Abstract
We cloned a cDNA for a gibberellin-induced ribonuclease (RNase) expressed in barley (Hordeum vulgare) aleurone and the gene for a second barley RNase expressed in leaf tissue. The protein encoded by the cDNA is unique among RNases described to date in that it contains a novel 23-amino acid insert between the C2 and C3 conserved sequences. Expression of the recombinant protein in tobacco (Nicotiana tabacum) suspension-cultured protoplasts gave an active RNase of the expected size, confirming the enzymatic activity of the protein. Analyses of hormone regulation of expression of mRNA for the aleurone RNase revealed that, like the pattern for α-amylase, mRNA levels increased in the presence of gibberellic acid, and its antagonist abscisic acid prevented this effect. Quantitative studies at early times demonstrated that cycloheximide treatment of aleurone layers increased mRNA levels 4-fold, whereas a combination of gibberellin plus cycloheximide treatment was required to increase α-amylase mRNA levels to the same extent. These results are consistent with loss of repression as an initial effect of gibberellic acid on transcription of those genes, although the regulatory pathways for the two genes may differ.
The plant hormone GA plays a key role in the regulation of expression of genes involved in many different plant functions (Huttly and Phillips, 1995). In conjunction with its antagonist ABA, GA3 controls the germination of monocotyledonous seeds, including the intensively studied events involved in its effects on the cereal aleurone layer (Jones and Jacobsen, 1991). Although the role of GA3 in inducing expression of the α-amylase gene families has received considerable and detailed attention in recent years, earlier experiments showed a connection between the presence of GA3 and the induction of numerous other hydrolytic enzymes, including RNase (Chrispeels and Varner, 1967) and DNase (Taiz and Starks, 1977; Brown and Ho, 1986; Brown et al., 1988). α-Amylase gene expression in cereal aleurone has been a model system for studying the effects of GA3 and ABA on transcription. To date all models for mechanisms by which GA3 increases and ABA suppresses transcription in cereal aleurone are derived from studies of α-amylase promoters (Gubler and Jacobsen, 1992; Huttly et al., 1992; Lanahan et al., 1992; Rogers and Rogers, 1992), although transient expression assays have demonstrated GA3 transcriptional effects on promoters from protease (Cejudo et al., 1992) and (1–3,1–4)-β-glucanase (Wolf, 1992), genes that are expressed in aleurone cells during germination.
Although RNases may be present in many different compartments in plant cells (Green, 1994), our interests are in RNases, as the one described here, that enter the secretory pathway. These RNases are homologous to the extracellular RNase T2 from the fungus Aspergillus oryzae (Kawata et al., 1988). In general, proteins enter the secretory pathway by cotranslational translocation into the ER lumen and will follow a default pathway of secretion to the cell exterior unless they contain structural determinants causing them to be retained or diverted to another organelle (Okita and Rogers, 1996). RNases secreted to the cell exterior include those with functions in self-incompatibility (Matton et al., 1994) and functions somehow related to wound responses (Ye and Droste, 1996). Alternatively, RNases could be sorted from the secretory pathway to lytic or protein storage vacuoles (Neuhaus and Rogers, 1998); RNase MC purified from gourd (Homordica charantia) seeds presumably would be an example of the latter (Ide et al., 1991). The function of RNases in vacuoles is engimatic because they should be separated in those organelles from potential substrate molecules in the cell cytoplasm or in the extracellular space. One potential role would be in autophagic vacuoles that are induced by Suc starvation, where cytoplasm and cytoplasmic organelles are engulfed and degraded (Aubert et al., 1996; Moriyasu and Ohsumi, 1996). A similar role might pertain for RNases that are induced by phosphate starvation or by tissue senescence (Taylor et al., 1993; Bariola et al., 1994; Köck et al., 1995; Dodds et al., 1996).
In germinating cereal grains RNases are thought to contribute to nutrient mobilization during digestion of the dead starchy endosperm (Green, 1994). Ingle and Hageman (1965) reported two RNase activities with different pH optima in the germinating seeds of corn and found that the increase in RNase activity required protein synthesis. Dry barley (Hordeum vulgare) seeds were the source for two RNases and a nuclease characterized by Pietrzak et al. (1980), who found sufficient similarity in the two analyzed RNases to result in identical reactions in a double immunodiffusion test and to show similar substrate specificities.
There are two separate classes of enzymes induced by GA treatment of aleurone layers that degrade RNA. The first is a Zn-containing metalloenzyme that digests both DNA and RNA (Brown and Ho, 1986, 1987); the second class includes at least two separate activities that appear to be RNA specific (Brown and Ho, 1986). Both activities were secreted from aleurone cells into the incubation medium, although the peaks of accumulation occurred after 72 h of hormone treatment (Brown and Ho, 1986). It is interesting that Chrispeels and Varner (1967) found that RNase activity induced by GA accumulated in aleurone layers for 24 h and then was progressively released into the medium. Release of the enzyme appeared to be an active process because it could be prevented by treatment with protein and RNA synthesis inhibitors (Chrispeels and Varner, 1967). It would be reasonable to expect secretion of the enzyme, where it could contribute to starchy endosperm degradation, but it is not clear where the enzyme might be sequestered before the time secretion occurs.
