Abstract
Background
African-Americans (AAs) with diabetes have high incidence rates of end-stage renal disease (ESRD) with associated high mortality. Genetic factors modulating the risk of mortality on dialysis are poorly understood. Meth ods: A genome-wide association study was performed in 610 AAs with type 2 diabetes (T2D) and ESRD on dialysis, using the Affymetrix 6.0 platform (868,155 SNPs). Time to death was assessed using Cox proportional hazards model adjusting for ancestry and other confounding variables. Cases were censored at kidney transplant or (if living) at study conclusion.
Results
Mean follow-up was 5.4 ± 3.5 years; 434 deaths were recorded. Five SNPs were associated with time to death at p < 1.00 × 10−6: rs2681019 (HR = 2.58, PREC = 8.00 × 10−8), rs815815 in CALM2 (HR = 1.51, PADD = 6.50 × 10−7), rs926392 (HR = 2.37, PREC = 4.80 × 10−7), and rs926391 (HR = 2.30, PREC = 7.30 × 10−7) near DHX35, and rs11128347 in PDZRN3 (HR = 0.57, PADD = 6.00 × 10−7). Other SNPs had nominal associations with time to death (p < 1.00 × 10−5).
Conclusion
Genetic variation may modify the risk of death on dialysis. SNPs in proximity to genes regulating vascular extracellular matrix, cardiac ventricular repolarization, and smoking cessation are associated with dialysis survival in AAs with T2D. These results warrant replication in other cohorts and races.
Key Words: African-Americans, Diabetes mellitus, Dialysis, Genome-wide association study, Survival
Introduction
Despite advances in technique and medications, alarmingly high mortality rates continue to be observed in the dialysis population, particularly in patients with diabetes (http://www.usrds.org; last accessed September, 2010). Factors associated with accelerated mortality in the dialysis population include traditional cardiovascular disease risk factors as well as non-traditional inflammatory markers [1]. Cardiovascular disease contributes to the largest proportion of deaths, related in part to pre-dialysis co-morbidity from diabetes, hypertension, and metabolic syndrome [2]. Derangements in mineral metabolism, anemia and hemoglobin variability, malnutrition, BMI and fat composition, residual renal function, dialysis access type, biocompatibility of dialysis membranes, intensity of pre-dialysis care, and frailty are independent determinants of life expectancy on dialysis [1].
Striking racial differences exist in the incidence and prevalence rates of end-stage renal disease (ESRD) and in mortality rates on dialysis (http://www.usrds.org; last accessed September, 2010). Compared with Caucasians, African-Americans (AAs) have a markedly increased risk of ESRD, yet manifest a paradoxical lower risk of death on dialysis [3,4]. The survival advantage in AAs with kidney failure stands in stark contrast to the higher prevalence of negative prognosticators of survival and the higher mortality rate observed in the general (non-renal disease) AA population [3,5].
Genetic factors have been demonstrated to underlie part of the risk for ESRD in AAs [6], and may also play a role in survival after initiation of dialysis. Several studies have explored genetic determinants for racial differences in atherosclerosis and heart failure mortality [7,8,9]. However, no such analysis has been performed in dialysis patients. In this study, an unbiased genome-wide association scan was performed to detect genetic variants associated with survival on dialysis in AAs with type 2 diabetes (T2D). Using genome-wide data from a local AA cohort with T2D [10], coupled with the dialysis survival span, we sought to detect genetic variants associated with survival on dialysis that may provide novel insights into the molecular mechanisms that underlie death in patients on dialysis and set the stage for future functional studies.
Research Design and Methods
Study Population and Measurements
Self-reported AAs initiating renal replacement therapy between February 1985 and September 2009 in eight ESRD Network 6 facilities comprised the study cohort. Participants were enrolled in a study of genetic factors in AAs with type 2 diabetic ESRD, and had ESRD clinically attributed to diabetic nephropathy on the Center for Medicare and Medicare Services 2728 form [11]. T2D was diagnosed with age at diabetes onset >30 years and/or use of oral hypoglycemic agents (not insulin alone) 1 year after onset. T2D-associated ESRD was diagnosed in the presence of either: (1) T2D preceding ESRD by >5 years, in the absence of other causes of nephropathy, (2) diabetic retinopathy, or (3) dipstick proteinuria ≥100 mg/dl (or 500 mg/day). Exclusion criteria included non-AA race and evidence of recovery of renal function or renal transplant within 6 months of initiating dialysis. Baseline measures of serum albumin and serum hemoglobin were obtained at the start of dialysis and BMI was computed. Proportions, mean values, and standard deviations were calculated for baseline variables.
