Abstract
The shade avoidance syndrome (SAS) allows plants to anticipate and avoid shading by neighbouring plants by initiating an elongation growth response. The phytochrome photoreceptors are able to detect a reduction in the red:far red ratio in incident light, the result of selective absorption of red and blue wavelengths by proximal vegetation. A shade-responsive luciferase reporter line (PHYB::LUC) was used to carry out a high-throughput screen to identify novel SAS mutants. The dracula 1 (dra1) mutant, that showed no avoidance of shade for the PHYB::LUC response, was the result of a mutation in the PHYA gene. Like previously characterized phyA mutants, dra1 showed a long hypocotyl in far red light and an enhanced hypocotyl elongation response to shade. However, dra1 additionally showed a long hypocotyl in red light. Since phyB levels are relatively unaffected in dra1, this gain-of-function red light phenotype strongly suggests a disruption of phyB signalling. The dra1 mutation, G773E within the phyA PAS2 domain, occurs at a residue absolutely conserved among phyA sequences. The equivalent residue in phyB is absolutely conserved as a threonine. PAS domains are structurally conserved domains involved in molecular interaction. Structural modelling of the dra1 mutation within the phyA PAS2 domain shows some similarity with the structure of the phyB PAS2 domain, suggesting that the interference with phyB signalling may be the result of non-functional mimicry. Hence, it was hypothesized that this PAS2 residue forms a key distinction between the phyA and phyB phytochrome species.
Keywords: Arabidopsis thaliana, light, luciferase, phytochrome, shade avoidance
Introduction
The light environment provides a wealth of information crucial for the development of plants, regulating germination, seedling establishment, architecture, and flowering time. Light reflected from neighbouring vegetation is depleted in red light (R) but remains relatively rich in far red light (FR). Plants are capable of perceiving a reduction in the R:FR ratio in incident light, indicative of potential vegetative shading. In species native to open habitats, such light triggers a phenomenon known as the shade avoidance syndrome (SAS): most noticeably characterized by a pronounced promotion of elongation growth causing a plant to overtop its neighbours, preventing the anticipated shading (Franklin, 2008; Martinez-Garcia et al., 2010).
Plants possess a range of photoreceptors capable of regulating growth so as to allow them to take maximum advantage of their situation. The phytochromes form the key to the SAS. Phytochromes exist in two reversibly photointerconvertible forms, an inactive, R-absorbing Pr form in which they are synthesized and an active, and an FR-absorbing Pfr form. Absorption of light causes an isomerization in the chromophore which, in turn, causes a change in conformation within the protein moiety (Rockwell et al., 2006). Some overlap, however, exists between the absorption spectra of the two forms, meaning that, even in monochromatic light, it is not possible to form a homogeneous pool of one form or the other. Instead, a dynamic equilibrium will exist between the two forms. Formation of the active Pfr form triggers a range of responses, most notably an inhibition of elongation growth and, thus, removal of Pfr in a light of a low R:FR ratio results in the promotion of elongation, characteristic of SAS.
The phytochrome family consists of five distinct proteins in eudicots, phyA–phyE. Each is a protein of 124 kDa consisting of a globular N-terminus and a linear C-terminus. The linear tetrapyrrole chromophore, phytochromobilin, is bound covalently by a cysteine in the N-terminus. While the N-terminus has been shown to be important mainly in light perception, the C-terminus is responsible for dimerization and signal transduction (Rockwell et al., 2006). Indeed, phytochromes exist as dimers, with two molecules, each with a chromophore, believed to be bound in an X-shaped configuration.
Several further subdomains within the phytochrome molecules have also been identified. The N-terminus consists of an amino-terminal extension segment (ATS), a Per/Arnt/Sim (PAS)-like domain (PLD), a cGMP phosphodiesterase/adenyl cyclase/FhlA (GAF) domain, and an eponymous phytochrome (PHY) subdomain (for a review, see Rockwell et al., 2006). The ATS inhibits dark reversion from Pfr to Pr and also stabilizes the conformation of Pfr. Along with the PLD this may also to be important in signal transduction (Oka et al., 2008). The N-terminal PLD and GAF domain fold into a groove which allows the binding of phytochromobilin, which occurs within the GAF domain. The PHY domain remains poorly understood but, along with the GAF domain, may contribute to light sensing (Oka et al., 2008). The C-terminus contains a large PAS-related domain (PRD) including two separate PAS domains, PAS1 and PAS2, and a histidine kinase-related domain. Within the PRD, a region known as the Quail-box (amino acids 714–731) is of particular interest in both phyA and phyB, containing the majority of known mutations. Sites for nuclear localization and for dimerization are also found within the C-terminal PRD (Rockwell et al., 2006).
The distinct phytochrome species show different expression patterns and functions. PhyA is by far the most abundant phytochrome in etiolated seedlings but is rapidly degraded upon conversion to the Pfr form. PhyA plays an important role in germination and de-etiolation in response to very low fluences of light [very low fluence response (VLFR)] or to very bright light of high fluence rates [high irradiance response (HIR)] (Franklin and Quail, 2010). In the VLFR the large pool of phyA in etiolated seedlings makes for a very sensitive ‘antenna‘, initiating photomorphogenesis even when the seedling in not in full light. In the HIR, FR, which maintains a small pool of the light-labile Pfr form, generates a persistent signal which increases with increasing fluence rate. This means that phyA is capable of triggering responses to FR wavelengths. Etiolated phyA null mutants show a complete insensitivity to FR for inhibition of hypocotyl elongation or promotion of cotyledon opening, indicating that phyA is the sole phytochrome mediating this response (Whitelam et al., 1993). PhyA levels in light-grown plants are ∼4% of those in etiolated seedlings (Sharrock and Clack, 2002). PhyA, however, continues to play a minor role throughout the life of the plant despite this. In particular, phyA is continually transcribed, meaning that any light conditions in which the Pfr form is not favoured will allow a re-accumulation of phyA (Clack et al., 1994; Bae and Choi, 2008).
PhyB–phyE are relatively light stable, with phyB becoming the major photoreceptor in light-grown seedlings. Like phyA, phyB–phyE also play a role in responses throughout the life of the plant. PhyB–phyE mediate low fluence responses (LFRs), which are typically activated by R but reversed by FR (Franklin and Quail, 2010). This R/FR reversibility means that these phytochromes are acutely sensitive to the R:FR ratio in the light environment and it is these phytochromes which are the key to SAS. Removal of light-stable phytochrome Pfr results in a removal of inhibition of elongation growth, causing a plant to overtop its neighbours, preventing any anticipated shading (Franklin, 2008; Martinez-Garcia et al., 2010). However, such a low R:FR ratio also activates phyA signalling. It is proposed that the low R:FR ratio allows re-accumulation of phyA which acts as a moderator of SAS by inhibiting elongation growth (Johnson et al., 1994; Yanovsky et al., 1995; Devlin et al., 2003).
Etiolated phyB null mutants show a severe loss of sensitivity to R for inhibition of hypocotyl elongation (Somers et al., 1991). PhyB is therefore identified as the major phytochrome mediating de-etiolation in R. Adult phyB mutant seedlings display a constitutively elongated, shade-avoiding phenotype (Lopez-Juez et al., 1992). PhyB is therefore identified as the major phytochrome mediating SAS. PhyD and phyE also play minor roles in this response but act redundantly with phyB (Devlin et al., 1998, 1999).
Molecular studies have revealed that phytochrome is cytoplasmic in the Pr form but production of Pfr results in a migration to the nucleus where it acts to regulate gene expression (Fankhauser and Chen, 2008). In the nucleus, phytochromes might interact with a number of phytochrome-interacting molecules, which play key roles in light responses, of which Phytochrome-interacting factor 3 (PIF3) was the original interactor to be characterized (Ni et al., 1998). PIF proteins are members of the basic helix–loop–helix (bHLH) family of transcription factors which play key roles in phytochrome signalling (for a review, see Castillon et al., 2007). Many of them have the ability to bind regulatory sequences of the genome (Martinez-Garcia et al., 2000; Huq et al., 2004; Hornitschek et al., 2009), hence providing a means to orchestrate a transcriptional network instrumental to transfer light conditions into the SAS morphological and physiological responses. Consistently, microarray analyses have identified dozens of shade-responsive genes (Devlin et al., 2003; Sessa et al., 2005), with a number of PHYTOCHROME RAPIDLY REGULATED (PAR) genes being identified as primary targets in a transcriptional cascade (Roig-Villanova et al., 2006). Several members of the PAR and PIF family have been genetically involved in the regulation of the SAS: five members of the homedomain-leucine zipper class II subfamily (ATHB2, ATHB4, and HAT1–HAT3), PAR1, PAR2, and PIF3-like 1 (PIL1) have been implicated in positive and/or negative aspects of SAS (Steindler et al., 1999; Salter et al., 2003; Roig-Villanova et al., 2006, 2007; Sorin et al., 2009). PIF4 and PIF5, which act as positive regulators of elongation growth in general (Nozue et al., 2007), were also demonstrated to play a key positive role (Lorrain et al., 2008). Another PIF relative, HFR1, acts to negatively regulate SAS by titrating out PIF4 and PIF5 (Sessa et al., 2005; Hornitschek et al., 2009). Two morphological-based screens have identified the involvement of auxin in SAS (Faigon-Soverna et al., 2006; Tao et al., 2008). Interestingly, PAR1 and PAR2 were shown to act as direct transcriptional repressors of auxin-responsive genes (Roig-Villanova et al., 2007), linking the above studies. Despite the success of these approaches, our understanding of phytochrome signalling in SAS is far from complete.
To complement this knowledge and identify novel SAS mutants, a molecular, high-throughput screen using an available shade-responsive luciferase reporter construct (Kozma-Bognar et al., 1999) was designed. The mutant dracula1 (dra1), a novel phyA mutant allele resulting in a decreased luciferase response to a low R:FR ratio, was identified. Based on protein structure modelling, it was proposed that the mutated residue, which is absolutely conserved in the PAS2 domain within each phytochrome species but which varies between phytochrome species, defines a key determinant of activity specific to those different phytochrome species.
Materials and methods
All data are representative of at least two independent experiments.
Plant materials and growth conditions
Seeds of Arabidopsis wild type, phyA-410, and phyB-464-19 containing PHYB::LUC (line ‘Ws-21a’) in the Ws-2 background were those described previously (Kozma-Bognar et al., 1999).
