Abstract
Two novel type I ribosome-inactivating proteins (RIPs) were found in the storage roots of Mirabilis expansa, an underutilized Andean root crop. The two RIPs, named ME1 and ME2, were purified to homogeneity by ammonium sulfate precipitation, cation-exchange perfusion chromatography, and C4 reverse-phase chromatography. The two proteins were found to be similar in size (27 and 27.5 kD) by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and their isoelectric points were determined to be greater than pH 10.0. Amino acid N-terminal sequencing revealed that both ME1 and ME2 had conserved residues characteristic of RIPs. Amino acid composition and western-blot analysis further suggested a structural similarity between ME1 and ME2. ME2 showed high similarity to the Mirabilis jalapa antiviral protein, a type I RIP. Depurination of yeast 26S rRNA by ME1 and ME2 demonstrated their ribosome-inactivating activity. Because these two proteins were isolated from roots, their antimicrobial activity was tested against root-rot microorganisms, among others. ME1 and ME2 were active against several fungi, including Pythium irregulare, Fusarium oxysporum solani, Alternaria solani, Trichoderma reesei, and Trichoderma harzianum, and an additive antifungal effect of ME1 and ME2 was observed. Antibacterial activity of both ME1 and ME2 was observed against Pseudomonas syringae, Agrobacterium tumefaciens, Agrobacterium radiobacter, and others.
For the past 3 years, we have been studying the biology of underutilized ARTCs (Flores and Flores, 1997; Flores et al., 1997). These species constitute an important staple for the people in the highlands of Peru, Bolivia, and Ecuador, and they are a possible source for novel bioactive phytochemicals and future novel food products (Johns et al., 1982; Flores and Flores, 1997). The ARTCs have great potential as new introduced crop species (Sperling and King, 1988), but they have been largely overlooked and poorly studied at the biological and biochemical levels. No fewer than nine plant families (Asteraceae, Basellaceae, Brassicaceae, Leguminoseae, Nyctaginaceae, Oxalidaceae, Solanaceae, Tropaeolaceae, and Umbelliferae) are represented among this crop complex, which is well adapted to cultivation at 6,000 to 14,000 feet above sea level and includes a fascinating reservoir of intraspecific and interspecific germ plasm biodiversity (National Research Council, 1989). One of the most neglected ARTCs is Mirabilis expansa, a relatively disease-resistant Andean crop that produces edible storage roots (Angeles Millones, 1996; Franco Pebe et al., 1996). The cultivation of this crop has been restricted to three small regions in Peru, Ecuador, and Bolivia (National Research Council, 1989), and its basic biology and agronomy are virtually unknown.
Storage organs such as tubers and roots synthesize proteins, the major function of which is to provide a store of N, S, and C (Shewry, 1995). Sprouting of such tissues may be accompanied by general proteolysis, during which structural and metabolic proteins are digested. In some cases, storage proteins have also been shown to exhibit antipest and antipathogen activity (Yeh et al., 1997). Because of the reported disease-resistant behavior of M. expansa (Angeles Millones, 1996), we decided to focus on the isolation of potential root defense proteins, with emphasis on RIPs. Our focus on RIPs was based on studies of Mirabilis jalapa (MacBride, 1951), a widely known ornamental plant also originally from South America and, like M. expansa, a member of the Nyctaginaceae. The storage roots of M. jalapa contain an RIP (MAP) with antiviral activity against important economically devastating viruses, such as tobacco mosaic virus, potato virus X, potato virus Y, and viroids such as potato spindle tuber viroid (Kubo et al., 1990; Kataoka et al., 1991, 1992; Vivanco, 1997). Root extracts of M. jalapa sprayed over potato plants conferred stable inhibition of viral infection (Vivanco, 1997). The viral inhibition was attributable to MAP and was based on its RIP activity and its biochemical stability.
RIPs are widely distributed among higher plants (Mehta and Boston, 1998). They inhibit protein synthesis by virtue of their N-glycosidase activity, selectively cleaving an adenine residue at a conserved site of the 28S rRNA (26S rRNA in yeast), such as the adenine-4324 of rat liver 28S rRNA (Endo and Tsurigi, 1988). This cleavage prevents the binding of elongation factor 2 (Stirpe et al., 1992), with the consequent arrest of protein synthesis. In addition to rRNA N-glycosidase activity, many RIPs display a variety of other biological and enzymatic activities. RIPs have broad-spectrum antiviral activity against RNA and DNA plant and animal viruses (Batelli and Stirpe, 1995). Furthermore, some RIPs have specific DNA-nuclease activity against supercoiled, covalently closed, circular plasmid DNA and single-stranded phage DNA (Roncuzzi and Gasperi-Campani, 1996). Some type I RIPs have been shown to inhibit fungal growth (Roberts and Selitrennikoff, 1986). Likewise, RIPs have been found to have insecticidal properties against coleopteran and lepidopteran species (Verma and Kymar, 1979; Gatehouse et al., 1990).
RIPs are compartmentalized in vacuoles and cell walls (Kataoka et al., 1991), which apparently allows the ribosome-inactivating activity to be sequestered from their own ribosomes. They may be released or induced in response to pathogen infection or injury. This apparent defense activity is usually coordinated with other defense proteins, such as chitinases (Leah et al., 1991), β-1,3 glucanases (Mauch et al., 1988a), and thaumatins (Hejgaard et al., 1991). Studies of trichosanthin, a type I RIP isolated from the roots of Trichosanthes kirilowii, suggest that full RIP expression is developmentally coordinated (Savary and Flores, 1994). However, a clear function of RIPs in plants has yet to be elucidated.
In this paper, we report the isolation and characterization of two type I RIPs, ME1 (27.5 kD) and ME2 (27 kD), from the storage roots of M. expansa. We also report antifungal and antibacterial activities of ME1 and ME2 against soil-borne microorganisms. Based on our findings, we suggest that ME1 and ME2 hold promise as candidates for broad-spectrum disease protection.
MATERIALS AND METHODS
Plant Material and Protein Extraction from Roots
Seeds from Mirabilis expansa obtained from the International Potato Center (Lima, Peru) were washed five times with sterile water and germinated on filter paper in a Petri dish at room temperature. The seeds were then transferred to pots and placed in the greenhouse. Five months after the seeds were planted, the storage roots were harvested and the total soluble proteins from the roots were extracted according to the method described by Savary and Flores (1994). Analyses were also conducted on storage roots collected by J.V. from the experimental station at the Universidad Nacional de Cajamarca (Peru). Storage roots were immersed in liquid N2, ground to a powder, and stored at −20°C until use. Root protein extracts were prepared by homogenizing 100 mg of ground root tissue per 1 mL of extraction buffer (25 mm NaPO4, pH 7.0, with 250 mm NaCl, 10 mm EDTA, 10 mm thiourea, 5 mm DTT, 1 mm PMSF, and 1.5% [w/v] polyvinylpolypyrrolidone). The solution was subsequently centrifuged for 20 min at 10,000g. The supernatant was incubated overnight at 4°C as a 20% (w/v) saturated ammonium sulfate solution under constant stirring. The supernatant was collected after centrifugation (10,000g for 20 min). Samples were dialyzed with two changes of 20 mm Hepes buffer (pH 8.0) containing 50 mm NaCl. Smaller sample volumes were desalted using Econopac 10DG columns (Bio-Rad). The protein solution was concentrated to 1 mg/mL using a Stirred Cell 8050 (Amicon, Beverly, MA) with a YM 10 membrane. Protein concentration was determined by the Bradford (1976) method using BSA as a standard and by using a laser densitometer (Ultrascan XL, LKB, Bromma, Sweden) to quantify individual proteins.