End Points and Data Collection
The primary end point was death from any cause. Survival time on dialysis was computed as the time between the start of dialysis and the date of death. Cases lost to follow-up or undergoing kidney transplantation were censored at their final follow-up date or transplant date, respectively. Follow-up in surviving participants ended on January 1, 2010. Data involving deaths were collected by means of primary cause of death documentation in the Death Notification Form (form 2746) required for every death occurring in the US ESRD population. Death events were categorized into 5 broad groups. Deaths coded as any of the following were defined as ‘cardiovascular’: atherosclerotic heart disease, cardiac arrest, cardiac arrhythmia, cardiomyopathy, congestive heart failure, acute myocardial infarction, valvular heart disease, ischemic stroke, or ischemic brain damage/anoxic encephalopathy due to cardiac event. Deaths coded as any of the following causes were classified as ‘infectious’: abdominal infection, perforated bowel, diverticulitis, gallbladder infection, endocarditis, pulmonary infection, or septicemia. Deaths ascribed to any of the following causes were grouped as ‘other’: accident related or unrelated to treatment, acidosis, cachexia, intracranial hemorrhage, chronic obstructive lung disease, cirrhosis, dementia (including dialysis dementia), Alzheimer's, gastrointestinal hemorrhage, hemorrhage from vascular access, hyperkalemia, malignant disease, viral hepatitis, pancreatitis, pulmonary embolus, or seizures. Withdrawal from dialysis and unknown cause of death constituted the 2 remaining groups.
Study approvals were obtained from the Wake Forest University Baptist Medical Center Institutional Review Board, and all subjects provided written informed consent.
Genotyping
DNA extraction from whole blood was performed using the PureGene system (Gentra Systems, Minneapolis, Minn., USA). Genotyping was performed at the Center for Inherited Disease Research using 1 μg of genomic DNA (diluted in 1× TE buffer and at 50 ng/μl) on Affymetrix Genome-Wide Human SNP array 6.0. DNA from cases was interleaved on 96-well master plates. To confirm sample identity, a SNP barcode (96 SNPs) was generated prior to genotyping on the Affymetrix array and confirmed on downstream released genotyping data. Genotypes were called using Birdseed version 2; APT 1.10.0 by grouping samples by DNA plate to determine the genotype cluster boundaries. All autosomal SNPs (n = 868,157) were included in the analysis, but classified on data quality with primary inference drawn from polymorphic SNPs (minor allele frequency > 0.05) with <5% missing data (Hardy-Weinberg Equilibrium p values >0.0001) resulting in a total of 832,357 SNPs for analysis. The average sample call rate was 99.16% for all autosomal SNPs. Forty-six blind duplicates were included in genotyping and had a concordance rate of 99.59%. In addition, one individual was removed whose self-reported gender was inconsistent with X chromosome genotype data. Relatedness was estimated using the identity-by-descent analysis implemented in the PLINK analysis software package (http://pngu.mgh.harvard.edu/purcell/plink/). One duplicate pair and 110 first-degree relative pairs were identified, and a single individual from each pair was retained for analysis based on the completeness of the phenotypic data.
Statistical Analysis
To account for admixture, ancestral allele frequencies were estimated from the results of the 70 ancestry informative markers genotyped in 44 Yoruba Nigerians and 39 European Americans. Individual ancestral proportions were generated for each subject using FRAPPE, an expectation maximization algorithm, under a two-population model [12].
To test for association between each SNP and survival, adjusting for covariates, a Cox proportional hazard semi-parametric regression model was computed. The proportional hazards assumption for each covariate was examined before allowing that covariate into the model. Time to all-cause death was tested for each SNP using dominant, recessive, and additive models. Primary inference was based on the additive genetic model unless significant departure from additivity was observed (p < 0.05). All genetic models were defined relative to the minor allele where the dominant model tests whether the presence of the minor allele influences survival, the additive model tests for a dose effect on the number of minor alleles (0, 1, or 2) and the recessive model tests whether 2 copies of the minor allele were associated with survival. The recessive and additive genetic models required at least 30 and 10 individuals homozygous for the minor allele, respectively. All analyses adjusted for population structure using the ancestral proportions noted above. Quantile-quantile (Q-Q) plots and test inflation factors were computed to assure that the experiment had appropriate family-wise type 1 error rates. Participants were censored when lost to follow-up due to transfer to another ESRD Network, receipt of a kidney transplant, or (if living) on January 1, 2010. Due to recent improvements in dialysis prescription and medications, factors with potential impact on dialysis survival introduced in approximately the year 2000, we adjusted the analysis using a binary variable denoting pre- and post-2000 initiation of dialysis. Model 1 adjusted for ancestry, age at start of dialysis, sex, BMI, pre-dialysis diabetes duration, and incident year of dialysis; model 2 adjusted for these factors plus serum albumin and hemoglobin at the initiation of dialysis. HR with 95% CIs are presented. We estimated study power using the Genetic Power Calculator; assuming dominant models with minor allele frequencies of 0.07, our sample size had 95% power to detect a minimal HR of 1.6 for death while on dialysis at α = 0.05 [13].