As a second hydrolytic enzyme induced by GA3 in barley aleurone, the RNase described here provides a means by which to test the universality of the conclusions based on the α-amylase model regarding the effects of GA on transcription. Two general models have been offered to explain how GA3 could increase transcription from α-amylase gene promoters. The first proposes that GA3 causes an increase in the abundance of mRNA for a Myb transcription factor, GAMyb, and synthesis of increased amounts of the GAMyb protein, which, in turn, specifically binds to the GARE and is responsible for the increase in transcription. In this role, GAMyb is proposed to be the “master regulator” of the system (Gubler et al., 1995, 1997). A second model proposes that a transcriptional repressor, HRT, interacts with the GARE sequences and prevents transcription in the absence of GA3: The primary action of that hormone is to cause loss of transcriptional repression, perhaps by physical displacement of the HRT protein (Raventós et al., 1998). This model would not exclude the possibility that additional transcription factors could then be recruited to the derepressed promoter by GA3 to further increase the level of expression. One advantage of the second model is that it would explain the antagonistic effects of ABA by proposing that ABA would stabilize the repressor on the promoter (Raventós et al., 1998). It will be important to test the α-amylase-derived models by discerning hormonal regulation of other genes expressed in aleurone cells, and results presented here represent a step toward that goal.
Here we describe the cloning of a cDNA for a GAR-RNase expressed in barley aleurone but not in leaf tissue. The predicted protein sequence of the RNase includes a canonical signal peptide; therefore, the enzyme should enter the secretory pathway. Its structure, however, is unique compared with other plant RNases because it contains a novel 23-amino acid insertion. Detailed studies of early times following treatment with GA3 and with the protein synthesis inhibitor Chx indicate the existence of two separate mechanisms that result in increases of mRNA for the GAR-RNase and for high-pI α-amylase.
MATERIALS AND METHODS
Cloning and rDNA Techniques
Methods for cloning, DNA sequencing, and blot hybridization were described previously (Khursheed and Rogers, 1988). A cDNA for a barley (Hordeum vulgare L.) RNase (pSR360) was isolated from a library prepared from GA3-treated aleurone layers of barley var Himalaya using Lambda-Zap (Stratagene). The library (a gift from Dr. John Mundy, University of Copenhagen, Denmark) was screened using a protein expression strategy to find proteins interacting with a multimer of a known α-amylase promoter cis-acting element sequence. This clone appeared to be positive in the screen, but sequencing revealed its homology to known RNases. A gene for a different RNase was recovered from a barley genomic library (var Igri) in a Lambda-Fix II vector (Stratagene) using the full-length RNase cDNA as a probe. Analysis of the protein sequence for structural features was performed using the BCM Protein Secondary Structure Prediction Program (accessible at http://dot.imgen.bcm.tmc.edu:9331/; Kim C. Worley, Human Genome Center, Baylor College of Medicine).
Southern-Blot Analysis
Southern-blot hybridization of restriction enzyme digests of DNA from barley cvs Morex and Steptoe was kindly performed by D. Kudrna and A. Kleinhofs (Department of Crop and Soil Sciences, Washington State University, Pullman), as described previously (Kleinhofs et al., 1993).
Northern-Blot Analysis
Preparation of total RNA from aleurone and leaves, electrophoresis, and blotting were as described previously using 20 μg of RNA per lane (Rogers, 1985). For repeat probes blots were boiled in 0.1× SSC and 0.1% SDS for 10 min between hybridizations. Probes included the full-length RNase cDNA, the high-pI α-amylase cDNA-coding sequence (Rogers, 1985), and the coding sequence for a PAPI present in aleurone, the abundance of which is not affected by either GA3 or ABA (Mundy and Rogers, 1986). In Chx experiments the inhibitor was added concurrently with hormone addition. Washing conditions included a final wash in 0.1× SSC and 0.1% SDS at 65°C for 30 min. Blots were analyzed using a phosphor imager (model 445SI, Molecular Dynamics, Sunnyvale, CA) with Imagequant software (Macintosh). Barley cv Himalaya seeds, 1996 and 1998 crops, were purchased from the Department of Agronomy, Washington State University.
Transient Expression in Tobacco Suspension-Cultured Protoplasts
For expression in protoplasts, the aleu-RNase cDNA was placed between the BamHI and SacI sites in pBI221 (Jefferson et al., 1987). Culture of tobacco (Nicotiana tabacum cv Xanthi) TxD cells, protoplast preparation, and transient expression of constructs under control of the cauliflower mosaic virus 35S promoter after electroporation were described previously (Holwerda et al., 1992). Control cells were electroporated with unmodified pBI221. Twenty-four hours after electroporation, protoplasts (approximately 0.4 mL packed volume) and medium (10 mL) were separated by centrifugation at 200g. Protoplasts were dissolved directly in 0.4 mL of 2× sample buffer, and proteins in 0.5 mL of medium were precipitated with acetone and taken up in 50 μL of sample buffer. The presence of RNase activity in each fraction was assayed after electrophoresis through an acrylamide gel containing RNA as described previously (Brown and Ho, 1986), except that the incubation buffer was 0.1 m sodium succinate, pH 5.5, 0.01 m KCl, and 1 mm Cys and the incubation temperature was 50°C.
