. Maher TM. Nicholson AG. Moffatt JD. Laurent GJ. Derian CK. Eickelberg O. Chambers RC. Increased local expression of coagulation factor X contributes to the fibrotic response in human and murine lung injury. J Clin Invest. 2009;119:2550–2563. doi: 10.1172/JCI33288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Haaften T. Byrne R. Bonnet S. Rochefort GY. Akabutu J. Bouchentouf M. Rey-Parra GJ. Galipeau J. Haromy A. Eaton F. Chen M. Hashimoto K. Abley D. Korbutt G. Archer SL. Thebaud B. Airway delivery of mesenchymal stem cells prevents arrested alveolar growth in neonatal lung injury in rats. Am J Respir Crit Care Med. 2009;180:1131–1142. doi: 10.1164/rccm.200902-0179OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aslam M. Baveja R. Liang OD. Fernandez-Gonzalez A. Lee C. Mitsialis SA. Kourembanas S. Bone marrow stromal cells attenuate lung injury in a murine model of neonatal chronic lung disease. Am J Respir Crit Care Med. 2009;180:1122–1130. doi: 10.1164/rccm.200902-0242OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.