In all experiments, seeds were sterilized in 30% bleach, 0.02% Triton X-100, stratified for 3 d in darkness at 4 °C before germination, and plants were grown at 21 °C. For analysis of PHYB::LUC expression and for quantitative real-time PCR (qRT-PCR) analysis, seeds were sown on Murashige and Skoog (MS) medium containing 2% sucrose. For analysis of PHYB::LUC expression in response to simulated shade, plants were germinated in constant white light (cool white fluorescent light, 50 μmol m−2 s−1) for 7 d then transferred to white light supplemented with FR (R:FR ratio of 0.02) for 2 h. Where R/FR reversibility was examined seedlings were subsequently returned to white light for 2 h. Supplementary FR was provided by arrays of FR LEDs (λ max 735 nm, Shinkoh Electronics). For analysis of PHYB::LUC expression in response to end of day far red light (EODFR), seedlings were grown in 12 h light/12 h dark cycles for 7 d prior to treatment. At the end of the light period on day 7, EODFR-treated seedlings were transferred to FR for 15 min, then returned to darkness for the remainder of 2 h. Control seedlings were immediately transferred to darkness for 2 h. Bioluminescence images were taken before and after transfer. For morphological measurements and protein assay, seeds were sown on MS medium without sucrose. For analysis of response to monochromatic light, following stratification, seeds were given a 2 h white light (50 μmol m−2 s−1) treatment to synchronize germination. The plates were returned to darkness for 24 h and then either maintained in darkness or transferred to R (13 μmol m−2 s−1), FR (16 μmol m−2 s−1), or blue light (B; 17 μmol m−2 s−1) for 3 d. Monochromatic R and B sources used here were those described previously by Lopez-Juez et al. (2007). Monochromatic FR was obtained by filtering the output from the FR LEDs through one layer each of blue no. 363 and deep orange no. 158 celluloid filters (Lee Filters, Andover, UK). For analysis of growth responses to a low R:FR ratio, seeds were grown under constant white light (50 μmol m−2 s−1) for 6 d then either maintained under the same conditions or transferred to white light supplemented with FR (R:FR ratio of 0.02) for 2 d. For adult plant analysis, plants were germinated and grown in soil under 16 h white light (120 μmol m−2 s−1), 8 h dark cycles. All light measurements were made using a StellarNet EPP2000-HR spectroradiometer.
Luciferase imaging
Following 6 d growth in constant white light, seedlings were sprayed with 1 mM d-luciferin dissolved in 0.01% Triton (1 ml per plate). After one further day in white light, bioluminescence measurements were made before and after low R:FR ratio treatment using a NightOwl Molecular Imager (Berthold Technologies, UK). Data were analysed by using Winlight image analysis software version 2.17 (Berthold Technologies, UK).
Mutagenesis and screening
For ethylmethane sulphonate (EMS) mutagenesis of wild-type seeds of the PHYB::LUC line Ws-21a, 2500 seeds were suspended in 15 ml of 0.1% Tween-20 for 15 min. The Tween-20 was then replaced with 15 ml of 0.3% EMS and the seeds were agitated overnight. Seeds were then rinsed in H2O and subjected to three further 1 h washes with H2O. Subsequently seeds were pipetted onto filter paper and stratified at 4 °C in darkness for 3 d before sowing in pots.
M2 seed was collected from 1800 EMS-mutagenized M1 seeds, in pooled batches of 12 M1 lines. A toal of 30 000 M2 seedlings were screened, two Petri plates of 100 M2 seeds from each pool. Bioluminescence was measured before and after a 2 h low R:FR ratio treatment. Seedlings showing a bioluminescence level and/or a response differing from the rest of the batch population (Z-test at a confidence level 0.01) were selected and transplanted to soil. The seeds were collected from these plants and the next generation was screened again using 50 seeds for each mutant line. Those showing variation versus a wild-type control (t-test at a confidence level 0.01) were selected as genuine mutants.
Measurement of hypocotyl elongation
Seedlings were laid out horizontally in rows on agar plates along with a scale marker. Seedlings were photographed and hypocotyls were measured on digital images using Scion Image software (Scion Corporation, USA). At least two repeats were performed.
Mutation mapping
The dra1 mutant was crossed to the Col-0 wild type and a mapping population was created by selecting the plants with the mutant phenotype in the F2 population. DNA was extracted from fresh, young leaves (∼50 mg of tissue) and ground for 10 s in a 1.5 ml micro-centrifuge tube using a plastic pestle. A 10–20 μl aliquot of 0.5 M NaOH was then added to the crushed tissue and the mixture incubated at 95 °C for 40 s. A 120 μl aliquot of TE buffer (10 mM TRIS-HCl, pH 7.5, 1 mM EDTA) was added to the sample. Following 1 min centrifugation at 13 000 rpm, 1.2 μl of supernatant was used for PCR.
For all PCRs, a 25 μl volume PCR was set up using the BioMix Red kit (Bioline, UK) according to the manufacturer's instructions. PCR was performed in an Eppendorf Mastercycler Gradient PCR machine (Eppendorf, Germany) or in a Techne Flexigene PCR machine (Techne, UK) using standard reaction conditions. Primers for polymorphisms used as markers were identified using the MarkerTracker repository for genetic markers running on the Bio-Array Resource at the University of Toronto (http://bar.utoronto.ca/markertracker/). A list of primers used for sequencing of the PHYA gene is provided in Supplementary Table S1 available at JXB online.
Where restriction enzyme digestion was required for cleaved amplified polymorphism (CAP) molecular markers, DNA produced from PCR was digested directly without any purification. A 25 μl aliquot of PCR product was used in a 60 μl reaction with 5 U of enzyme and incubated for 2–4 h at the recommended temperature and buffer concentration for each enzyme. Enzymes were obtained from Promega (UK) or New England Biolabs (UK). Products were analysed following separation by gel electrophoresis, compared with a DNA size marker, HyperLadder V (Bioline, UK).
Analysis of gene expression
Following growth and treatment, seedlings (∼100 mg) were collected and frozen in liquid nitrogen. Total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen, UK). Possible DNA contamination in extracted RNA was removed by on-column DNase digestion with RNase-free DNase (Qiagen, UK) for 20 min. The quantity of RNA was determined by measurement of the absorbance at 260 nm in an Eppendorf Bio-photometer (Eppendorf, Germany). RNA was checked for DNA contamination by PCR which was designed to produce different product lengths due to the presence or absence of an intron.
Synthesis of cDNA was carried out using a cDNA synthesis kit (Bioline, UK) employing recommended reaction conditions. Original cDNA was diluted 10 times with diethylpyrocarbonate (DEPC)-treated water, and 1 μl of diluted cDNA was used for qPCR.
qPCR was performed using a SensiMix NoRef DNA kit (Quantace, UK) in a Qiagen Rotor gene 6000 (Qiagen, UK). PCR was performed for 40 cycles of 95 °C for 20 s, 60 °C for 30 s, and 72 °C for 30 s, following an initial enzyme activation step at 95 °C for 10 min. A standard dilution series was prepared of cDNA from the wild type after shade treatment. The sample cDNA was diluted 10 times prior to qPCR and the standard and sample cDNAs were amplified in the same PCR run. A standard curve for each amplified gene was plotted from the critical threshold (Ct) data of the standard dilution series, and quantitation of expression in samples was read from a standard curve using Qiagen Rotor-gene 1.7.65 software. Gene expression was normalized to the β-actin-2 housekeeping gene (At3g18780). In order to confirm that the PCR was specific and that the PCR product length is unique, the melting curve of each PCR product was analysed over a temperature range from 72 °C to 95 °C. A list of primers used for qPCR is provided in Supplementary Table S1 at JXB online.
Protein extraction and immunoblotting
For crude protein extracts, 100 mg of seedlings were collected under a dim green safelight and snap-frozen in liquid nitrogen. Seedlings were ground using a chilled pestle and mortar before 200 μl of extraction buffer (50% ethylene glycol, 0.1 M TRIS, 0.14 M ammonium sulphate, 10 mM EDTA, 1 mM phenylmethylsulphonyl fluoride) was added. Samples were centrifuged at 4 °C for 20 min and the supernatant was recovered. The protein concentration was determined by Bradford protein assay (Bio-Rad, UK). Samples were mixed with SDS–PAGE sample buffer (Laemmli, 1970) and heated at 100 °C for 3 min before transfer to ice.
Electrophoresis and immunoblotting were carried out according to the method of Devlin et al. (1992) with the following modifications. A 6% SDS–polyacrylamide (Laemmli, 1970) gel was prepared. A 25 μg aliquot of each sample was loaded in each lane along with the protein molecular markers (Prestained Page Ruler™, Fermentas, UK). Proteins were transferred to a PVDF membrane (Immobilon-P, Millipore UK), pre-wet in methanol, using a Bio-Rad Transblot Mini Trans-Blot Electrophoretic Transfer Cell (Biorad, UK) according to the manufacturer's instructions. The membrane was blocked in 5% (w/v) milk protein in TBST buffer (20 mM TRIS pH 7.5, 150 mM NaCl, 0.05% Tween) for 1 h at room temperature. Imunoblotting was carried out in TBST buffer containing 5% (w/v) milk powder with 3 μg ml−1 antibody at 4 °C in an incubation bag overnight with slow shaking. Following the primary antibody incubation, the membrane was washed three times with TBST for 10 min. Horseradish peroxidase-labelled secondary antibody (Promega, UK) was diluted 1:20 000 in 2.5% (w/v) milk protein/TBST and was added to the membrane and incubated for 1 h. The membrane was washed three times in TBST buffer. A Super Signal West Pico chemiluminescent substrate kit (Thermo Scientific, UK) was used for the development of the blot according to the manufacturer's instructions.
Following phytochrome detection, the blot was stained with Coomassie total protein stain (2 g l−1 Coomassie, 10% acetic acid, and 50% methanol) for 5 min and then destained with 7% acetic acid and 10% methanol.
AA01 anti-phyA monoclonal antibodies and BA02 anti-phyB monoclonal antibodies were a kind gift of Professor Akira Nagatani (Kyoto University, Japan).
Results
Identification of a phytochrome-responsive luciferase reporter line
With the goal of taking a fresh approach to the study of SAS, a high-throughput in vivo molecular-level screen in young seedlings of Arabidopsis was designed. The aim was to use a shade-responsive luciferase reporter to isolate potentially novel SAS mutants on the basis of a molecular rather than a morphological defect in light responsivity. For that purpose, the PHYB::LUC reporter line in the Ws-2 ecotype background developed by Kozma-Bognar et al. (1999) was used.
