Abstract
Mammalian meiosis is a process that allows diploid progenitor germ cells to produce haploid gametes after proceeding through 2 rounds of cell divisions. The first division (MI) is unique and results in the separation of homologous chromosomes, while the second division (MII) leads to the separation of sister chromatids similar to a somatic cell division. However, the mechanisms by which meiotic cells regulate their 2 very different cell divisions are not well understood. We postulated a role for epigenetic chromatin modifications in regulating these processes. We found prior to the onset of MI that pericentromeric heterochromatic regions, which are enriched with histone H3K9me2 throughout meiosis, become enriched at late pachytene with H3S10ph and at diplotene with H4K5ace and H4K16ace, but remain underacetylated at other sites examined. RNA polymerase II, which is clearly excluded from pericentromeric heterochromatin at pachytene, becomes exclusively associated with these regions from diplotene to MI. By contrast, pericentromeric heterochromatic regions at MII are not engaged by RNA polymerase II nor enriched with H3S10ph. Furthermore, we found DICER to localize exclusively to pericentromeric heterochromatin at MI, but not MII. These results are significant since they suggest: (1) that distinct chromatin modifications differentiate the 2 meiotic divisions; (2) a role for repetitive DNA elements and RNAi in mammalian meiosis; (3) H3K9me2 is not sufficient to block RNA polymerase II elongation through heterochromatin, and (4) H3S10ph provides a ‘binary switch’ to activate transcription in heterochromatin.
Key Words: DICER, Histone modifications, Non-coding RNA, RNAi, Spermatogenesis
Introduction
Mammalian meiosis occurs in both the male and female germline during spermatogenesis and oogenesis, respectively, and it is critical for the propagation of species [Clermont, 1972]. The division of a meiotic cell is distinct from the division of a somatic cell since each meiotic cell undergoes 2 consecutive divisions to produce 4 haploid cells; by contrast, each somatic cell undergoes one division to produce 2 diploid cells. Prior to both meiosis and mitosis, germ and somatic cells, respectively, replicate their DNA to double their original chromosomal content. In a somatic cell, DNA is replicated in the S phase of the cell cycle prior to mitosis to produce a tetraploid (4N) cell which can be divided to 2 diploid (2N) cells. By contrast, DNA of pre-meiotic cells (2N) is replicated prior to entry into meiosis to produce a tetraploid (4N) state (fig. 1); after entry into meiosis, meiotic cells undergo 2 consecutive cell divisions that reduce a tetraploid (4N) state of a cell to a haploid (1N) [Clermont, 1972]. The first meiotic division (metaphase I or MI) during meiosis (fig. 1B) is unique since it results in the separation of homologous chromosomes but not sister chromatids. By contrast, the second meiotic division (MII) results in the separation of sister chromatids similar to mitosis in somatic cells (fig. 1) [Suja and Barbero, 2009]. However, since both these divisions take place during meiosis, meiotic cells must have developed a mechanism to allow the separation of homologous chromosomes during MI while keeping sister chromatids tethered together. Also, these cells must be able to reverse these factors prior to MII to allow the separation of sister chromatids. The mechanisms that regulate the first and second meiotic divisions are not well understood. Several meiosis-specific cohesion complexes have been implicated in regulating the first and second meiotic divisions, reviewed in Suja and Barbero [2009]; however, the exact mechanism of this process is still to be uncovered. There are several meiosis-specific cohesion proteins that are only expressed in germ cells, and form several different cohesion complexes during meiosis that are targeted to different regions of the chromosomes [Suja and Barbero, 2009]. While further studies are needed to delineate the role of different cohesion complexes in meiosis, the targeting of these complexes to specific chromosomal regions has been largely ignored. One intriguing possibility is local chromatin structure, which is influenced by several factors including epigenetic modifications, and non-coding RNAs may play a role in this process [Mattick, 2001; Bernstein and Allis, 2005; Sugiyama et al., 2005].
