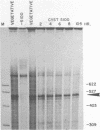
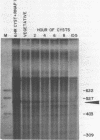
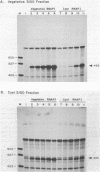
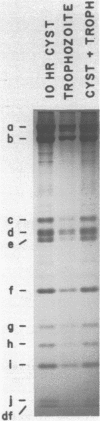
Abstract
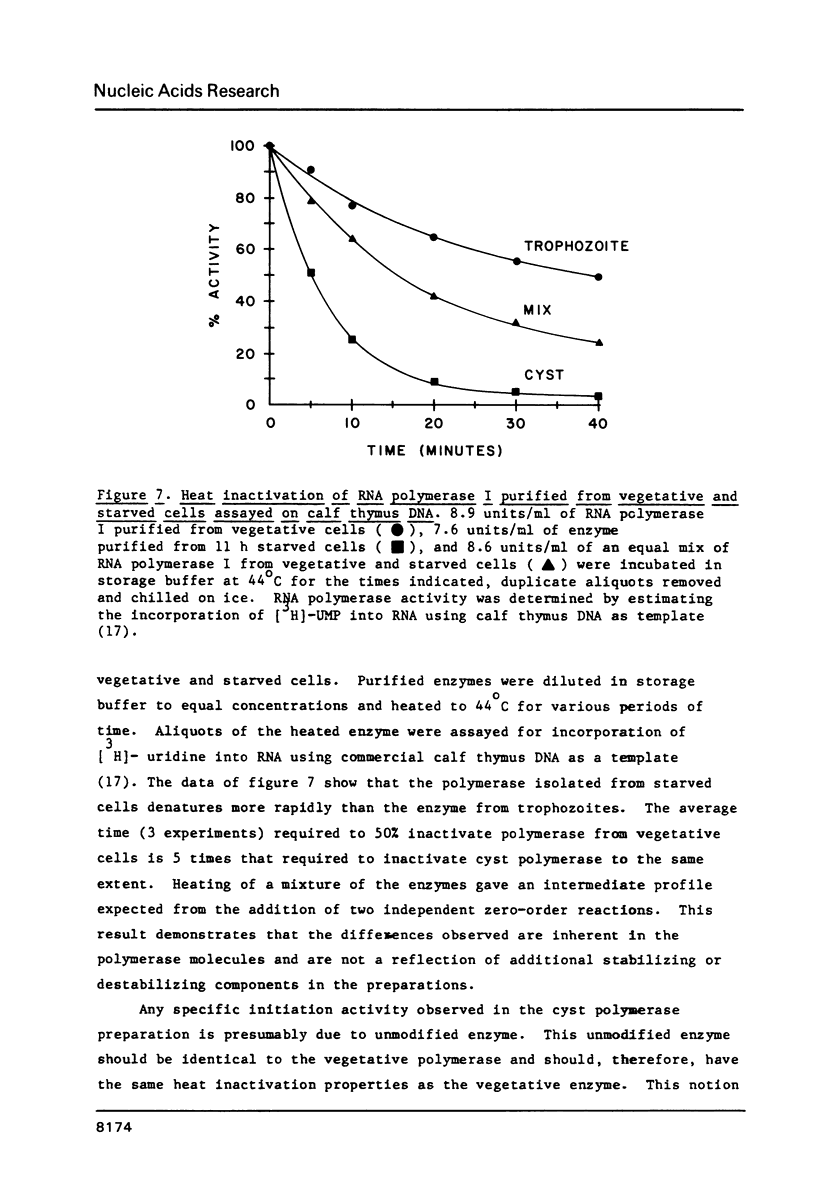
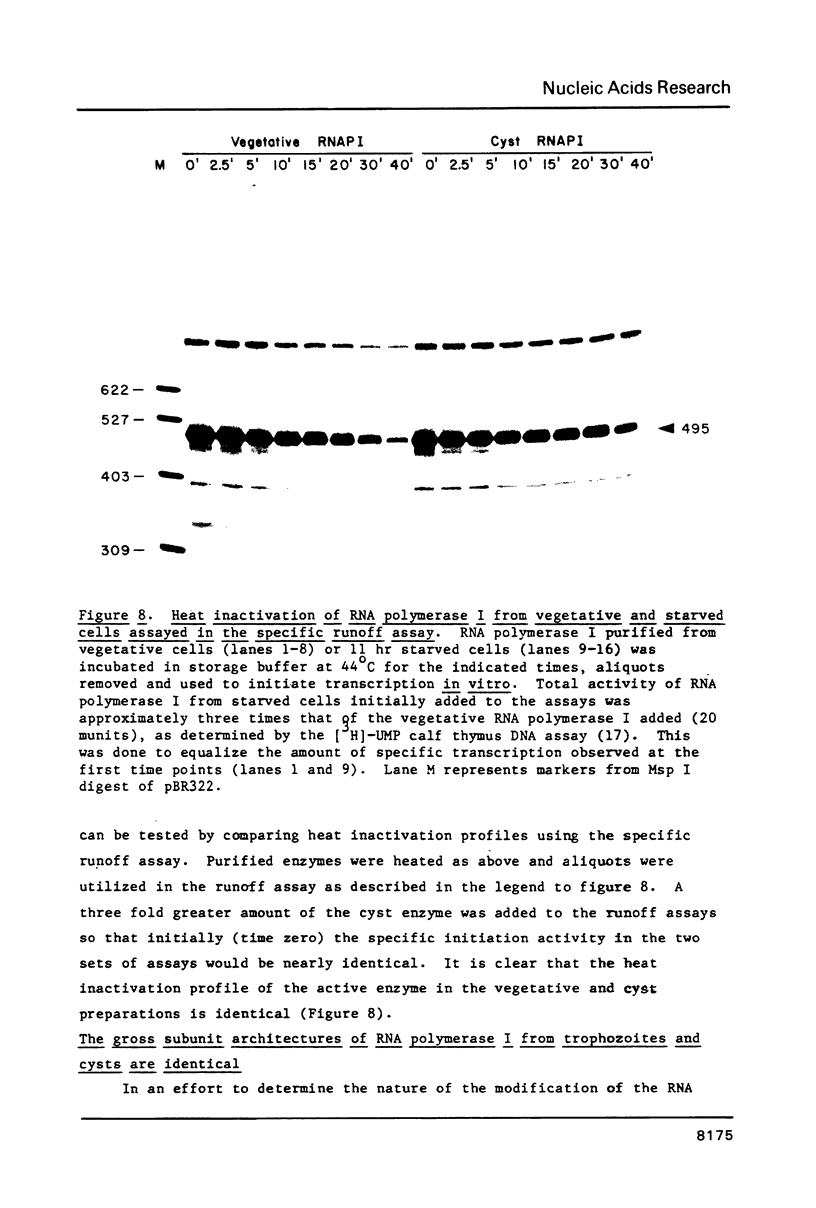
We have utilized a cell-free transcription system from Acanthamoeba castellanii to test the functional activity of RNA polymerase I and transcription initiation factor I (TIF-I) during developmental down regulation of rRNA transcription. The results strongly suggest that rRNA transcription is regulated by modification, probably covalent, of RNA polymerase I: (1) The level of activity of TIF-I in extracts from transcriptionally active and inactive cells is constant. (2) The number of RNA polymerase I molecules in transcriptionally active and inactive cells is also constant. (3) In contrast, though the specific activity of polymerase I on damaged templates remains constant, both crude and purified polymerase I from inactive cells have lost the ability to participate in faithful initiation of rRNA transcription. (4) Polymerase I purified from transcriptionally active cells has the same subunit architecture as enzyme from inactive cells. However, the latter is heat denatured 5 times faster than the active polymerase.