HPLC
A 4.6- × 100-mm column (1.66-mL volume) was packed with Poros 20 HS cation-exchange perfusion medium (Perkin-Elmer-Applied Biosystems) and used in conjunction with a 600E HPLC system equipped with a photodiode-array detector (model 990, Waters). Equilibration, loading, and washing were carried out in 25 mm Hepes, pH 8.0, containing 50 mm NaCl. The target proteins were eluted with a linear gradient of 30 column volumes (approximately 50 mL) from 50 to 200 mm NaCl at a flow rate of 5 mL/min. Individual peaks were collected and concentrated by ultrafiltration using a Stirred Cell 8050 (Amicon). Protein purity and peak size were confirmed by SDS-PAGE.
Reverse-phase HPLC was used to prepare proteins for N-terminal sequencing and for antiserum production. Individual peaks collected during the cation-exchange step were further separated on a 4.6- × 100-mm column packed with Poros R2 reverse-phase perfusion medium. The column was equilibrated with 0.1% (v/v) trifluoroacetic acid containing 10% (v/v) acetonitrile at a flow rate of 5 mL/min. Proteins were eluted through a 50-column-volume linear gradient from 10% to 60% acetonitrile (in 0.1% trifluoroacetic acid). All chromatographic separations were performed at room temperature.
Amino Acid Analysis and N-Terminal Sequencing
Amino acid analysis and composition were determined using an analyzer (model 420H, Perkin-Elmer-Applied Biosystems), according to the methodology described by Tarr (1986). The N-terminal-sequence analysis was performed on a protein sequencer (model 477A) equipped with an analyzer (model 120A, Perkin-Elmer-Applied Biosystems) at the Hershey Medical Center (The Pennsylvania State University, University Park). The standard Edman degradation procedure was used as described by Allen (1981).
Electrophoresis and Western-Blot Analysis
SDS-PAGE was performed with 13.5% or 15% (v/v) acrylamide discontinuous gels (Laemmli, 1970) using an electrophoresis cell (Mini-Protean II, Bio-Rad), according to the manufacturer's instructions. A low-molecular-mass (14–66 kD) protein-marker kit (Sigma) was used to determine approximate protein sizes. Proteins were visualized with Coomassie brilliant blue G-250 (Calbiochem, La Jolla, CA) or zinc staining (Bio-Rad). Proteins were electroblotted to Immobilon-P PVDF membranes (Millipore) with a Bio-Rad Mini-Trans electrotransfer cell for 1 h at 150 V (constant voltage), using 10 mm 3-(cyclohexylamino)propanesulfonic acid (pH 11.0 with NaOH) and 10% (v/v) methanol-transfer buffer (LeGendre and Matsudaira, 1989). Membranes were developed with the Promega secondary antibody-alkaline phosphatase detection system, according to the manufacturer's instructions. An antiserum titer of 1:5000 was used for all experiments.
IEF
The pI of purified ME1 and ME2 was estimated by IEF using an Ampholine PAG plate (pH 3.5–9.5; Pharmacia) with high-range pI marker proteins (Pharmacia) stained with Coomassie blue.
Antibodies
Polyclonal antibodies were produced in male New Zealand White rabbits against reverse-phase HPLC-purified ME1 and ME2. Three injections were given 20 d apart, to a total of 300 μg of each protein. The proteins were injected together with Freund's adjuvant. After clot removal, the serum was stored at −40°C. Antibodies were further purified according to the method described by Olmsted (1986). The procedure was carried out at the Centralized Biological Laboratories (The Pennsylvania State University).
rRNA Depurination Assay
The assay was conducted according to the method of Tumer et al. (1997). Yeast ribosomes (50 μg) were incubated with 100 ng of ME1, ME2, ME1 plus ME2, and PAP at 30°C for 30 min. The reaction was stopped by the addition of 0.1% SDS. RNA was extracted from yeast ribosomes, incubated on ice for 30 min with 1 m aniline acetate, pH 4.5, and precipitated with ethanol. RNA was electrophoresed in a 4.5% urea-polyacrylamide gel and stained with ethidium bromide.
Antimicrobial Assays
Antifungal activity of M. expansa root proteins was demonstrated in a radial growth-inhibition assay adapted from the method of Schlumbaum et al. (1986). A fungal plug was placed in the center of a potato-dextrose-agar plate. Sterile paper discs were placed into punched wells in the agar around the outside of the plate. Protein solutions ranging from 0.5 to 100 μg were filter sterilized—using 0.22-μm filters (Ultrafree-MC Durapore, Millipore). Various amounts of protein were pipetted into the wells, and sterile buffer was used to bring the final volume to 100 μL. The Petri plates were incubated in the dark at 23°C. Antifungal activity was observed as a crescent-shaped zone of inhibition at the mycelial front. The effect on fungal growth was expressed qualitatively, according to the procedure of Schlumbaum et al. (1986).
Antibacterial activity was screened using an inhibition-halo-plate assay. Bacterial cultures were inoculated into liquid Luria-Bertani medium and placed onto a shaker at 23°C for 2 to 3 d. The A600 of bacterial cultures was measured and adjusted to an optical density of 0.2 for the antibacterial experiments. One hundred microliters of bacterial suspension was placed onto a sterile plate containing Luria-Bertani medium and subsequently spread over the entire surface of the plate. A sterile paper disc was placed into punched wells in the agar in the middle of the plate. Protein solutions were filter-sterilized using 0.22-μm filters (Ultrafree-MC Durapore, Millipore) and applied to the paper discs. The Petri plates were incubated in a dark room at 23°C for 24 h. Antibacterial activity was measured as the radius of inhibition from the border of the protein-impregnated disc.