Results
A total of 647 participants were evaluated for inclusion. Of these, 5 recovered renal function, 4 had temporary discontinuation of dialysis, 2 were transplanted within 6 months of dialysis onset, and 26 did not have the date of first dialysis recorded; these patients were withdrawn. The remaining 610 subjects were included in the final analysis. Table 1 summarizes the baseline characteristics of participants. Hemodialysis was the predominant initial mode of renal replacement therapy (93% of patients), and we observed an overall 10% rate of modality conversion from any initial to an alternative renal replacement modality during the period of follow-up. After mean follow-up of 5.4 ± 3.5 years, 434 deaths, 44 kidney transplants, and 2 cases lost to follow-up were recorded (table 2). The mean survival on dialysis for the 434 subjects who died was 5.2 ± 3.1 years. A total of 130 subjects remained alive at the last follow-up. Table 2 summarizes the major causes of death. Sex, BMI, incident albumin, incident hemoglobin, and admixture were not individually significantly associated with survival, while age at dialysis inception (5-year HR = 1.2; p < 0.0001) and incident year of dialysis (dialysis vintage, 5-year HR = 1.16; p= 0.0008) were significantly associated with mortality (data not shown).
Table 1.
Demographics at dialysis inception
| n | Value | |
|---|---|---|
| Age, years | 610 | 59±11 |
| Females | 610 | 279 (45) |
| BMI | 475 | 29.6±6.8 |
| Pre-dialysis diabetes duration, years | 524 | 20±10 |
| Duration of follow-up, years | 610 | 5.4±3.5 |
| Undergoing hemodialysis | 567 (93) | |
| Hemoglobin, g/dl | 481 | 9.8±5.4 |
| Serum albumin, g/dl | 451 | 3.1±0.6 |
| ESRD Network | 610 | |
| 3 | 3 (0.5) | |
| 4 | 3 (0.5) | |
| 5 | 2 (0.3) | |
| 6 | 597 (98) | |
| 7 | 1 (0.1) | |
| 8 | 2 (0.3) | |
| 10 | 1 (0.1) | |
| 14 | 1 (0.1) |
Data presented as n (%) or means ± SD. Conversion factor for hemoglobin and serum album (g/dl to g/l) was ×10.
Table 2.
Events at follow-up
| n (%) | |
|---|---|
| Deaths | 434 (71) |
| Cardiovascular death | 236 (54) |
| Infection | 80 (18) |
| Withdrawal from dialysis | 26 (6) |
| Other | 39 (9) |
| Unknown | 53 (12) |
| Transplants | 44 (7) |
| Switched dialysis modality | 63 (10) |
| Censored1 | 176 (29) |
Patients transplanted (n = 44), lost to follow-up (n = 2), or alive (n = 130).
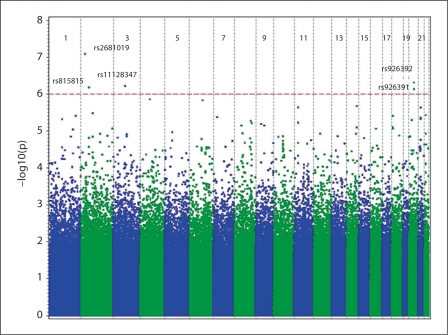
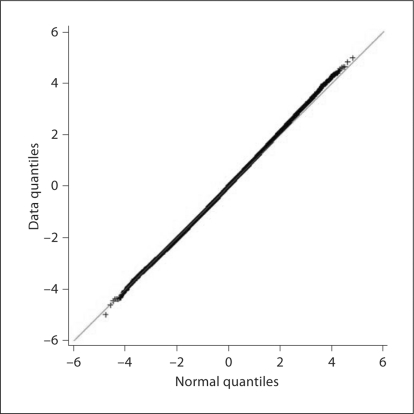
A number of genomic regions showed evidence of association with survival at p < 1.00 × 10−5, with 5 of the landmark SNPs associated at p < 1.00 × 10−6 (fig. 1). Overall, there was no evidence of inflation of the tests of association, and there was appropriate fit to the expected overall distribution of association (fig. 2).
Fig. 1.
GWAS results for death on dialysis. The genome-wide distribution of –log10 p values from the adjusted trend is shown across the chromosomes. The dotted line indicates the genome-wide significance threshold (p < 1 × 10−6). Annotated are the top five SNPs with genome-wide significance for dialysis survival at p < 1 × 10−6.
Fig. 2.
Q-Q plot from the genome-wide association with dialysis survival analysis. The black bold line represents the observed p values; the red line is the expected line under the null distribution.