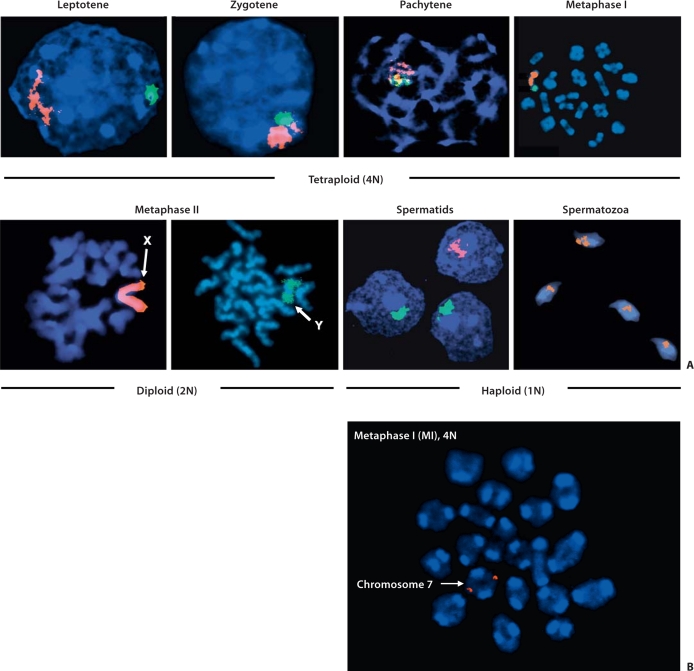
Fig. 1.
Stages of mammalian male meiosis. A Spermatogenesis initiates from spermatogonial cells (not shown) which undergo continuous cell division in post-pubertal males. Daughter cells replicate their DNA (4N) prior to entering meiosis (pre-leptotene). At late leptotene and zygotene, homologous chromosomes begin to pair to form fully synapsed chromosomes by pachytene. At diplotene, dysynapsis of homologous chromosomes takes place. Thereafter, chromosomes begin to condense to prepare for the first meiotic division (MI) (B), which leads to the separation of homologous chromosomes. Note that homologous chromosomes are joined by their telomeric end (red dots: FISH probes for chr. 7 telomere) with the pericentromeric regions (strong DAPI staining) holding sister chromatids together. The second meiotic division MII (2N) leads to separation of sister chromatids similar to a somatic cell division. Round spermatids (1N) undergo extensive loss of cytoplasm and chromatin condensation to form spermatozoa (1N). Whole chromosome paints were used to identify the X (pink/orange) and Y (yellow/green) chromosomes.
Epigenetic regulation by chromatin-modifying enzymes plays key roles in gene expression, nuclear architecture, chromosome dynamics, meiotic recombination, and other processes [Rideout et al., 2001; Arney et al., 2002; Crosio et al., 2002; Hake et al., 2005; Khalil and Driscoll, 2006]. Many chromatin-modifying complexes can modify tails of core histones (H4, H3, H2A, and H2B) by acetylation or methylation at lysine residues and phosphorylation at serine residues [Strahl and Allis, 2000; Turner, 2002]. Numerous studies have now shown that distinct patterns of histone modifications mark euchromatin and heterochromatin. While euchromatin is enriched with H3 and H4 acetylation, H3K36me3 (histone H3 lysine 36 trimethylation), H3K79me2 and H3K4me3; pericentromeric heterochromatin is enriched with H3K9me2, H3K9me3, H3K27me1, and H4K20me3 [Rice and Allis, 2001; Richards and Elgin, 2002; Khalil et al., 2004; Schotta et al., 2004; Craig, 2005]. These observations have been recently validated using high-resolution genome-wide studies with ChIP-Sequence technology: chromatin immunoprecipitation followed by deep sequencing [Barski et al., 2007; Mikkelsen et al., 2007].
While chromatin modifications regulate gene expression at the transcriptional level, post-transcriptional gene regulation can be achieved by several mechanisms including RNA interference (RNAi). RNAi is a conserved mechanism of gene regulation whereby small interfering RNAs (siRNAs) lead to the degradation of mRNAs which have a complementary sequence [Fire et al., 1998]. Several recent studies using yeast genetics have now shown that core components of the RNAi pathway (Dicer, Argonaute and RNA-dependent RNA polymerase) are involved in the formation and maintenance of heterochromatin [Andersen and Panning, 2003; Verdel et al., 2004; Martienssen et al., 2005; Matzke and Birchler, 2005; Sugiyama et al., 2005]. Heterochromatin is enriched with repetitive elements that become transcribed in both the sense and antisense direction, these transcripts form dsRNAs that are processed by DICER [Reinhart and Bartel, 2002; Murchison et al., 2005; Sugiyama et al., 2005]. By unknown mechanisms, these dsRNA recruit histone-modifying enzymes to maintain heterochromatin integrity [Volpe et al., 2002; Partridge et al., 2007; Peng and Karpen, 2007]. Furthermore, the DICER knockout is an embryonic lethal in mice [Bernstein et al., 2003] and mouse embryonic cells that lack DICER fail to properly form pericentromeric heterochromatin (PCH) which leads to chromosome segregation defects [Reinhart and Bartel, 2002; Fukagawa et al., 2004; Murchison et al., 2005]. Recently, a conditional knockout of DICER in mouse germ cells demonstrated that DICER is required for male fertility and proper meiotic progression of germ cells [Maatouk et al., 2008].