RESULTS
Sequence Information
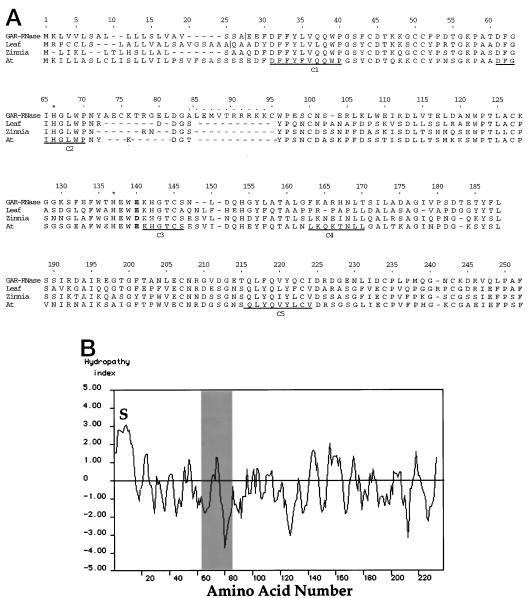
The predicted protein sequence encoded by pSR360 is clearly that of an RNase, as shown by its homology to other known plant RNases (Fig. 1A; numbering is for reference only and does not represent residue numbers for any of the sequences). Accordingly, we have identified the clone as GAR-RNase (accession no. AF000939). Based on the amino acid sequence, the expected molecular mass of the mature GAR-RNase protein, minus the signal peptide, is 24.8 kD and the expected pI is 5.14, making it likely to correspond to the 25-kD RNase identified by Brown and Ho (1986). GAR-RNase contains the five highly conserved RNase regions (Green, 1994; Fig. 1A, C1–C5), as well as the two pairs of Cys residues (positions 100 and 127 and 207 and 234) that form disulfide linkages characteristic of T2 RNases in fungi (Kawata et al., 1988). Other highly conserved amino acids include the third active-site His (position 137) and the Glu (position 140, bold print; Green, 1994). GAR-RNase contains an insert of 23 amino acids, with a third pair of Cys residues, which is not found in previously described RNases and has no detectable homology to any sequences in the current database. The unique region does contain a central DGA, which is common to other sequences, and a hydrophobic region, followed by a strongly hydrophilic one (Fig. 1B). The residues indicated with dots above have a high likelihood of forming an α-helix (data not presented). Its hydrophilic nature indicates that the helical structure is likely to be on the protein surface. By homology, the position of the insert corresponds to residue 52 of RNase Rh (Kurihara et al., 1992), which, according to its crystal structure, is on the surface of the molecule.
Figure 1.
Sequence analysis. A, A comparison is presented of predicted amino acid sequences of barley aleurone RNase (GAR-RNase) (accession no. AF000939) and the leaf RNase (Leaf) encoded by the gene isolated from a barley genomic library (accession no. AF000940) and known sequences from the database for RNases of zinnia (accession no. U19924; Zinnia) and Arabidopsis (accession no. U05206; At). Numbering begins with the initial Met. Asterisks indicate active-site His residues, and conserved Glu is shown in bold. Five highly conserved regions are underlined and labeled C1 to C5. A 23-amino acid insert unique to GAR-RNase is present from residues 73 to 95. Dots above residues indicate a region that is predicted to form an α-helix. Vertical lines show positions of predicted signal peptide cleavage. B, Kyte-Doolittle hydropathy plot of the predicted protein sequence of GAR-RNase cDNA (Kyte and Doolittle, 1982). The area highlighted in gray is the 23-amino acid insert unique to GAR-RNase. Numbering of amino acids is the same as in A. S, Hydrophobic region corresponding to signal peptide.
We also identified a barley genomic clone encoding a different RNase (accession no. AF000940). The predicted sequence of the protein encoded by this gene is also presented in Figure 1A (Leaf). As shown from the sequence comparison, it lacks the insert found in the cDNA and is more closely related to the zinnia and Arabidopsis RNases than is GAR-RNase. When compared with GAR-RNase, there is only 41% sequence identity, indicating that the cDNA and the gene encode different members of the RNase family. As shown below, this RNase is expressed in leaf but not aleurone tissue. The predicted size of the RNase gene product is 22.4 kD, which is somewhat smaller than the 25.9-kD size determined by Lantero and Klosterman (1973) for their purified leaf RNase. In addition to the sequences shown in Figure 1A, the GenBank database contains an expressed sequence tag clone from barley (accession no. L43983), which represents a partial RNase cDNA containing a large deletion 5′ to the C5 region and an extended 3′ region (data not shown). This presumably represents a third, entirely different type of barley RNase. The predicted signal peptide cleavage sites for both GAR-RNase and the leaf RNase, based on the ExPASy program (Heinrik et al., 1997), are indicated on Figure 1A by vertical lines.
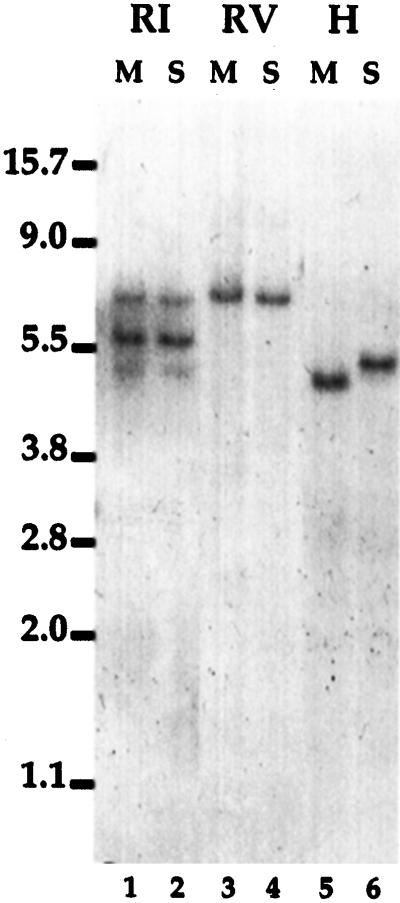
The results of Southern-blot hybridizations using the cDNA as a probe and washing at high stringency indicate that this RNase belongs to a small gene family with one or two other genes closely related to that encoding the cDNA. This is illustrated in Figure 2, where restriction digests of DNA from barley cvs Morex (lanes M) and Steptoe (lanes S) are presented. Digestions with EcoRV and HindIII (lanes 3–6), enzymes that do not cut within the coding sequence of the cDNA, gave single hybridizing bands for both DNAs. In contrast, digestion with EcoRI, an enzyme that cuts within the cDNA-coding sequence, gave three bands with the same pattern in both DNAs: a faint band <5 kb, a strong band of approximately 6 kb, and an intermediate band of approximately 7 kb (lanes 1 and 2). Since two bands could be explained by the EcoRI site in the cDNA sequence, the third presumably is derived from another, homologous gene. This estimate is consistent with the identification of two RNases expressed in aleurone (Brown and Ho, 1986).