After adjustment for ancestry, age at dialysis, sex, BMI, diabetes duration, and incident year factor (model 1), 30 SNPs provided evidence for association (p < 1.00 × 10−5), 12 were located in 10 genes (table 3), and 18 SNPs were located in intergenic regions (table 4). One of the top hits (rs2412980, chr22q12.2, HR = 1.65, p = 3.67 × 10−6) was located in LOC729980, a gene encoding a protein with unknown function. Two of the top hits (rs6546886, chr2p13.1, HR = 2.13, p = 3.28 × 10−6; rs9921518, chr16q12.2, HR = 2.11, p = 8.62 × 10−6) were located in regions of reported copy number variation. Of note, several SNPs clustered around A Disintegrin And Metalloproteinase with Thrombospondin Motifs (ADAMTS) and Iroquois (IRX) genes (rs6816344, chr4q13.3, HR = 1.7, p = 1.37 × 10−6; rs1452093, chr21q21.3, HR = 1.59, p = 2.31 × 10−6; rs9977499, chr21q21.3, HR = 1.57, p = 4.36 × 10−6; rs1817114, chr21q21.3, HR = 1.57, p = 4.36 × 10−6; rs2830881, chr6q22.31, HR = 1.57, p = 4.67 × 10−6; and rs9921518, chr16q12.2, HR = 2.11, p = 8.62 × 10−6).
Table 3.
Summary results for top gene SNPs associated with all-cause death on dialysis
| Rank | SNP | Chromosome | Position | Gene | Name | Function | MAF | Best model | HR | CI | p value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | rsl 1128347 | 3p13 | 73702252 | PDZRN3 | PDZ domain containing ring finger 3 | intronic with no known function | 11 | additive | 0.54 | 0.43-0.69 | 6.00 × 107 |
| 4 | rs815815 | 2p21 | 47252569 | CALM2 | calmodulin 2 (phos-phorylase kinase, delta) | intronic enhancer | 19 | additive | 1.51 | 1.28-1.77 | 6.50 × 107 |
| 9 | rs7111546 | 11p14.3 | 22786334 | GAS2 | growth arrest-specific 2 | intronic enhancer | 44 | recessive | 1.72 | 1.37-2.16 | 2.30 × 106 |
| 12 | rs2412980 | 22q12.2 | 28922070 | LOC729980 | hypothetical LOC729980 | N/A | 9 | additive | 1.65 | 1.33-2.04 | 3.67 × 106 |
| 14 | rs8098064 | 18p11.23 | 8199270 | PTPRM | protein tyrosine phosphatase, receptor type, M | intronic with no known function | 26 | additive | 1.42 | 1.22-1.65 | 3.87 × 106 |
| 19 | rsl7110736 | 1p22.1 | 94235358 | ABCA4 | ATP-binding cassette, subfamily A (ABC1), member 4 | intronic enhancer | 35 | recessive | 2.08 | 1.52-2.86 | 4.80 × 106 |
| 20 | rs4814615 | 20p12.1 | 17305574 | PCSK2 | proprotein convertase subtilisin/kexin type 2 | intronic enhancer | 19 | dominant | 1.59 | 1.30-1.95 | 5.02 × 106 |
| 21 | rs712022 | 11pl4.3 | 22799732 | SVIP | small VCP/p97-interacting protein | downstream with no known function | 47 | dominant | 0.62 | 0.51-0.76 | 5.96 × 106 |
| 23 | rs2439312 | 8pl2 | 32531902 | NRG1 | neuregulin 1 | intronic enhancer | 24 | recessive | 2.36 | 1.62-3.45 | 7.08 × 106 |
| 25 | rs7243299 | 18p11.23 | 7745772 | PTPRM | protein tyrosine phosphatase, receptor type, M | intronic enhancer | 10 | additive | 1.63 | 1.31-2.03 | 8.01 × 106 |
| 27 | rs9953514 | 18p11.23 | 7821266 | PTPRM | protein tyrosine phosphatase, receptor type, M | intronic with no known function | 9 | dominant | 1.74 | 1.36-2.22 | 8.56 × 106 |
| 28 | rsl 1676855 | 2q37.2 | 235564911 | SH3BP4 | SH3-domain binding protein 4 | promoter/regulatory region | 33 | additive | 0.7 | 0.60-0.82 | 8.58 × 106 |
Results ordered by p values and adjusted for ancestry, age at start of dialysis, gender, BMI, pre-dialysis diabetes duration, and incident year of dialysis. Chromosome positions are based on NCBI build 36_3. HR, CI, and p values are shown for the best-fit model. MAF = Minor allele frequency.
Table 4.