To determine if there is a link between chromatin modifications and the RNAi pathway in the regulation of meiosis, we examined several histone modifications and the RNAi core enzyme Dicer during male meiosis. In the present manuscript we show that a distinct pattern of histone modifications distinguish pericentromeric heterochromatin during the first and second meiotic divisions of mammalian meiosis. Remarkably, both RNA polymerase II and DICER co-localize to pericentromeric heterochromatin at the diplotene-MI transition, but not at other stages of meiosis. Collectively, our results suggest that pericentromeric regions are transcribed during MI, but not MII and pave the ground for functional studies to determine the role of the RNAi pathway and chromatin modifications in meiotic divisions, which could be a part of a complex mechanism that involves other structural proteins such as cohesions.
Materials and Methods
Cells Used in This Study
Human lymphoblasts were grown in RPMI media containing 10% fetal bovine serum in a 5% CO2 incubator. Mouse germ cells were isolated from testes after the mice were sacrificed according to an approved Institutional Animal Care and Use Committee (IACUC) protocol by the University of Florida College of Medicine.
Immunocytochemistry
This was performed as previously described [Khalil et al., 2004]. Both human and mouse somatic cells and mouse germ cells were cytospun onto microscope slides using a Shandon 4 cytospin. The cells were washed in KCM buffer (120 mM KCl, 20 mM NaCl, 10 mM Tris·HCl pH 8, 0.5 M EDTA, 0.1% Triton X-100) for 5 min prior to incubation with the primary antibody at room temperature for 1 h. Slides were washed 3 times in KCM buffer prior to incubation with secondary antibody for 30 min at room temperature. Slides were finally washed in KCM buffer (3×) prior to fixation in 10% formalin at room temperature for 10 min. Cells were stained with propidium iodide and visualized using an Olympus fluorescence microscope. Images were captured using an Applied Imaging CytoVision system. Antibodies for H3K9me2, H3K4me3 and H3S10ph were purchased from Abcam (Cambridge, UK). Antibodies for H4K5ace, H4K8ace, H4K12ace, H4K16ace were purchased from Upstate (Now Millipore). An antibody for RNA polymerase II (H5) was purchased from Covance (USA). Several hundred cells within the same slide, as well as on other slides, were examined for each antibody. Each experiment was repeated 3 or more times to ensure reproducibility of the results. Positive labeling of the antibodies at the various stages was only considered if it was observed in over 95% of the cells examined.
Fluorescence in situ Hybridization
DNA in situ hybridization was performed on immunostained slides using DNA whole chromosome paints for the mouse X and Y chromosomes (Cambio, UK). Briefly, slides were dehydrated in an ethanol series, denatured in 70% formamide, 30% 2× SSC for 5 min at 65°C before incubation with denatured chromosomal specific DNA probes overnight at 42°C. The slides were washed in 0.4× SSC at 45°C for 2 min and in 2× SSC, 0.1% NP-40 for 1 min. Chromosomal DNA was stained blue by 4′,6-diamidino-2-phenylindole (DAPI) and red by propidium iodide (PI). Images were captured as described above.
Results
Histone Modifications of Pericentromeric Heterochromatin during Male Meiosis
In this report we conducted our experiments in male mice and therefore the focus of this report will be on spermatogenesis. Mammalian spermatogenesis initiates from type A spermatogonia that undergo multiple rounds of mitotic divisions to produce type B spermatogonia which enter the meiotic phase by the formation of pre-leptotene primary spermatocytes [Bellve et al., 1977]. During prophase I of meiosis each spermatocyte progresses through a series of cytologically identifiable stages known as (i) leptotene (homologous chromosomes are not paired), (ii) zygotene (initiation of pairing between homologs), (iii) pachytene (complete pairing of homologous autosomes, X and Y form a sex body and only pair at their pseudoautosomal region), (iv) diplotene (desynapsis of homologs), (v) diakinesis (condensation of chromosomes in preparation for the first meiotic division (MI)) (fig. 1A, B). After the first meiotic division (MI) which results in the separation of homologs, but not sister chromatids, secondary spermatocytes undergo a second division (MII) to separate sister chromatids and produce haploid round spermatids (fig. 1A). Round spermatids undergo morphological changes to produce mature sperms through a process known as spermiogenesis [Clermont, 1972].
