Abstract
Here, we demonstrate that the reduction in leaf K+ observed in a mutant previously identified in an ionomic screen of fast neutron mutagenized Arabidopsis thaliana is caused by a loss-of-function allele of CPR5, which we name cpr5-3. This observation establishes low leaf K+ as a new phenotype for loss-of-function alleles of CPR5. We investigate the factors affecting this low leaf K+ in cpr5 using double mutants defective in salicylic acid (SA) and jasmonic acid (JA) signalling, and by gene expression analysis of various channels and transporters. Reciprocal grafting between cpr5 and Col-0 was used to determine the relative importance of the shoot and root in causing the low leaf K+ phenotype of cpr5. Our data show that loss-of-function of CPR5 in shoots primarily determines the low leaf K+ phenotype of cpr5, though the roots also contribute to a lesser degree. The low leaf K+ phenotype of cpr5 is independent of the elevated SA and JA known to occur in cpr5. In cpr5 expression of genes encoding various Cyclic Nucleotide Gated Channels (CNGCs) are uniquely elevated in leaves. Further, expression of HAK5, encoding the high affinity K+ uptake transporter, is reduced in roots of cpr5 grown with high or low K+ supply. We suggest a model in which low leaf K+ in cpr5 is driven primarily by enhanced shoot-to-root K+ export caused by a constitutive activation of the expression of various CNGCs. This activation may enhance K+ efflux, either indirectly via enhanced cytosolic Ca2+ and/or directly by increased K+ transport activity. Enhanced shoot-to-root K+ export may also cause the reduced expression of HAK5 observed in roots of cpr5, leading to a reduction in uptake of K+. All ionomic data presented is publically available at www.ionomicshub.org.
Introduction
Potassium (K+) is a macronutrient essential for normal plant growth and development. It participates in numerous physiological processes including regulation of enzyme activity, cell expansion, stomata movement and defence towards pathogens [1], [2]. Numerous mechanisms are known to be involved in K+ homeostasis, and many K+ channels and transporters have been identified. In plants K+ is highly mobile. Its movement into the xylem is driven by transpiration, and in the phloem by the specific requirements of tissues and organs [3], [4]. Environmental conditions also regulate the movement of K+. For example, under drought stress the concentration of K+ in the xylem decreases, possibly because of the closure of stomatal pores induced by abscisic acid (ABA) [5].
The effects of low K+ have been investigated in some depth, and it has emerged that jasmonic acid (JA) plays a central role in the response of plants to K+ shortage [6], [7]. Nevertheless, whether the application of K+ is beneficial or not in conferring resistance towards pathogens is controversial [8]. At the molecular level the role of K+ is clearer. Plant-pathogen interactions cause an increase in cytosolic Ca2+ triggering anion channel activation, plasma membrane depolarization, activation of K+ permeable efflux channels leading to enhanced K+ efflux and the initiation of the hypersensitive response (HR) [9], [10].
In this study, we describe the characterization of an A. thaliana mutant with a 10–30% reduction in leaf K+ which was previous identified in an ionomic screen of fast neutron mutagenized plants [11]. Genetic analysis revealed this mutant to be a new null allele of CPR5, a gene originally identified in two independent screens for altered response to pathogens [12], [13]. cpr5 has a high content of salicylic acid (SA) and shows constitutive expression of pathogenesis related genes (PR), as well as plant defensin PDF1.2 which is a marker of the JA-dependent pathway. Therefore, it has been suggested the CPR5 is a negative regulator of local defence response to pathogens [14]. CPR5 is also implicated in cell senescence [15]–[17], cell proliferation and trichome development [18], cell wall biogenesis [19] redox balance [20], and water relations via enhanced ABA sensitivity [21]. Here, we show that CPR5 is also associated with K+ homeostasis possibly via modulation of expression of various CNGCs and HAK5.
Materials and Methods
Plant growth and mutant screening
Fast neutron-mutagenized M2 A. thaliana seeds were purchased from Lehle Seeds (Round Rock, TX) and plants screened for their leaf elemental profile by ICP-MS [11]. Seeds of Col-0 (CS6000) and cpr5-2 (CS3770) were provided by the Arabidopsis Biological Resource Center (The Ohio State University).
For non axenic conditions, plants were grown in pots containing moist soil (Scotts Potting Medium, Scotts-Sierra Horticultural Products Company, Marysville, OH) in a climate-controlled room (temperature 19–22°C, day-night; humidity 60%; photoperiod 10–14 hours light-dark; light intensity 100±10 µmol m−2 sec−1) and bottom watered at regular intervals with a solution containing 0.25× Hoagland's macro and micronutrients [11].
For plants grown in axenic conditions, surface sterilized seeds were stratified at 4°C in the dark for five days before sowing. To measure the expression of HAK5 and AKT1 plants were grown for 2 weeks on solidified medium containing 1/20th MS salts accordingly to Cheong et al., [22] and containing 20 mM or 100 µM of KCl, and in a second experiment on a minimal medium without NH4 + [23] with 10 g L−1 UltraPure sucrose (Sigma) and solidified with 10 g L−1 pure agarose (Molecular Biology Grade, Research Products International Corp.), which contains negligible amounts of K+ (approximately 8 µg Kg−1 as determined by ICP-MS analysis). K+ was added as KCl at the final concentration of 10, 50 and 100 µM and the pH adjusted to 5.8 with Ca(OH)2. K+ content in root and shoot was measured in plants grown for 2 weeks on solidified medium containing 1/20th MS salts accordingly to Cheong et al., [22] and containing 20 mM of KCl.
Genetic analysis
cpr5-3 was outcrossed to A. thaliana Landsberg erecta (Ler-0) accession, F1 seeds were planted on 0.5× MS medium solidified with 10 g L−1 of agar and seedlings visually screened for hyponastic and early yellowing cotyledons. All seedlings of F1 generation looked wild-type and were transferred to soil and grown to produce F2 seeds. F2 plants grown on soil were visually selected for small size and yellow early senescing leaves and used to map the mutation with a positional cloning approach with single strand length polymorphism (SSLP) markers.