Summary results for top intergenic SNPs associated with all-cause death on dialysis
| Rank | SNP | Chromosome | Position | Upstream gene and distance, kb | Downstream gene and distance, kb | MAF | Best model | HR | CI | p value |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs2681019 | 2p24.1 | 23041010 | KLHL29 (420) | none within 500 kb | 28 | rec. | 2.58 | 1.82-3.66 | 8.00 × 108 |
| 2 | rs926392 | 20q12 | 37123879 | LOC339568 (151) | DHX35 (22) | 29 | rec. | 2.37 | 1.69-3.33 | 4.80 × 107 |
| 5 | rs926391 | 20q12 | 37123900 | LOC339568 (151) | DHX35 (22) | 30 | rec. | 2.3 | 1.65-3.20 | 7.30 × 107 |
| 6 | rs6816344 | 4q13.3 | 74002269 | COX18 (137) | ADAMTS3 (348) | 49 | rec. | 1.7 | 1.37-2.12 | 1.37 × 106 |
| 7 | rs594442 | 6q16.1 | 93904606 | EPHA7 (101) | none within 500 kb | 5 | add. | 1.99 | 1.50-2.64 | 1.47 × 106 |
| 8 | rsl009170 | 14q32.12 | 91706467 | SLC24A4 (152) | CPSF2 (6) | 43 | rec. | 0.5 | 0.38-0.67 | 2.10 × 106 |
| 10 | rsl452093 | 21q21.3 | 27666228 | NCRNA00113 (350) | ADAMTS5 (404) | 23 | dom. | 1.59 | 1.31-1.94 | 2.31 × 106 |
| 11 | rs6546886 | 2p13.1 | 74099286 | TET3 (27) | DGUOK (59) | 29 | rec. | 2.13 | 1.55-2.93 | 3.28 × 106 |
| 13 | rsl497828 | 1q41 | 215593648 | GPATCH2 (76) | ESRRG (215) | 37 | add. | 0.71 | 0.61-0.82 | 3.85 × 106 |
| 15 | rs17364464 | 7pl5.3 | 22480579 | IL<5 (252) | RAPGEF5 (117) | 8 | add. | 1.76 | 1.38-2.25 | 4.19 × 106 |
| 16 | rs9977499 | 21q21.3 | 27656869 | NCRNA00113 (359) | ADAMTS5 (3995) | 24 | dom. | 1.57 | 1.29-1.91 | 4.36 × 106 |
| 17 | rsl817114 | 21q21.3 | 27661425 | NCRNA00113 (355) | ADAMTS5 (400) | 24 | dom. | 1.57 | 1.29-1.91 | 4.36 × 106 |
| 18 | rs2830881 | 21q21.3 | 27657679 | NCRNA00113 (358) | ADAMTS5 (396) | 24 | dom. | 1.57 | 1.29-1.91 | 4.67 × 106 |
| 22 | rsl7232789 | 8q24.13 | 122691682 | HAS2 (2) | none within 500 kb | 37 | dom. | 0.63 | 0.52-0.77 | 6.09 × 106 |
| 24 | rs6560517 | 9q21.13 | 78227991 | GCNT1 (18) | RPSAP9 (23) | 29 | dom. | 0.64 | 0.53-0.78 | 7.09 × 106 |
| 26 | rs4904947 | 14q32.12 | 92041916 | RIN3 (7) | SLC24A4 (9) | 5 | add. | 1.9 | 1.43-2.52 | 8.03 × 106 |
| 29 | rs9921518 | 16q12.12 | 53051926 | TRX5 (470) | IRX3 (174) | 28 | rec. | 2.11 | 1.52-2.93 | 8.62 × 106 |
| 30 | rsl6844716 | 1q32.1 | 197623140 | none within 500 kb | none within 500 kb | 9 | add. | 1.65 | 1.32-2.07 | 9.09 × 106 |
SNPs ordered by p values and ranked in relation to the intragenic SNPs. Results are adjusted for ancestry, age at start of dialysis, gender, BMI, pre-dialysis diabetes duration, and incident year of dialysis. Chromosome positions are based on NCBI build 36_3. MAF = Minor allele frequency. HR, CI, and p values are shown for the best-fit model.
Additional covariate adjustments in model 2 (incident serum albumin and hemoglobin) maintained comparable HRs, but diminished the p values modestly (data not shown).
Discussion
This report represents the first genome-wide search for variation influencing dialysis survival in AAs with T2D. We found several alleles (30 SNPs) that had strong statistical associations (p < 5.00 × 10−6) with survival on dialysis, even after adjustment for important covariates known to impact dialysis survival (model 1). The lack of change in the HR with modest reduction of statistical significance (10-fold drop) in model 2 reflected a reduction in statistical power due to the reduced sample size resulting from a lack of measured covariates in some participants. Of the top 30 SNPs, some indicated a negative effect on survival by association with a higher propensity for death (HR > 1.0), while other SNPs indicated a protective effect by association with a lower rate of death (HR < 1.0). These associations tag potentially interesting genomic regions and the functional qualities of the top SNPs relating to death on dialysis are unknown, but may prove to be important based on their detection using unbiased methodologies. Replication, subsequent fine mapping, and functional studies are necessary to more accurately narrow down regions of interest and establish loci as influencing survival. Genome-wide association study (GWAS) results from the entire cohort have been entered in the Database of Genotypes and Phenotypes (dbGaP) [10].
The top SNPs correlating with dialysis survival were located primarily in or near genes with mechanistic roles that can be categorized in 3 major groups: regulation of extracellular matrix (ECM) composition and turnover, myocardial cell development and repolarization, and neurobiological regulation of smoking cessation. At this stage, these results are hypothesis-generating and require replication in other cohorts and races. If replicated, they merit experimental analysis to define the molecular pathways that impact survival.