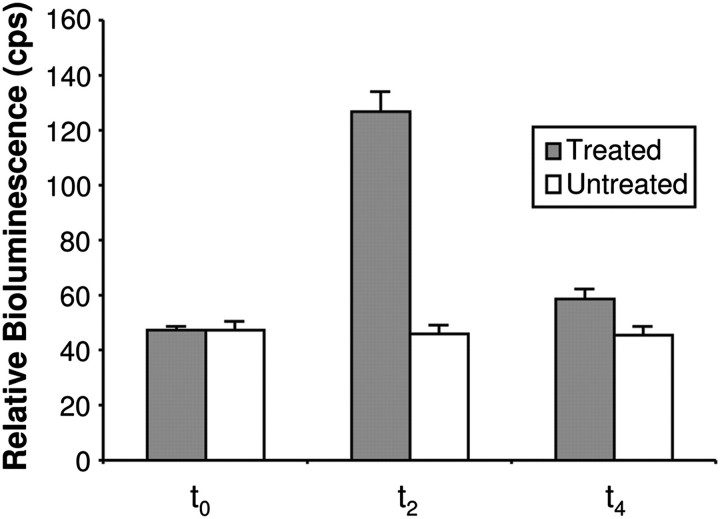
Previous work has shown that 7-day-old white light-grown seedlings of Arabidopsis show strong gene expression responses to a reduced R:FR ratio (simulated shade) (Devlin et al., 2003). Transcriptomic analysis of this response to the R:FR ratio in Arabidopsis revealed the PHYB gene to be an R:FR ratio-responsive gene, showing an increase in expression as part of the SAS (Devlin et al., 2003). The luciferase activity in response to a reduced R:FR ratio in seedlings of the PHYB::LUC reporter line was then tested. Seven-day-old white light-grown seedlings were imaged in a photon counting camera before being transferred to white light supplemented with FR (simulated shade) for 2 h, after which a second luciferase image was taken. The PHYB::LUC seedlings showed a clear luciferase signal after 7 d in constant white light. More importantly, they showed a marked increase in luciferase expression in response to a reduced R:FR ratio, tracking that previously seen for the transcript level. Subsequent return of treated seedlings to high R:FR ratio light reversed the increase in expression, confirming that this is a phytochrome-mediated response. In contrast, PHYB::LUC expression in untreated seedlings remained constant during this period of time (Fig. 1).
Fig. 1.
PHYB::LUC expression shows an R:FR-reversible increase in response to shade. Seven-day-old white light-grown seedlings were imaged (t0), transferred to simulated shade for 2 h before a second image was taken (t2), and then returned to white light for 2 h when a third image was taken (t4). Control seedlings were maintained in white light. Data represent mean bioluminescence measurements relative to t0 ±SE from at least 22 seedlings.
Isolation of SAS mutants using a luciferase reporter screen
Seeds of the PHYB::LUC line were treated with EMS to generate a mutagenized population of 1800 M1 lines. A total of 30 000 M2 plants were screened from within these, evenly covering the 1800 M1 lines. Seedlings were screened in batches of 200 M2 seeds, each batch sown from seed collected from a pool of 12 M1 lines. Seeds were sown evenly spaced at a density of 100 seeds on a 10 cm square Petri plate (two plates per batch of M2 seeds). Following release from stratification, seedlings were screened after exactly 7 d in constant white light in order to avoid any circadian variation in PHYB expression. Seedlings were imaged before (t0) and after a 2 h reduced R:FR ratio treatment (t2). For each plate of 100 seedlings, those individually showing an increased or decreased relative change in LUC expression in response to a reduced R:FR ratio [(t2–t0)/t0] relative to the batch population were selected for further analysis. Seedlings were selected as putative mutants where this aberrant LUC expression response showed a z-score of –3.29 or less, or of ≥3.29 based on the mean response of the screened population of 100 seedlings on each Petri plate. This constitutes a significant difference (P <0.001).
Following first round screening, 217 putative mutants were isolated and grown for seed. A population of 20 M3 seed collected from each line was used for a second round of screening to confirm the heritability of the mutant phenotype compared with seed of the original PHYB::LUC line. Populations consistently showing a significant difference in response to a reduced R:FR ratio (P <0.001) were flagged for further analysis. Twenty-nine putative mutants were retained after this second round as genuine mutants, and larger batches of 50 M3 seeds from each population were then analysed in a third round of screening. Fourteen lines showing a significant difference in SAS in response to a reduced R:FR ratio with a P-value of <0.001 at this stage were selected for further study.
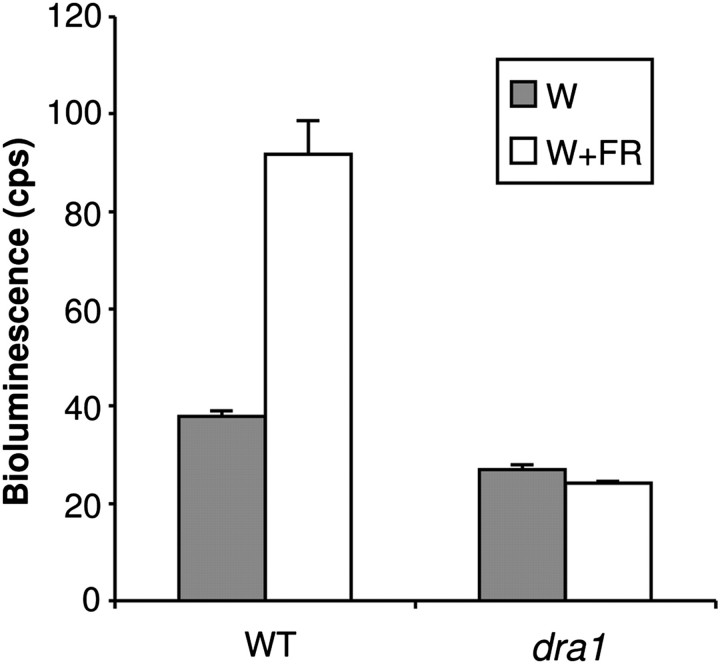
One mutant in particular was selected for more in-depth analysis. This mutant was named dracula 1 (dra1) because it showed a substantially reduced avoidance of shade in terms of the LUC expression response (Fig. 2). Inhibition of hypocotyl elongation in response to monochromatic R, B, and FR was examined in etiolated seedlings of this mutant line. Similarly, the hypocotyl elongation response to a reduced R:FR ratio was examined in established seedlings. The aim here was to identify any pleiotropic effect on morphology that may also be caused by the mutation, since defective morphological light responses during de-etiolation under monochromatic light in addition to those in response to simulated shade might suggest a more general defect in phytochrome signalling. M3 seedlings of dra1 showed normal de-etiolation under B but were observed to show reduced sensitivity to R and a severely reduced sensitivity to FR for inhibition of hypocotyl elongation. In contrast to the LUC response, M3 seedlings of dra1 also showed a greatly increased hypocotyl elongation in response to a reduced R:FR ratio (see below for a full phenotypic analysis of the backcrossed line).
Fig. 2.
dra1 shows a greatly reduced PHYB::LUC expression response to shade. Seven-day-old white light (W)-grown seedlings of the wild type (WT) and dra1 were transferred to simulated shade (W+FR) for 2 h. Data represent mean bioluminescence measurements ±SE from at least 27 seedlings.
Co-segregation analysis of dra1 phenotypes
A backcross of dra1 was performed to the ‘wild-type’ PHYB::LUC line to examine co-segregation of the mutant's morphological and LUC expression phenotypes. The long hypocotyl in FR was used as the initial phenotype for selection in order to test co-segregation. Seedlings of the F2 population from the backcross of dra1 with the parental ‘wild-type’ PHYB::LUC line showed a 3:1 ratio of wild type to long hypocotyl phenotypes under FR, indicating this to be the result of a single, recessive mutation. A number of wild-type and long hypocotyl seedlings from this population were selected and grown for seed. More than 50 independent F3 lines were tested, each the offspring of F2 individuals selected on the basis of a long hypocotyl in FR. All individuals within each F3 population displayed a reduced LUC response to shade, indicating that the long hypocotyl in FR phenotype is 100% linked to the dra1 reduced LUC response to shade. Supplementary Fig. S1A at JXB online shows LUC response data for seven of these lines. Similarly, the long hypocotyl in FR phenotype was found to be 100% linked to the long hypocotyl in R in >50 independent F3 lines. Supplementary Fig. S1B shows data for hypocotyl length in R and FR for individuals from the same seven F3 lines as above. Thus, all three phenotypes are highly likely to be the result of the same mutation.
For ease of selection, the long hypocotyl in FR phenotype was therefore used to perform one additional round of backcrossing and selection of the dra1 mutant to clean up the mutant line with respect to other possible induced mutations elsewhere in the genome. All the following phenotypic and molecular analyses were performed with the twice-backcrossed line.
Physiological analysis of mutant lines
An examination of SAS responses was carried out in the twice-backcrossed line. Seedlings were grown for 5 d in constant white light before being transferred to white light supplemented with additional FR for another 2 d. Control seedlings were maintained in constant white light. In contrast to luciferase activity, dra1 seedlings also showed a greatly increased hypocotyl elongation in response to a reduced R:FR ratio (Fig. 3A), in agreement with the observations with the M3 seedlings.
Fig. 3.
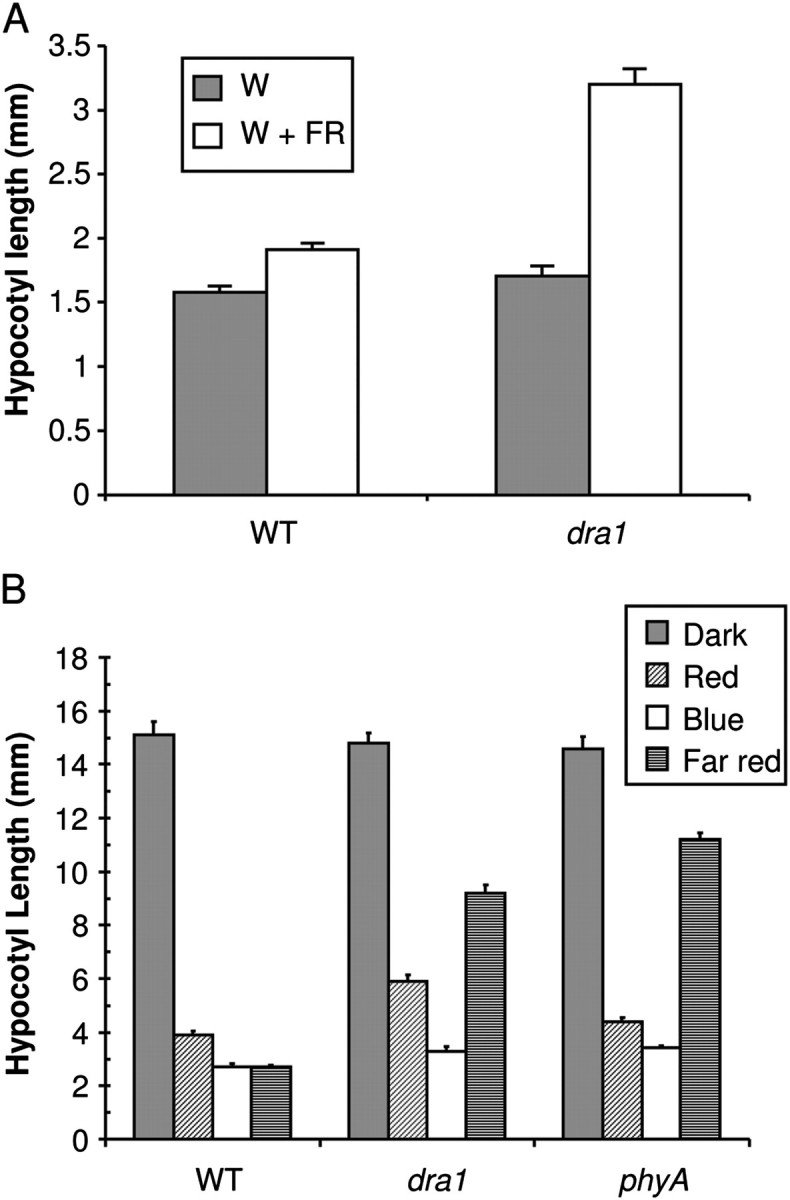
dra1 displays altered phytochrome responses. (A) dra1 shows aberrant elongation responses to shade. Five-day-old seedlings of the wild type (WT) and dra1 were either maintained in white light (W) or transferred to simulated shade (W+FR) for 2 d. Data represent mean hypocotyl length ±SE for 20 seedlings. (B) dra1 displays a long hypocotyl in red and far red light. One-day-old etiolated seedlings of the wild type (WT), dra1, and phyA-410 were either maintained in darkness or transferred to monochromatic red, blue, or far red light for 3 d. Data represent the mean hypocotyl length ±SE from at least 20 seedlings.