Quantitative real-time PCR
Total RNA was extracted with the Qiagen RNeasyPlant Mini Kit (http://www.qiagen.com) from five weeks old plants grown on soil or from two week old plants grown on plates. DNase digestion on column was performed to eliminate possible contamination with DNA. Two micrograms of total RNA were used as a template to synthesize first-strand cDNA with SuperScript VILO cDNA Synthesis Kit (InvitrogenLife Technologies, http://www.invitrogen.com). Quantitative real-time PCR was performed with the SYBR Green reagent mix in a StepOnePlus instrument according to the manufacturer's instructions (Applied Biosystems, California, USA). The expression of K+ channel/transporter genes was detected with the following set of primers: HAK5, 147 (5′-TGCTGATCTAGGTCACTTCAGTGTT-3′) and 148 (5′-AAAGCAGGATATGCGACACATG-3′); AKT1, 170 (5′-TCTAAATTGTGTTCTTCTTCTGTTGGA-3′) and 171 (5′-CCTTCCGCGTCTCTGCAA-3′); CNGC19, 137 (5′-TTCTCACTTGGTGCCTCTCTTCT-3′) and 138 (5′-AATCCCTTTGGTGGCATCTTT-3′); CNGC10, 139 (5′-TCATCATTGATCTACTCTCTATCCTTCCT-3′) and 140 (5′-TGGTTGACGCTTGGAATAACG-3′); CNGC12, 156 (5′-CAACGAACATTCAGGTTATACTCACA-3′) and 157 (5′-GCCGCTTGAATGAAGAATGC-3′); CNGC20, 160 (5′-TGCCTCGAACGCTCTTCTG-3′) and 161 (5′-CCTTTGATGGCATCCTTATCCT-3′); CNGC11, 196 (5′-GATAAAAACATGAATCTTCAGAGGAGAA-3′) and 197 (5′-CTAACACTTTTCAATTTTCCATCAACTC-3′). As an internal reference the expression of PP2A (At1g13320) was used since it showed stable expression throughout the experimental series of development, shoot and root abiotic stress, hormones, nutrient stress, light and biotic stress [24], all of which are affected by loss-of-function of CPR5. In addition to PP2A the expression of HAK5 and AKT1 was also normalized to UBQ10 (At4g05320). UBQ10 also shows stable expression in cpr5. The average value from real-time PCR measurements from at least three independent biological replicates was used to evaluate transcript abundance. Biological replicates were composed of tissues harvested from between 5 and 10 plants for analysis of shoot tissue, and between 25 and 30 plants for analysis of root tissue, from plants grown on plates. For plants grown in soil 2–3 leaves from at least 3 individual plants were used. Steady state mRNA levels were calculated relative to a reference gene and presented based on the 2−ΔCt method [25].
Determination of the elemental content of plant tissues
Plants grown in soil were non-destructively sampled by removing 1–2 leaves (approximately 3 mg dry weight), rinsed with 18 MΩ water, placed into Pyrex digestion tubes and dried at 92°C for 20 hours. Alternatively, shoots and roots were harvested from plants grown on 1/20th MS medium modified accordingly to Cheong et al. [22] as previously described. After cooling, all samples were digested with 0.7 mL concentrated nitric acid (OmniTrace, VWR) and diluted to 6.0 mL with 18 MΩ water. Acid used for digestion was spiked with gallium (Ga) to act as an internal standard to control for errors in dilution, variations in sample introduction, and plasma stability in the ICP-MS instrument. Sample sets also contained analytical blanks, standard reference material (NIST SRM 1547) digested in the same manner as the plant samples, and quantitative calibration standards. Calibration standards and standard reference material samples were included at the beginning and end of the sample sets to control for drift during the analysis. Samples were introduced into an inductively coupled plasma mass spectrometer (ICP-MS) (Elan DRCe, PerkinElmer) and analyzed for Li, B, Na, Mg, P, K, Ca, Mn, Fe, Co, Ni, Cu, Zn, As, Se, Mo and Cd. All samples were normalized to calculated weights, as determined with an iterative algorithm using the best-measured elements, the weights of the seven weighed samples, and the solution concentrations, detailed at www.ionomicshub.org.
Determination of SA content
Total SA content was quantified in leaves of five weeks old plants using a Waters Alliance HPLC system equipped with Millenium software, 2695 Separation Module, 2475 Fluorescence Detector, and 2996 Photodiode array detector. A Nova-Pack C-18 column was used with a flow rate and methanol gradient as described previously [26]. SA (Sigma; catalogue no. S-6271) was used to develop the standard curve for quantification.
Grafting
cpr5-2 and Col-0 seeds were germinated in the dark on 0.5× MS plates containing 1 mL L−1 of MS Vitamins (Caisson Laboratories, Inc.), 3 mg L−1 Benomyl (methyl 1-(butylcarbamoyl)-2-benzimidazolecarbamate; Sigma), 0.04 mg L−1 BA (6-benzylaminopurine; Sigma), 0.02 mg L−1 IAA (indole acetic acid; Sigma), and 12 g L−1 agar. Plates were held vertically and after seven days seedlings were grafted as previously described [27] and then grown for an additional seven days on plates before transfer into soil. Plants were grown for a further four weeks in soil before being analyzed for their elemental content. Plants that from a visual inspection showed adventitious roots coming from the shoot above the graft were excluded from the experiment.
Statistical analysis
ANOVA was conducted using the software CoStat 6.2 (CoHort Software, CA, USA). Separation of means was performed using LSD test at P = 0.05 significance level.
Results
Mutant identification
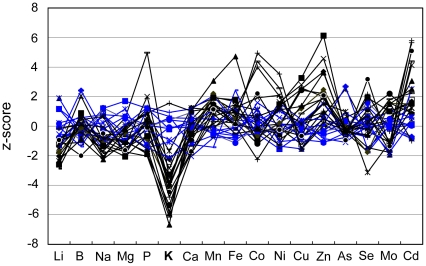
Fast neutron mutagenized A. thaliana Col-0 plants were previously screened for altered leaf elemental composition [11]. In this screen 12645 was identified as a low K+ mutant with a reduction in leaf K+ of approximately 20% compared to wild-type Col-0 (raw data are available at www.ionomicshub.org, experimental tray 229) (Fig. 1). In addition, 12645 was also smaller in size than wild-type Col-0 and developed symptoms of hypersenescence in cotyledons and mature leaves. Moreover, cotyledons of mutant plants were hyponastic when seeds were germinated in soil, as well as in plates.
Figure 1. Leaf ionomic profile of cpr5-3 (12645).
The concentration of each element was measured in leaves of five weeks old plants of both 12645 (M3 generation) (black lines; N = 21) and wild type Col-0 (blue lines; N = 12) plants. For each element given on the abscissa the correspondent z-score value is given on the ordinate. The z-score represents the number of standard deviations of the wild-type Col-0 each plant differs from the mean of the wild-type Col-0 (raw data are available at www.ionomicshub.org, experimental tray 743).
Genetic analysis
To map and define the Mendelian character of the mutation, 12645 was out-crossed to Ler-0, and F1 and F2 populations scored for mutant like plants. All F1 plants from this cross looked wild-type, while in the F2 small plants with early senescent leaves segregated with a ratio of 3:1 (wild-type to mutant phenotype), indicating that 12645 is inherited as a single recessive nuclear mutation. Small hypersenescent plants from the F2 population also showed reduced K+ content (38.3 µg g−1 dry weight) compared to those that looked wild-type (42.8 µg g−1 dry weight), indicating that the traits of early senescence and small size co-segregate with reduced leaf K+ (P<0.001).