Full text
PDF



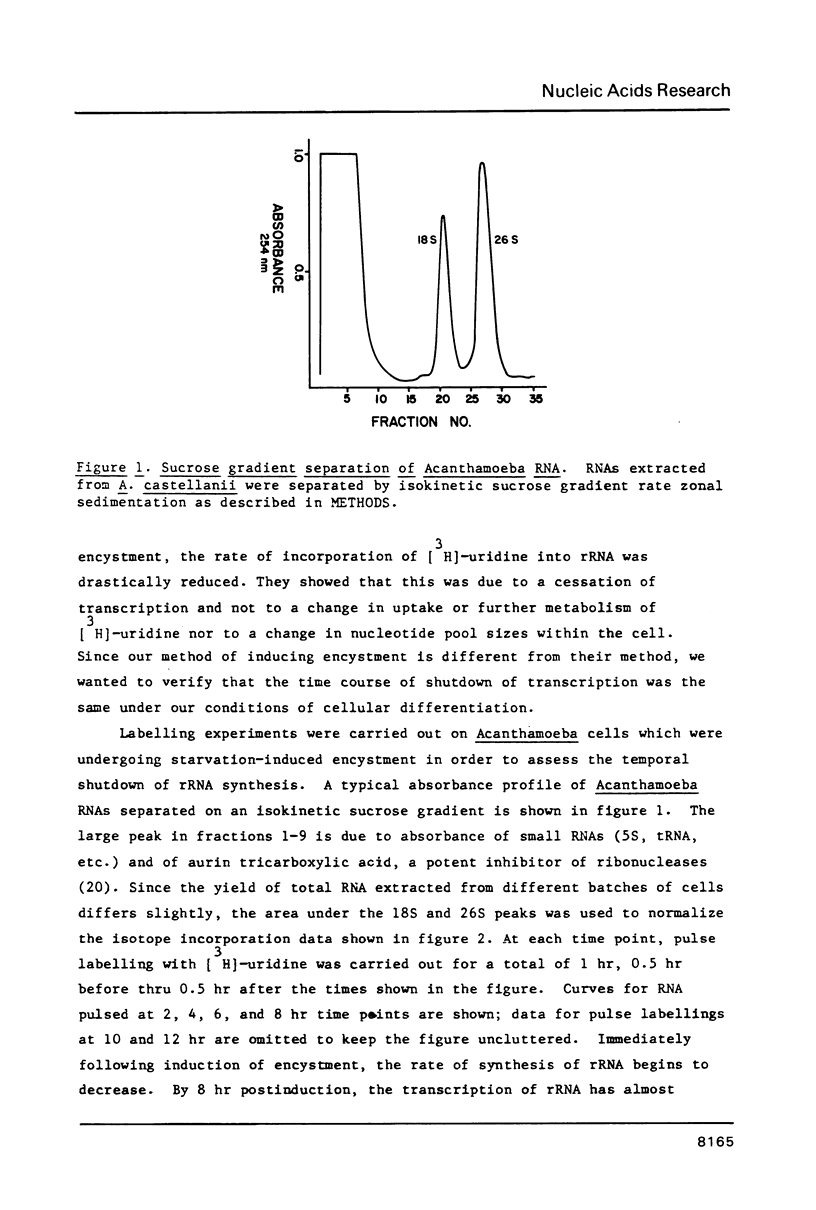
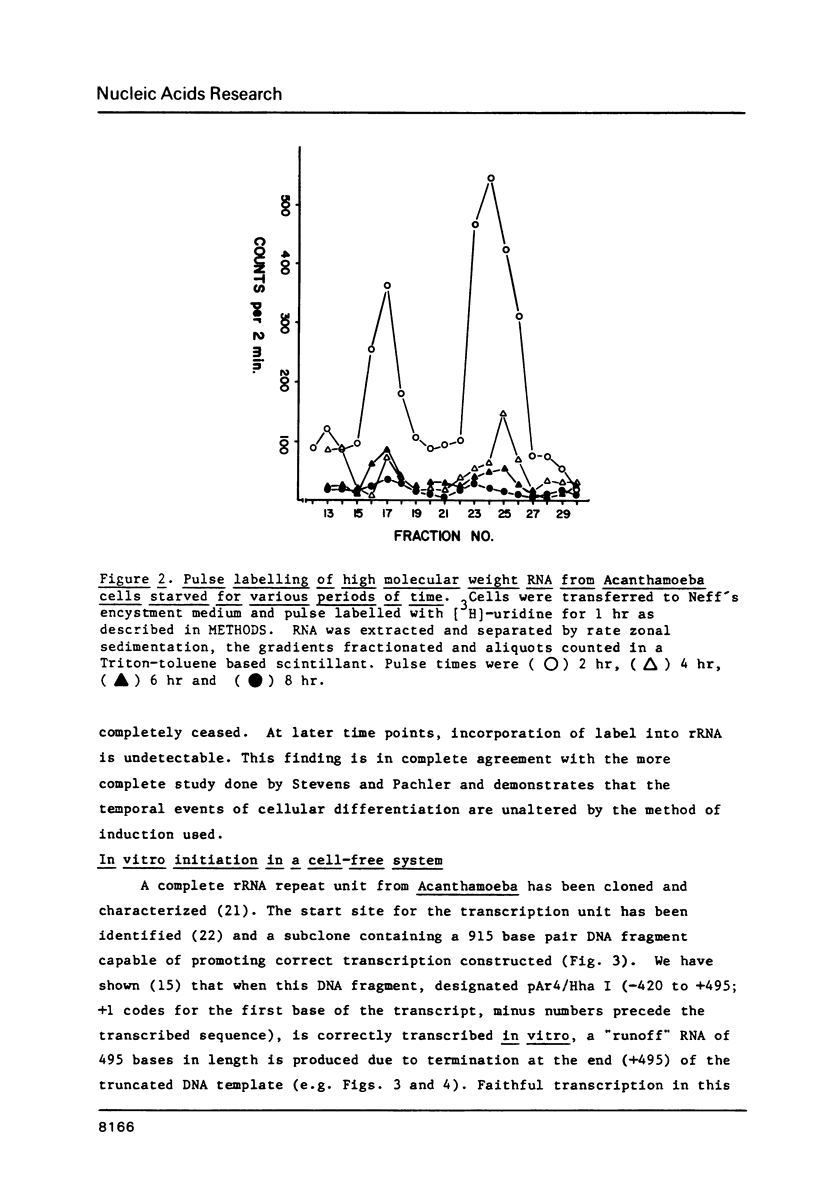
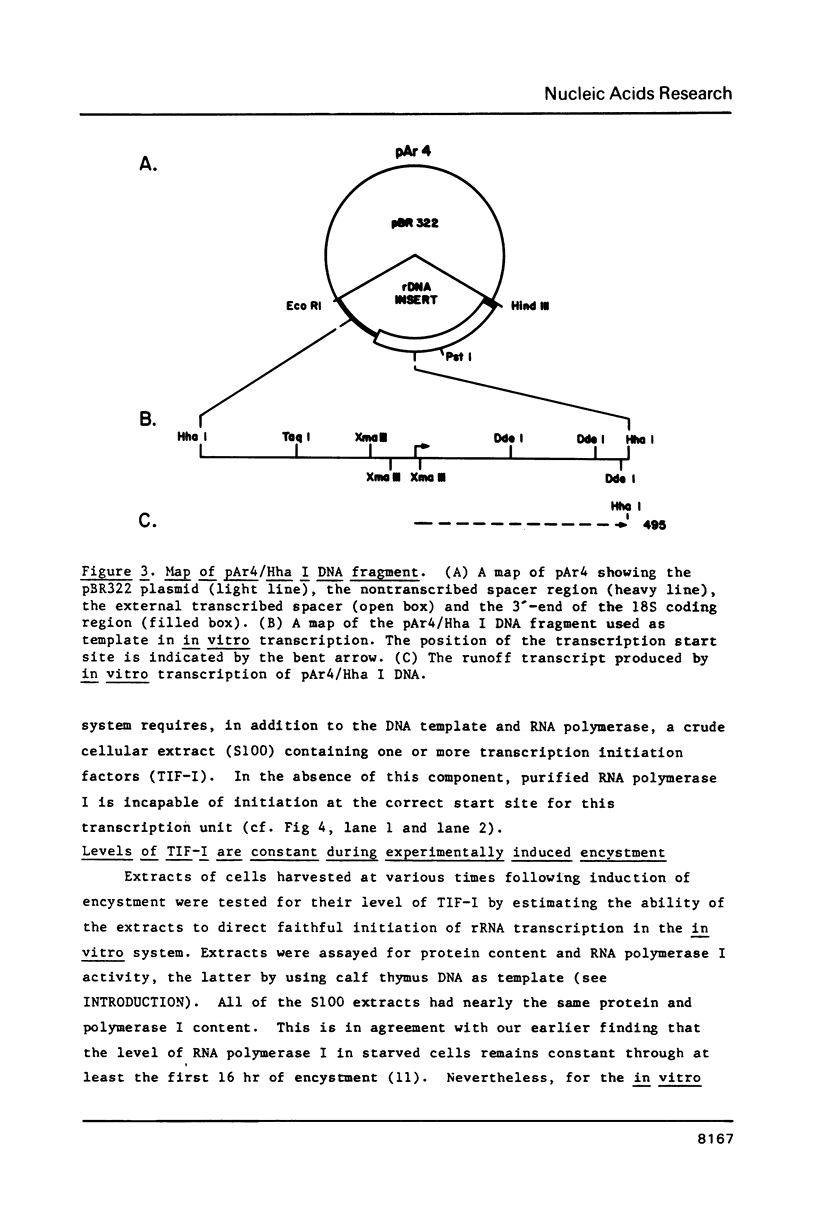


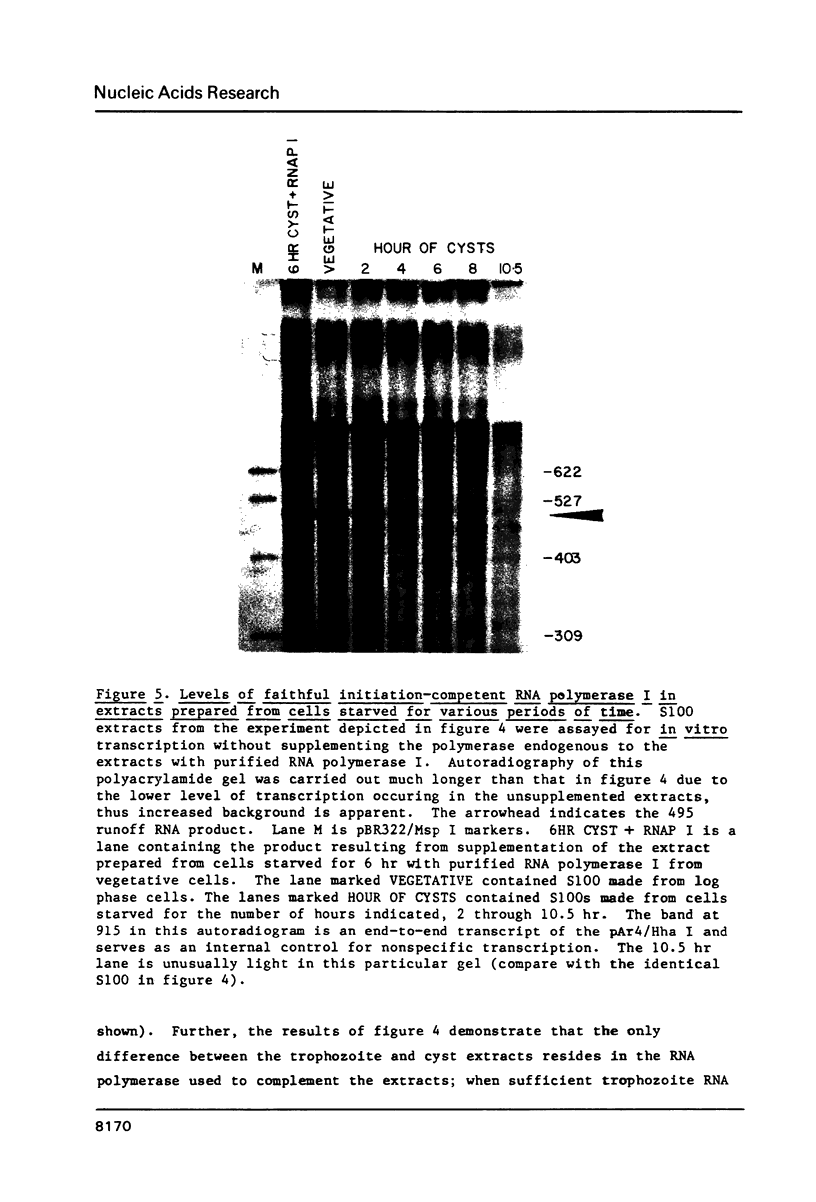







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Bréant B., Buhler J. M., Sentenac A., Fromageot P. On the phosphorylation of yeast RNA polymerases A and B. Eur J Biochem. 1983 Feb 1;130(2):247–251. doi: 10.1111/j.1432-1033.1983.tb07143.x. [DOI] [PubMed] [Google Scholar]
- Cizewski V., Sollner-Webb B. A stable transcription complex directs mouse ribosomal RNA synthesis by RNA polymerase I. Nucleic Acids Res. 1983 Oct 25;11(20):7043–7056. doi: 10.1093/nar/11.20.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessio J. M., Harris G. H., Perna P. J., Paule M. R. Ribosomal ribonucleic acid repeat unit of Acanthamoeba castellanii: cloning and restriction endonuclease map. Biochemistry. 1981 Jun 23;20(13):3822–3827. doi: 10.1021/bi00516a024. [DOI] [PubMed] [Google Scholar]
- D'Alessio J. M., Perna P. J., Paule M. R. DNA-dependent RNA polymerases from Acanthamoeba castellanii. Comparative subunit structures of the homogeneous enzymes. J Biol Chem. 1979 Nov 25;254(22):11282–11287. [PubMed] [Google Scholar]
- Detke S., Paule M. R. DNA-dependent RNA polymerase I from Acanthamoeba castellanii: comparison of the catalytic properties and subunit architectures of the trophozoite and cyst enzymes. Arch Biochem Biophys. 1978 Jan 30;185(2):333–343. doi: 10.1016/0003-9861(78)90175-3. [DOI] [PubMed] [Google Scholar]