The following bacterial strains were obtained from a collection of field isolates maintained in the laboratory of Dr. Leland S. Pierson, III, at the University of Arizona in Tucson: Bacillus subtilis 613R, Bacillus thuringiensis Gnatrol, Clavibacter michigenensis subsp. nebraskensis CN74-1, Agrobacterium radiobacter K84, Agrobacterium tumefaciens C58, Bacillus cepacea Deny, Escherichia coli ESS, Erwinia amylovora, Erwinia carotovora ATCC 15713, Pseudomonas aureofaciens 30-84, Pseudomonas fluorescens 2-79, Pseudomonas syringae B, Pseudomonas syringae pv phaseolicola, Pseudomonas putida Qd8, Ralstonia solanacearum, Serratia marcescens, Xanthomonas campestris pv versicatoria, Streptomyces griseovivides Mycostop, and Rhizobium leguminosarum. Erwinia herbicola was isolated from pea seedling roots by Dr. Lindy Brigham at the University of Arizona. Agrobacterium rhizogenes ATCC 15834 was maintained by Dr. Hector E. Flores at The Pennsylvania State University. Xanthomonas campestris pv pelargonii was obtained from the collection of Dr. Gary Moorman at The Pennsylvania State University.
Microscopy
The inhibitory halo was observed on a stereoscopic microscope (model SMZ-U, Nikon), and the antifungal effect was confirmed in vivo (see Fig. 8). The effect of the basic M. expansa proteins was studied in hyphae growing on agar, using a modification of the method described by Arlorio et al. (1992). Fungal hyphae from the center of the actively growing plates (healthy) and from the front of the inhibition zone adjacent to the wells (unhealthy) were mounted on glass slides and stained with lacto-phenol blue. The samples were destained with distilled water before viewing with bright-field microscopy using an inverted microscope (Diaphot-TMD, Nikon).
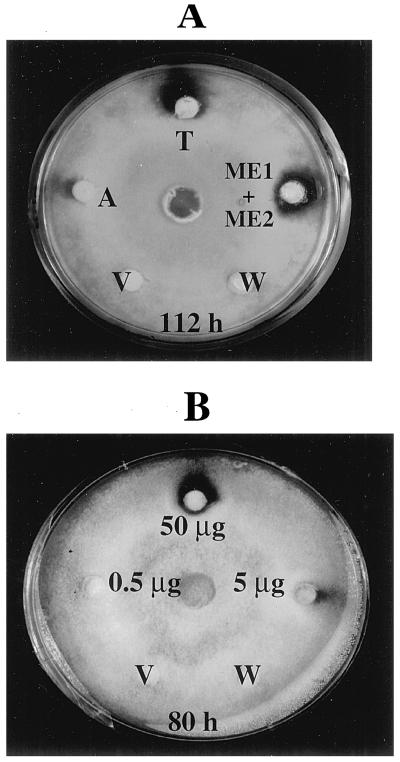
Figure 8.
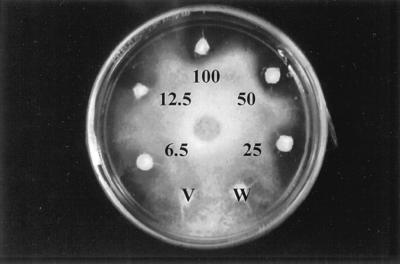
A, Antifungal activity of M. expansa proteins against T. reesei. Fifty micrograms of total storage-root protein (T), ME1 and ME2 together, and acidic (unretained) proteins (A) were applied to the discs and assayed for antifungal activity, as described in Methods. B, Antifungal activity of the purified ME1 and ME2 against T. reesei. Numbers indicate different concentrations of ME1 and ME2. The controls consisted of boiled M. expansa total storage-root proteins (V) and 25 mm Hepes buffer, pH 8.0 (W).
RESULTS
Purification of ME1 and ME2
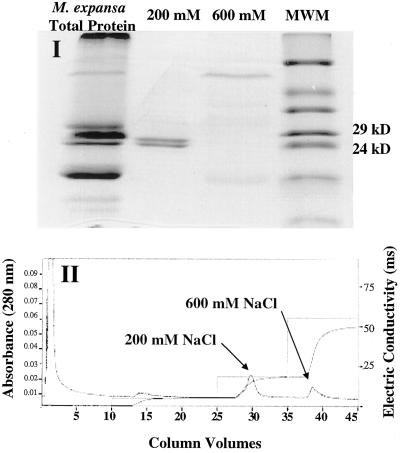
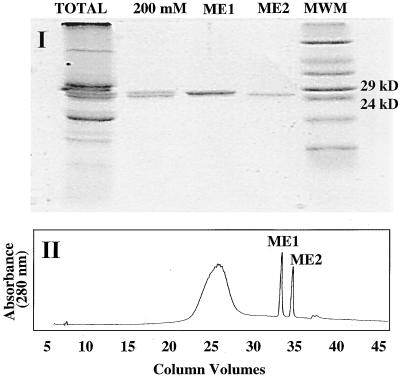
The root proteins of M. expansa were buffer exchanged to pH 8.0 (see Methods), which facilitates the enrichment and separation of strongly basic proteins, such as RIPs, by cation-exchange chromatography. Two major basic proteins, with molecular masses of 27 and 27.5 kD, were resolved through cation-exchange perfusion chromatography and eluted through a 200 mm NaCl step gradient. As shown in Figure 1, these two proteins eluted close to each other, indicating a similarity in charge. Reverse-phase chromatography was performed to further purify the proteins for amino acid composition analysis and sequencing. This technique yielded individual bands of essentially pure protein, as confirmed by SDS-PAGE (Fig. 2). The two proteins were named ME1 (27.5 kD) and ME2 (27 kD). A cation-exchange perfusion chromatography linear gradient (0–150 mm NaCl) was also used to resolve ME1 and ME2 under nondenaturing conditions, and these fractions were used individually and in combination to determine their biological activities. The perfusion medium greatly enhanced the speed and resolution of our separations and enabled us to purify milligram quantities of ME1 and ME2 in 15 min.
Figure 1.
Cation-exchange perfusion chromatography of the total storage-root proteins from M. expansa. I, SDS-PAGE of the fractions collected by cation-exchange perfusion chromatography. II, Cation-exchange perfusion chromatogram. Samples for electrophoresis were prepared as described in Methods. Fifteen and five micrograms were loaded onto the gel for total proteins and fractions, respectively. Approximately 33 mg of total protein was injected into the column, and 6.1 mg was recovered in the 200 mm NaCl fraction. This separation was repeated several times to obtain sufficient protein separation for further characterization.
Figure 2.
Isolation of ME1 and ME2 from the 200 mm cation-exchange fraction by reverse-phase column chromatography, as described in Methods. I, SDS-PAGE of the proteins separated after the reverse-phase HPLC step. Approximately 2 μg of protein was loaded per lane. II, Reverse-phase perfusion chromatogram. Approximately 300 μg of protein from the 200 mm NaCl fraction was injected into the column, and 100 μg of each fraction was collected.