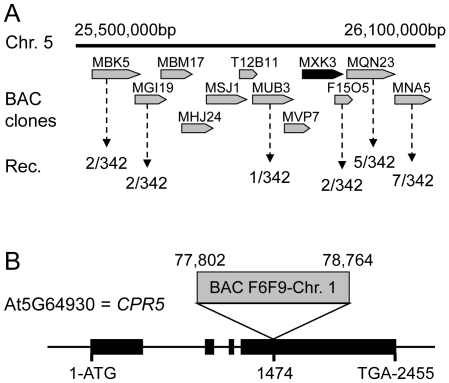
The chromosomal position of the 12645 mutation was determined using a positional cloning approach with SSLP markers in 171 F2 plants from the outcross with Ler-0. The recombinant population scored for markers in the region between nucleotides 25,500,000 and 26,100,000 on chromosome 5 revealed that only one plant had Ler-0 alleles on the BAC clone MUB3 and two plants on F15O5, which placed the mutation on the BAC clone MXK3 (Fig. 2A). DNA sequencing of this region revealed that 12645 contains a 972 base pair insertion at nucleotide 1474 in the fourth exon of the At5g64930 gene (Fig. 2B). This insertion originated from nucleotides 77,802–78,764 on chromosome 1, a region that does not contain any annotated loci. A. thaliana mutants carrying recessive loss-of-function alleles of At5g64930 have been previously and independently identified in screens for constitutive expression of pathogen resistance, and the early appearance of hypersenescence symptoms, and named cpr5-1 [13], cpr5-2 [12] and hys1 [17]. Based on the previous nomenclature we renamed 12645 as cpr5-3. As expected CPR5 expression is lost in cpr5-3 (Fig. S1A). All cpr5 mutants (cpr5-1, cpr5-2 and cpr5-3) were grown together, along with the wild-type Col-0 and the concentration of Li, B, Na, Mg, P, S, K, Ca, Mn, Fe, Co, Ni, Cu, Zn, As, Se, Mo, Cd in leaves of each genotype determined by ICP-MS (n = 10–22 replicate plants per genotype). Of all the elements measured only K+ was found to be significantly different between the wild-type Col-0 and all the cpr5 alleles (P<0.005 after Bonferroni correction for multiple testing). Wild-type Col-0 had a leaf K+ concentration of 40,067±6222 µg g−1 dry weight, compared to 27,490±3575 µg g−1 dry weight for cpr5-1, 29,751±2616 µg g−1 dry weight for cpr5-2, and 27,324±2502 µg g−1 dry for cpr5-3 (raw data are available at www.ionomicshub.org, experimental tray 1020).
Figure 2. Mapping and gene structure of cpr5-3 (12645).
A. BAC clones covering the cpr5-3 (12645) locus. Frequency of recombination is expressed as number of recombinant chromosomes over the total number of examined chromosomes. B. 972 bp insertion from BAC clone F6F9 from chromosome 1 inserted at nucleotide 1474 in the fourth exon of At5g64930 gene. Exons are shown by boxes and introns by lines.
Low leaf K+ in cpr5 is not dependent on SA or JA, and it is a unique trait of cpr5 among lesion mimic mutants
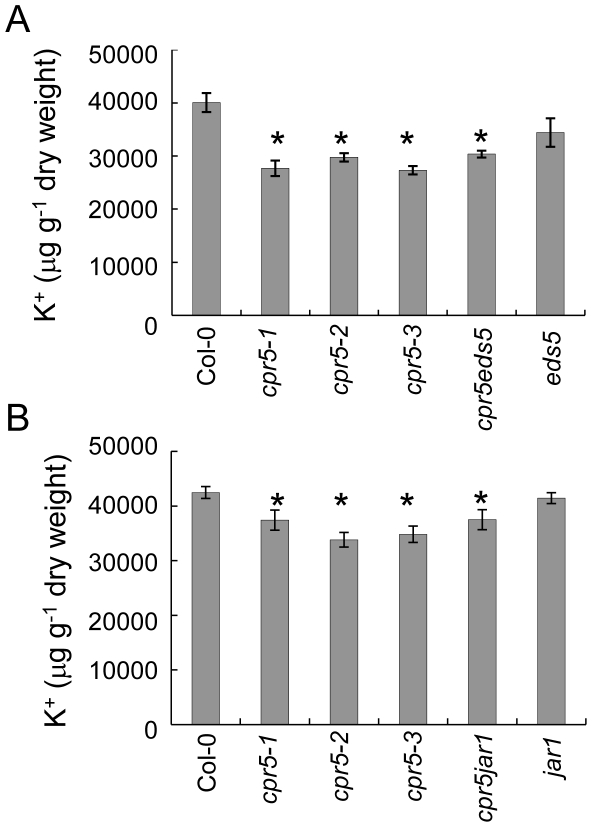
CPR5 acts as a negative regulator of local resistance to pathogens by partially repressing the accumulation of SA, so that cpr5 mutants have elevated SA content [13], [14], which we also confirmed to be the case for cpr5-3 (Fig. S1C). This raised the question, whether low leaf K+ in cpr5 results from the constitutively high SA in this mutant. To address this we measured K+ content in three different cpr5 alleles, in the single eds5-1 mutant, and in the double mutant cpr5eds5 prepared from cpr5-1 and eds5-1 [14]. The mutation in eds5-1 suppresses the biosynthesis of SA and in the double mutant cpr5eds5 the elevated SA content of cpr5-1 is returned to the level of the wild-type [14], [28], [29]. As shown in Fig. 3A, K + content in eds5-1 leaves is slightly reduced but not significantly different from Col-0 whereas in cpr5eds5, which has low SA but carries the cpr5-1 mutation, the content of K+ is similar to that of single cpr5 alleles (raw data are available at www.ionomicshub.org, experimental tray 1020).
Figure 3. K+ content in salicylic acid and jasmonic acid defective mutants.
K+ (µg g-1 of dry weight) was measured in leaves of five weeks old plants of wild-type Col-0, cpr5-1, cpr5-2, cpr5-3 and (A) eds5 and cpr5eds5, and (B) jar1 and cpr5jar1. Wild-type Col-0 was used as a control. Values represent means of independent biological replicates (between 10 and 22 plants per genotype). Error bars represent standard errors. An asterisk (*) indicates values significantly different from the mean of the wild-type Col-0 at 99% confidence interval (raw data are available at www.ionomicshub.org; experimental trays 838, 839, 1020 and 1840).
We also followed a similar approach to determine if the low leaf K+ of cpr5 is dependent on the elevated JA in this mutant [16]. The jasmonate resistant 1 (jar1) mutant is unable to conjugate JA with isoleucine to form the active jasmonoyl-L-isoleucine form of JA that is required to elicit the JA response [30], and therefore this mutant is insensitive to JA [31]. Fig. 3B shows that the double mutant cpr5jar1, prepared from cpr5-1 and jar1-1 [14], and single allelic mutants cpr5-1, cpr5-2 and cpr5-3 all share a similar reduction of leaf K+ which is not observed in the single jar1-1 mutant (raw data are available at www.ionomicshub.org; experimental tray 1840). From this we conclude that the low leaf K+ of cpr5 is not dependent on JA signalling. Taken together these experiments support the conclusion that the reduced leaf K+ of cpr5 is independent of both SA and JA.