De-etiolation under monochromatic light was also examined in seedlings of dra1. One-day-old etiolated seedlings were transferred to R, FR, or B for 3 d, after which hypocotyl lengths were observed. dra1 seedlings showed normal de-etiolation under B but were observed to show reduced sensitivity to R and a severely reduced sensitivity to FR for inhibition of hypocotyl elongation (Fig. 3B). This phenotype is suggestive of a defect in both phyA and phyB signalling, as previously observed in the hy1 and hy2 loss-of-phytochromobilin chromophore mutants (Koornneef et al., 1980). However, hy1 and hy2 mutants both show a pale, spindly phenotype, a small rosette, and attenuated SAS responses (Chory et al., 1989; Halliday et al., 1994). In contrast, dra1 seedlings showed quite a normal adult phenotype in white light (Supplementary Fig. S2 at JXB online) and an exaggerated elongation response to shade, suggesting that this may be a novel phenotype caused by the molecular lesion found in the PHYA gene of dra1
Molecular analysis of dra1
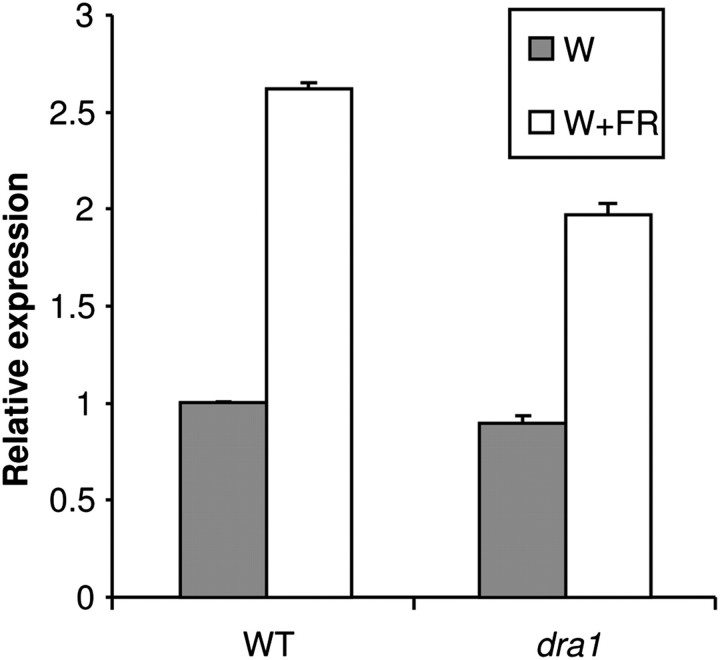
The response of the endogenous PHYB transcript to simulated shade was examined in a dra1 line using qRT-PCR. Total RNA was extracted from 1-week-old constant white light-grown seedlings of the wild type and dra1 mutants either maintained for 2 h in white light or subjected to a low R:FR ratio treatment. As expected, the PHYB message was increased ∼2.5-fold in wild type seedlings in response to simulated shade, closely mirroring the increase in LUC expression seen under these conditions. The dra1 mutant showed a very slightly reduced PHYB transcript level relative to the wild type in white light but showed a significantly reduced increase in PHYB transcript level compared with the wild type in response to shade (Fig. 4). However, the magnitude of the reduction was not as dramatic as that seen for the bioluminescence phenotype. Nonetheless, both early molecular responses to simulated shade were significantly reduced in dra1 seedlings (P <0.000001).
Fig. 4.
dra1 shows a reduced PHYB expression response to shade. Seven-day-old white light (W)-grown seedlings of the wild type (WT) and dra1 were transferred to simulated shade (W+FR) for 2 h. Relative PHYB expression was measured by quantitative RT-PCR, normalized to the ACT2 control gene. Data represent the mean ±SE from five independent batches of 20 seedlings.
Cloning and characterization of the mutant gene
The easy-to-score long hypocotyl in FR phenotype was also used to select dra1 mutant seedlings from the F2 of a mapping cross between the M3 dra1 line and a wild-type plant of the Col-0 ecotype. Identification of markers polymorphic between ecotypes Ws-2 and Col-0 in the region of the top of chromosome 1 revealed the mutation to lie between markers N1-3041125-EcoRI (Nordborg et al., 2005) at 3 041 125 bp and NGA63 (Bell and Ecker, 1994) at 3 224 463 bp (exact location updated). This location is distinct from the location of the PHYB::LUC transgene on chromosome 2 (L. Kozma-Bognar, personal communication), indicating that this was not a mutation within the transgene. The region between 3 041 125 bp and 3 224 463 bp of chromosome 1 contains the PHYA gene and, as the dra1 mutant showed a defect in phyA signalling, the PHYA gene in dra1 was therefore sequenced to check for mutations. Sequencing revealed a G to A substitution at 3625 bp downstream of the ATG within the PHYA genomic sequence. This would result in a G to E substitution at amino acid 773 of the phyA protein. This also results in the loss of a BtsCI restriction enzyme digest site, providing a convenient restriction fragment length polymorphism useful for identification of the dra1 mutation. The G to E substitution at amino acid 773, a previously uncharacterized mutation, lies within the C-terminal region of the phyA protein.
The LUC response to shade of the phyA-410 mutant carrying PHYB::LUC was examined to determine whether this loss of response is allele specific or a universal effect of phyA deficiency. Seven-day-old light-grown seedlings of the phyA-410 mutant, conversely, showed an increase in luciferase activity in response to shade (Fig. 5). This suggests that the lack of response in dra1 is allele specific.
Fig. 5.
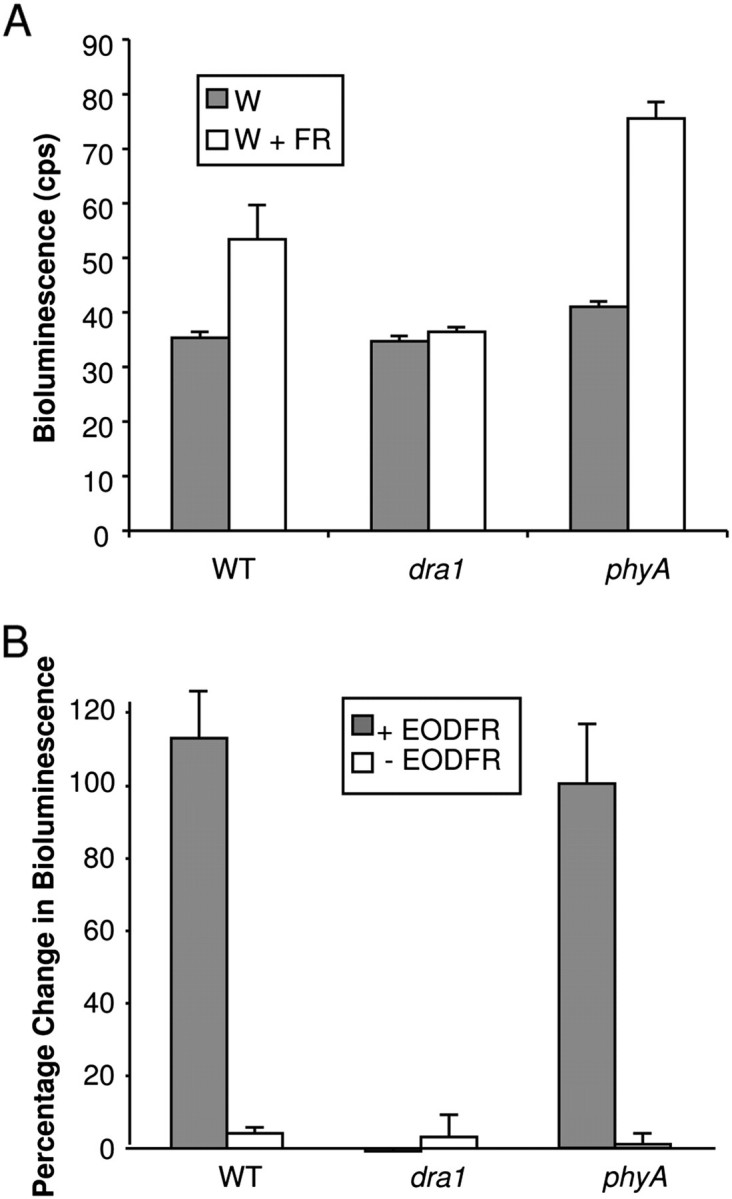
phyA-410 shows an enhanced PHYB::LUC response to shade but not end of day far red. (A) Seven-day-old white light (W)-grown seedlings of the wild type (WT), dra1, and phyA-410 were transferred to simulated shade (W+FR) for 2 h. Data represent mean bioluminescence measurements ±SE from 20 seedlings. (B) Seven-day-old W-grown seedlings of the WT, dra1, and phyA-410, grown in 12 h light/12 h dark cycles, were transferred at the end of the light period to far red light for 15 min, then returned to darkness for the remainder of 2 h (+EODFR). Control seedlings were immediately transferred to darkness for 2 h (–EODFR). Bioluminescence images were taken before and after transfer. Data represent the mean percentage change in bioluminescence ±SE from 20 seedlings.