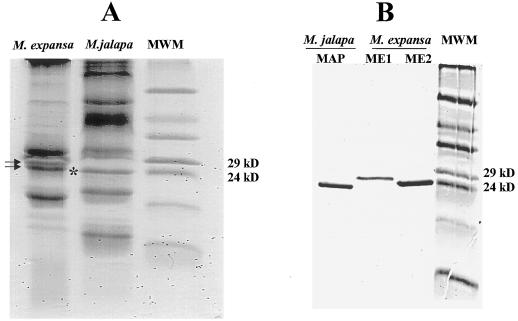
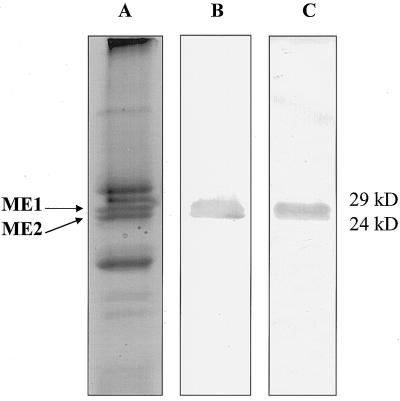
The patterns of total root-soluble proteins of M. jalapa and M. expansa were compared by SDS-PAGE. Species-specific patterns were observed in both species, including common and unique major proteins. It was clear that MAP, a type I RIP from M. jalapa, migrated at the same protein range (24–29 kD) as ME1 and ME2 from M. expansa (Fig. 3A). MAP was purified to homogeneity and compared with ME1 and ME2. As shown in Figure 3B, ME2 was similar in size to MAP (27 kD). Densitometric determination of the total storage-root proteins of M. expansa revealed that ME1 and ME2 account for almost 20% of the soluble proteins (data not shown).
Figure 3.
A, Comparison of the total root-protein patterns from M. expansa and M. jalapa. The arrows indicate ME1 (top arrow) and ME2 (bottom arrow) in M. expansa, and the asterisk indicates MAP in M. jalapa. B, Comparison of the RIP from M. jalapa (MAP) and the two proteins isolated from M. expansa (ME1 and ME2). Samples were extracted as described in Methods. About 15 and 3 μg of total and individual proteins, respectively, were loaded.
Characterization of ME1 and ME2
ME1 and ME2 are strongly basic proteins with pI values greater than pH 10.0, as determined by IEF-PAGE (data not shown). Amino acid composition analysis of ME1 and ME2 indicated a similar distribution of amino acids, with the exception of Pro, Ala, and Phe (Table I). ME1 had more Pro and less Ala and Phe than ME2, and the amino acid composition of ME1 and ME2 showed a strong similarity with that of MAP. ME1 and ME2 appear to share distinctive features among RIPs, such as low Cys and His levels and high Lys content.
Table I.
Amino acid composition of ME1 and ME2
| Amino Acid | ME1 | ME2 | MAP | PAP-S | Petroglaucin |
|---|---|---|---|---|---|
| Asp | 33 | 30 | 27 | 34 | 26 |
| Ser | 14 | 13 | 19 | 15 | 14 |
| His | 1 | 1 | 0 | 2 | 1 |
| Thr | 25 | 28 | 27 | 14 | 19 |
| Pro | 12 | 8 | 8 | 12 | 9 |
| Val | 13 | 16 | 14 | 16 | 18 |
| 1/2Cys | 0 | 0 | 2 | 2 | 0 |
| Leu | 23 | 20 | 21 | 27 | 26 |
| Lys | 23 | 22 | 23 | 24 | 23 |
| Glu | 15 | 16 | 20 | 29 | 24 |
| Gly | 15 | 15 | 12 | 18 | 18 |
| Arg | 12 | 10 | 7 | 14 | 14 |
| Ala | 26 | 31 | 20 | 16 | 24 |
| Tyr | 8 | 7 | 11 | 10 | 5 |
| Met | 3 | 3 | 4 | 6 | 4 |
| Ile | 14 | 15 | 20 | 20 | 15 |
| Phe | 10 | 15 | 13 | 7 | 9 |
| Trp | 0 | 0 | 2 | 14 | 0 |
Amino acid composition of ME1 and ME2, MAP (Wong et al., 1992), PAP-S (Barbieri et al., 1982), and Petrocoptis glaucifolia (petroglaucin) (Arias et al., 1992).
The N-terminal amino acid sequences of ME1 and ME2 were determined and compared with each other and with those from other RIPs (Fig. 4). A significant matching with conserved hydrophobic amino acids was found in the N-terminal region with 11 RIPs reported to date (Funatsu et al., 1991). Furthermore, the ME1 and ME2 amino acid residues Tyr-16 (Y) and Phe-19 (F) aligned perfectly with the consensus sequences found in the same positions for all of the examined RIPs. Our data suggest homology between ME1 and ME2, as well as between ME2 and MAP. Antisera were produced for each protein, from which monospecific antibodies were prepared and used for western-blot analysis (Olmsted, 1986). Each antiserum was reactive with both proteins in the total root-protein extract (Fig. 5). In other experiments in which the individual purified proteins were blotted, the antisera were cross-reactive between proteins (data not shown), indicating structural homology.
Figure 4.
Comparison of the N-terminal sequences from RIPs. RIC, Ricin A-chain from Ricinus communis; RCA, agglutinin from R. communis; ABR, abrin A-chain from Abrus precatorius; LUFa and LUFb, luffin-a and luffin-b from Luffa cylindrica; TRI, trichosanthin from T. kirilowii; MOM, momordin from Momordica charantia; SAP, saporin from Saponaria officinalis; and BAR, barley translation inhibitor from barley (Funatsu et al., 1991). Shaded boxes and boldface characters represent two amino acids that are conserved in all RIPs currently reported. Shaded boxes and lightface characters symbolize homology regions among ME1, ME2, and MAP. The asterisk shows the presence of conserved amino acids with polar R groups (hydrophobic).
Figure 5.
Western-blot analysis of ME1 and ME2. A, SDS-PAGE of the total storage-root proteins from M. expansa stained with Coomassie blue. B and C, Western-blot analysis of M. expansa total storage-root proteins using ME1 antibody (B) and ME2 antibody (C). SDS-PAGE, electrobloting, and western-blot development were conducted as described in Methods.
rRNA Depurination Assay
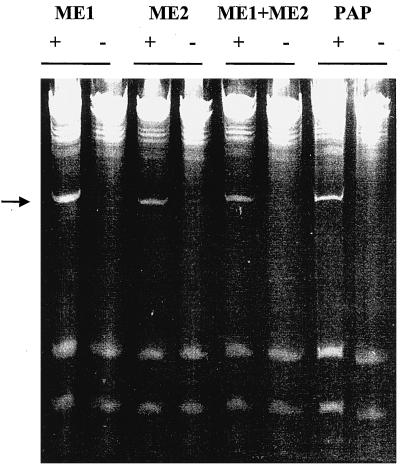
RIP-mediated depurination of the large rRNA renders the RNA sugar-phosphate backbone susceptible to hydrolysis at the depurination site (Endo and Tsurugi, 1988). When depurinated rRNA is treated with aniline, cleavage occurs at the depurinated site and a small 367-nucleotide fragment (in the yeast 26S rRNA) is released (Stirpe et al., 1986). Yeast ribosomes were individually incubated with ME1, ME2, ME1 and ME2 together, and PAP, a type I RIP, as a control. RNA was extracted, treated with aniline, and analyzed by gel electrophoresis. As shown in Figure 6, ME1 and ME2 depurinated the 26S rRNA and released the 367-nucleotide fragment upon treatment with aniline. The combination of ME1 and ME2 also produced the same fragment. These results demonstrate that ME1 and ME2 are RIPs.