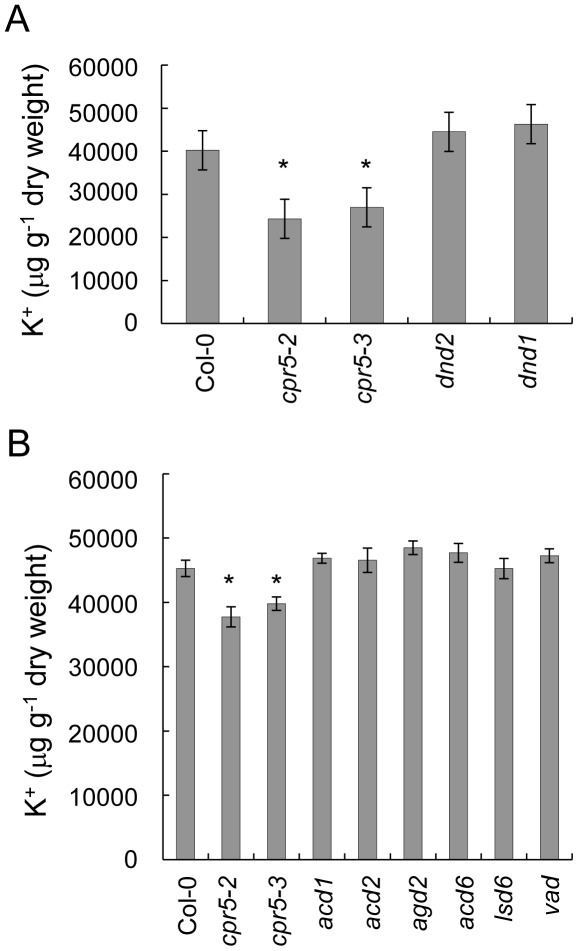
cpr5 is easily distinguishable from wild-type plants from the presence of necrotic and chlorotic spots on the leaves, a trait which characterizes plants infected by bacteria and/or fungi, as well as mutants with a constitutively active response to pathogens. Hypothesizing that low leaf K+ was associated with the response to pathogens, we measured K+ content in leaves of lesion mimic mutants (LMM) selected from the two major classes of initiation (dnd1, dnd2, agd2, acd6, lsd6) and propagation (acd1, acd2, vad1) of lesion mutants [32]. None of the lesion mimic mutants tested showed reduced leaf K+ (Fig. 4A, B). From this we conclude that the low leaf K+ observed in cpr5 is not a result of the presence of lesions.
Figure 4. K+ content in lesion mimic mutants.
K+ (µg g−1 of dry weight) content was measured in leaves of five weeks old plants of cpr5-1, cpr5-2, cpr5-3 and (A) initiation of lesion mimic mutants dnd1 and dnd2, and (B) initiation of lesion mimic agd2, acd6, lsd6, and propagation of lesion mimic mutants acd1, acd2, vad1. Wild-type Col-0 was used as a control. Values represent means of independent biological replicates (between 10 and 22 plants per genotype). Error bars represent standard errors. An asterisk (*) indicates values significantly different from wild-type Col-0 at 99% confidence interval (raw data are available at www.ionomicshub.org; experimental trays 1770 and 1771).
Low leaf K+ in cpr5 is primarily driven by the shoot but roots also play a role
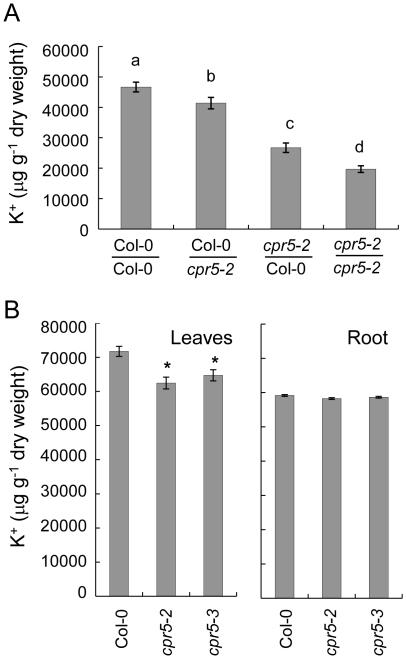
Low K+ in cpr5 leaves could be caused by reduced uptake from roots, impaired root-to- shoot translocation, or enhanced shoot-to-root circulation through the phloem. CPR5 is equally expressed in both roots and leaves (Fig. S1B) making it possible that CPR5 could contribute to K+ homeostasis in either organ. Therefore, we performed reciprocal grafting of cpr5 and wild-type Col-0 to determine in which tissue cpr5 exerts its influence. Shoots of cpr5-2 were grafted onto wild-type Col-0 roots and vice versa, grafted plants allowed to grow for four weeks in soil and the K+ content measured in leaves. When cpr5-2 shoots were grafted on wild-type Col-0 roots we observed the K+ content in leaves to be significantly reduced by 43% compared to self-grafted Col-0 (Fig. 5A). Moreover, plants with cpr5-2 shoot and wild-type Col-0 root also retained the hypersenescence phenotype of chlorotic and necrotic spots observed on leaves of non-grafted cpr5-2 plants. In contrast, plants with wild-type Col-0 shoots grafted on cpr5-2 roots showed only an 11% reduction in leaf K+ and were indistinguishable from Col-0 in respect to symptoms of hypersenescence. Interestingly, self-grafted cpr5-2 plants showed the lowest leaf K+ of all the grafting types tested, with a reduction in leaf K of 58%. These results suggest that loss-of-function of CPR5 in shoots plays a primary role in the reduced leaf K+ phenotype of cpr5, but loss-of-function of CPR5 in roots also exerts a lesser yet significant influence.
Figure 5. K+ content in cpr5 root and leaves and in cpr5 grafted plants.
A. K+ (µg g-1 of dry weight) in leaves of grafted plants transferred to soil after grafting and grown for four weeks. Values represent means of independent measurements performed on 19 to 32 different plants per graft combination. Error bars show standard errors. Bars with different letters are significantly different at P = 0.05 level (LSD test). Col-0/Col-0, self-grafted wild-type Col-0; Col-0/cpr5-2, wild-type Col-0 shoot grafted on cpr5-2 root; cpr5-2/Col-0, cpr5-2 shoot grafted on wild-type Col-0 root; cpr5-2/cpr5-2, self-grafted cpr5-2. B. K+ (µg g−1 of dry weight) in leaves and roots of wild-type Col-0, cpr5-2 and cpr5-3 plants grown for two weeks on solidified 1/20th MS medium supplemented with 20 mM of K+. Data represents the mean of 10 measurements of a pool of 10 to 15 shoots or roots per genotype grown on 15 different plates. Error bars represent standard errors. An asterisk (*) indicates values significantly different from the mean of wild-type Col-0 at P = 0.005.
To further understand the role of roots versus shoots in the low leaf K+ phenotype of cpr5 we measured the K+ content of shoot and root tissue of plants grown on solidified MS medium in plates. This experiment revealed no significant difference in the root concentration of K+ between cpr5 and Col-0, while cpr5 shoots retained the low K+ phenotype observed in plants grown in soil (Fig. 5B).