The LUC response to EODFR was also examined in dra1 and phyA-410. EODFR treatment comprises a pulse of FR at the end of a light period in plants grown in light–dark cycles and is more specific to examining the effect of phyB Pfr removal without triggering the moderating effects of phyA HIR which requires more prolonged irradiation. Here wild-type and phyA-410 seedlings behaved identically, but dra1 seedlings showed no response to EODFR (Fig. 5b). The allele-specific effect of the phyA mutation in dra1 in suppressing PHYB::LUC expression would therefore appear to be constitutive, not dependent on phyA activation.
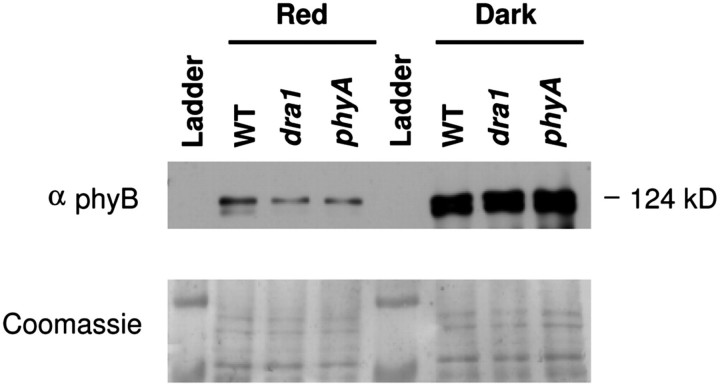
Measurement of PHYA mRNA via qRT-PCR revealed only a minor reduction in PHYA message in the dra1 mutant (Fig. 6A) while measurement of phyA protein levels in dark-grown seedlings by western blotting, likewise, showed no significant difference in phyA protein levels (Fig. 6B). This suggests that the mutation results in the production of a defective phyA protein. The phyA-410 mutant was also analysed. The phyA-410 mutation, similarly, results in a defective phyA protein and showed only a slight decrease in phyA protein levels (Fig. 6B). Despite the mutation in dra1, the phyA protein produced was still degraded following 3 d in R in conditions where the labile Pfr form would be expected to form, indicating that it is photoactive (Fig. 6B). This, therefore, suggests a signalling rather than a light perception defect, consistent with other C-terminal mutations of phytochrome molecules (Wagner and Quail, 1995).
Fig. 6.
dra1 shows normal levels of PHYA message and phyA protein. (A) Relative PHYA expression in 7-day-old white light (W)-grown seedlings of the wild type (WT) and dra1. PHYA expression was measured by quantitative RT-PCR, normalized to the ACT2 control gene. Data represent the mean ±SE from five independent batches of 20 seedlings. (B) One-day-old etiolated seedlings were either maintained in darkness or transferred to monochromatic red light for 3 d, at which point protein was extracted. Upper panel: western blot of extracts of the WT, dra1, and phyA-410 probed with phyA-specific monoclonal antibodies. Lower panel: Coomassie-stained blot showing equal loading.
dra1/phyAG773E shows a novel allele-specific defect in R signalling
A long hypocotyl in monochromatic FR, a wild-type hypocotyl elongation in monochromatic R, and an exaggerated elongation response to shade are well established as being consistent with a loss of phyA function (Whitelam et al., 1993; Johnson et al., 1994; Yanovsky et al., 1995; Sessa et al., 2005). However, such dramatically reduced sensitivity to R for inhibition of hypocotyl elongation in de-etiolation has not been observed in previous phyA mutant alleles. For comparison, 1-day-old etiolated seedlings of the phyA-410 null mutant carrying PHYB::LUC were grown alongside dra1 for 3 d in the above de-etiolation experiments in monochromatic light. As expected, both mutants showed the long hypocotyl in FR phenotype (Fig. 3B). It is noticeable that dra1 hypocotyls were significantly shorter under FR than phyA-410 null mutant hypocotyls, indicating that the phyAG773E mutation results in only a partial loss of phyA function. However, the phyA-410 null mutant showed a wild-type hypocotyl length in R, confirming that the long hypocotyl in R was a specific defect of the phyAG773E mutation in the dra1 allele.
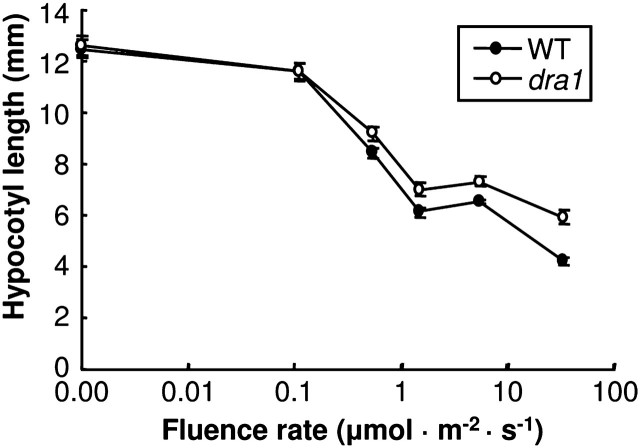
The fluence rate dependence of the reduced inhibition of hypocotyl elongation in R was further investigated using a fluence rate response curve. One-day-old etiolated seedlings were transferred to one of a range of fluence rates of R for 3 d, after which hypocotyl lengths were observed. dra1 seedlings were observed to show reduced sensitivity to R for inhibition of hypocotyl elongation at fluence rates as low as 0.5 μmol m−2 s−1. The discrepancy between the wild type and dra1 became greater as the fluence rate increased (Fig. 7).
Fig. 7.
dra1 displays a reduced sensitivity to red light over a range of fluence rates. One-day-old etiolated seedlings of the wild type (WT) and dra1 were either maintained in darkness or transferred to one of a range of fluence rates of monochromatic red light for 2 d. Data represent the mean hypocotyl length ±SE from at least 20 seedlings.
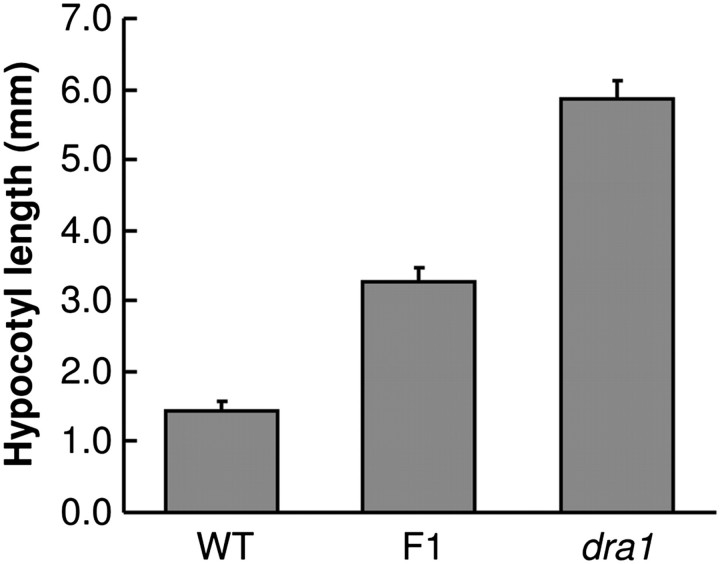
The dominance of the novel long hypocotyl in R phenotype in the dra1 mutant was tested by examination of hypocotyl length in R in F1 seedlings of the backcross of dra1 with the wild-type PHYB::LUC line. F1 seedlings displayed an intermediate hypocotyl length under R, indicating this to be a gain-of-function effect, probably showing dosage dependence and, hence, is the result of a partially dominant mutation (Fig. 8).
Fig. 8.
The long hypocotyl in red light of dra1 is a dose-dependent gain-of-function phenotype. One-day-old etiolated seedlings of the wild type, dra1, and the F1 of a cross between the two were transferred to monochromatic red light for 3 d. Data represent the mean hypocotyl length ±SE from at least 11 seedlings.
A significant long hypocotyl phenotype in R is indicative of a deficiency in phyB signalling (Somers et al., 1991). Although PHYB transcript levels are unaffected in white light in dra1, a possible reason for a loss of sensitivity to R caused by the phyAG773E mutation may have been a secondary effect causing a reduction in phyB protein levels. An examination of phyB protein levels was therefore carried out in seedlings of wild-type, dra1/phyAG773E, and phyA-401 mutant seedlings grown in darkness or in R. In darkness, levels of phyB were similar in all three lines. Wild-type, dra1, and phyA-401 seedlings showed a significant decrease in phyB protein in response to R but, following growth in R, both dra1/phyAG773E and phyA-401 seedlings contained slightly less phyB protein than the wild type (Fig. 9). Therefore, the slightly lower level of phyB protein may be a common feature of phyA mutants. The phyA-410 mutant does not show any measurable defect in the de-etiolation response to monochromatic R, meaning that it seems unlikely that the R phenotype of dra1/phyAG773E is simply due to the observed decrease in phyB protein levels.
Fig. 9.
dra1 shows normal levels of phyB protein. One-day-old etiolated seedlings were either maintained in darkness or transferred to monochromatic red light for 3 d, at which point protein was extracted. Upper panel: western blot of extracts of the wild type (WT), dra1, and phyA-410 with phyB-specific monoclonal antibodies. Lower panel: Coomassie-stained gel showing equal loading.
It was considered that the allele-specific loss of responsivity to a low R:FR ratio might also be a feature of this phyB signalling defect. However, the loss of the LUC response phenotype was found to be recessive, suggesting that it is primarily caused by the similarly recessive phyA signalling deficiency rather than the gain-of-function phyB signalling defect (Supplementary Fig. S3 at JXB online).
Modelling of wild-type and mutant phyA structures
The mutation in dra1 lies in the PAS2 domain of the phyA protein. Residue 773 lies between the first two β-strands of the PAS domain (Supplementary Fig. S4 at JXB online), a domain shown to be necessary for downstream signal transduction (Quail et al., 1995; Park et al., 2000). Although analysis of the predicted effect of the mutation on secondary structure indicates that the PAS domain would still fold normally (Supplementary Fig. S4), the most likely reason for a partial loss of phyA function in this case would therefore seem to be a disruption of an aspect of the phyA signalling pathway.
It is interesting that while the structure of the PAS2 domain around this residue is conserved in all phytochromes, the mutated residue is absolutely conserved as a G in all phyA sequences from a range of higher plants, while the equivalent residue in the phyB PAS2 domain is absolutely conserved as a T in all published phyB/phyD sequences (Supplementary Table S2 at JXB online). This suggests that this residue may make a key contribution to the distinct characteristics of phyA or phyB. The unusual R signalling defect resulting from the dra1/phyAG773E mutation was speculated to be the result of a gain of ability of phyAG773E to interfere with some aspect of the phyB signalling pathway by mimicking the phyB PAS2 domain. Therefore, a predictive modelling package was used to examine the effect of the phyAG773E mutation on the shape of the PAS2 domain of the phyA molecule, and this was compared with the shape of the PAS2 domain of the phyB molecule. The sequence was initially mapped onto similar PAS domains for which definitive structural data already exist. The best structural homologue to the sequences examined here was identified using mGenThreader which assigns the fold (McGuffin and Jones, 2003). mGenThreader identified the redox sensor domain of Azotobacter vinelandii NifL corresponding to the PDB code 2gj3 (Key et al., 2007) for these sequences with a medium confidence. This structure was then used as a template to model the structures corresponding to the sequences using the modelling package Modeller (Marti-Renom et al., 2000), using the alignment provided by mGenThreader. Ten models were generated for each structure and the best one was selected on the basis of the DOPE score.