To gain insights into the role of histone modifications in meiotic cell divisions, we began by immunostaining the various stages of mouse meiosis with antibodies against specific histone modifications. We found the pericentromeric regions at the pachytene stage of meiosis to be devoid of histone H4 acetylation at all the lysine residues examined (lysine 5, 8, 12 and 16). However, by diplotene the pericentromeric regions become enriched with H4K16ace and H4K5ace, but remain underacetylated at H4K8 and H4K12 (fig. 2 and data not shown). While H4K16ace is maintained at the pericentromeric regions at MI and MII, H4K5 is only maintained at MI, but is lost in MII. We also examined H3K9me2 and found this modification to be enriched at the pericentromeric regions at pachytene, diplotene, MI, MII, anaphase II and round spermatids (fig. 2A, B). In males, the X and Y chromosomes become inactivated during meiosis and become enriched with H3K9me2 along with the pericentromeric regions (fig. 2A, B) [Khalil and Driscoll, 2006]. As expected, H3K9ace was not detectable at the pericentromeric regions since H3K9me and H3K9ace are mutually exclusive. H3K4me3, a histone modification that correlates strongly with transcriptional activity in euchromatin [Khalil and Driscoll, 2006, 2007; Bernstein et al., 2006; Barski et al., 2007], was not detectable at the pericentromeric regions either in meiosis or in somatic cells. Collectively, these results demonstrate that while the pericentromeric regions are enriched with a repressive histone modification (H3K9me2), there are some histone modifications of active chromatin (H4K5ace and H4K16ace) that spread to those regions suggesting a unique form of regulation of pericentromeric regions during meiosis.
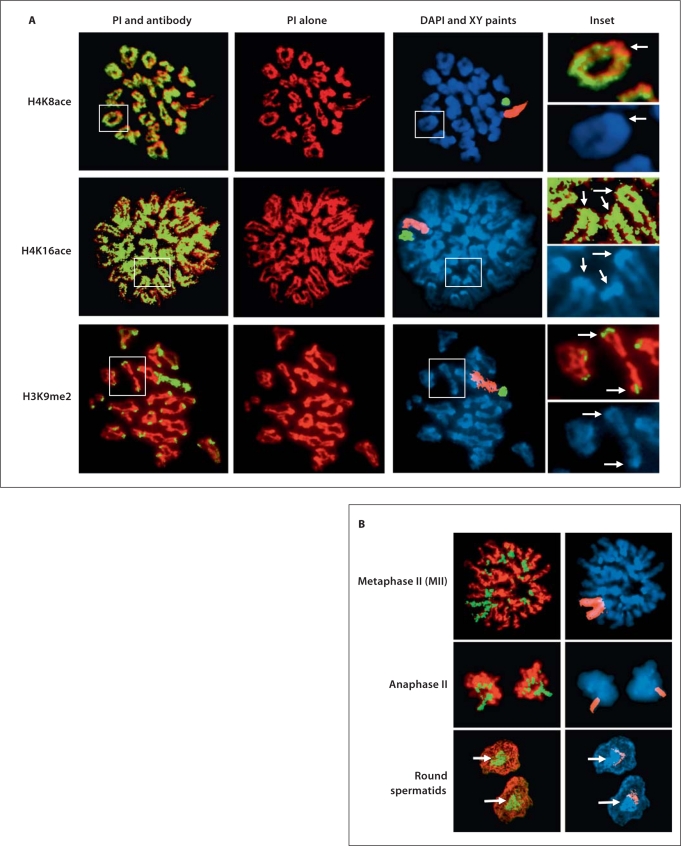
Fig. 2.
Histone modifications of pericentromeric heterochromatin during meiosis. A Diplotene-MI spermatocytes immunostained with antibodies against various histone modifications (FITC). The same cells are shown with propidium iodide (PI) staining alone (red) and again the same cells are shown in DAPI staining (blue) and X (pink) and Y (green) chromosome paints. H4K8ace is present on the arms of all the chromosomes (except the X and Y), but excluded from the pericentromeric heterochromatin. By contrast, H4K16ace is present on all the arms of the chromosomes and the pericentromeric regions. H3K9me2 is enriched at the pericentromeric regions and the X and Y chromosomes. The insets and arrows highlight the observations noted above. B H3K9me2 continues to be enriched at the pericentromeric heterochromatin as well as the X and Y chromosomes at MII and anaphase II. In round spermatids, H3K9me is lost from the X and Y chromosomes but remains enriched at the pericentromeric heterochromatin regions (arrows). Only the X chromosome (pink) is shown in these panels. Note: For each antibody we examined several hundred cells. We also repeated each experiment at least 3 times and we consistently observed similar staining patterns in both technical and biological replicates. This is also true for all staining patterns in other figures.