- Detke S., Paule M. R. DNA-dependent RNA polymerase III from Acanthamoeba castellanii: comparison of the catalytic properties of the trophozoite and cyst enzymes. J Protozool. 1979 May;26(2):319–323. doi: 10.1111/j.1550-7408.1979.tb02788.x. [DOI] [PubMed] [Google Scholar]
- Detke S., Paule M. R. DNA-dependent RNA polymerases from Acanthamoeba castellanii. Multiple forms of the class III enzyme and levels of activity of the polymerase classes during encystment. Biochim Biophys Acta. 1978 Aug 23;520(1):131–138. doi: 10.1016/0005-2787(78)90014-x. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983 Mar;32(3):669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- Grummt I., Roth E., Paule M. R. Ribosomal RNA transcription in vitro is species specific. Nature. 1982 Mar 11;296(5853):173–174. doi: 10.1038/296173a0. [DOI] [PubMed] [Google Scholar]
- Grummt I. Specific transcription of mouse ribosomal DNA in a cell-free system that mimics control in vivo. Proc Natl Acad Sci U S A. 1981 Feb;78(2):727–731. doi: 10.1073/pnas.78.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallick R. B., Chelm B. K., Gray P. W., Orozco E. M., Jr Use of aurintricarboxylic acid as an inhibitor of nucleases during nucleic acid isolation. Nucleic Acids Res. 1977 Sep;4(9):3055–3064. doi: 10.1093/nar/4.9.