Abstract
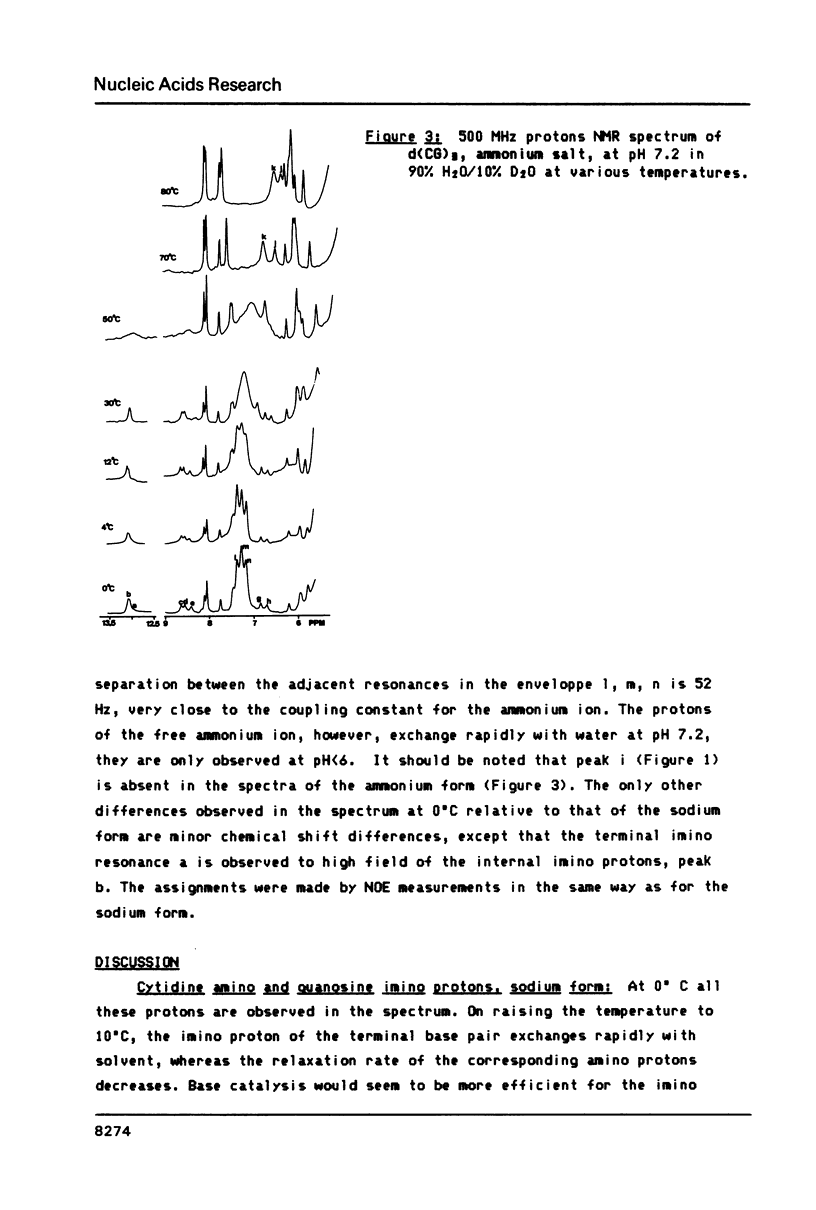
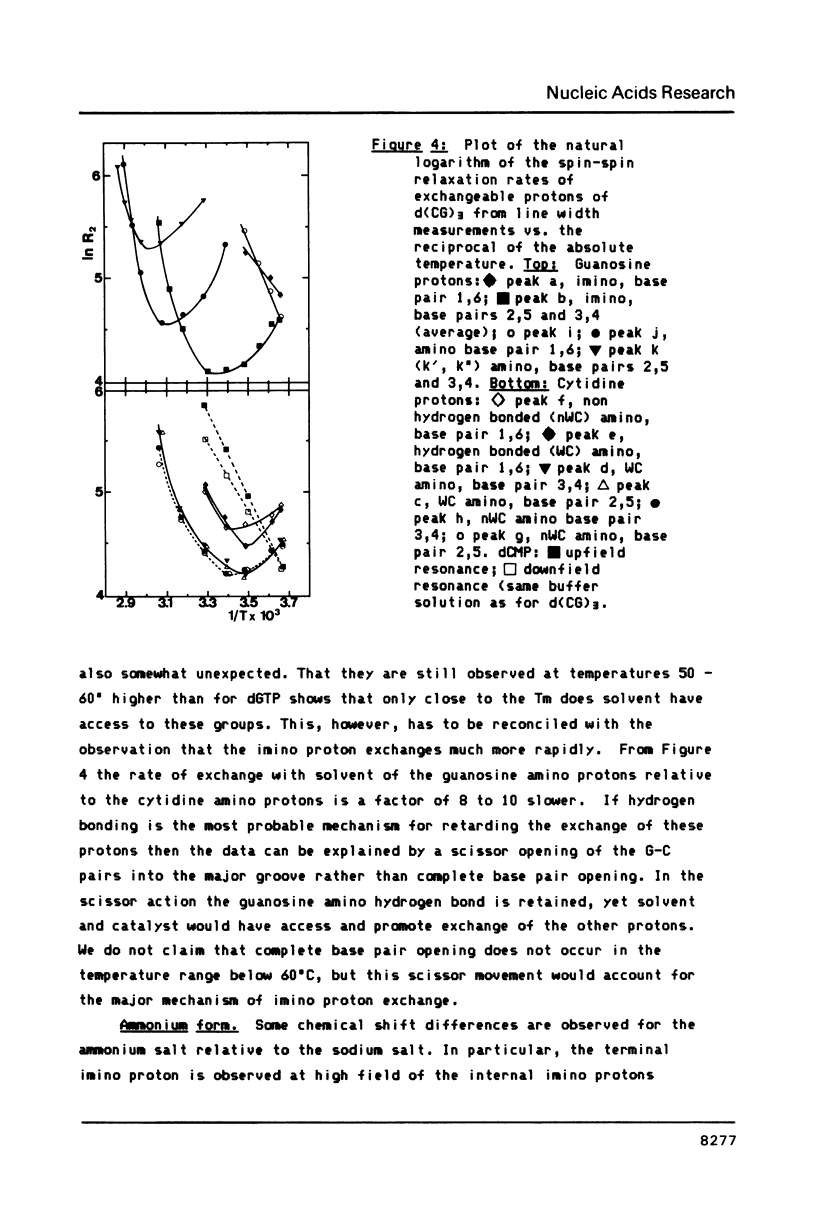
All exchangeable protons in a short DNA helix, d(CG)3 sodium salt, have been studied by proton nuclear magnetic resonance. The cytidine and guanosine amino protons have been assigned for the first time. As a function of temperature the cytidine amino protons and the imino protons behave very similarly, their relaxation is dominated by exchange with solvent above 30 degrees C. The guanosine amino protons, however, show that helix opening can only be described by a multistate model. The most rapid process observed is probably a twist about the helix axis which lengthens or breaks the guanosine amino hydrogen bond and allows rotation of the amino group. The second fastest process is a scissor opening into the major groove which gives rise to solvent exchange with the imino and cytidine amino protons. The slowest process observed is the complete base pair opening in which the guanosine amino protons also exchange with solvent. For the ammonium salt of the oligonucleotide, a specific ammonium ion complex is observed which at low temperature may catalyze exchange of the guanosine amino protons with the protons of the ammonium ion, but retards exchange with solvent. The complex appears to be specific for the sequence d(CpG).
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clore G. M., Gronenborn A. M. A nuclear-Overhauser-enhancement study of the solution structure of a double-stranded DNA undecamer comprising a portion of the specific target site for the cyclic-AMP-receptor protein in the gal operon. Sequential resonance assignment. Eur J Biochem. 1984 May 15;141(1):119–129. doi: 10.1111/j.1432-1033.1984.tb08166.x. [DOI] [PubMed] [Google Scholar]
- Early T. A., Kearns D. R., Hillen W., Wells R. D. A 300- and 600-MHz proton nuclear magnetic resonance investigation of a 12 base pair deoxyribonucleic acid restriction fragment: relaxation behavior of the low-field resonances in water. Biochemistry. 1981 Jun 23;20(13):3756–3764. doi: 10.1021/bi00516a014. [DOI] [PubMed] [Google Scholar]