Figure 6.
Depurination of yeast ribosomes in vitro. Ribosomes were isolated and incubated with ME1 and ME2 individually, ME1 and ME2 together, and PAP as described in Methods. rRNA was extracted, treated with aniline, separated on a 4.5% urea-polyacrylamide gel, and stained with ethidium bromide. The presence (+) or absence (−) of aniline is denoted. The arrow shows the presence of the diagnostic 367-nucleotide cleavage product of rRNA.
Antifungal Activity of ME1 and ME2
To explore the biological activity of ME1 and ME2, we used a standard antifungal assay (Schlumbaum et al., 1986). Growth of Trichoderma harzianum treated with the total root soluble proteins of M. expansa was inhibited, indicating antifungal activity. As shown in Figure 7, inhibitory activity was observed at doses as low as 6.5 μg of total protein. The antifungal activity of ME1 and ME2 was also tested against an array of plant fungal pathogens. As shown in Table II, the antifungal action was more evident against the hyphomycete fungi, such as Verticillium dahliae, Alternaria solani, Fusarium oxysporum solani, Fusarium proliferatum, Trichoderma reesei, and T. harzianum. Based on these observations, we focused our studies on T. reesei, an indicator organism commonly used to study antimicrobial activity (Roberts and Selitrennikoff, 1986; Schlumbaum et al., 1986; Arlorio et al., 1992). A time-course experiment for antifungal activity was performed using 50 μg of total root soluble protein, acidic root protein, and ME1 and ME2 together (Fig. 8A). The results showed strong antifungal activity of the total protein and the purified ME1 and ME2 for up to 112 h of incubation. The acidic proteins showed only weak antifungal activity, but this could be the result of minor contamination by ME1 or ME2 during the cation-exchange chromatography separation.
Figure 7.
Antifungal activity of M. expansa total storage-root proteins against T. harzianum. Numbers indicate the concentrations (in micrograms) of proteins applied to the discs and tested for antifungal activity, as described in Methods. The controls consisted of boiled M. expansa total storage-root proteins (V) and 25 mm Hepes buffer, pH 8.0 (W).
Table II.
Antifungal activity of ME1 and ME2
| Tested Fungal Pathogens | Inhibition of Fungal Growth |
|---|---|
| Oomycetes | |
| Pythium irregulare | + |
| Pythium ultimum | − |
| Phytophtohora drechsleri | + |
| Phytophthora megasperma | − |
| Phytophtohora palmivora | − |
| Phytophthora crytoptogea | − |
| Phytophthora cinnamomi | − |
| Hyphomycetes | |
| V. dahliae | + |
| A. solani | ++ |
| F. oxysporum solani | + |
| F. proliferatum | + |
| T. harzianum | +++ |
| T. reesei | +++ |
| Agonomycetes | |
| Rhizoctonia solani | − |
Antifungal activity of ME1 and ME2 against various fungal pathogens. About 50 μg of purified protein in 100 μL of 25 mm phosphate buffer, pH 70, was used for determination of antifungal activity. The effect on fungal growth is expressed in qualitative terms, according to the method of Schlumbaum et al. (1986), in which +++ stands for strong inhibition, + for just detectable inhibition, and − for no inhibition.
T. reesei was treated with various doses of ME1 and ME2. As little as 0.5 μg of protein showed inhibitory activity after 24 h of incubation (data not shown). Zones of inhibition were dose dependent. As shown in Figure 8B, the most prominent inhibition zones were obtained with 50 and 5 μg after 80 h of incubation. Fungal inhibition persisted after 3 weeks of incubation in the dark growth chamber at 23°C and after the plates were transferred to the cold room (4°C) for 2 more weeks. The individual effects of ME1 and ME2 separated by cation-exchange chromatography were also monitored against T. reesei growth, revealing an additive inhibitory effect of ME1 and ME2 (data not shown). The inhibitory activity of ME1 was higher than that of ME2.
Evaluation of the Effect of ME1 and ME2 on T. reesei Hyphae Growth at the Microscopic Level
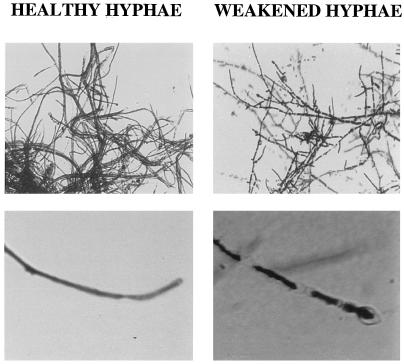
The inhibitory halo caused by ME1 and ME2 was evaluated microscopically to analyze the antifungal effect in detail. Comparison by light microscopy of the healthy hyphae growing at the center of the plate and the weakened hyphae at the border of the inhibition halo showed extensive septum formation and thinning of the unhealthy hyphae (Fig. 9). Enlarged tips were also seen on the unhealthy hyphae, showing a marked swelling and lysis (Fig. 9).
Figure 9.
Microscopic analysis of the antifungal effect in T. reesei. Comparison of T. reesei healthy hyphae growing at the middle of the Petri dish and weakened hyphae growing at the border of the inhibition halo.
Antibacterial Activity of ME1 and ME2
Because the two M. expansa root proteins were isolated from underground organs, the inhibitory activity of ME1 and ME2 was tested against 27 soil-borne bacterial species (Table III), and inhibitory activity was found against 8 of them. Among these bacteria there were nonpathogenic species, such as B. subtilis, R. leguminosarum, and S. marcescens, and pathogenic species, including P. syringae, A. tumefaciens, A. rhizogenes R100nal, X. campestris pv versicatoria, and E. carotovora. These bacteria include important plant pathogens that cause severe diseases, such as wilts, crown galls, leaf spots, and soft rots. It is interesting that S. marcescens, one of the bacteria most susceptible to ME1 and ME2, can be a human pathogen under certain conditions (von Graevenitz, 1980). To our knowledge, this is the first time that in vitro antibacterial activity has been reported for type I RIPs against bacterial plant pathogens. Previous studies performed by Kataoka et al. (1991) showed that E. coli expressing MAP displayed a reduced growth rate, which is consistent with our antibacterial assays. A. rhizogenes inoculations were also performed on M. expansa plants to obtain “hairy-root” cultures (data not shown). However, no infection occurred. Thus, it is possible that A. rhizogenes was inhibited by ME1 and ME2 activity.
Table III.