Figure 2.
Southern blot probed with GAR-RNase cDNA. DNA from barley cvs Morex (lanes M) and Steptoe (lanes S) was digested with EcoRI (RI; lanes 1 and 2), EcoRV (RV; lanes 3 and 4), or HindIII (H; lanes 5 and 6), electrophoresed, transferred to a membrane, and hybridized with the GAR-RNase cDNA probe. Sizes of molecular mass markers (in kb) are indicated to the left.
Enzymatic Activity of GAR-RNase Expressed as a Recombinant Protein
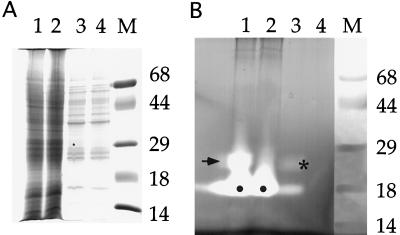
We used transient expression in tobacco suspension-cultured protoplasts to establish that the GAR-RNase protein, as predicted from its sequence, was an enzymatically active RNase. An assay was used with which RNase activity is determined in situ after denaturation of proteins with SDS and electrophoresis through an acrylamide gel containing RNA (Brown and Ho, 1986). Activity is shown as cleared bands when RNA remaining in the gel is stained after incubation. As shown in Figure 3B, lane 2, control protoplasts transfected with plasmid pBI221 expressing Escherichia coli GUS contained an RNase activity that migrated at approximately 18 kD (indicated by a dot), whereas medium from those protoplasts (lane 4) lacked detectable activity. In contrast, protoplasts transfected with the GAR-RNase construct (lane 1) not only had the approximately 18-kD activity (dot) but also a similar amount migrating at approximately 26 kD (arrow). In addition, a lesser amount of this approximately 26-kD activity was present in the medium from these protoplasts (asterisk). The fact that a similar proportion of the endogenous, approximately 18-kDa activity was also present in the medium indicates that both may have resulted from a small amount of protoplast lysis. As shown in Figure 3A, the amounts of protein in the protoplast extracts (lanes 1 and 2) and in the medium samples (lanes 3 and 4) for the two sets were similar. Therefore, the presence of the approximately 26-kD RNase activity was not an artifact of unequal loading but was, instead, due to specific expression of GAR-RNase in the test cells. Although the approximately 26-kD size of the RNase activity is an estimate limited by the fact that intramolecular disulfide bonds were not reduced before electrophoresis of the samples, it fits well with the size of GAR-RNase predicted from the cDNA sequence.
Figure 3.
Expression of recombinant GAR-RNase in tobacco suspension-cultured protoplasts. A, Coomassie blue-stained gel from SDS-PAGE. B, RNase activity gel. Lanes M, Molecular mass markers with size in kD to the right. Lanes 1, Cell extract from protoplasts expressing GAR-RNase; lanes 2, extract from protoplasts expressing E. coli GUS; lanes 3, medium from GAR-RNase-expressing cells; and lanes 4, medium from control cells. Dots indicate approximately 18-kD RNase activity in both cell extracts, arrow indicates approximately 26-kD RNase activity in extract from GAR-RNase-expressing cells, and asterisk indicates approximately 26-kD RNase activity in medium from GAR-RNase-expressing cells.
Analysis of mRNA Expression Patterns
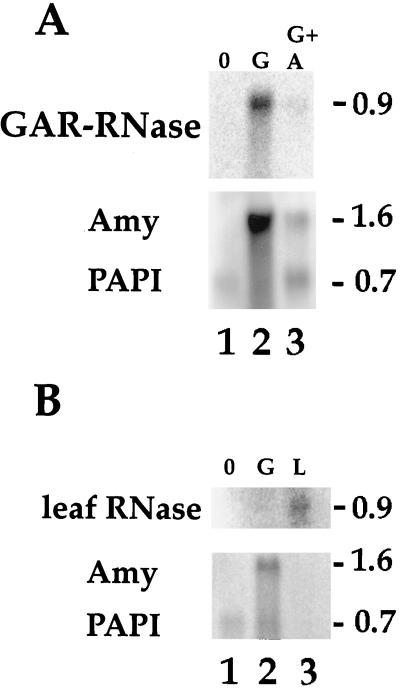
We used aleurone-specific PAPI mRNA as an internal standard, because its abundance in aleurone is not altered with hormone treatments (Mundy and Rogers, 1986). Blots were sequentially hybridized with an RNase probe and then combined α-amylase and PAPI cDNAs; therefore, the expression patterns of RNase and α-amylase mRNAs could be compared directly. Northern blots prepared from barley aleurone total RNA (Fig. 4A) showed that mRNA for GAR-RNase was at essentially undetectable levels in untreated layers (lane 1), as was true for high-pI α-amylase mRNA (Rogers, 1985). Neither probe hybridized to leaf RNA (data not shown). In a manner similar to high-pI α-amylase mRNA, the GAR-RNase mRNA level was substantially increased after 24 h of exposure to 10−6 m GA3 (lane 2), and incubation with 10−5 m ABA reduced the GA3-mediated mRNA increase by >80% for both (lane 3). The presence of similar amounts of 0.7 kb of PAPI mRNA in each of the lanes indicates that the differences observed for GAR-RNase and α-amylase hybridizations in the different samples could not be explained by RNA gel-loading variations. The specificity of the pattern observed for GAR-RNase mRNA was confirmed by using the RNase genomic clone as a control. A second northern blot (Fig. 4B) was probed with an approximately 600-bp SalI fragment containing a portion of the coding sequence of the leaf RNase gene. These results demonstrate that this RNase was expressed in leaf (lane 3) but not in aleurone (lanes 1 and 2) and the results establish the tissue specificity of expression of each of the gene types.