RNA Polymerase II Becomes Enriched at Pericentromeric Heterochromatin at Diplotene
Since we observed histone modifications of active chromatin (H4K5ace and H4K16ace) that spread to the pericentromeric regions in meiosis, we wanted to determine if these modifications are correlated with a change in transcriptional status. To accomplish this, we utilized an antibody that recognizes the phosphorylated form of RNA polymerase II at serine 2 (Covance) which is indicative of transcriptional elongation [Ahn et al., 2004; Sims et al., 2004a]. We found RNA polymerase II to be excluded from pericentromeric regions at pachytene, but remarkably becomes exclusively associated with pericentromeric regions at the diplotene stage of meiosis (fig. 3). This association is maintained at diakinesis and MI, but is lost by the second meiotic division (MII). In round spermatids, the pericentromeric regions of autosomes come in close proximity to form a dense structure known as the chromocenter (fig. 4A, arrows). We co-stained round spermatids with both RNA polymerase II (red) (fig. 4B) and H3K4me3 (green) (fig. 4C) and found both to be largely excluded from the chromocenter but strongly co-localize in euchromatic regions (fig. 4D). To determine if RNA polymerase II localization to pericentromeric regions only occurs in diplotene-MI or if it also occurs in somatic cells, we examined human (fibroblasts and lymphoblasts) and mouse (fibroblasts) somatic interphase and metaphase spreads. We did not observe RNA polymerase II at the pericentromeric regions in any of these somatic cells. Collectively, these results show that RNA polymerase II is associated with pericentromeric heterochromatin at MI, but not at MII or in somatic cells indicating a novel role for these regions during the first meiotic division.
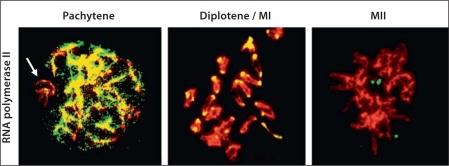
Fig. 3.
RNA polymerase II distribution during meiosis. RNA polymerase II (phosphorylated at serine 2) (FITC) is present at the chromosomes (propidium iodide stain) at pachytene with the exception of the X and Y chromosomes (sex body) (arrow), but excluded from the pericentromeric heterochromatin regions. By diplotene-MI, RNA polymerase II becomes almost exclusively associated with the pericentromeric heterochromatin (PCH) regions, but this association is not observed at MII.
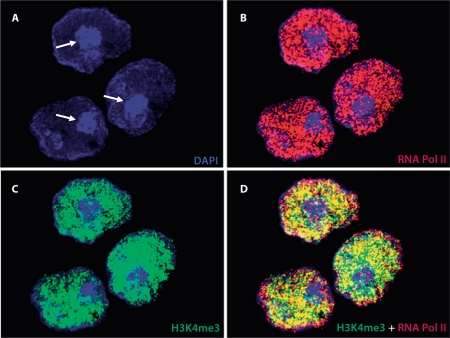
Fig. 4.
RNA polymerase II and H3K4me3 are excluded from PCH in round spermatids. A DAPI-stained round spermatids showing the densely stained chromocenter (arrows) which is composed of pericentromeric heterochromatin. B RNA polymerase II (phosphorylated at serine 2, shown in red) is present throughout round spermatids, but excluded from pericentromeric heterochromatin regions. C H3K4m3 (shown in green), a strong marker of active transcription, is also excluded from pericentromeric heterochromatin. D A merged image of all panels.