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda B. M., Roeder R. G. Association of a 5S gene transcription factor with 5S RNA and altered levels of the factor during cell differentiation. Cell. 1980 Nov;22(1 Pt 1):119–126. doi: 10.1016/0092-8674(80)90160-9. [DOI] [PubMed] [Google Scholar]
- Kohorn B. D., Rae P. M. Accurate transcription of truncated ribosomal DNA templates in a Drosophila cell-free system. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1501–1505. doi: 10.1073/pnas.79.5.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- Learned R. M., Smale S. T., Haltiner M. M., Tjian R. Regulation of human ribosomal RNA transcription. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3558–3562. doi: 10.1073/pnas.80.12.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned R. M., Tjian R. In vitro transcription of human ribosomal RNA genes by RNA polymerase I. J Mol Appl Genet. 1982;1(6):575–584. [PubMed] [Google Scholar]
- Matsui T., Segall J., Weil P. A., Roeder R. G. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J Biol Chem. 1980 Dec 25;255(24):11992–11996. [PubMed] [Google Scholar]
- Miller K. G., Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981 Nov;27(1 Pt 2):165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- Mishima Y., Financsek I., Kominami R., Muramatsu M. Fractionation and reconstitution of factors required for accurate transcription of mammalian ribosomal RNA genes: identification of a species-dependent initiation factor. Nucleic Acids Res. 1982 Nov 11;10(21):6659–6670. doi: 10.1093/nar/10.21.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y., Yamamoto O., Kominami R., Muramatsu M. In vitro transcription of a cloned mouse ribosomal RNA gene. Nucleic Acids Res. 1981 Dec 21;9(24):6773–6785. doi: 10.1093/nar/9.24.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paule M. R., Iida C. T., Perna P. J., Harris G. H., Brown Shimer S. L., Kownin P. Faithful initiation of ribosomal RNA transcription from cloned DNA by purified RNA polymerase I. Biochemistry. 1984 Aug 28;23(18):4167–4172. doi: 10.1021/bi00313a025. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Brown D. D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner J. B., Goldsmith M. R., Hamkalo B. A. Chromosome organization during male meiosis in Bombyx mori. Chromosoma. 1981;82(3):341–351. doi: 10.1007/BF00285760. [DOI] [PubMed] [Google Scholar]
- Rattner J. B., Saunders C., Davie J. R., Hamkalo B. A. Ultrastructural organization of yeast chromatin. J Cell Biol. 1982 Apr;93(1):217–222. doi: 10.1083/jcb.93.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall J., Matsui T., Roeder R. G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980 Dec 25;255(24):11986–11991. [PubMed] [Google Scholar]