Figure 4.
Hormone effects on mRNA expression. A, Hybridization with the GAR-RNase probe. Total RNA was isolated from barley aleurone layers treated for 24 h with no hormone (0; lane 1), with GA3 (G; lane 2), or with GA plus ABA (G + A, lane 3). Following electrophoresis, RNA was transferred to nitrocellulose and blots were probed sequentially. First, the blot was probed with the full-length GAR-RNase cDNA. After the signal was collected by phosphor imager analysis, the blot was stripped and probed with a mixture of high-pI α-amylase cDNA (Amy) and PAPI probes. Sizes (in kb) are indicated to the right of each panel. B, Hybridization with barley leaf RNase probe. Total RNA blots were prepared and probed as for Figure 3A, except that here the RNase probe was an approximately 600-bp SalI fragment of coding sequence from the RNase gene. Lane 1, No hormone (0); lane 2, 24 h with GA3 (G); and lane 3, leaf RNA (L). Sizes of mRNA (in kb) are indicated to the right of each panel.
Chx Effects
We wanted to investigate mechanisms that could contribute to the GA3-induced increase in mRNA documented in Figure 4A. In other hormonally regulated systems, Chx has been used to test for the presence of a transcriptional repressor (Theologis, 1986; Franco et al., 1990; Ballas et al., 1995). If the half-life of a repressor protein is short enough, inhibition of protein synthesis by Chx should result in depletion of repressor before other positive transcription factors are depleted. As repressor is depleted, transcription should increase. Therefore, aleurone layers were incubated untreated or treated with GA3, Chx, or Chx plus GA3, and the relative amounts of GAR-RNase and high-pI α-amylase mRNAs were measured.
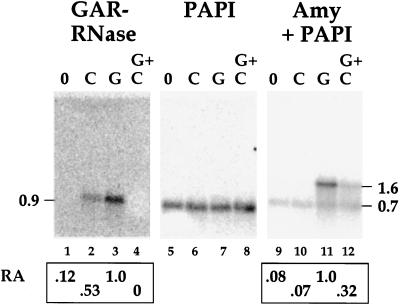
Results are presented in Figure 5 for aleurone layers incubated for 6 h under various conditions. In this experiment the same blot was hybridized sequentially with GAR-RNase, PAPI, and high-pI α-amylase probes; the blot was not stripped between hybridizations. Amounts of hybridization for the two probes were expressed as relative abundance. These numbers were corrected for variation in loading based on hybridization to the PAPI cDNA probe, using phosphor imager analysis. Little hybridization for the approximately 0.9-kb GAR-RNase was present in control RNA (Fig. 5, lane 1); in contrast, strong hybridization signals were obtained from Chx-treated (lane 2) and GA3-treated (lane 3) layers, where the signals represented 4- and 8-fold increases over the control, respectively. Surprisingly, little or no hybridization was obtained from the Chx plus GA3-treated layers (lane 4). The latter result was not an artifact of unequal sample loading or transfer to the membrane because the signal for the approximately 0.7-kb PAPI mRNA hybridization, positioned immediately below the position of GAR-RNase, was essentially equal in all of the samples (lanes 5–8). For high-pI α-amylase, little hybridization was detected in control (lane 9) and Chx-treated (lane 10) samples. GA3 treatment (lane 11) caused a 12-fold increase in mRNA over the control level, and mRNA also increased 4-fold over the control in the Chx plus GA3-treated sample (lane 12). These results were obtained with 1998 harvest seeds, and similar results were also obtained with 1996 harvest seeds.
Figure 5.
Effect of Chx on mRNA expression patterns. A blot carrying 20 μg/lane of total aleurone RNA from de-embryonated half-seeds not treated (0; lanes 1, 5, and 9), treated with 50 μm Chx (C; lanes 2, 6, and 10), treated with 10−6 m GA (G; lanes 3, 7, and 11), or treated with GA plus Chx (GC; lanes 4, 8, and 12) for 6 h was probed sequentially with cDNAs for GAR-RNase, PAPI, and high-pI α-amylase (Amy). After the blot was washed at high stringency, images were captured and quantitated with a phosphor imager. The signals for PAPI in each lane were used to correct for differences in loading. RA, Relative abundance, where the value obtained with GA was set to 1.0.
We analyzed the different results obtained with GAR-RNase and high-pI α-amylase as follows. Chx treatment was effective because it resulted in an increase in GAR-RNase mRNA; therefore, the negative result with α-amylase (lane 10) is a true difference and not the result of inadequate treatment. Similarly, the Chx treatment was sufficient to prevent GA3 induction of GAR-RNase (lane 4); therefore, the presence of a substantial signal in the Chx plus GA3 lane for α-amylase (lane 12) is not simply due to inadequate Chx treatment, but, instead, means that GA3 has an effect on α-amylase mRNA accumulation when synthesis of new protein is blocked. These results are consistent with the conclusion that loss of a short-lived protein affected mRNA levels for both GAR-RNase and high-pI α-amylase but by different mechanisms. For GAR-RNase, loss of the protein was sufficient to induce mRNA accumulation, and addition of the GA signal blocked that process. In contrast, loss of a short-lived protein alone was not sufficient to cause accumulation of α-amylase mRNA, but loss of a protein coupled with the effects of the GA signal transduction pathway was.