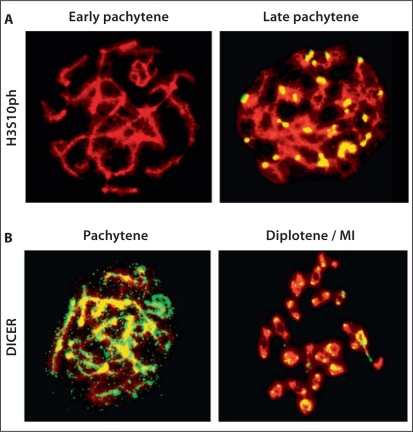
Since we found the pericentromeric regions to be enriched with H3K9me2 (a mark of inactive chromatin), and the phosphorylated form of RNA polymerase II at serine 2 (which is indicative of the elongation phase of transcription) [Orphanides and Reinberg, 2000; Ahn et al., 2004; Sims et al., 2004a, 2004b], we therefore postulated that this could be an example of the ‘binary switch hypothesis’ [Fischle et al., 2003]. This hypothesis states that the phosphorylation of a serine residue could influence the effect of a repressive methylation mark on a neighboring lysine residue [Nowak and Corces, 2004]. Therefore we examined the various stages of meiosis with an antibody against H3S10ph. We found H3S10ph to be largely absent from early pachytene, but become enriched at the pericentromeric regions by the late pachytene stage of meiosis (fig. 5A). This enrichment was also observed at diakinesis and MI, but not at MII. This is consistent with RNA polymerase II localization at diplotene-MI, but not at MII. In metaphases of somatic cells and at MII of meiosis H3S10ph is present at the chromosome arms, as well as the pericentromeric regions with equal intensity. These results suggest that H3S10ph is induced by late pachytene to ‘neutralize’ the effects of H3K9me2 and allow RNA polymerase II to elongate through these regions.
Fig. 5.
Distribution of H3S10ph and DICER at percentromeric heterochromatin in meiosis. A Early- and late-pachytene spermatocytes immunostained with an antibody for H3S10ph. H3S10ph is absent in early pachytene but becomes enriched at the pericentromeric heterochromatin regions by the late-pachytene stage. B DICER is enriched across all the chromosomes at pachytene but becomes almost exclusively associated with the pericentromeric heterochromatin regions by diplotene.
DICER Is Enriched at Pericentromeric Heterochromatin at Diplotene-MI
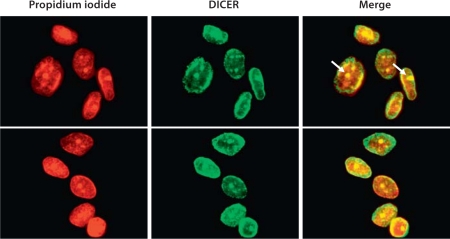
Since we found the phosphorylated form of RNA polymerase II at serine 2 to be localized to pericentromeric heterochromatin from diplotene-MI, we postulated that potential transcripts from these regions, which are mostly repetitive DNA elements, can form double-stranded RNA and enter the RNAi pathway. Previous studies in yeast have shown that transcripts from pericentromeric heterochromatin can indeed form double-stranded RNA that become processed by DICER [Volpe et al., 2002; Hall et al., 2003; Sugiyama et al., 2005]. To investigate whether a similar phenomenon may also occur in mammals, we used an antibody for DICER to examine its localization during mouse meiosis. We found DICER to be enriched on chromosomes arms (including the XY body) at pachytene. However, DICER becomes almost exclusively associated with the pericentromeric regions at diplotene-MI stage of meiosis, but not at MII (fig. 5B). It is important to point out that our method of preparing chromosome spreads for immunofluorescence requires a hypotonic solution and a cytospin, which disrupts the cytoplasm of cells, and therefore we do not detect the cytoplasmic fraction of DICER in our preparations. We also examined human somatic cells (lymphoblasts) (also subjected to a hypotonic solution and cytospun onto slides) and found DICER to show nuclear localization with strong staining in the nuclear periphery as well as a strong staining of the nucleolus (fig. 6, arrows). However, we did not detect DICER during metaphase of somatic cells. Our data show that DICER is also localized to the nucleus of somatic cells similar to what have been observed in yeast [Volpe et al., 2002; Hall et al., 2003; Sugiyama et al., 2005] as well as to pericentromeric heterochromatin during MI of meiosis. Collectively, our results suggest that pericentromeric heterochromatin is transcribed at specific stages of mouse meiosis (diplotene-MI) and these potential transcripts may be processed by DICER into small RNAs.
Fig. 6.
DICER shows nuclear localization in human somatic cells. Disruption of cell cytoplasm by a cytospin centrifugation reveals nuclear localization of DICER (FITC) in human somatic cells (DNA stained red with PI). DICER is mostly localized to perinuclear region and the nucleolus (arrows).