The 3D structures also predict that the PAS2 domain containing the mutated sequence in dra1/phyAG773E would fold normally. The mutated residue is revealed to be at the entrance to the PAS pocket. PAS pockets are common structurally conserved domains involved in ligand binding or protein–protein interaction (Pellequer et al., 1998), with the ligand binding within the pocket. The G773E mutation would replace the minimal side chain of glycine with the more extensive side chain of glutamate, and the model predicts that this side chain would stick out into the pocket (Fig. 10A, B). It is interesting that the predicted model of the phyB PAS2 domain predicts that the conserved threonine at this point in phyB PAS2 sequences would similarly leave a relatively large side chain protruding out into the mouth of the pocket (Fig. 10C). This is, therefore, consistent with the proposal that the cause of the interference of the dra1/phyAG773E phyA protein with the phyB signalling pathway is a degree of mimicry.
Fig. 10.
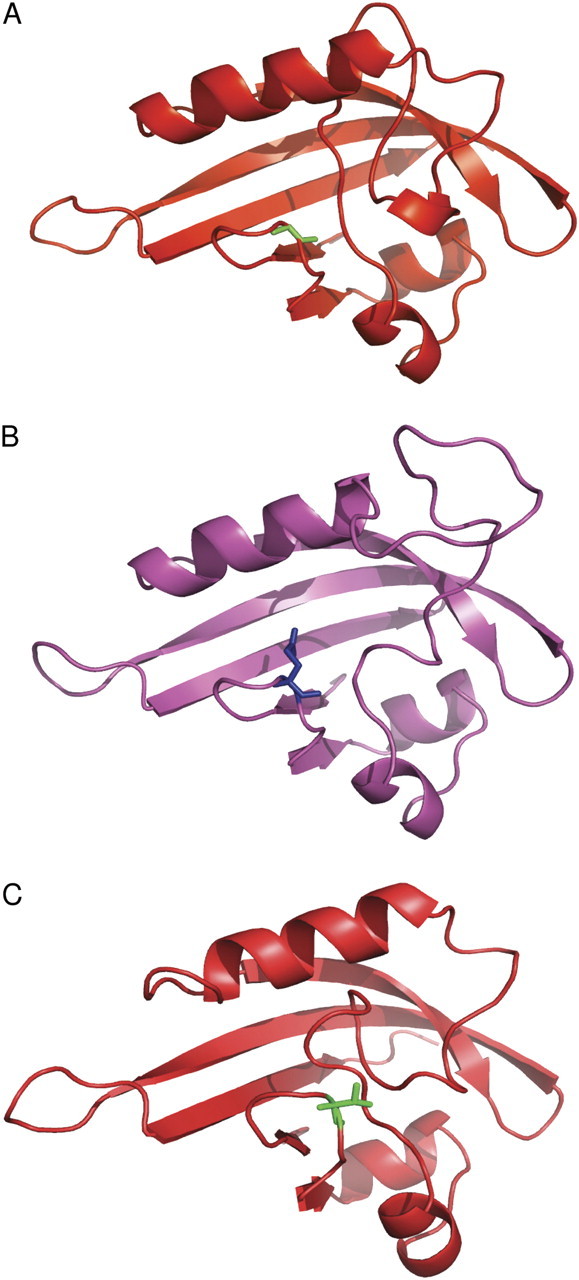
3D modelling suggests some similarity between the PAS2 domains of dra1 phyA and wild-type phyB. Cartoon representations of 3D models generated by the ‘Modeller’ package, using the redox sensor domain of Azotobacter vinelandii NifL as a template. (A) The wild-type phyA PAS2 domain structure. Residue 773 (glycine) of the phyA molecule, labelled in green in stick notation, does not obstruct the PAS pocket. (B) The dra1 mutant phyA PAS2 domain structure. Residue 773 (glutamate) of the phyA molecule, labelled in blue in stick notation, protrudes across the PAS pocket. (C) The phyB PAS2 domain structure. Residue 808 (threonine) of the phyB molecule, labelled in blue in stick notation and equivalent to residue 773 of the phyA molecule, also protrudes across the PAS pocket.
Discussion
A novel, high-throughput molecular screen to identify SAS mutants
Luciferase reporters have been used previously in screening for mutants defective in responses to a number of regulators of gene expression such as the circadian clock (Millar et al., 1995), UV light (Jackson et al., 1995), and jasmonate (Ellis and Turner, 2001). However, no previous attempts have been made to use this technology for screening of SAS or even phytochrome signalling mutants. The present screen builds on previous work identifying R:FR ratio-responsive genes in established, 1-week-old seedlings (Devlin et al., 2003). At this stage, luciferase bioluminescence can be accurately measured in individuals whilst still being small enough to allow screening of high numbers of seedlings growing on Petri plates (Millar et al., 1995).
The PHYB promoter was used as a regulator of LUC expression. PHYB message is strongly responsive to the R:FR ratio (Devlin et al., 2003), making it a good reporter for a SAS screening. The ecological reason for the dramatic increase in PHYB expression in response to a reduced R:FR ratio is unclear. Such conditions trigger SAS as a result of a shift in the Pr:Pfr equilibrium in favour of Pr. The conversion of phyB Pfr to the inactive Pr form removes an inhibition of elongation growth. If phyB levels subsequently increased as a result of increased PHYB expression, this would increase levels of both Pr and Pfr. Although the equilibrium between the two would be unchanged, the increased phyB Pfr level would act to moderate elongation growth, perhaps preventing excessive elongation. However, it remains to be seen whether the increase in PHYB message results in a concomitant increase in phyB protein. Previous analysis of phyB expression has shown a circadian rhythm in PHYB transcription and in levels of PHYB message. However, measurement of phyB protein has failed to show any such pattern being replicated at the protein level (Kozma-Bognar et al., 1999). The previously generated PHYB::LUC line, Ws-21a, created by Kozma-Bognar et al. (1999) was used to confirm that the increase in PHYB message in response to a low R:FR ratio was reflected at the level of transcription. The 2.5-fold change in luciferase bioluminescence in response to a low R:FR ratio very closely followed a similar magnitude of change previously observed in message levels (Devlin et al., 2003). This finding is also in agreement with the observation of Hall et al. (2002) who analysed the response of this line to EODFR treatment which also depletes phytochrome Pfr.
Screening identified a novel phyA mutant showing dominant-negative suppression of phyB signalling
Following EMS mutagenesis of this line, a screen of 30 000 M2 lines yielded 217 putative response mutants, 14 of which showed an inherited phenotype. These included mutants showing increased and decreased luciferase response but, curiously, none was identified showing a constitutively high luciferase activity signal. Such constitutive shade-avoiding mutants might be expected as the phyB mutant itself showed this pattern of behaviour (data not shown). The 14 most extreme mutants were examined further. Approximately half of these mutants showed some physiological phenotype associated with more generally defective light signalling. However, in many of the mutants showing physiological defects, these defects were minor and would very probably be missed by standard morphological screens.
A mutant showing an extreme but previously unreported phenotype was the focus of this work. dra1 was so named because it showed a greatly reduced LUC response to simulated shade. However, in contrast to the reduced LUC response to a low R:FR ratio, dra1 showed an enhanced hypocotyl elongation response to a low R:FR ratio (Fig. 3A). dra1 also showed an elongated hypocotyl following de-etiolation in either R or FR, but not under B (Fig. 3B). It remains a slim possibility that these phenotypes may be the result of very tightly linked mutations, but the fact that these phenotypes showed 100% linkage in >50 independent F3 lines strongly suggests that the phenotypes are all a result of a single mutation. Such a phenotype has previously been observed in the hy1 and hy2 chromophore biosynthesis mutants of Arabidopsis, but as adult plants these mutants show a pale, elongated appearance, with small rosettes (Chory et al., 1989), which was not the case for dra1 (Supplementary Fig. S2 at JXB online). A number of pef mutants were identified some time ago showing deficiency in response to R and FR for hypocotyl elongation (Ahmad and Cashmore, 1996); however, the genes disrupted in these mutants have not been identified.
The dra1 mutation results in an amino acid change from glycine to glutamate at position 773 near the beginning of the PAS2 domain of the phyA protein. This constitutes a previously uncharacterized phyA mutation. Analysis of the PHYA transcript shows that the PHYA gene is expressed at levels only slightly below that in the wild type, while western blotting shows that the levels of phyA protein are comparable with those in the wild type (Fig. 6A, B). However, the mutant showed greatly reduced sensitivity to monochromatic FR for inhibition of hypocotyl elongation (Fig. 2), a response solely controlled by phyA (Whitelam et al., 1993), and indicative of a severe loss of phyA function. The fact that some inhibition of hypocotyl elongation in FR was observed in the phyAG773E mutant, however, suggests that there is not a complete loss of phyA function. In addition, the phyAG773E mutant showed an exaggerated hypocotyl elongation response to a low R:FR ratio-simulated shade (Fig. 3A), typical of a phyA mutant (Johnson et al., 1994). Analysis of the change in PHYB::LUC expression in response to shade in the previously characterized phyA-410 mutant revealed that this phyA mutant also showed an extreme response to shade for this PHYB::LUC response. However, this would suggest that the originally observed loss-of-PHYB::LUC response phenotype in phyAG773E is not a standard result of phyA deficiency but rather an allele-specific phenotype. The PHYB::LUC defect in phyAG773E co-segregates with the long hypocotyl in FR phenotype and is recessive in nature, suggesting that it is associated with the specific disruption of phyA. It is clear from the phyA-410 mutant that phyA does normally play a role in suppressing PHYB transcription, and it would appear that this role has somehow been enhanced by the mutation in phyAG773E. Such enhancement was also observed to overcome the EODFR response where only 15 min of FR light was given to dra1 seedlings prior to transfer to darkness, suggesting that this is a constitutive effect not dependent on phyA activation. That one aspect of phyA signalling is enhanced while another is diminished would suggest that the two functions, inhibition of hypocotyl elongation and inhibition of PHYB expression, involve distinct mechanisms possibly involving distinct binding partners.