DISCUSSION
Here we present the protein sequences for two new barley RNases and characterize the patterns of expression at the mRNA level for each. The RNase cDNA and gene from which the protein sequences were derived provide additional tools with which to increase our understanding of both the roles of RNases in many phases of plant development and interaction of the plant hormones GA3 and ABA in regulating gene expression.
The role of S1-RNases in self-incompatability in various Nicotiana species has been the subject of intense recent study (McClure et al., 1989, 1990). The sequences of the two RNases presented here fall into a class different from S1- RNases (Green, 1994); the enzymes in this class have been termed “S-like RNases” to indicate their different functions and sequence divergence from S-RNases (Taylor et al., 1993). Phosphate starvation has been shown to induce expression of a number of different S-like RNases (Taylor et al., 1993; Bariola et al., 1994; Köck et al., 1995; Dodds et al., 1996), and it is likely that these enzymes contribute to a phosphate starvation rescue system in plants (Dodds et al., 1996). Because GAR-RNase is expressed in aleurone during germination, it may contribute to digestion of RNA in the dead starchy endosperm cells and thereby may function to help mobilize Pi for the developing embryo. Although the protein has a predicted signal peptide and almost certainly is translocated into the ER during synthesis, we have not yet obtained antibodies to GAR-RNase and therefore cannot comment about its possible intracellular or secreted location in aleurone cells. The fact that most of the GAR-RNase activity expressed in the tobacco protoplasts was intracellular may indicate that it is sorted to vacuoles, as well as possibly being secreted.
The characterization of a wound-induced RNase cDNA in zinnia (Ye and Droste, 1996) suggests an additional, defensive task for which plants might elicit RNase activity. The leaf RNase encoded by the barley genomic sequence is closely related to this type of RNase and may have a similar function. Comparison of the protein sequence encoded by GAR-RNase cDNA with those for other RNases reveals a 23-amino acid insert, which, exclusive of the conserved DGA core, appears to be unique among presently known RNases. The insertion is predicted, using the crystal structure of the Rhizopus niveus RNase Rh as a model (Kurihara et al., 1992), to be located on the surface of the molecule. The insertion contains two Cys residues; because the other Cys residues in the protein correspond to invariant residues known to participate in intramolecular disulfide bonds (Green, 1994), we predict that the two Cys residues in this insertion would be linked together. This would isolate the insertion in a looped domain with the disulfide bridge at the base. Analysis of the possible structure of this insertion indicates that one-half of the insertion would likely form an α-helix, which should protrude from the surface of the molecule. In the future, it will be of considerable interest to learn how the insertion affects the mechanism of action of this aleurone-specific RNase. Similarly, structural analysis of the protein encoded by the genomic sequence described here may prove useful in understanding what role RNases play in plant wound response.
As previously observed for most other GA3-regulated genes (Huttly and Phillips, 1995), mRNA levels for GAR-RNase were greatly increased in aleurone layers treated with GA3, and this increase was largely prevented by the simultaneous presence of ABA. Similar to GA3/ABA regulation of α-amylase gene expression, it is likely that the primary action of these hormones in regulating GAR-RNase gene expression is at the transcriptional level. However, we recently isolated a genomic clone for GAR-RNase and assayed promoter function in the particle bombardment transient expression system (Lanahan et al., 1992; Rogers and Rogers, 1992; Rogers et al., 1994). The promoter has three potential GARE sequences within −500 nucleotides but lacks all other sequences found in amylase GA-response complexes. Its promoter, assayed as either a −500- or a −3000-nucleotide construct, gives very little transcription above baseline, and such a small amount of response to GA3 that assays 6 times longer than usual (Rogers and Rogers, 1992) are required to detect it (data not presented). Stepwise truncation of the promoter and mutation of potential GARE sequences did not change this result. At present we do not know how GA3 regulates GAR-RNase mRNA abundance.
In this regard, the effects of Chx observed in our studies may help in understanding mechanisms for transcriptional regulation. Chx treatment of aleurone layers caused a 4-fold increase in GAR-RNase mRNA over a 6-h incubation, under conditions in which GA3 treatment gave an 8-fold increase. In addition, Chx plus GA3 caused a 4-fold increase in high-pI α-amylase mRNA abundance under conditions in which GA3 treatment alone gave a 12-fold increase. These results in general provide support for a recently proposed model by which GA3/ABA might regulate transcription (Raventós et al., 1998), where loss of repression, an early result of GA3 treatment, would allow a certain level of transcription, whereas later effects of GA signaling would increase transcription by recruiting positive factors to the promoter. Our data indicate that loss of a short-lived protein by inhibiting protein synthesis with Chx, in either the absence (for GAR-RNase) or the presence (for high-pI α-amylase), results in increased abundance of both RNase and α-amylase mRNA. These results would be consistent with a model in which loss of a short-lived transcriptional repressor allowed a certain level of transcription from the gene promoters, but we cannot exclude other possible mechanisms, such as effects on mRNA stability.
The effects of protein synthesis inhibitors on hormone-regulated expression of plant genes was first described and carefully studied with certain auxin-regulated genes (Theologis, 1986; Franco et al., 1990; Ballas et al., 1995). That system differs greatly from the one described here. Auxin induces transcription of those genes within minutes, and the combination of auxin plus Chx caused “superinduction” of the mRNAs. These observations indicated that all of the factors necessary for maximal transcription were present within the cells before auxin treatment. In contrast, in the aleurone system used here, increased mRNA levels were detected only after several hours of treatment with GA3 (data not shown), and 6 h were required to define the Chx effect. It is likely that Chx treatment over this prolonged period, in contrast to the short times used in the auxin experiments, would also cause depletion of somewhat longer-lived positive transcription factors. The effects seen from Chx treatment would reflect the relative half-lives of a repressor versus positive factors. If the half-life of the repressor was only moderately less than that of the positive factors, a result similar to what we present here would be observed. This may explain why in previous studies with a less-sensitive hybridization system (Muthukrishnan et al., 1983) an effect with Chx was not observed.