Putative Atherogenic SNPs
Accelerated atherosclerosis, a multi-factorial process involving inflammation, altered matrix turnover, and composition in vascular walls is often present in patients with diabetes and kidney disease [14]. Proteases from the ADAMTS family regulate ECM turnover in atherosclerotic plaque [15], and hyaluronic acid (encoded by HAS2 gene) is a prominent constituent of the ECM in atherosclerotic vascular lesions [16]. Our analysis found several SNPs clustered near the ADAMTS5 gene (rs6816344, rs1452093, rs9977499, rs1817114, rs2830881) and a single SNP located near the HAS2 gene (rs17232789) to have significant correlations with survival. These SNPs may impact the rate of atherosclerosis, either pre-dialysis or on dialysis. Further studies are needed to replicate these findings and delineate their pathophysiology.
Putative Arrhythmogenic SNPs
Sudden cardiac death due to arrhythmia is the leading cause of death in patients on dialysis. Several genes are known to control myocardial development and repolarization. The protein tyrosine phosphatase receptor type M (PTPRM) gene and PDZ domain containing the ring finger 3 (PDZRN3) gene play important roles in myocardial development, myocardial cell resistance to ischemia-reperfusion injury, and electrolyte-induced depolarization [17,18]. Calmodulin regulates intracellular calcium homeostasis in myocardial and vascular endothelial cells, and Ca2+/calmodulin kinase complex plays a major role in ventricular repolarization [19]. Polymorphisms in the calmodulin 2 (CALM2) gene (rs815815), PTPRM gene (rs8098064, rs7243299, rs9953514), and PDZRN3 gene (rs11128347) were among the top associations with dialysis survival, suggesting a role for differential myocardial cell response to triggers of cell depolarization. Experimental studies of the morphologic and functional impacts of genomic variation detected in this study could identify arrhythmogenic pathways in myocardial cells that contribute to survival on dialysis.
Smoking Cessation Genes
Association was detected between dialysis survival and polymorphisms in and near genes thought to predict successful smoking cessation: PDZRN3, PTPRM, neuregulin 1 (NRG1), SH3-domain binding protein 4 (SH3BP4), kelch-like 29 (KLHL29), DEAH (Asp-Glu-Ala-His) box polypeptide 35 (DHX35), ATP-binding cassette sub-family A member 4 (ABCA4), ephrins receptor A7 (EPHA7), proprotein convertase subtilisin/kexin type 2 (PCSK2), tet oncogene family member 3 (TET3), G patch domain-containing 2 (GPATCH2), estrogen-related receptor gamma (ESRRG), Rap guanine nucleotide exchange factor 5 (RAPGEF5), and Ras and Rab interactor 3 (RIN3) [20,21]. Unfortunately, our dataset lacks information pertaining to smoking history.
Unique strengths of this GWAS include the novel phenotype (dialysis survival) and use of an unbiased genetic approach in a population enriched for cardiovascular disease risk factors. Previous genetic studies of dialysis survival included mainly Caucasian and non-diabetic individuals, and applied a priori single gene analysis (e.g. Klotho, CCR5, Fetuin A) [22,23,24]. The current GWAS did not reveal polymorphisms in or near these genes, potentially due to race-dependent and environment-conducive genetic variation. These confounding factors were minimized in our analysis by virtue of homogenous ethnicity and cause of ESRD.
GWAS analyses have been applied in large study cohorts, including the Wellcome Trust Case Control Consortium (WTCCC) and Atherosclerosis Risk in Communities (ARIC) studies, to detect genetic variants associated with cardiovascular events. These analyses detected several SNPs associated with incident heart failure and coronary heart disease in African-ancestry populations, but there was no overlap with risk variants in our study [7,9]. This is not surprising, since the mechanisms of cardiovascular death in dialysis patients are likely to differ from those in the general population. Since few studies with ESRD patients have undergone GWAS, it will be necessary to evaluate dialysis survival as a phenotype in other GWAS. Although the genetic mechanisms underlying various etiologies of death on dialysis may differ, we elected not to perform separate analyses by cause of death, since the sample sizes were small and inconsistencies might exist in the attributed cause of death. However, it is clear that the majority of deaths in our cohort were related to cardiovascular events – an expected observation.
Limitations of this study include (1) a relatively small cohort, (2) inability to adjust for being on hemodialysis versus peritoneal dialysis, (3) inability to assess the type of hemodialysis access used, (4) inability to ascertain that T2D was the actual cause of ESRD in clinically diagnosed subjects due to lack of kidney biopsies. An inherent limitation of the study represents the fact that the statistical association only tags regions of interest. Thus, replication and fine mapping studies will be necessary. Frequent changes in dialysis modality and access type are commonly observed, making adjustment for these factors difficult. The possibility of false-positive results relating to our relatively small sample size cannot be excluded; however, we detected suggestive evidence of genome-wide association. An unadjusted p value of <1 × 10−8 is regarded as significant in genome wide association studies. Several SNPs in this discovery cohort had p values <1 × 10−6; it is possible that these may play important roles in survival on dialysis or may be false-positive results. A replication cohort to validate these signals and exclude genomic noise is critical. The relatively large numbers of deaths was not unexpected in this high-risk sample of subjects with diabetes and ESRD. Known biological connections between the associated genes and their pathways suggest a plausible mechanism for effects on survival.