The mutation in phyAG773E occurs at a very highly conserved glycine residue within the PAS2 domain. The PAS domain is a structurally rather than sequence conserved domain. It would appear that sequence can vary greatly while still creating the same folding structure typifying a PAS pocket (Pellequer et al., 1998). Alignment of all available phyA sequences revealed that, while the residues constituting much of the PAS2 domain vary from one plant species to another, the glycine residue is unchanged, suggesting an absolute requirement for glycine at this position in phyA species (Supplementary Table S2 at JXB online). Structural predictions suggest that the larger side chain of the substituting glutamate of the mutation in phyAG773E would protrude across the entrance of the PAS pocket where the minimal side chain of glycine would create no such obstruction (Fig. 10). It is easy to envisage that this would interfere with the ability of the PAS domain to bind to a target molecule, accounting for a loss of phyA function. Consistent with this being a defect in signalling rather than perception, the phyA protein present in darkness in the phyAG773E mutant shows normal R-induced degradation (Fig. 6B).
The phyA N-terminal domain with an attached green fluorescent protein (GFP) fusion in transgenic plants showed constitutive nuclear localization of the fusion protein. However, this construct was not sufficient to rescue a phyA mutant (Mateos et al., 2006), confirming the importance of the C-terminus in signal transduction. Furthermore, the phyA PAS1 and PAS2 domains have both, specifically, been found to be important in signalling transduction (Quail et al., 1995; Park et al., 2000). There are also previously reported phyA mutants in the PAS2 domain. PhyA-108 (G768D) and phyA-302 (E768K) are both missense, loss-of-function mutations which produce spectrally active phyA (Xu et al., 1995; Parks et al., 1996), but no dominant-negative interference with phyB signalling was observed in either mutant. Interestingly, phyA-302 (E768K) affects phyA nuclear localization (Yanovsky et al., 2002), providing one possible, speculative cause for the loss of activity in the phyAG773E mutant.
The phyAG773E mutant shows a novel, additional R phenotype not typical of previously characterized phyA mutants, even those similarly resulting from mutations causing a specific loss of phyA signalling as opposed to photoperception. The phyAG773E mutant fails to de-etiolate properly under R, displaying a long hypocotyl in R more typical of a partial loss of phyB function (Figs 3B, 7). A role for phyA in de-etiolation in R has been previously demonstrated by Mazzella et al. (1997) and by Franklin et al. (2007), but in the former case only in the absence of phyB and in the latter case only at photon irradiances much greater than those used here. The phyB photoreceptor is almost entirely responsible for de-etiolation responses to R at photon irradiances used here (Aukerman et al., 1997; Franklin et al., 2003; Monte et al., 2003). This significant deficiency in response to R in the phyAG773E mutant therefore indicates an additional defect in phyB signalling. The dominance of this phenotype is suggestive of a dominant-negative interference with phyB signalling by the mutant phyAG773E molecule. Several examples have been recorded of dominant-negative effects mediated by a mutated phyA on a functional phyA protein (Fry et al., 2002; Weller et al., 2004), but only one example of a dominant-negative effect of a mutant phyA on phyB signalling has been observed previously. This was witnessed in experiments overexpressing truncated phyA. Here oat phyA truncated at residue 617 or missing residues 617–686, overexpressed in Arabidopsis, resulted in a similar loss of sensitivity to R (Boylan et al., 1994), a result interpreted as a consequence of the interference of the overexpressed mutant phyA on the endogenous phytochrome signalling machinery. Similarly, the mechanism by which such dominant-negative interference of mutant phyA on phyB signalling could occur in dra1 can be speculated upon. One possible cause centres on the extremely high level of conservation of the mutated residue. The equivalent residue in the phyB PAS2 domain, Thr808, is also extremely highly conserved; however, the residue is conserved as a threonine in all available phyB sequences (Supplementary Table S2 at JXB online). This further suggests that the residue may form a key distinguisher of phytochrome species-specific signalling. Furthermore, this threonine in phyB projects a long side chain into the mouth of the PAS pocket in a manner similar to that seen in the phyAG773E mutant. It was hypothesized that the phyAG773E mutant phyA protein may, therefore, mimic phyB to some extent and bind non-functionally to normal phyB interactors, perhaps titrating them out, and reducing the effectiveness of phyB signalling as a result. Speculating further, similarity between the mutant phyAG773E protein and phyB could be even greater if the Thr808 in phyB were to be phosphorylated. Glutamate is known to be a functional mimic of phosphorylated threonine. However, no evidence currently exists that Thr808 is phosphorylated in phyB.
The phyB PAS2 domain has not been notably implicated in direct signalling, but more in nuclear localization and dimerization, both essential for phyB signal transduction. Using phyB–GFP fusions, Matsushita et al. (2003) showed that the N-terminus of phyB alone is sufficient for function once in the nucleus. A phyB–GFP fusion was engineered to dimerize and locate to the nucleus and actually induced higher photosensitivity than full-length phyB. In addition, this fusion is able to rescue a phyB mutant phenotype. Furthermore, Chen et al. (2005) demonstrated that the phyB PRD interacts directly with the N-terminal Bilin Lyase and PHY domains to regulate nuclear accumulation. The mutant phyAG773E protein may, therefore, bind a component normally responsible for phyB nuclear translocation following conversion to the Pfr form. Alternatively, rather than binding a phyB interactor, the mutant phyAG773E protein may form non-functional dimers with the endogenous phyB. PhyB is believed to form primarily homodimers (Wagner and Quail, 1995), but additional phyB heterodimers have recently been detected. Myc6-tagged phyB has been used to co-immunoprecipitate phyC, phyD, and phyE, while myc6-phyD co-precipitates phyB and phyE (Sharrock and Clack, 2004). However, phyA–phyB heterodimers were not detected by this method, suggesting that they are not a normal functional aspect of phytochrome signalling.
The fact that a deficiency in phyB signalling is not observed in established seedlings of the phyAG773E mutant is also consistent with the defect being associated with the effect of the mutant phyA molecule. PhyA protein is degraded in R or white light, and any effect of the mutant phyA would, therefore, be quickly lost, allowing normal phyB signalling. PhyA protein has been shown to be undetectable after 7 d of growth in white light. However, it is still detectable after 24 h of R irradiation (Sharrock and Clack, 2002), meaning that, if the long hypocotyl in R is a result of loss of phyB signalling, this loss would only be manifest to a large extent within the first day or so of R irradiation. Nonetheless, even such a brief loss of phyB signalling would be capable of causing a difference between mutant and wild-type seedlings (Parks and Spalding, 1999). That no phenotype is observed after a similar duration of growth in white light presumably reflects the ability of cryptochrome blue light photoreceptors to substitute for the reduced phyB action during de-etiolation in white light. In support of this, it has been demonstrated previously that phyB mutants have a much more severe phenotype in R than when R is mixed with B (Casal and Mazzella, 1998).
Conclusion
In conclusion, a novel, high-throughput, molecular screen was used to identify SAS signalling mutants successfully. A phyA mutant causing a dominant-negative inhibition of phyB signalling was identified. The phyAG773E mutation results in the production of a photoactive phyA protein, severely disrupted in several aspects of phyA signalling. The mutation also causes a significant reduction in phyB signalling in etiolated seedlings. It is hypothesized that the mutant phyAG773E protein interferes with normal phyB signalling either by non-functional interaction with a phyB signalling intermediate, thus titrating it out, or by forming non-functional dimers with phyB itself.
Although speculative, it is hoped that this hypothesis may provide a clue for further investigation into the poorly understood distinctions that define differences in the roles and functionality of the various phytochrome species.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. The reduced PHYB::LUC expression response to shade phenotype and the long hypocotyl in red light phenotype co-segregate with the long hypocotyl in far red light phenotype of dra1.
Figure S2. Adult phenotype of dra1. Five-week-old plants of the wild type and dra1 grown in light/dark cycles (16 h/8 h).
Figure S3. The reduced PHYB::LUC expression response to shade of dra1 is a recessive phenotype.
Figure S4. The mutation in the phyA PAS2 domain in dra1 does not alter the predicted secondary structure. The secondary structure for the phyA PAS2 domain (amino acids 764–876) from the wild type and the dra1 mutant was predicted using Jnet software (http://www.compbio.dundee.ac.uk/). E and H indicate stretches of β-sheet and α-helix, respectively. The mutated residue is highlighted.
Table S1. Primers for amplifying and sequencing PHYA in Ws-2 and dra1, and primers for qPCR.
Table S2. Blast–Clustal W alignments of Arabidopsis phytochrome protein sequences corresponding to amino acids 765–785 of Arabidopsis phyA.
Acknowledgments
Seeds of Arabidopsis wild type, phyA-410, and phyB-464-19 containing PHYB::LUC (line ‘Ws-21a’) in the Ws-2 background were kindly provided by Professor Andrew Millar, University of Edinburgh. We would like to thank Dr Laszlo Kozma-Bognar (BRC, Hungary) for sequencing work in order to confirm the position of the PHYB::LUC transgene. AA01 anti-phyA monoclonal antibodies and AB01 anti-phyB monoclonal antibodies were a kind gift of Professor Akira Nagatani (Kyoto University). This work was supported by The Royal Society and the Spanish CSIC (2004GB0016 to PFD and JFM-G); the University of London Central Research Fund (AR/CRF/B to PFD); and the Generalitat de Catalunya (Xarba, 2009-SGR697) and the Ministerio de Ciencia e Innovación (MICINN)-FEDER (BIO2005-00154, CSD2007-00036, and BIO2008-00169) to JFM-G. IR-V was the recipient of a pre-doctoral fellowship from the MICINN. XW was the recipient of a KC Wong studentship.