Expression of Cyclic Nucleotide Gated Channels is elevated in shoots of cpr5
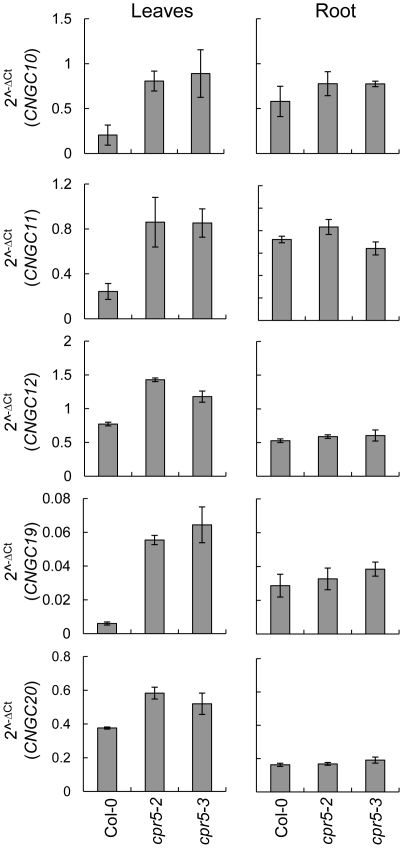
Based on the importance of the shoot in driving the reduced leaf K+ in cpr5 we investigated expression of Cyclic Nucleotide Gated Channels (CNGCs) that may be directly or indirectly involved in K+ efflux during the response to pathogens in A. thaliana. An initial survey of transcriptional data publically available in experiment 175 on Genevestigator [33] revealed that the expression of numerous CNGCs was up-regulated in cpr5 leaves. We confirmed the differences initially observed in the database using qRT-PCR, and performed similar measurements in root tissue. Our analysis revealed that steady state levels of CNGC10, CNGC11, CNGC12, CNGC19 and CNGC20 mRNA are all elevated in leaves of both cpr5-2 and cpr5-3 compared to wild-type Col-0 (Fig. 6). Analogous measurements performed in roots did not revealed any substantial differences from wild-type Col-0 for the cpr5 mutants (Fig. 6).
Figure 6. Expression of CNGCs in cpr5.
Steady state levels of CNGC10, CNGC11, CNGC12, CNGC19, CNGC20 mRNA in leaves and roots of cpr5-2, cpr5-3 and wild-type Col-0 evaluated by qRT-PCR. Seedlings were germinated and grown on 1/20th MS medium supplemented with 20 mM KCl. After two-week of growth shoot and root tissue was harvested and RNA extract for qRT-PCR. PP2A (At1g13320) was used as an endogenous reference gene for normalization across samples, and data presented as 2−ΔCt. Error bars represent standard deviations calculated following [29].
Expression of HAK5 encoding a high affinity K+ transporter is reduced in roots of cpr5
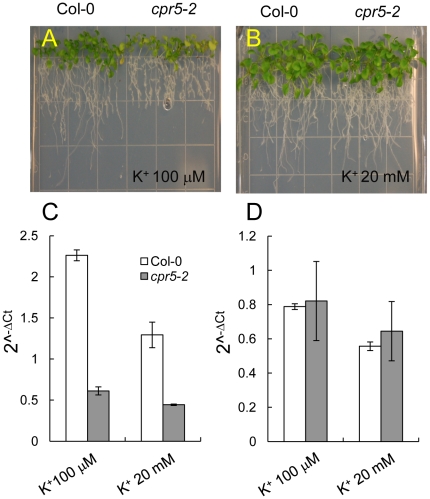
The K+ transporter HAK5 is known to contributes to K+ uptake in A. thaliana primarily at low K+ supply (0–0.25 mM) [23], [34]–[37], whereas the K+ channel AKT1 [38], [39] contributes to K+ uptake at both low and intermediate K+ supply (0.01–5 mM) [35]–[37], [40]. Above 5 mM external K+ the transport processes involved in K+ uptake are currently undefined [36]. Given the importance of both HAK5 and AKT1 in K+ uptake in A. thaliana we used qRT-PCR to quantify the steady state levels of HAK5 and AKT1 mRNA in cpr5 roots to test if altered expression of these genes may be involved in the low leaf K+ phenotype we observe in cpr5. Steady state levels of AKT1 mRNA in cpr5 where found to be the same as wild-type Col-0 after growth on medium supplemented with either high (20 mM) or low (100 µM) K+ (Fig. 7C). A slight increase in AKT1 mRNA was observed in wild-type Col-0 plants grown on medium supplemented with 100 µM K+ compared to 20 mM K+. Enhanced expression of AKT1 was not previously observed [41], though this is possibly due to the fact that the previous authors used RT-PCR to determine expression of AKT1. Interestingly, steady state levels of HAK5 mRNA were observed to be reduced in cpr5 compared to wild-type Col-0 grown in medium with either high and low K+ supply (Fig. 7B). Further, the increase in HAK5 mRNA observed in wild-type Col-0 grown on medium containing 100 µM K+ was essentially abolished in cpr5 (Fig. 7B). Interestingly, root growth of cpr5 on medium supplemented with 100 µM K+ was found to be reduced compared to wild-type (Fig. 7A). However, in medium supplemented with 20 mM K+ growth of cpr5 and wild-type was similar (Fig. 7A).
Figure 7. Expression of HAK5 and AKT1 in roots of cpr5.
Seeds were germinated on solidified 0.5× MS salts, and after five days seedlings transferred to solidified 1/20th MS medium supplemented with either 100 µM (A) or 20 mM KCl (B). Pictures were taken after two weeks of growth. Steady state levels of HAK5 (C) and AKT1 (D) mRNA were quantified using qRT-PCR in roots of wild-type Col-0 (white bars) and cpr5-2 (gray bar). RNA was extracted from roots of three independent samples generated from between 25 and 30 plants per plates. UBQ10 (At4g05320) was used as an endogenous reference gene for normalization across the samples, and data presented as 2−ΔCt. Error bars represent standard deviations calculated following [29].
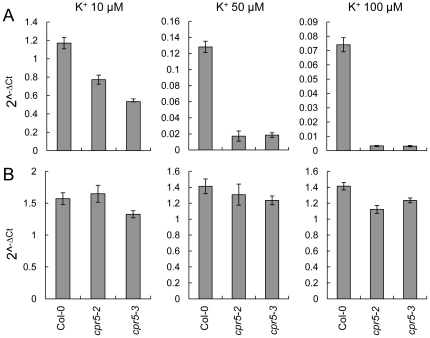
To confirm and extend these results we performed a more detailed dose response experiment with solidified medium supplemented with 10, 50, and 100 µM KCl lacking NH4 + given that NH4 + is known to suppress activity [38], [39] and expression [23] of HAK5. Pure agarose was used to solidify the growth medium to avoid any extra K+ supply [42]. In wild-type Col-0 root steady state levels of HAK5 mRNA were observed to increase as external K+ was reduced (Fig. 8A), as expected [34]. In roots of cpr5-2 and cpr5-3 steady state levels of HAK5 mRNA were also increased as the external K+ concentration was reduced (Fig. 8A). However, steady state levels of HAK5 mRNA in cpr5-2 and cpr5-3 were consistently lower then wild-type Col-0 at 10, 50 and 100 µM K+ in the growth medium (Fig. 8A). In agreement with our previous observations no consistent differences in expression of AKT1 between wild-type Col-0 and cpr5 were observed (Fig. 8B).