ACKNOWLEDGMENTS
We thank Kay Walker-Simmons for her gift of biologically active ABA, Lynn Holappa for valuable assistance with preparation of northern blots, and Liwen Jiang for transfecting tobacco suspension-cultured protoplasts. Special thanks are due to Dave Kudrna and Andy Kleinhofs for providing the Southern-blot data.
Abbreviations:
- Chx
cycloheximide
- GARE
GA response element
- GAR-RNase
GA-regulated RNase
- PAPI
lipid transfer protein
Footnotes
This research was supported by the Department of Energy (grant no. DE-FG 95ER 20165).
LITERATURE CITED
- Aubert S, Gout E, Bligny R, Marty-Mazars D, Barrieu F, Alabouvette J, Marty F, Douce R. Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: control by the supply of mitochondria with respiratory substrates. J Cell Biol. 1996;133:1251–1263. doi: 10.1083/jcb.133.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Wong L-M, Malcolm K, Theologis A. Two auxin-responsive domains interact positively to induce expression of the early indoleacetic acid-inducible gene PS-IAA4/5. Proc Natl Acad Sci USA. 1995;92:3483–3487. doi: 10.1073/pnas.92.8.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J. 1994;6:673–685. doi: 10.1046/j.1365-313x.1994.6050673.x. [DOI] [PubMed] [Google Scholar]
- Brown PH, Ho T-HD. Barley aleurone layers secrete a nuclease in response to gibberellic acid. Plant Physiol. 1986;82:801–806. doi: 10.1104/pp.82.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PH, Ho T-HD. Biochemical properties and hormonal regulation of barley nuclease. Eur J Biochem. 1987;168:357–364. doi: 10.1111/j.1432-1033.1987.tb13427.x. [DOI] [PubMed] [Google Scholar]
- Brown PH, Mecham RP, Ho T-HD. Hormonal regulation of barley nuclease: investigation using a monoclonal antibody. Plant Cell Environ. 1988;11:747–753. [Google Scholar]
- Cejudo FJ, Ghose TK, Stabel P, Baulcombe DC. A gibberellin-regulated gene from wheat with sequence homology to cathepsin B of mammalian cells. Plant J. 1992;2:937–948. [PubMed] [Google Scholar]
- Chrispeels MJ, Varner JE. Gibberellic acid-enhanced synthesis and release of α-amylase and ribonuclease by isolated barley aleurone layers. Plant Physiol. 1967;42:398–406. doi: 10.1104/pp.42.3.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Clarke AE, Newbigin E. Molecular characterisation of an S-like RNase of Nicotiana alata that is induced by phosphate starvation. Plant Mol Biol. 1996;31:227–238. doi: 10.1007/BF00021786. [DOI] [PubMed] [Google Scholar]
- Franco AR, Gee MA, Guilfoyle TJ. Induction and superinduction of auxin-responsive mRNAs with auxin and protein synthesis inhibitors. J Biol Chem. 1990;265:15845–15849. [PubMed] [Google Scholar]
- Green PJ. The ribonucleases of higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:421–445. [Google Scholar]
- Gubler F, Jacobsen JV. Gibberellin-responsive elements in the promoter of a barley high-pI α-amylase gene. Plant Cell. 1992;4:1435–1441. doi: 10.1105/tpc.4.11.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV. Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell. 1995;7:1879–1891. doi: 10.1105/tpc.7.11.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Watts RJ, Kalla R, Jacobsen JV. GAMyb. A transcription factor mediating gibberellin-regulated gene expression in aleurone cells of barley (abstract no. 1493) Plant Physiol. 1997;114:S-286. [Google Scholar]
- Heinrik N, Englebrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engin. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Holwerda BC, Padgett HS, Rogers JC. Proaleurain vacuolar targeting is mediated by short contiguous peptide interactions. Plant Cell. 1992;4:307–318. doi: 10.1105/tpc.4.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttly AK, Phillips AL. Gibberellin-regulated plant genes. Physiol Plant. 1995;95:310–317. [Google Scholar]
- Huttly AK, Phillips AL, Tregear JW. Localization of cis elements in the promoter of a wheat α-Amy2 gene. Plant Mol Biol. 1992;19:903–911. doi: 10.1007/BF00040523. [DOI] [PubMed] [Google Scholar]
- Ide H, Kimura M, Arai M, Funatsu G. The complete amino acid sequence of ribonuclease from the seeds of bitter gourd (Momordica charantia) FEBS Lett. 1991;284:161–164. doi: 10.1016/0014-5793(91)80675-s. [DOI] [PubMed] [Google Scholar]
- Ingle J, Hageman RH. Metabolic changes associated with the germination of corn. II. Nucleic acid metabolism. Plant Physiol. 1965;40:48–53. doi: 10.1104/pp.40.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanaugh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RL, Jacobsen JV. Regulation of synthesis and transport of secreted proteins in cereal aleurone. Int Rev Cytol. 1991;126:49–88. doi: 10.1016/s0074-7696(08)60682-8. [DOI] [PubMed] [Google Scholar]
- Kawata Y, Sakiyama F, Tamaoki H. Amino acid sequence of ribonuclease T2 from Aspergillus oryzae. Eur J Biochem. 1988;176:683–697. doi: 10.1111/j.1432-1033.1988.tb14331.x. [DOI] [PubMed] [Google Scholar]