Antibacterial activity of ME1 and ME2
| Bacterial Strain | Inhibition Zone |
|---|---|
| mm | |
| B. subtilis G13R | 2.5 ± 0.15 |
| B. cepacea Deny | 0.0 |
| B. thuringernesis Gnatrol | 0.0 |
| P. syringae B | 1.0 ± 0.1 |
| P. syringae pv phaseolicola | 0.0 |
| P. aureofaciens 30-84 | 0.0 |
| P. fluorescens PFS rpos- | 0.0 |
| P. putida Qd8 | 0.0 |
| P. fluorescens 2-79 | 0.0 |
| R. solanacearum | 0.0 |
| A. radiobacter K84 | 0.0 |
| A. tumefaciens C58 | 1.0 ± 0.11 |
| A. rhizogenes ATCC 15834 | 1.0 ± .09 |
| C. michigenensis subsp nebraskensis | 0.0 |
| F. carotovora ATCC 15713 | 2.0 ± 0.12 |
| E. amylovora | 0.0 |
| E. herbicola | 0.0 |
| X. campestris pv vesicatoria | 1.0 ± 0.11 |
| X. campestris pv peligonii | 0.0 |
| R. leguminosarum | 2.0 ± 0.2 |
| E. coli ESS | 0.0 |
| S. griseovivides Mycostop | 0.0 |
| S. marcescens | 2.5 ± 0.1 |
Antibacterial activity of ME1 and ME2 against various root-tor bacteria. Approximately 50 μg of purified protein in 100 μL of 25 mm phosphate buffer, pH 70), was used for determination of antibacterial activity. These experiments were repeated three times.
DISCUSSION
We have isolated and characterized two major basic proteins (ME1 and ME2) that constitute about 20% of the total soluble proteins from the storage roots of M. expansa. ME1 and ME2 showed antimicrobial activity against various fungal and bacterial species, including some important plant pathogens. Based on amino acid composition, N-terminal sequence analysis, and enzyme activity, we conclude that ME1 and ME2 are type I RIPs. Both proteins share features common to RIPs, such as low Cys and His levels and high Lys content. Highly conserved hydrophobic amino acid residues were also observed at the N-terminal regions of ME1 and ME2 (Funatsu et al., 1991), as was the presence of conserved amino acids with polar R groups (hydrophobic), such as Met (M), Ile (I), and Val (V) (Fig. 4). It is well established that hydrophobic residues stabilize the interaction between nucleic acids and proteins by intercalating between bases of DNA or RNA (Frankel et al., 1989). The molecular-weight and N-terminal sequence similarities between ME2 and MAP suggest the presence of RIPs among the genus Mirabilis, similar to the widespread content of RIPs in the genus Trichosanthes (Ng et al., 1992, 1993).
RIPs are widely represented in taxonomically distinct plant families, such as Asparagaceae, Caryophyllaceae, Cucurbitaceae, Euphorbiaceae, Nyctaginaceae, Phytolaccaceae, and Poaceae (Mehta and Boston, 1998). Some plant species show a higher constitutive content of RIPs than others (Stirpe and Barbieri, 1986; Stirpe et al., 1992; Mehta and Boston, 1998). It has been speculated that RIPs were originally used by all plant species as a primary defense response (Stirpe et al., 1992).
ME1 and ME2 showed strong antifungal activity against an array of pathogenic and nonpathogenic fungi, such as P. irregulare, V. dahliae, A. solani, F. oxysporum solani, F. proliferatum, and T. reesei (Table II); the hyphomycetes was the most susceptible fungal group. It has been suggested that as little as one RIP molecule per cell is capable of shutting down protein synthesis (Stirpe and Barbieri, 1986). However, to reach the ribosomes, RIPs must penetrate the target cell. Anatomic features such as gaps, natural openings, and damaged tissue may facilitate RIP penetration. According to Roberts et al. (1986), a barley RIP was able to penetrate T. reesei through gaps in cell membranes. The antifungal activity of ME1 and ME2 on T. reesei growth could be attributable to the penetration of these two proteins into the hyphae by the same mechanism. It has also been suggested that some RIPs may have a role as chitinases (Remi Shih et al., 1997). In vivo RIPs may play a synergistic role with other defense-related proteins such as chitinases (Broekaert et al., 1989) and β-1,3 glucanases (Kombrick et al., 1988; Mauch et al., 1988b); the latter proteins may break down fungal cell walls and thus facilitate RIP entrance into the cell. We found that ME1 and ME2 showed antibacterial activity against important plant pathogens. The mechanism by which ME1 and ME2 inhibit certain bacteria remains unknown. In addition to ME1 and ME2 antimicrobial activity, preliminary results suggest antiviral activity associated with these two proteins (data not shown). Preventive applications of ME1 and ME2 over Gomphrena globosa leaves inhibited potato virus X infection. Nonetheless, toxicity was observed with higher concentrations of ME1 and ME2.
It is intriguing that ME1 and ME2 have an additional role as storage proteins, because they constitute 20% of the total storage-root proteins in M. expansa. This hypothesis is in accordance with studies of trichosanthin, a root-specific RIP from T. kirilowii, in which trichosanthin accumulation was associated with the onset of root secondary growth (Savary and Flores, 1994). In fact, maximum levels of trichosanthin (>25% total soluble root protein) were observed in fully developed tuberous roots. The main storage protein of sweet potato roots, sporamin, may function in response to root pests as well (Yeh et al., 1997). Patatin, the major tuber storage protein of potato, has been reported to have lipid acyl hydrolase activity and to inhibit the growth of southern corn rootworm and western corn rootworm when fed to them in an artificial diet (Strickland et al., 1995). Thus, we may speculate that more than a few major proteins found in underground storage organs have evolved more than one function.
The majority of RIPs reported so far have been isolated from leaves, stems, and seeds. To date, the isolation of root-specific RIPs has been restricted to MAP, trichosanthin, and related proteins from Cucurbitaceae species (Maragonore et al., 1985; Savary and Flores, 1994; Mehta and Boston, 1998). Preliminary investigations using tissue printing suggest the localization of ME1 and ME2 in the epidermis of the storage root, as well as higher concentrations at the site of emergence of lateral roots (data not shown), consistent with their role as pathogen/pest defense proteins. PAP, a type I RIP, and PAP mutants have been genetically engineered into potato and tobacco plants. The transformants have been shown to express resistance to a broad spectrum of viruses and to fungal pathogens as well, such as R. solani (Lodge et al., 1993; Zoubenko et al., 1997). Our data suggest the role of ME1 and ME2 as broad-spectrum defense proteins in M. expansa and their potential use in the development of transgenic pathogen-resistant crops. Alternatively, their antimicrobial and antiviral activity could be used in simple crop-protection methods in low-input agricultural systems, such as the spraying of root extracts on leaves of various crops to prevent or control pathogen infection (Vivanco, 1997).