Discussion
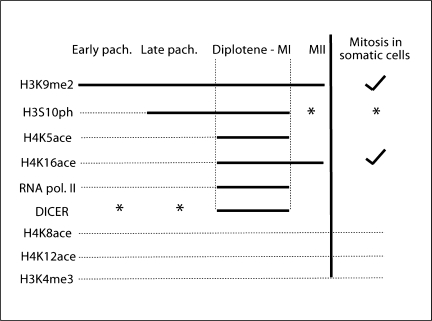
Mammalian meiosis is a complex and unique process with multiple steps. Appropriate segregation of chromosomes during MI and MII is crucial to producing haploid cells with the correct number of chromosomes. We have demonstrated in this study that distinct epigenetic modifications at pericentromeric heterochromatin differentiate the 2 meiotic divisions, MI and MII. While MI has a distinct combination of histone and epigenetic modifications, MII and somatic metaphases have similar patterns (fig. 7). The first meiotic division (MI) is unique to meiosis since meiotic cells are the only cells that require the segregation of homologous chromosomes. However, MII leads to the separation of sister chromatids similar to mitosis in somatic cells.
Fig. 7.
Summary of modifications at pericentromeric heterochromatin (PCH). (1) H3K9me2 is enriched at PCH throughout meiosis and in somatic cells (check mark indicates a similar localization is observed in somatic cells). (2) H3S10ph is absent in early pachytene from all chromosomal regions; however, it becomes enriched at PCH from late pachytene to diplotene. At MII and in somatic cells, H3S10ph is present on the arms of chromosomes and PCH with equal intensity (this is marked by ∗). (3) H4K5ace and H4K16ace are enriched at PCH from diplotene-MI. Only H4K16ace remains enriched at PCH at MII. (4) RNA polymerase II is only enriched at PCH from diplotene to MI. (5) DICER is present at both the arms of the chromosomes and PCH with equal intensity (∗) in early and late pachytene. DICER is enriched at PCH at diplotene and MI. (6) H4K8ace, H4K12ace and H3K4me3 are not detectable at PCH throughout meiosis or in somatic cells.
It has previously been shown that methylation of H3K9 by Clr4, a histone methyltransferase homologous to SUV39H1 in mammals, acts upstream of DICER in the establishment of centromeric heterochromatin in fission yeast [Partridge et al., 2007]. Our observations that H3K9me2 is enriched on heterochromatin during meiosis prior to DICER and RNA polymerase II further support this finding in yeast and suggest that the order of events leading to heterochromatin formation may be conserved between yeast and mammals. Furthermore, it indicates that mammalian spermatogenesis which has cytologically distinguishable stages is a powerful experimental system to study the interaction between the RNAi pathway and chromatin modifications.
H3K9me2 has been classified as a mark of inactive chromatin, although one report found this modification to also be present in the body of active genes [Vakoc et al., 2005]. Our observations indicate that although H3K9me2 is enriched at pericentromeric heterochromatin, this enrichment is not sufficient to block RNA polymerase II localization to heterochromatin. Also, since H3S10ph becomes enriched at pericentromeric heterochromatin prior to RNA polymerase II, we postulate that H3S10ph may provide a ‘binary switch’ to activate transcription in heterochromatin during meiosis [Lo et al., 2000; Nowak and Corces, 2004].
Pericentromeric heterochromatin regions are mostly composed of repetitive DNA elements with little known about their function. The localization of RNA polymerase II and epigenetic modifications of active chromatin suggest that these elements are transcribed and play a crucial role during specific stages of meiosis (diplotene-MI). Also, since DICER becomes enriched at pericentromeric heterochromatin, it is possible that these transcripts are processed by DICER, suggesting a role of the RNAi pathway in mammalian meiosis. It has been previously shown that a mutation in the second largest subunit of RNA polymerase II results in the loss of heterochromatin histone modifications (e.g. H3K9me) and siRNA production in fission yeast [Kato et al., 2005]. Also, RNA-dependent RNA polymerase (Rdp1) localizes to pericentromeric heterochromatin along with RITS and DICER in fission yeast [Sugiyama et al., 2005]. Abolishing the catalytic activity of Rdp1 results in mitotic chromosome segregation defects and in the absence of siRNA production from pericentromeric heterochromatin [Sugiyama et al., 2005].
Recently, a conditional knockout of DICER in mouse germ cells has demonstrated that DICER is essential for male fertility and proper meiotic progression [Maatouk et al., 2008]. The exact mechanism by which DICER disruption leads to meiotic defects is not known. Our current findings would suggest that the disruption of DICER might lead to chromosome segregation defects. Emerging evidence also suggests a role of RNAi pathway in heterochromatin formation in mammalian stem cells. For example, mouse ES cells that lack DICER fail to localize Xist and H3K27me3 to the inactive X chromosome [Ogawa et al., 2008]. However, it is worth noting that another independent study in mouse ES cells suggested that DICER may be dispensable for Xist and H3K27me3 localization to the inactive X chromosome [Kanellopoulou et al., 2009]. Further studies will be needed to address these questions. Nevertheless, our current findings add further evidence to the important role for repetitive DNA elements and the RNAi pathway in mammalian meiosis, in agreement with previous observations [Andersen and Panning, 2003].