- Spindler S. R., Duester G. L., D'Alessio J. M., Paule M. R. A rapid and facile procedure for the preparation of RNA polymerase I from Acanthamoeba castellanii. Purification and subunit structure. J Biol Chem. 1978 Jul 10;253(13):4669–4675. [PubMed] [Google Scholar]
- Stevens A. R., Pachler P. F. RNA synthesis and turnover during density-inhibited growth and encystment of Acanthamoeba castellanii. J Cell Biol. 1973 May;57(2):525–537. doi: 10.1083/jcb.57.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M. E., Holland M. J. RNA polymerase I-dependent selective transcription of yeast ribosomal DNA. Identification of a new cellular ribosomal RNA precursor. J Biol Chem. 1983 Mar 10;258(5):3242–3250. [PubMed] [Google Scholar]
- Wandelt C., Grummt I. Formation of stable preinitiation complexes is a prerequisite for ribosomal DNA transcription in vitro. Nucleic Acids Res. 1983 Jun 11;11(11):3795–3809. doi: 10.1093/nar/11.11.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil P. A., Luse D. S., Segall J., Roeder R. G. Selective and accurate initiation of transcription at the Ad2 major late promotor in a soluble system dependent on purified RNA polymerase II and DNA. Cell. 1979 Oct;18(2):469–484. doi: 10.1016/0092-8674(79)90065-5. [DOI] [PubMed] [Google Scholar]