Glossary
Abbreviations
- SAS
shade avoidance syndrome
- R
red light
- FR
far red light
- B
blue light
- PAS
Per/Arnt/Sim
References
- Ahmad M, Cashmore AR. The pef mutants of Arabidopsis thaliana define lesions early in the phytochrome signaling pathway. The Plant Journal. 1996;10:1103–1110. doi: 10.1046/j.1365-313x.1996.10061103.x. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA. A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. The Plant Cell. 1997;9:1317–1326. doi: 10.1105/tpc.9.8.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Annual Review of Plant Biology. 2008;59:281–311. doi: 10.1146/annurev.arplant.59.032607.092859. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Boylan M, Douglas N, Quail PH. Dominant negative suppression of Arabidopsis photoresponses by mutant phytochrome A sequences identifies spatially discrete regulatory domains in the photoreceptor. The Plant Cell. 1994;6:449–460. doi: 10.1105/tpc.6.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Mazzella MA. Conditional synergism between cryptochrome 1 and phytochrome B is shown by the analysis of phyA, phyB, and hy4 simple, double, and triple mutants in Arabidopsis. Plant Physiology. 1998;118:19–25. doi: 10.1104/pp.118.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends in Plant Sciences. 2007;12:514–521. doi: 10.1016/j.tplants.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Chen M, Tao Y, Lim J, Shaw A, Chory J. Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Current Biology. 2005;15:637–642. doi: 10.1016/j.cub.2005.02.028. [DOI] [PubMed] [Google Scholar]
- Chory J, Peto CA, Ashbaugh M, Saganich R, Pratt L, Ausubel F. Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. The Plant Cell. 1989;1:867–880. doi: 10.1105/tpc.1.9.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Molecular Biology. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC. Phytochrome E influences internode elongation and flowering time in Arabidopsis. The Plant Cell. 1998;10:1479–1487. doi: 10.1105/tpc.10.9.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Robson PR, Patel SR, Goosey L, Sharrock RA, Whitelam GC. Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiology. 1999;119:909–915. doi: 10.1104/pp.119.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Rood SB, Somers DE, Quail PH, Whitelam GC. Photophysiology of the elongated internode (ein) mutant of Brassica rapa. ein mutant lacks a detectable phytochrome B-like polypeptide. Plant Physiology. 1992;100:1442–1447. doi: 10.1104/pp.100.3.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA. A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiology. 2003;133:1617–1629. doi: 10.1104/pp.103.034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Turner JG. The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. The Plant Cell. 2001;13:1025–1033. doi: 10.1105/tpc.13.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faigon-Soverna A, Harmon FG, Storani L, Karayekov E, Staneloni RJ, Gassmann W, Mas P, Casal JJ, Kay SA, Yanovsky MJ. A constitutive shade-avoidance mutant implicates TIR-NBS-LRR proteins in Arabidopsis photomorphogenic development. The Plant Cell. 2006;18:2919–2928. doi: 10.1105/tpc.105.038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chen M. Transposing phytochrome into the nucleus. Trends in Plant Sciences. 2008;13:596–601. doi: 10.1016/j.tplants.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Franklin KA. Shade avoidance. New Phytologist. 2008;179:930–944. doi: 10.1111/j.1469-8137.2008.02507.x. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Allen T, Whitelam GC. Phytochrome A is an irradiance-dependent red light sensor. The Plant Journal. 2007;50:108–117. doi: 10.1111/j.1365-313X.2007.03036.x. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Davis SJ, Stoddart WM, Vierstra RD, Whitelam GC. Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. The Plant Cell. 2003;15:1981–1989. doi: 10.1105/tpc.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. Journal of Experimental Botany. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry RC, Habashi J, Okamoto H, Deng XW. Characterization of a strong dominant phytochrome A mutation unique to phytochrome A signal propagation. Plant Physiology. 2002;130:457–465. doi: 10.1104/pp.005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Kozma-Bognar L, Bastow RM, Nagy F, Millar AJ. Distinct regulation of CAB and PHYB gene expression by similar circadian clocks. The Plant Journal. 2002;32:529–537. doi: 10.1046/j.1365-313x.2002.01441.x. [DOI] [PubMed] [Google Scholar]
- Halliday KJ, Koornneef M, Whitelam GC. Phytochrome B and at least one other phytochrome mediate the accelerated flowering response of Arabidopsis thaliana L. to low red/far-red ratio. Plant Physiology. 1994;104:1311–1315. doi: 10.1104/pp.104.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO Journal. 2009;28:3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Al Sady B, Hudson M, Kim C, Apel K, Quail PH. Phytochrome interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science. 2004;305:1937–1941. doi: 10.1126/science.1099728. [DOI] [PubMed] [Google Scholar]
- Jackson JA, Fuglevand G, Brown BA, Shaw MJ, Jenkins GI. Isolation of Arabidopsis mutants altered in the light-regulation of chalcone synthase gene expression using a transgenic screening approach. The Plant Journal. 1995;8:369–380. doi: 10.1046/j.1365-313x.1995.08030369.x. [DOI] [PubMed] [Google Scholar]
- Johnson E, Bradley M, Harberd NP, Whitelam GC. Photoresponses of lightgrown phyA mutants of arabidopsis. Phytochrome A is required for the perception of daylength extensions. Plant Physiology. 1994;105:141–149. doi: 10.1104/pp.105.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key J, Hefti M, Purcell EB, Moffat K. Structure of the redox sensor domain of Azotobacter vinelandii NifL at atomic resolution: signaling, dimerization, and mechanism. Biochemistry. 2007;46:3614–3623. doi: 10.1021/bi0620407. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Rolf E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Zeitschrift für Planzenphysiologie. 1980;100:147–160. [Google Scholar]
- Kozma-Bognar L, Hall A, Adam E, Thain SC, Nagy F, Millar AJ. The circadian clock controls the expression pattern of the circadian input photoreceptor, phytochrome B. Proceedings of the National Academy of Sciences, USA. 1999;96:14652–14657. doi: 10.1073/pnas.96.25.14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lopez-Juez E, Bowyer JR, Sakai T. Distinct leaf developmental and gene expression responses to light quantity depend on blue-photoreceptor or plastid-derived signals, and can occur in the absence of phototropins. Planta. 2007;227:113–123. doi: 10.1007/s00425-007-0599-7. [DOI] [PubMed] [Google Scholar]
- Lopez-Juez E, Nagatani A, Tomizawa K, Deak M, Kern R, Kendrick RE, Furuya M. The cucumber long hypocotyl mutant lacks a light-stable PHYB-like phytochrome. The Plant Cell. 1992;4:241–251. [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. The Plant Journal. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annual Review of Biophysics and Biomolecular Structure. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Galstyan A, Salla-Martret M, Cifuentes-Esquivel N, Gallemi M, Bou-Torrent J. Regulatory components of shade avoidance syndrome. In: Kader J-C, Delseny M, editors. Advances in botanical research. Vol. 53. London: Elsevier; 2010. pp. 65–116. [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Mateos JL, Luppi JP, Ogorodnikova OB, Sineshchekov VA, Yanovsky MJ, Braslavsky SE, Gartner W, Casal JJ. Functional and biochemical analysis of the N-terminal domain of phytochrome A. Journal of Biological Chemistry. 2006;281:34421–34429. doi: 10.1074/jbc.M603538200. [DOI] [PubMed] [Google Scholar]
- Matsushita T, Mochizuki N, Nagatani A. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature. 2003;424:571–574. doi: 10.1038/nature01837. [DOI] [PubMed] [Google Scholar]
- Mazzella MA, Magliano TMA, Casal JJ. Dual effect of phytochrome A on hypocotyl growth under continuous red light. Plant, Cell and Environment. 1997;20:261–267. [Google Scholar]
- McGuffin LJ, Jones DT. Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics. 2003;19:874–881. doi: 10.1093/bioinformatics/btg097. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Carré IA, Strayer CA, Chua N-H, Kay SA. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science. 1995;267:1161–1163. doi: 10.1126/science.7855595. [DOI] [PubMed] [Google Scholar]
- Monte E, Alonso JM, Ecker JR, Zhang Y, Li X, Young J, Austin-Phillips S, Quail PH. Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. The Plant Cell. 2003;15:1962–1980. doi: 10.1105/tpc.012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman J, Quail P. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix–loop–helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biology. 2005;3 doi: 10.1371/journal.pbio.0030196. e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- Oka Y, Matsushita T, Mochizuki N, Quail PH, Nagatani A. Mutant screen distinguishes between residues necessary for light-signal perception and signal transfer by phytochrome B. PLoS Genetics. 2008;4 doi: 10.1371/journal.pgen.1000158. e1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CM, Bhoo SH, Song PS. Inter-domain crosstalk in the phytochrome molecules. Seminars in Cell and Developmental Biology. 2000;11:449–456. doi: 10.1006/scdb.2000.0200. [DOI] [PubMed] [Google Scholar]
- Parks BM, Quail PH, Hangarter RP. Phytochrome A regulates red-light induction of phototropic enhancement in Arabidopsis. Plant Physiology. 1996;110:155–162. doi: 10.1104/pp.110.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Spalding EP. Sequential and coordinated action of phytochromes A and B during Arabidopsis stem growth revealed by kinetic analysis. Proceedings of the National Academy of Sciences, USA. 1999;96:14142–14146. doi: 10.1073/pnas.96.24.14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellequer JL, Wager-Smith KA, Kay SA, Getzoff ED. Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proceedings of the National Academy of Sciences, USA. 1998;95:5884–5890. doi: 10.1073/pnas.95.11.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D. Phytochromes: photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signalling mechanisms. Annual Review of Plant Biology. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou J, Sorin C, Devlin PF, Martinez-Garcia JF. Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiology. 2006;141:85–96. doi: 10.1104/pp.105.076331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portoles S, Rodriguez-Concepcion M, Martinez-Garcia JF. Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO Journal. 2007;26:4756–4767. doi: 10.1038/sj.emboj.7601890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC. Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature. 2003;426:680–683. doi: 10.1038/nature02174. [DOI] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes and Development. 2005;19:2811–2815. doi: 10.1101/gad.364005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Clack T. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiology. 2002;130:442–456. doi: 10.1104/pp.005389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Clack T. Heterodimerization of type II phytochromes in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2004;101:11500–11505. doi: 10.1073/pnas.0404286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Sharrock RA, Tepperman JM, Quail PH. The hy3 long hypocotyl muntant of Arabidopsis is deficient in phytochrome B. The Plant Cell. 1991;3:1263–1274. doi: 10.1105/tpc.3.12.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C, Salla-Martret M, Bou-Torrent J, Roig-Villanova I, Martinez-Garcia JF. ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. The Plant Journal. 2009;59:266–277. doi: 10.1111/j.1365-313X.2009.03866.x. [DOI] [PubMed] [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I. Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development. 1999;126:4235–4245. doi: 10.1242/dev.126.19.4235. [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Quail PH. Mutational analysis of phytochrome B identifies a small COOH-terminal-domain region critical for regulatory activity. Proceedings of the National Academy of Sciences, USA. 1995;92:8596–8600. doi: 10.1073/pnas.92.19.8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Batge SL, Smith JJ, Kerckhoffs LH, Sineshchekov VA, Murfet IC, Reid JB. A dominant mutation in the pea PHYA gene confers enhanced responses to light and impairs the light-dependent degradation of phytochrome A. Plant Physiology. 2004;135:2186–2195. doi: 10.1104/pp.103.036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. Phytochrome A null mutants of arabidopsis display a wild-type phenotype in white light. The Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Parks BM, Short TW, Quail PH. Missense mutations define a restricted segment in the C-terminal domain of phytochrome A critical to its regulatory activity. The Plant Cell. 1995;7:1433–1443. doi: 10.1105/tpc.7.9.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Casal JJ, Whitelam GC. Phytochrome A, phytochrome B and HY4 are involved in hypocotyl growth responses to natural radiation in Arabidopsis: weak de-etiolation of the phyA mutant under dense canopies. Plant, Cell and Environment. 1995;18:788–794. [Google Scholar]
- Yanovsky MJ, Luppi JP, Kirchbauer D, et al. Missense mutation in the PAS2 domain of phytochrome A impairs subnuclear localization and a subset of responses. The Plant Cell. 2002;14:1591–1603. doi: 10.1105/tpc.000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.