Figure 8. Expression of HAK5 and AKT1 in roots of cpr5 grown in the absence of NH4 +.
Wild-type Col-0, cpr5-2 and cpr5-3 seedlings were grown on solidified defined medium without NH4 + and supplemented with different concentrations of KCl (10, 50 and 100 µM). RNA was extracted from roots of two week old plants and steady state levels of HAK5 (A) and AKT1 (B) quantified using qRT-PCR. Data represents the mean of four biological replicates each composed of a pool of 5 to 10 plants. PP2A (At1g13320) was used as an endogenous reference gene for normalization across samples, and data presented as 2−ΔCt. Error bars represent standard deviations calculated following [29].
Discussion
The data presented here establishes that the wild-type allele of CPR5 is required to maintain normal K+ homeostasis in A. thaliana Col-0. We observed that loss-of-function alleles of CPR5 show a consistent and specific reduction in leaf K+ of 10–30% in plants grown in both soil or on solidified MS medium with high K+ supply. Further, this defect leads to reduced growth at low K+ supply. Genetic analysis confirmed that this reduction in leaf K+ is not dependent on the elevated SA or JA known to occur in cpr5. Further, genetic analysis confirmed that reduced leaf K+ is not a feature of lesion mimic mutants in general. Through reciprocal grafting we establish that the reduced leaf K+ observed in cpr5 is primarily driven by the shoot (74%), with the root playing a significant but smaller role (19%). Interestingly, the presence of the cpr5 allele in both roots and shoots is required to produce the full low leaf K+ phenotype, suggesting that feedback between both organs is needed. In investigating the factors that in cpr5 cause the reduction of leaf K+, we surveyed the experiment AT-175 performed on cpr5 shoots (www.genevestigator.ethz.ch) and this revealed that genes belonging to the CNGC family are highly expressed in cpr5. Using qRT-PCR we confirmed that CNGC10, CNGC11, CNGC12, CNGC19, CNGC20 are indeed highly expressed in leaves, but not roots, of cpr5. The expression of CNGC1 and CNGC13 which showed a milder increase in the AT-175 array did not show any difference when expression was analyzed by qRT-PCR. CNCG2 and CNGC4, null mutants of which (dnd1 and dnd2) have enhanced disease resistance, also did not show any misregulation when analyzed by qRT-PCR. In the interaction between plant and pathogens the recognition of factors of avirulence by the host plant induces fluxes of Ca2+ that initiate the immune response and leads to enhanced K+ efflux [10]. CNGCs are believed to be the channels that deliver the Ca2+ signal required for pathogen recognition [43], and dnd1, dnd2 and the gain of function chimeric CNGC11/12 mutant cpr22 strongly support this conclusion [44]–[50]. Moreover, heterologous expression of specific CNGCs provides direct evidence of Ca2+ transport activity [51], [52]. It is therefore possible that in cpr5 the constitutively high level of expression of CNGCs we observe leads to a persistent activation of this Ca2+ initiated signalling cascade that in turn leads to constitutively enhanced K+ efflux [10]. Such an enhanced K+ efflux could increase shoot-to-root K+ export, and may explain the leaf-driven portion of the reduced leaf K+ observed in cpr5. The transporters/channels involved in this K+ efflux are currently unknown. However, these is evidence that CNGC10 transports K+ [53], and steady state levels of CNGC10 mRNA are elevated in cpr5, making it possible that CNGC10 could play a role in the proposed enhanced K+ efflux and shoot-to-root export in cpr5.
HAK5 encodes the primary high affinity root K+ transporter in A. thaliana [23], [34]–[37], and its expression in wild-type Col-0 roots is induced at low K+ supply [34]. Significantly, cpr5 roots show a consistent reduction in the steady state levels of HAK5 mRNA under both high and low K+ supply. Though HAK5 primarily plays a role in K+ uptake at low K+ supply, it is possible that the reduced steady state levels of HAK5 mRNA in cpr5 roots we observe even at high K+ supply is responsible for the small (18%) but significant contribution of roots to the reduced leaf K+ phenotype of cpr5. It is also interesting to speculate that expression of HAK5 in cpr5 roots is suppressed by an increased flux of K+ arriving in the roots from the shoots driven by elevated expression of CNGCs in shoots.
Based on sequence analysis CPR5 encode a membrane protein with five transmembrane domains and a nuclear localization signal (NLS). CPR5 appears to localize to the nucleus [54] where it has been proposed to be targeted to the inner nuclear membrane [19], [54]. It has been further suggested that proteolytic cleavage can release the nucleosolic domain of CPR5 from the membrane to allow its participation in transcriptional processes [19], [54]. Recent studies have identified the transcription factors TPR1 and EDS1 that direct repression of expression of CNGC2 and CNGC4 [55], [56], and we speculate that CPR5 may play a similar role in the nucleus to negatively regulate the expression of the various CNGCs affected on the cpr5 mutant.
In summary, we have identified a new low leaf K+ phenotype for cpr5 that is primarily driven by the shoot, and is independent of the elevated levels of SA and JA found in this mutant. We suggest that the reduced leaf K+ of cpr5 is caused by the elevated expression of various CNGCs in shoots and the reduced expression of HAK5 in roots, driving both an enhanced K+ export from shoots and a reduced K+ uptake in roots. Our observation of altered K+ homeostasis in cpr5 may also be an important piece of evidence linking the function of CPR5 as a negative regulator of local defence responses to pathogens with the role K+ efflux plays in these responses; likely through the direct or indirect regulation of CNGCs by CPR5.
Supporting Information
CPR5 expression and salicylic acid (SA) levels in A. thaliana leaves and roots of wild-type Col-0 and cpr5. A. Steady state levels of CPR5 mRNA in wild-type Col-0 and cpr5-3 quantified using qRT-PCR. RNA was extracted from leaves of five-week old plants grown in soil. Data represents the mean of measurements of four independent biological replicates. Each biological replicate consisted of 2-3 leaves from individual plants. Errors bars represent standard deviation. B. Steady state levels of CPR5 mRNA in root and shoots of wild-type Col-0 quantified using qRT-PCR. RNA was extracted from shoots and roots of two week old wild-type Col-0 plants grown on 0.5× MS media solidified with 1% agar (w/v). Values represent mean of measurements from at least three independent replicates. Errors bars represent standard deviation. Steady state mRNA levels (A & B) are presented as 2−ΔCt. UBQ10 (At4g05320) was used as an endogenous reference gene for normalization across samples. C. SA content (mg g−1 of fresh weight) in leaves of wild-type Col-0, cpr5-2 and cpr5-3. Data represents the mean of three independent leaf samples harvested from individual plants grown in soil for five weeks. Error bars represent the standard error.