Conclusion
In summary, a GWAS employing 868,155 SNPs identified 30 genetic variants that appeared to influence the risk of death on dialysis in AAs with T2D and ESRD. These results highlight several important pathways and processes involved in cardiac and ECM physiology, and provide insights into the potential genetic architecture of dialysis survival as a polygenic trait. This is a hypothesis-generating GWAS that presents a preliminary set of SNPs associated with survival in AA ESRD patients with T2D. In the future, GWAS in other racial groups and dialysis cohorts, replication of these findings, fine-mapping and functional studies of these loci in myocardial, vascular, and/or neuronal cells will be necessary to validate these results and determine the role played by these polymorphisms in the pathogenesis of dialysis survival.
Disclosure Statement
The authors report no conflicts of interest.
Acknowledgements
We wish to thank the patients and the Southeastern Kidney Council, Inc./ESRD Network 6 for their participation. This work was supported by NIH grants R01 DK066358 (D.W.B.), R01 DK053591 (D.W.B.), R01 HL56266 (B.I.F.), R01 DK070941 (B.I.F.) and in part by the General Clinical Research Center of the Wake Forest University School of Medicine grant M01 RR07122.
References
- 1.Zoccali C, Tripepi G, Mallamaci F. Predictors of cardiovascular death in ESRD. Semin Nephrol. 2005;25:358–362. doi: 10.1016/j.semnephrol.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Goodkin DA, Bragg-Gresham JL, Koenig KG, Wolfe RA, Akiba T, Andreucci VE, Saito A, Rayner HC, Kurokawa K, Port FK, Held PJ, Young EW. Association of comorbid conditions and mortality in hemodialysis patients in Europe, Japan, and the United States: the Dialysis Outcomes and Practice Patterns Study (DOPPS) J Am Soc Nephrol. 2003;14:3270–3277. doi: 10.1097/01.asn.0000100127.54107.57. [DOI] [PubMed] [Google Scholar]
- 3.Jha AK, Shlipak MG, Hosmer W, Frances CD, Browner WS. Racial differences in mortality among men hospitalized in the Veterans Affairs health care system. JAMA. 2001;285:297–303. doi: 10.1001/jama.285.3.297. [DOI] [PubMed] [Google Scholar]
- 4.Newsome BB, McClellan WM, Coffey CS, Allison JJ, Kiefe CI, Warnock DG. Survival advantage of black patients with kidney disease after acute myocardial infarction. Clin J Am Soc Nephrol. 2006;1:993–999. doi: 10.2215/CJN.01251005. [DOI] [PubMed] [Google Scholar]
- 5.Frankenfield DL, Rocco MV, Frederick PR, Pugh J, McClellan WM, Owen WF., Jr Racial/ethnic analysis of selected intermediate outcomes for hemodialysis patients: results from the 1997 ESRD Core Indicators Project. Am J Kidney Dis. 1999;34:721–730. doi: 10.1016/s0272-6386(99)70399-9. [DOI] [PubMed] [Google Scholar]
- 6.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bressler J, Folsom AR, Couper DJ, Volcik KA, Boerwinkle E. Genetic variants identified in a European genome-wide association study that were found to predict incident coronary heart disease in the atherosclerosis risk in communities study. Am J Epidemiol. 2010;171:14–23. doi: 10.1093/aje/kwp377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrison AC, Felix JF, Cupples LA, Glazer NL, Loehr LR, Dehghan A, Demissie S, Bis JC, Rosamond WD, Aulchenko YS, Wang YA, Haritunians T, Folsom AR, Rivadeneira F, Benjamin EJ, Lumley T, Couper D, Stricker BH, O'Donnell CJ, Rice KM, Chang PP, Hofman A, Levy D, Rotter JI, Fox ER, Uitterlinden AG, Wang TJ, Psaty BM, Willerson JT, van Duijn CM, Boerwinkle E, Witteman JC, Vasan RS, Smith NL. Genomic variation associated with mortality among adults of European and African ancestry with heart failure: the cohorts for heart and aging research in genomic epidemiology consortium. Circ Cardiovasc Genet. 