- Khursheed B, Rogers JC. Barley α-amylase genes: quantitative comparison of steady-state mRNA levels from individual members of the two different families expressed in aleurone cells. J Biol Chem. 1988;263:18953–18960. [PubMed] [Google Scholar]
- Kleinhofs A, Kilian A, Saghai Maroof MA, Biyashev RM, Hayes P, Chen FQ, Lapitan N, Fenwick A, Blake TK, Kanazin V and others. A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor Appl Genet. 1993;86:705–712. doi: 10.1007/BF00222660. [DOI] [PubMed] [Google Scholar]
- Köck M, Löffler A, Abel S, Glund K. cDNA structure and regulatory properties of a family of starvation-induced ribonucleases from tomato. Plant Mol Biol. 1995;27:477–485. doi: 10.1007/BF00019315. [DOI] [PubMed] [Google Scholar]
- Kurihara H, Mitsui Y, Ohgi K, Irie M, Mizuno H, Nakamura KT. Crystal and molecular structure of RNase Rh, a new class of microbial ribonuclease from Rhizopus niveus. FEBS Lett. 1992;306:189–192. doi: 10.1016/0014-5793(92)80997-u. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lanahan MB, Ho T-HD, Rogers SW, Rogers JC. A gibberellin response complex in cereal α-amylase gene promoters. Plant Cell. 1992;4:203–211. doi: 10.1105/tpc.4.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantero OJ, Klosterman HJ. Purification and properties of barley leaf ribonuclease. Phytochemistry. 1973;12:775–784. [Google Scholar]
- Matton DP, Nass N, Clarke AE, Newbigin E. Self-incompatibility: how plants avoid illegitimate offspring. Proc Natl Acad Sci USA. 1994;91:1992–1997. doi: 10.1073/pnas.91.6.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Gray JE, Anderson MA, Clarke AE. Self-incompatability in Nicotiana alata involves degradation of pollen rRNA. Nature. 1990;347:757–760. [Google Scholar]
- McClure BA, Haring V, Ebert PR, Anderson MA, Simpson RJ, Sakiyama F, Clarke AE. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature. 1989;342:955–958. doi: 10.1038/342955a0. [DOI] [PubMed] [Google Scholar]
- Moriyasu Y, Ohsumi Y. Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol. 1996;111:1233–1241. doi: 10.1104/pp.111.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy J, Rogers JC. Selective expression of a probable amylase/protease inhibitor in barley aleurone cells: comparison to the barley amylase/subtilisin inhibitor. Planta. 1986;169:51–63. doi: 10.1007/BF01369775. [DOI] [PubMed] [Google Scholar]
- Muthukrishnan S, Chandra GR, Maxwell ES. Hormonal control of α-amylase gene expression in barley: studies using a cloned cDNA probe. J Biol Chem. 1983;258:2370–2375. [PubMed] [Google Scholar]
- Neuhaus JM, Rogers JC. Sorting of proteins to vacuoles in plant cells. Plant Mol Biol. 1998;38:127–144. [PubMed] [Google Scholar]
- Okita TW, Rogers JC. Compartmentation of proteins in the endomembrane system of plant cells. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:327–350. doi: 10.1146/annurev.arplant.47.1.327. [DOI] [PubMed] [Google Scholar]
- Pietrzak M, Cudny H, Maluszynski M. Purification and properties of two ribonucleases and a nuclease from barley seeds. Biochim Biophys Acta. 1980;614:102–112. doi: 10.1016/0005-2744(80)90171-0. [DOI] [PubMed] [Google Scholar]
- Raventós D, Skriver K, Schlein M, Karnahl K, Rogers SW, Rogers JC, Mundy J. HRT, a novel zinc finger, transcriptional repressor from barley. J Biol Chem. 1998;273:23313–23320. doi: 10.1074/jbc.273.36.23313. [DOI] [PubMed] [Google Scholar]
- Rogers JC. Two barley α-amylase gene familes are regulated differently in aleurone cells. J Biol Chem. 1985;260:3731–3738. [PubMed] [Google Scholar]
- Rogers JC, Lanahan MB, Rogers SW. The cis-acting gibberellin response complex in high-pI α-amylase gene promoters: requirement of a coupling element for high-level transcription. Plant Physiol. 1994;105:151–158. doi: 10.1104/pp.105.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JC, Rogers SW. Definition and functional implications of gibberellin and abscisic acid cis-acting hormone response complexes. Plant Cell. 1992;4:1443–1451. doi: 10.1105/tpc.4.11.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L, Starks JE. Gibberellic acid enhancement of DNA turnover in barley aleurone cells. Plant Physiol. 1977;60:182–189. doi: 10.1104/pp.60.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CB, Bariola PA, DelCardayré SB, Raines RT, Green PJ. RNS2: a senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc Natl Acad Sci USA. 1993;90:5118–5122. doi: 10.1073/pnas.90.11.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologis A. Rapid gene regulation by auxin. Annu Rev Plant Physiol. 1986;37:407–438. [Google Scholar]
- Wolf N. Structure of the genes encoding Hordeum vulgare (1–3,1–4)-β-glucanase isoenzymes I and II and functional analysis of their promoters in barley aleurone protoplasts. Mol Gen Genet. 1992;234:33–42. doi: 10.1007/BF00272342. [DOI] [PubMed] [Google Scholar]
- Ye ZH, Droste DL. Isolation and characterization of cDNAs encoding xylogenesis-associated and wound-induced ribonucleases in Zinnia elegans. Plant Mol Biol. 1996;30:697–709. doi: 10.1007/BF00019005. [DOI] [PubMed] [Google Scholar]