Acknowledgement
This work was supported by grants from the NIH (RO1 HD36417 to D.J.D. and T32 AR007603-07 to A.M.K.), the Hayward Foundation and the Children's Miracle Network.
References
- Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- Andersen AA, Panning B. Epigenetic gene regulation by noncoding RNAs. Curr Opin Cell Biol. 2003;15:281–289. doi: 10.1016/s0955-0674(03)00041-3. [DOI] [PubMed] [Google Scholar]
- Arney KL, Bao S, Bannister AJ, Kouzarides T, Surani MA. Histone methylation defines epigenetic asymmetry in the mouse zygote. Int J Dev Biol. 2002;46:317–320. [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bellve AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Clermont Y. Kinetics of spermatogenesis in mammals: Seminiferous epithelium cycle and spermatogonial renewal. Physiol Rev. 1972;52:198–236. doi: 10.1152/physrev.1972.52.1.198. [DOI] [PubMed] [Google Scholar]
- Craig JM. Heterochromatin – many flavours, common themes. Bioessays. 2005;27:17–28. doi: 10.1002/bies.20145. [DOI] [PubMed] [Google Scholar]
- Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, et al. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian aurora kinases. Mol Cell Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, et al. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Hake SB, Garcia BA, Kauer M, Baker SP, Shabanowitz J, et al. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc Natl Acad Sci USA. 2005;102:6344–6349. doi: 10.1073/pnas.0502413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Noma K, Grewal SI. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci USA. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Dimitrov SD, Chen X, Colin C, et al. X chromosome inactivation in the absence of DICER. Proc Natl Acad Sci USA. 2009;106:1122–1127. doi: 10.1073/pnas.0812210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Driscoll DJ. Histone H3 lysine 4 dimethylation is enriched on the inactive sex chromosomes in male meiosis but absent on the inactive X in female somatic cells. Cytogenet Genome Res. 2006;112:11–15. doi: 10.1159/000087508. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Driscoll DJ. Trimethylation of histone H3 lysine 4 is an epigenetic mark at regions escaping mammalian X inactivation. Epigenetics. 2007;2:114–118. doi: 10.4161/epi.2.2.4612. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Boyar FZ, Driscoll DJ. Dynamic histone modifications mark sex chromosome inactivation and reactivation during mammalian spermatogenesis. Proc Natl Acad Sci USA. 2004;101:16583–16587. doi: 10.1073/pnas.0406325101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to GCN5-mediated acetylation at lysine 14. Mol Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- Maatouk DM, Loveland KL, McManus MT, Moore K, Harfe BD. Dicer1 is required for differentiation of the mouse male germline. Biol Reprod. 2008;79:696–703. doi: 10.1095/biolreprod.108.067827. [DOI] [PubMed] [Google Scholar]
- Martienssen RA, Zaratiegui M, Goto DB. RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe. Trends Genet. 2005;21:450–456. doi: 10.1016/j.tig.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Mattick JS. Non-coding RNAs: The architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Sun BK, Lee JT. Intersection of the RNA interference and X-inactivation pathways. Science. 2008;320:1336–1341. doi: 10.1126/science.1157676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. RNA polymerase II elongation through chromatin. Nature. 2000;407:471–475. doi: 10.1038/35035000. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Debeauchamp JL, Kosinski AM, Ulrich DL, Hadler MJ, Noffsinger VJ. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol Cell. 2007;26:593–602. doi: 10.1016/j.molcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: Rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- Rideout WM, 3rd, Eggan K, Jaenisch R. Nuclear cloning and epigenetic reprogramming of the genome. Science. 2001;293:1093–1098. doi: 10.1126/science.1063206. [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: The short and long of it. Genes Dev. 2004a;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Mandal SS, Reinberg D. Recent highlights of RNA-polymerase-II-mediated transcription. Curr Opin Cell Biol. 2004b;16:263–271. doi: 10.1016/j.ceb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci USA. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suja JA, Barbero JL. Cohesin complexes and sister chromatid cohesion in mammalian meiosis. Genome Dyn. 2009;5:94–116. doi: 10.1159/000166622. [DOI] [PubMed] [Google Scholar]
- Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine- 9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]