- Early T. A., Kearns D. R., Hillen W., Wells R. D. A 300-MHz proton nuclear magnetic resonance investigation of deoxyribonucleic acid restriction fragments: dynamic properties. Biochemistry. 1981 Jun 23;20(13):3764–3769. doi: 10.1021/bi00516a015. [DOI] [PubMed] [Google Scholar]
- Feigon J., Denny W. A., Leupin W., Kearns D. R. Proton nuclear magnetic resonance investigation of the conformation and dynamics in the synthetic deoxyribonucleic acid decamers d(ATATCGATAT) and d(ATATGCATAT). Biochemistry. 1983 Dec 6;22(25):5930–5942. doi: 10.1021/bi00294a037. [DOI] [PubMed] [Google Scholar]
- Feigon J., Wright J. M., Denny W. A., Leupin W., Kearns D. R. Application of multiple-pulse H-NMR techniques to the study of two synthetic DNA decamers. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):207–217. doi: 10.1101/sqb.1983.047.01.026. [DOI] [PubMed] [Google Scholar]
- Frechet D., Cheng D. M., Kan L. S., Ts'o P. O. Nuclear Overhauser effect as a tool for the complete assignment of nonexchangeable proton resonances in short deoxyribonucleic acid helices. Biochemistry. 1983 Oct 25;22(22):5194–5200. doi: 10.1021/bi00291a020. [DOI] [PubMed] [Google Scholar]
- Hilbers C. W., Patel D. J. Proton nuclear magnetic resonance investigations of the nucleation and propagation reactions associated with the helix-coil transition of d-ApTpGpCpApT in H2O solution. Biochemistry. 1975 Jun 17;14(12):2656–2660. doi: 10.1021/bi00683a015. [DOI] [PubMed] [Google Scholar]
- Kan L. S., Borer P. N., Ts'o P. O. Conformation and interaction of short nucleic acid double-stranded helices. II. Proton magnetic resonance studies on the hydrogen-bonded NH-N protons of ribosyl ApApGpCpUpU helix. Biochemistry. 1975 Nov 4;14(22):4864–4869. doi: 10.1021/bi00693a013. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Fratini A. V., Drew H. R., Dickerson R. E. Ordered water structure around a B-DNA dodecamer. A quantitative study. J Mol Biol. 1983 Jan 5;163(1):129–146. doi: 10.1016/0022-2836(83)90033-5. [DOI] [PubMed] [Google Scholar]