(TIF)
Acknowledgments
The authors wish to acknowledge Brett Lahner for ICP-MS analysis and Hyeong Cheol Park for salicylic acid determination. We wish to acknowledge Xinniang Dong's laboratory for providing seeds of cpr5-1, eds5-1 and cpr5eds5 and Fred Ausubel for giving us permission to use them. We also acknowledge Shannon Clarke and Luis Mur for providing seeds of cpr5jar1.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants to D.E.S. from the US National Science Foundation Arabidopsis 2010 and Plant Genome Research programs (IOS 0419695 and DBI 0701119). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Marschner H. Mineral nutrition of higher plants: New York: Academic Press. 2002. pp. 299–312.
- 2.Ashley MK, Grant M, Grabov A. Plant responses to potassium deficiencies: a role for potassium transport proteins. J Exp Bot. 2006;57:425–436. doi: 10.1093/jxb/erj034. [DOI] [PubMed] [Google Scholar]
- 3.Grignon C, Sentenac H. pH and ionic conditions in the apoplast. Annu Rev Plant Biol. 1991;42:103–128. [Google Scholar]
- 4.White P. The regulation of K+ influx into roots of rye (Secale cereale L.) seedlings by negative feedback via the K+ flux from shoot to root in the phloem. J Exp Bot. 1997;48:2063–2073. [Google Scholar]
- 5.Shabala S. Transport from root to shoot. In: Yeo A, Flowers T, editors. Plant solute transport. Blackwell Publishers; 2007. pp. 214–234. [Google Scholar]
- 6.Armengaud P, Breitling R, Amtmann A. The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol. 2004;136:2556–2576. doi: 10.1104/pp.104.046482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armengaud P, Sulpice R, Miller AJ, Stitt M, Amtmann A, et al. Multilevel analysis of primary metabolism provides new insights into the role of potassium nutrition for glycolysis and nitrogen assimilation in Arabidopsis roots. Plant Physiol. 2009;150:772–785. doi: 10.1104/pp.108.133629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amtmann A, Troufflard S, Armengaud P. The effect of potassium nutrition on pest and disease resistance in plants. Physiol Plantarum. 2008;133:682–691. doi: 10.1111/j.1399-3054.2008.01075.x. [DOI] [PubMed] [Google Scholar]
- 9.Jabs T, Tschope M, Colling C, Hahlbrock K, Scheel D. Elicitor-stimulated ion fluxes and O2- from the oxidative burst are essential components in triggering defense gene activation and phytoalexin synthesis in parsley. Proc Natl Acad Sci U S A. 1997;94:4800–4805. doi: 10.1073/pnas.94.9.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeworutzki E, Roelfsema MRG, Anschütz U, Krol E, Elzenga JTM, et al. Early signaling through the Arabidopsis pattern recognition receptors FLS2 and EFR involves Ca2+-associated opening of plasma membrane anion channels. Plant J. 2010;62:367–378. doi: 10.1111/j.1365-313X.2010.04155.x. [DOI] [PubMed] [Google Scholar]
- 11.Lahner B, Gong J, Mahmoudian M, Smith EL, Abid KB, et al. Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nature Biotech. 2003;21:1215–1221. doi: 10.1038/nbt865. [DOI] [PubMed] [Google Scholar]
- 12.Boch J, Verbsky M, Robertson T, Larkin J, Kunkel B. Analysis of resistance gene-mediated defense responses in Arabidopsis thaliana plants carrying a mutation in CPR5. Mol Plant Microbe In. 1998;11:1196–1206. [Google Scholar]
- 13.Bowling S, Clarke J, Liu Y, Klessig D, Dong X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X. Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell. 2000;12:2175–2190. doi: 10.1105/tpc.12.11.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jing H, Sturre M, Hille J, Dijkwel P. Arabidopsis onset of leaf death mutants identify a regulatory pathway controlling leaf senescence. Plant J. 2002;32:51–63. doi: 10.1046/j.1365-313x.2002.01400.x. [DOI] [PubMed] [Google Scholar]
- 16.Jing HC, Anderson L, Sturre MJ, Hille J, Dijkwel PP. Arabidopsis CPR5 is a senescence-regulatory gene with pleiotropic functions as predicted by the evolutionary theory of senescence. J Exp Bot. 2007;58:3885–3894. doi: 10.1093/jxb/erm237. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida S, Ito M, Nishida I, Watanabe A. Identification of a novel gene HYS1/CPR5 that has a repressive role in the induction of leaf senescence and pathogen-defense responses in Arabidopsis thaliana. Plant J. 2002;29:427–437. doi: 10.1046/j.0960-7412.2001.01228.x. [DOI] [PubMed] [Google Scholar]
- 18.Kirik V, Bouyer D, Schobinger U, Bechtold N, Herzog M, et al. CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein. Curr Biology. 2001;11:1891–1895. doi: 10.1016/s0960-9822(01)00590-5. [DOI] [PubMed] [Google Scholar]
- 19.Brininstool G, Kasili R, Simmons LA, Kirik V, Hulskamp M, et al. Constitutive Expressor Of Pathogenesis-related Genes5 affects cell wall biogenesis and trichome development. BMC Plant Biol. 2008;8:58. doi: 10.1186/1471-2229-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jing HC, Hebeler R, Oeljeklaus S, Sitek B, Stuhler K, et al. Early leaf senescence is associated with an altered cellular redox balance in Arabidopsis cpr5/old1 mutants. Plant Biol. 2008;10(Suppl 1):85–98. doi: 10.1111/j.1438-8677.2008.00087.x. [DOI] [PubMed] [Google Scholar]
- 21.Gao G, Zhang S, Wang C, Yang X, Wang Y, et al. Arabidopsis CPR5 Independently Regulates Seed Germination and Postgermination Arrest of Development through LOX Pathway and ABA Signaling. PLoS One. 2011;6:e19406. doi: 10.1371/journal.pone.0019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, et al. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 2007;52:223–239. doi: 10.1111/j.1365-313X.2007.03236.x. [DOI] [PubMed] [Google Scholar]
- 23.Qi Z, Hampton CR, Shin R, Barkla BJ, White PJ, et al. The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J Exp Bot. 2008;59:595–607. doi: 10.1093/jxb/erm330. [DOI] [PubMed] [Google Scholar]
- 24.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Freeman JL, Garcia D, Kim D, Hopf A, Salt DE. Constitutively elevated salicylic acid signals glutathione-mediated nickel tolerance in Thlaspi nickel hyperaccumulators. Plant Physiol. 2005;137:1082–1091. doi: 10.1104/pp.104.055293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, et al. Natural variants of AtHKT1 enhance Na accumulation in two wild populations of Arabidopsis. PLoS Genetics. 2006;2:e210. doi: 10.1371/journal.pgen.0020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nawrath C, Metraux J. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers E, Ausubel F. Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell. 1997;9:305–316. doi: 10.1105/tpc.9.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staswick PE. JAZing up jasmonate signaling. Trends Plant Sci. 2008;13:66–71. doi: 10.1016/j.tplants.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 31.Staswick PE, Su W, Howell SH. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. PNAS. 1992;89:6837–6840. doi: 10.1073/pnas.89.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorrain S, Vailleau F, Balagué, Roby D. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 2003;8:263–271. doi: 10.1016/S1360-1385(03)00108-0. [DOI] [PubMed] [Google Scholar]