As stated above, M. expansa is an underutilized root crop, the storage roots of which are used as a food source. Presumably, ME1 and ME2 become denatured during the cooking and roasting process, thus allowing the root to be eaten. However, some type I RIPs, such as PAP, are able to resist boiling without denaturing (Irvin, 1995). Our findings thus warrant detailed studies of the potential toxicity or beneficial use of M. expansa roots to humans (Moore, 1988). The study of new RIPs, such as those reported in this paper, may lead to a better understanding of their biology and diversity. Efforts are currently under way to isolate cDNA and genomic clones for ME1 and ME2 and to define their potential role in root-pathogen interactions.
ACKNOWLEDGMENTS
We thank Drs. N. Tumer and K. Hudak for assistance with the depurination assay. We also thank Dr. S. Kang, Ms. P. Michaels, and Ms. D. Needle for editing suggestions in the preparation of the manuscript.
Abbreviations:
- ARTCs
Andean root and tuber crops
- MAP
Mirabilis jalapa antiviral protein
- ME1 and ME2
Mirabilis expansa proteins 1 and 2
- PAP
pokeweed antiviral protein
- RIP
ribosome-inactivating protein
Footnotes
This work was supported by the McKnight Foundation.
LITERATURE CITED
- Allen G. Sequencing of proteins and peptides. In: Work TS, Burdon RH, editors. Laboratory Techniques in Biochemistry. Amsterdam: Elsevier; 1981. , 1981, pp 299–318. [Google Scholar]
- Angeles Millones E (1996) Cultivo del Chago. Instituto Nacional de Investigacion Agraria, Lima
- Arias FJ, Angeles Rojo M, Ferreras JM, Iglesias JM, Iglesias R, Munoz R, Rocher A, Mendez E, Barbieri L, Girbes J. Isolation and partial characterization of new ribosome-inactivating proteins from Petrocoptis glaucifolia (Lag.) Boiss. Planta. 1992;186:532–540. doi: 10.1007/BF00198033. [DOI] [PubMed] [Google Scholar]
- Arlorio M, Ludwing A, Boller T, Bonfante P. Inhibition of fungal growth by plant chitinases and β-1,3 glucanases: a morphological study. Protoplasma. 1992;171:34–43. [Google Scholar]
- Barbieri L, Aron GM, Irvin JD, Stirpe F. Purification and partial characterization of another form of the antiviral protein from the seeds of Phytolacca americana L. (pokeweed) Biochem J. 1982;203:55–59. doi: 10.1042/bj2030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batelli MG, Stirpe F. Ribosome-inactivating proteins from plants. In: Chessin M, DeBorde D, Zipf A, editors. Antiviral Proteins in Higher Plants. Boca Raton, FL: CRC Press; 1995. pp. 39–64. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Broekaert WF, Van Parijs J, Leyns F, Joos H, Peumans WJ. A chitin-binding lectin from stinging nettle rhizomes with antifungal properties. Science. 1989;245:1100–1102. doi: 10.1126/science.245.4922.1100. [DOI] [PubMed] [Google Scholar]
- Endo Y, Tsurigi K. RNA N-glycosidase activity of ricin A-chain: mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J Biol Chem. 1988;262:8128–8130. [PubMed] [Google Scholar]
- Flores HE, Brigham LA, Vivanco JM (1997) The future of radical biology? Connecting roots, people, and scientists. In HE Flores, JP Lynch, D Eissenstat, eds, Radical Biology: Advances and Perspectives on the Function of Plant Roots. American Society of Plant Physiologists, Rockville, MD, pp 320–339
- Flores HE, Flores T (1997) Biology and biochemistry of underground plant storage organs. In T Johns, J Romeo, eds, Functionality of Food Phytochemicals. Plenum Press, New York, pp 113–132
- Franco Pebe S, Uceda Vejarano J (1996) El Chago o Yuca Inca (Mirabilis expansa) Raiz Andina en Peligro de Extincion. Instituto Nacional de Investigacion Agraria, Cajamarca, Peru
- Frankel A, Schlossman D, Welsh P, Hertler A, Withers D, Johnston S. Selection and characterization of ricin toxin A-chain mutations in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:415–420. doi: 10.1128/mcb.9.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funatsu G, Islam MR, Minami Y, Sung-Sil K, Kimura M. Conserved amino acid residues in ribosome-inactivating proteins from plants. Biochimie. 1991;73:1157–1161. doi: 10.1016/0300-9084(91)90160-3. [DOI] [PubMed] [Google Scholar]
- Gatehouse AMR, Barbieri L, Stirpe F, Croy RRD. Effects of ribosome inactivating proteins on insect development: differences between Lepidoptera and Coleoptera. Entomol Exp Appl. 1990;54:43–51. [Google Scholar]
- Hejgaard J, Jacobsen S, Svendsen I. Two antifungal thaumatin-like proteins from barley grain. FEBS Lett. 1991;291:127–131. doi: 10.1016/0014-5793(91)81119-s. [DOI] [PubMed] [Google Scholar]
- Irvin JD. Antiviral proteins from Phytolacca. In: Chessin M, DeBorde D, Zipf A, editors. Antiviral Proteins in Higher Plants. Boca Raton, FL: CRC Press; 1995. pp. 65–94. [Google Scholar]
- Johns T, Kitts WD, Newsome F, Towers GHN. Anti-reproductive and other medicinal effects of Tropaeolum tuberosum. J Ethnopharmacol. 1982;5:149–161. doi: 10.1016/0378-8741(82)90040-x. [DOI] [PubMed] [Google Scholar]
- Kataoka J, Habuka N, Miyano M, Masuta C, Koiwai A. Adenine depurination and inactivation of plant ribosomes by an antiviral protein of Mirabilis jalapa (MAP) Plant Mol Biol. 1992;20:1111–1119. doi: 10.1007/BF00028897. [DOI] [PubMed] [Google Scholar]
- Kataoka J, Habuka N, Miyano M, Takanami Y, Koiwai A. DNA sequence of Mirabilis antiviral protein (MAP), a ribosome inactivating protein with antiviral property, from Mirabilis jalapa L. and its expression in E. coli. J Biol Chem. 1991;266:8426–8430. [PubMed] [Google Scholar]
- Kombrick E, Schroeder M, Hahlbrock K. Several “pathogenesis-related” proteins in potato are 1,3-β-glucanases and chitinases. Proc Natl Acad Sci USA. 1988;85:782–786. doi: 10.1073/pnas.85.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo S, Ikeda T, Imaizumi S, Takanami Y, Mikami Y. A potent plant virus inhibitor found in Mirabilis jalapa L. Ann Phytopathol Soc Jpn. 1990;56:481–487. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leah R, Tommerup H, Svendensen I, Mundy J. Biochemical and molecular characterization of three barley seed proteins with antifungal activity. J Biol Chem. 1991;266:1564–1573. [PubMed] [Google Scholar]