2010;3:248–255. doi: 10.1161/CIRCGENETICS.109.895995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith NL, Felix JF, Morrison AC, Demissie S, Glazer NL, Loehr LR, Cupples LA, Dehghan A, Lumley T, Rosamond WD, Lieb W, Rivadeneira F, Bis JC, Folsom AR, Benjamin E, Aulchenko YS, Haritunians T, Couper D, Murabito J, Wang YA, Stricker BH, Gottdiener JS, Chang PP, Wang TJ, Rice KM, Hofman A, Heckbert SR, Fox ER, O'Donnell CJ, Uitterlinden AG, Rotter JI, Willerson JT, Levy D, van Duijn CM, Psaty BM, Witteman JC, Boerwinkle E, Vasan RS. Association of genome-wide variation with the risk of incident heart failure in adults of European and African ancestry: a prospective meta-analysis from the cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium. Circ Cardiovasc Genet. 2010;3:256–266. doi: 10.1161/CIRCGENETICS.109.895763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonough CW, Palmer ND, Hicks PJ, Roh BH, An SS, Cooke JN, Hester JM, Wing MR, Bostrom MA, Rudock ME, Lewis JP, Talbert ME, Blevins RA, Lu L, Ng MCY, Sale MM, divers J, Langefeld CD, Freedman BI, Bowden DW. A genome-wide association study for diabetic nephropathy genes in African Americans. Kidney Int. 2011;79:563–572. doi: 10.1038/ki.2010.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowden DW, Colicigno CJ, Langefeld CD, Sale MM, Williams A, Anderson PJ, Rich SS, Freedman BI. A genome scan for diabetic nephropathy in African Americans. Kidney Int. 2004;66:1517–1526. doi: 10.1111/j.1523-1755.2004.00915.x. [DOI] [PubMed] [Google Scholar]
- 12.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 13.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 14.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 15.Salter RC, Ashlin TG, Kwan AP, Ramji DP. ADAMTS proteases: key roles in atherosclerosis? J Mol Med. 2010;88:1203–1211. doi: 10.1007/s00109-010-0654-x. [DOI] [PubMed] [Google Scholar]
- 16.Bot PT, Pasterkamp G, Goumans MJ, Strijder C, Moll FL, de Vries JP, Pals ST, de Kleijn DP, Piek JJ, Hoefer IE. Hyaluronic acid metabolism is increased in unstable plaques. Eur J Clin Invest. 2010;40:818–827. doi: 10.1111/j.1365-2362.2010.02326.x. [DOI] [PubMed] [Google Scholar]
- 17.Koop EA, Lopes SM, Feiken E, Bluyssen HA, Van DV, Voest EE, Mummery CL, Moolenaar WH, Gebbink MF. Receptor protein tyrosine phosphatase mu expression as a marker for endothelial cell heterogeneity; analysis of RPTPmu gene expression using LacZ knock-in mice. Int J Dev Biol. 2003;47:345–354. [PubMed] [Google Scholar]
- 18.Ko JA, Kimura Y, Matsuura K, Yamamoto H, Gondo T, Inui M. PDZRN3 (LNX3, SEMCAP3) is required for the differentiation of C2C12 myoblasts into myotubes. J Cell Sci. 2006;119:5106–5113. doi: 10.1242/jcs.03290. [DOI] [PubMed] [Google Scholar]
- 19.Maier LS, Bers DM, Brown JH. Calmodulin and Ca2+/calmodulin kinases in the heart – physiology and pathophysiology. Cardiovasc Res. 2007;73:629–630. doi: 10.1016/j.cardiores.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE, David SP, Niaura R, Lerman C. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry. 2008;65:683–693. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhl GR, Drgon T, Johnson C, Ramoni MF, Behm FM, Rose JE. Genome wide association for smoking cessation success in a trial of precessation nicotine replacement. Mol Med. 2010;16:513–526. doi: 10.2119/molmed.2010.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman DJ, Afkarian M, Tamez H, Bhan I, Isakova T, Wolf M, Ankers E, Ye J, Tonelli M, Zoccali C, Kuro-o M, Moe O, Karumanchi SA, Thadhani R. Klotho variants and chronic hemodialysis mortality. J Bone Miner Res. 2009;24:1847–1855. doi: 10.1359/JBMR.090516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muntinghe FL, Verduijn M, Zuurman MW, Grootendorst DC, Carrero JJ, Qureshi AR, Luttropp K, Nordfors L, Lindholm B, Brandenburg V, Schalling M, Stenvinkel P, Boeschoten EW, Krediet RT, Navis G, Dekker FW. CCR5 deletion protects against inflammation-associated mortality in dialysis patients. J Am Soc Nephrol. 2009;20:1641–1649. doi: 10.1681/ASN.2008040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stenvinkel P, Wang K, Qureshi AR, Axelsson J, Pecoits-Filho R, Gao P, Barany P, Lindholm B, Jogestrand T, Heimburger O, Holmes C, Schalling M, Nordfors L. Low fetuin-A levels are associated with cardiovascular death: impact of variations in the gene encoding fetuin. Kidney Int. 2005;67:2383–2392. doi: 10.1111/j.1523-1755.2005.00345.x. [DOI] [PubMed] [Google Scholar]