- Mandal C., Kallenbach N. R., Englander S. W. Base-pair opening and closing reactions in the double helix. A stopped-flow hydrogen exchange study in poly(rA).poly(rU). J Mol Biol. 1979 Dec 5;135(2):391–411. doi: 10.1016/0022-2836(79)90443-1. [DOI] [PubMed] [Google Scholar]
- McConnell B., Seawell P. C. Amino protons of cytosine. Chemical exchange, rotational exchange, and salt-induced proton magnetic resonance chemical shifts of aqueous 2',3'-cyclic cytidine monophosphate. Biochemistry. 1973 Oct 23;12(22):4426–4434. doi: 10.1021/bi00746a020. [DOI] [PubMed] [Google Scholar]
- McConnell B. The amino 1H resonances of oligonucleotide helices: d(CGCG). J Biomol Struct Dyn. 1984 Jun;1(6):1407–1421. doi: 10.1080/07391102.1984.10507528. [DOI] [PubMed] [Google Scholar]
- Pardi A., Morden K. M., Patel D. J., Tinoco I., Jr Kinetics for exchange of imino protons in the d(C-G-C-G-A-A-T-T-C-G-C-G) double helix and in two similar helices that contain a G . T base pair, d(C-G-T-G-A-A-T-T-C-G-C-G), and an extra adenine, d(C-G-C-A-G-A-A-T-T-C-G-C-G). Biochemistry. 1982 Dec 7;21(25):6567–6574. doi: 10.1021/bi00268a038. [DOI] [PubMed] [Google Scholar]
- Patel D. J. d-CpCpGpG and d-GpGpCpC self-complementary duplexes: Nmr studies of the helix-coil transition. Biopolymers. 1977 Aug;16(8):1635–1656. doi: 10.1002/bip.1977.360160804. [DOI] [PubMed] [Google Scholar]
- Raszka M., Kaplan N. O. Association by hydrogen bonding of mononucleotides in aqueous solution. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2025–2029. doi: 10.1073/pnas.69.8.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raszka M. Mononucleotides in aqueous solution: proton magnetic resonance studies of amino groups. Biochemistry. 1974 Oct 22;13(22):4616–4622. doi: 10.1021/bi00719a023. [DOI] [PubMed] [Google Scholar]
- Wong Y. P., Kearns D. R., Reid B. R., Shulman R. G. Investigation of exchangeable protons and the extent of base pairings in yeast phenylalanine transfer RNA by high resolution nuclear magnetic resonance. J Mol Biol. 1972 Dec 30;72(3):725–740. doi: 10.1016/0022-2836(72)90187-8. [DOI] [PubMed] [Google Scholar]