- LeGendre N, Matsudaira PT (1989) Purification of proteins and peptides by SDS-PAGE. In PT Matsudaira, ed, A Practical Guide to Protein and Peptide Purification for Microsequencing. Academic Press, New York, pp 49–69
- Lodge JK, Kaniewski JK, Tumer NE. Broad-spectrum virus resistance in transgenic plants expressing pokeweed antiviral protein. Proc Natl Acad Sci USA. 1993;90:7089–7093. doi: 10.1073/pnas.90.15.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacBride JF (1951) Flora of Peru: Euphorbiaceae, Spurge Family. Chicago Field Museum of Natural History, Chicago, pp 543–544
- Maragonore JM, Joseph J, Bailey MC. Purification and characterization of trichosanthin: homology to the ricin A chain and implications as to mechanism of abortifacient activity. J Biol Chem. 1985;262:11628–11633. [PubMed] [Google Scholar]
- Mauch F, Hadwiger LA, Boller T. Antifungal hydrolases in pea tissue. I. Purification and characterization of two chitinases and two β-1,3 glucanases differentially regulated during development in response to fungal infection. Plant Physiol. 1988a;87:325–333. doi: 10.1104/pp.87.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch F, Mauch-Mani B, Boller T. Antifungal hydrolases in pea tissue. II. Inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. Plant Physiol. 1988b;88:936–942. doi: 10.1104/pp.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta AD, Boston RS (1998) Ribosome-inactivating proteins. In J Bailey-Serres, DR Gallie, eds, A Look Beyond Transcription: Mechanisms Determining mRNA Stability and Translation in Plants. American Society of Plant Physiologists, Rockville, MD, pp 145–152
- Moore M (1988) Medicinal Plants of the Mountain West. Museum of New Mexico Press, Santa Fe
- National Research Council (1989) Lost Crops of the Incas. National Academy Press, Washington, DC, pp 22–123
- Ng TB, Chan WY, Yeung HW. Proteins with abortifacient, ribosome inactivating, immunomodulatory, antitumor and anti-AIDS activities from Cucurbitaceae plants. Gen Pharmacol. 1992;23:575–590. doi: 10.1016/0306-3623(92)90131-3. [DOI] [PubMed] [Google Scholar]
- Ng TB, Shaw PC, Yeung HW, Ho WKK. Immunological relatedness of ribosome-inactivating proteins from the Cucurbitaceae family. Biochem Mol Biol Int. 1993;31:447–453. [PubMed] [Google Scholar]
- Olmsted JB. Analysis of cytoskeletal structure using blot-purified monospecific antibodies. Methods Enzymol. 1986;134:467–472. doi: 10.1016/0076-6879(86)34112-0. [DOI] [PubMed] [Google Scholar]
- Remi Shih NR, McDonald KA, Jackman AP, Girbes T, Iglesias R. Bifunctional plant defense enzymes with chitinase and ribosome inactivating activities from Trichosanthes kirilowii cell cultures. Plant Sci. 1997;130:145–150. [Google Scholar]
- Roberts WK, Selitrennikoff CP. Isolation and characterization of two antifungal proteins from barley. Biochim Biophys Acta. 1986;880:161–170. doi: 10.1016/0304-4165(86)90076-0. [DOI] [PubMed] [Google Scholar]
- Roncuzzi L, Gasperi-Campani A. DNA-nuclease activity of the single chain ribosome inactivating proteins dianthin 30, saporin 6 and gelonin. FEBS Lett. 1996;392:16–20. doi: 10.1016/0014-5793(96)00776-4. [DOI] [PubMed] [Google Scholar]
- Savary BJ, Flores HE. Biosynthesis of defense-related proteins in transformed root cultures of Trichosanthes kirilowii Maxim. var japonicum (Kitam.) Plant Physiol. 1994;106:1195–1204. doi: 10.1104/pp.106.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumbaum A, Mauch F, Vogeli U, Boller T. Plant chitinases are potent inhibitors of fungal growth. Nature. 1986;324:365–367. [Google Scholar]
- Shewry PR. Plant storage proteins. Biol Rev. 1995;70:375–426. doi: 10.1111/j.1469-185x.1995.tb01195.x. [DOI] [PubMed] [Google Scholar]
- Sperling CR, King SR (1988) Andean tuber crops: worldwide potential. In J Janick, JE Simon, eds, Advances in New Crops: Research, Development, Economics. Oregon Timber Press, Portland, OR, pp 428–435
- Stirpe F, Barbieri L. Ribosome-inactivating proteins up to date. FEBS Lett. 1986;195:1–8. doi: 10.1016/0014-5793(86)80118-1. [DOI] [PubMed] [Google Scholar]
- Stirpe F, Barbieri L, Batelli MG, Soria M, Lappi DA. Ribosome inactivating proteins from plants: present status and future prospects. Bio/Technology. 1992;10:405–412. doi: 10.1038/nbt0492-405. [DOI] [PubMed] [Google Scholar]
- Strickland JA, Orr GL, Walsh TA. Inhibition of Diabrotica larval growth by patatin, the lipid hydrolase from potato tubers. Plant Physiol. 1995;109:667–674. doi: 10.1104/pp.109.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr GE (1986) In J Shively, ed, Methods in Protein Characterization. Human Press, New York, pp 155–194
- Tumer NE, Hwang D-J, Bonness M. C-terminal deletion mutant of pokeweed antiviral protein inhibits viral infection but does not depurinate ribosomes. Proc Natl Acad Sci USA. 1997;94:3866–3871. doi: 10.1073/pnas.94.8.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma HN, Kymar V. Prevention of potato plants from viruses and insect vectors. J Indian Potato Assoc. 1979;6:157–161. [Google Scholar]
- Vivanco JM (1997) Efecto inhibitorio de los extractos de Mirabilis jalapa en contra de PVX, PVY y PSTVd. Thesis. Universidad Nacional Agraria La Molina, Lima
- von Graevenitz A. Human diseases due to Serratia: infection and colonization with Serratia. In: von Graevenitz A, Rubin SJ, editors. The Genus Serratia. Boca Raton, FL: CRC Press; 1980. pp. 167–186. [Google Scholar]
- Wong R, Ng TB, Chan SH, Dong TX, Yeung HW. Characterization of Mirabilis jalapa antiviral protein: a ribosome inactivating protein from Mirabilis jalapa L. Biochem Int. 1992;28:585–593. [PubMed] [Google Scholar]
- Yeh K, Chen J, Lin M, Chen Y, Lin C. Functional activity of sporamin from sweet potato (Ipomoea batatas Lam.): a storage protein with trypsin inhibitor activity. Plant Mol Biol. 1997;33:565–570. doi: 10.1023/a:1005764702510. [DOI] [PubMed] [Google Scholar]
- Zoubenko O, Uckun F, Hur Y, Chet I, Tumer N. Plant resistance to fungal infection by nontoxic pokeweed antiviral protein mutants. Nat Biotechnol. 1997;15:992–996. doi: 10.1038/nbt1097-992. [DOI] [PubMed] [Google Scholar]