- 33.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis Microarray Database and Analysis Toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gierth M, Maser P, Schroeder JI. The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 2005;137:1105–1114. doi: 10.1104/pp.104.057216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubio F, Nieves-Cordones M, Aleman F, Martinez V. Relative contribution of AtHAK5 and AtAKT1 to K+ uptake in the high-affinity range of concentrations. Physiol Plantarum. 2008;134:598–608. doi: 10.1111/j.1399-3054.2008.01168.x. [DOI] [PubMed] [Google Scholar]
- 36.Rubio F, Alemán F, Nieves-Cordones M, Martínez V. Studies on Arabidopsis athak5, atakt1 double mutants disclose the range of concentrations at which AtHAK5, AtAKT1 and unknown systems mediate K uptake. Physiol Plant. 2010;139:220–228. doi: 10.1111/j.1399-3054.2010.01354.x. [DOI] [PubMed] [Google Scholar]
- 37.Pyo YJ, Gierth M, Schroeder JI, Cho MH. High-Affinity K+ transport in Arabidopsis: AtHAK5 and AKT1 are vital for seedling establishment and postgermination growth under low-potassium conditions. Plant Physiol. 2010;153:863–875. doi: 10.1104/pp.110.154369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirsch RE, Lewis BD, Spalding EP, Sussman MR. A role for the AKT1 potassium channel in plant nutrition. Science. 1998;280:918–921. doi: 10.1126/science.280.5365.918. [DOI] [PubMed] [Google Scholar]
- 39.Spalding EP, Hirsch RE, Lewis DR, Qi Z, Sussman MR, et al. Potassium uptake supporting plant growth in the absence of AKT1 channel activity: Inhibition by ammonium and stimulation by sodium. J Gen Physiol. 1999;113:909–918. doi: 10.1085/jgp.113.6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu J, Li H, Chen L, Wang Y, Liu L, et al. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell. 2006;125:1347–1360. doi: 10.1016/j.cell.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 41.Lagarde D, Basset M, Lepetit M, Conejero G, Gaymard F, et al. Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 1996;9:195–203. doi: 10.1046/j.1365-313x.1996.09020195.x. [DOI] [PubMed] [Google Scholar]
- 42.Jain A, Poling MD, Smith AP, Nagarajan VK, Lahner B, et al. Variations in the composition of gelling agents affect morphophysiological and molecular responses to deficiencies of phosphate and other nutrients. Plant Physiol. 2009;150:1033–1049. doi: 10.1104/pp.109.136184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moeder W, Urquhart W, Ung H, Yoshioka K. The role of cyclic nucleotide-gated ion channels in plant immunity. Mol Plant. 2010;4:442–452. doi: 10.1093/mp/ssr018. [DOI] [PubMed] [Google Scholar]
- 44.Balague C, Lin B, Alcon C, Flottes G, Malmstrom S, et al. HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell. 2003;15:365–379. doi: 10.1105/tpc.006999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clough SJ, Fengler KA, Yu IC, Lippok B, Smith RK, Jr, et al. The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. PNAS. 2000;97:9323–9328. doi: 10.1073/pnas.150005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jurkowski GI, Smith RK, Jr, Yu IC, Ham JH, Sharma SB, et al. Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol Plant Microbe In. 2004;17:511–520. doi: 10.1094/MPMI.2004.17.5.511. [DOI] [PubMed] [Google Scholar]
- 47.Yoshioka K, Kachroo P, Tsui F, Sharma SB, Shah J, et al. Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J. 2001;26:447–459. doi: 10.1046/j.1365-313x.2001.2641039.x. [DOI] [PubMed] [Google Scholar]
- 48.Yoshioka K, Moeder W, Kang HG, Kachroo P, Masmoudi K, et al. The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. Plant Cell. 2006;18:747–763. doi: 10.1105/tpc.105.038786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ali R, Zielinski R, Berkowitz G. Expression of plant cyclic nucleotide-gated cation channels in yeast. J Exp Bot. 2006;57:125–138. doi: 10.1093/jxb/erj012. [DOI] [PubMed] [Google Scholar]
- 50.Guo KM, Babourina O, Christopher DA, Borsics T, Rengel Z. The cyclic nucleotide-gated channel, AtCNGC10, influences salt tolerance in Arabidopsis. Physiol Plantarum. 2008;134:499–507. doi: 10.1111/j.1399-3054.2008.01157.x. [DOI] [PubMed] [Google Scholar]
- 51.Frietsch S, Wang Y-F, Sladek C, Poulsen LR, Romanowsky SM, et al. A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. Proc Natl Acad Sci U S A. 2007;104:14531–14536. doi: 10.1073/pnas.0701781104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leng Q, Mercier RW, Yao W, Berkowitz GA. Cloning and first functional characterization of a plant cyclic nucleotide-gated cation channel. Plant Physiol. 1999;121:753–761. doi: 10.1104/pp.121.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, Borsics T, Harrington HM, Christopher DA. Arabidopsis AtCNGC10 rescues potassium channel mutants of E. coli, yeast and Arabidopsis and is regulated by calcium/calmodulin and cyclic GMP in E. coli. Funct Plant Biol. 2005;32:643–653. doi: 10.1071/FP04233. [DOI] [PubMed] [Google Scholar]
- 54.Perazza D, Laporte F, Balagué C, Chevalier F, Remo S, et al. Plant Physiol; 2011. GeBP/GPL transcription factors regulate a subset of CPR5-dependent processes. doi: 10.1104/111.179804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, et al. Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. pollen Proc Natl Acad Sci U S A. 2010;107:13960–13965. doi: 10.1073/pnas.1002828107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.García AV, Blanvillain-Baufumé S, Huibers RP, Wiermer M, Li G, et al. Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog. 2010;6:e1000970. doi: 10.1371/journal.ppat.1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CPR5 expression and salicylic acid (SA) levels in A. thaliana leaves and roots of wild-type Col-0 and cpr5. A. Steady state levels of CPR5 mRNA in wild-type Col-0 and cpr5-3 quantified using qRT-PCR. RNA was extracted from leaves of five-week old plants grown in soil. Data represents the mean of measurements of four independent biological replicates. Each biological replicate consisted of 2-3 leaves from individual plants. Errors bars represent standard deviation. B. Steady state levels of CPR5 mRNA in root and shoots of wild-type Col-0 quantified using qRT-PCR. RNA was extracted from shoots and roots of two week old wild-type Col-0 plants grown on 0.5× MS media solidified with 1% agar (w/v). Values represent mean of measurements from at least three independent replicates. Errors bars represent standard deviation. Steady state mRNA levels (A & B) are presented as 2−ΔCt. UBQ10 (At4g05320) was used as an endogenous reference gene for normalization across samples. C. SA content (mg g−1 of fresh weight) in leaves of wild-type Col-0, cpr5-2 and cpr5-3. Data represents the mean of three independent leaf samples harvested from individual plants grown in soil for five weeks. Error bars represent the standard error.
(TIF)