Abstract
The leptin gene has received intensive attention and scientific investigation for its importance in energy homeostasis and reproductive regulation in mammals. Furthermore, study of the leptin gene is of crucial importance for public health, particularly for its role in obesity, as well as for other numerous physiological roles that it plays in mammals. In the present work, we report the identification of novel leptin genes in 4 species of Cetacea, and a comparison with 55 publicly available leptin sequences from mammalian genome assemblies and previous studies. Our study provides evidence for positive selection in the suborder Odontoceti (toothed whales) of the Cetacea and the family Phocidae (earless seals) of the Pinnipedia. We also detected positive selection in several leptin gene residues in these two lineages. To test whether leptin and its receptor evolved in a coordinated manner, we analyzed 24 leptin receptor gene (LPR) sequences from available mammalian genome assemblies and other published data. Unlike the case of leptin, our analyses did not find evidence of positive selection for LPR across the Cetacea and Pinnipedia lineages. In line with this, positively selected sites identified in the leptin genes of these two lineages were located outside of leptin receptor binding sites, which at least partially explains why co-evolution of leptin and its receptor was not observed in the present study. Our study provides interesting insights into current understanding of the evolution of mammalian leptin genes in response to selective pressures from life in an aquatic environment, and leads to a hypothesis that new tissue specificity or novel physiologic functions of leptin genes may have arisen in both odontocetes and phocids. Additional data from other species encompassing varying life histories and functional tests of the adaptive role of the amino acid changes identified in this study will help determine the factors that promote the adaptive evolution of the leptin genes in marine mammals.
Introduction
Leptin is an adipose tissue-derived circulating hormone with multiple functions pivotal in regulating energy homeostasis and reproductive functions in mammals [1], [2], [3], [4], [5], [6], [7]. Increases in leptin signal levels act on the central nervous system to inhibit food intake and/or regulate energy expenditure to maintain constancy in adipose mass [1], [4]. Mutations in the leptin gene are associated with a myriad of hormonal and metabolic alterations [1], [8]. In humans and mice, leptin deficiency has been shown to cause obesity and diabetes [1], [9], [10], [11], [12].
Evidence of rapid sequence evolution in leptin genes has been documented in seals [13], plateau pikas [14], [15] and primates [16], [17], [18], [19], [20], [21], [22], [23], [24]. It has been hypothesized that leptin function in these mammals may have undergone modification [13], [22]. For example, Siltberg and Liberles (2002) proposed that positive selection in primate leptin may be driven by dietary and/or reproductive changes during the evolution of primates relative to other mammals [22]. Therefore, the study of leptin genes in mammals encompassing different evolutionary histories will contribute to our understanding of other physiological roles of leptin.
Marine mammals are a diverse group of 120 species including cetaceans (whales, dolphins, and porpoises), sirenians (manatees and dugong), and pinnipeds (true seals, sea lions and walrus) [25]. Compared with their terrestrial counterparts, marine mammals are remarkable and evolutionarily significant in terms of their adaptation to an aquatic environment, yet there have been few studies of the genetic changes underlying adaptation to an aquatic lifestyle [26], [27], [28]. Interestingly, a recent study by Hammond et al. (2005) revealed a substantially larger number of amino acid substitutions in seal leptin sequences than in other mammals, and also demonstrated unusual expression of leptin in the seal lung [13]. They proposed a role for leptin in seal respiratory physiology in addition to its common role in energy balance as in other mammals. These observations raised important questions about (1) whether excessive amino acid substitutions in seal leptin were due to positive selection and (2) our lack of information about the evolutionary history of leptin genes in other marine mammalian lineages.
Therefore, we sequenced leptin genes from four cetacean species representing three families of the suborder Odontoceti (toothed whales) and one family of the suborder Mysticeti (baleen whales). In addition, 55 publicly available leptin sequences from mammalian genome assemblies and previous studies, including 3 pinniped species from the families Otariidae (sea lions) and Phocidae (seals), were included in our analyses to address the issue. Since leptin exerts its physiological effects on energy balance via direct binding to the leptin receptor (LPR) [2], [10], [29], we analyzed publicly available 24 LPR gene sequences to investigate whether leptin and its have receptor co-evolved.
Results
Leptin and LPR Gene Sequences
We obtained the complete coding sequences of leptin genes (501 bp) from all four cetaceans (See the Materials and Methods for the species list). They all show features typical of leptin, including an amino-terminal signal peptide of 21 amino acids and a mature protein of 146 amino acids, with four α-helices (helices A–D) and a distorted helix E in the CD loop. The amino acid sequences of the mature peptides encoded by these genes are shown in Figure S1.
In addition to these newly determined cetacean leptin genes, the amino acid sequences of the leptin and LPR genes that are obtained from database searches also have characteristic features of leptin (as described above; Figure S1) and the LPR (Figure S2). The LPR coding sequences span 3480–3501 bp, and include codons for the signal peptide (66 bp) and the mature peptide (3414–3435 bp) consisting of extracellular, transmembrane, and intracellular regions (Figure S2).
Phylogenetic Analysis
Neighbor-Joining (NJ) trees of the leptin and LPR genes are shown in Figure S3 and Figure S4, respectively. Maximum-parsimony and Bayesian analyses yielded similar tree topologies and levels of nodal support (See Figure S3 and Figure S4). Both leptin and LPR gene trees supported the monophyly of each of the eight placental mammalian orders included here, but they did not resolve the relationships among these orders, possibly due to the small number of nucleotide sites used.
Positive Selection in Leptin Genes of Cetacea and Pinnipedia
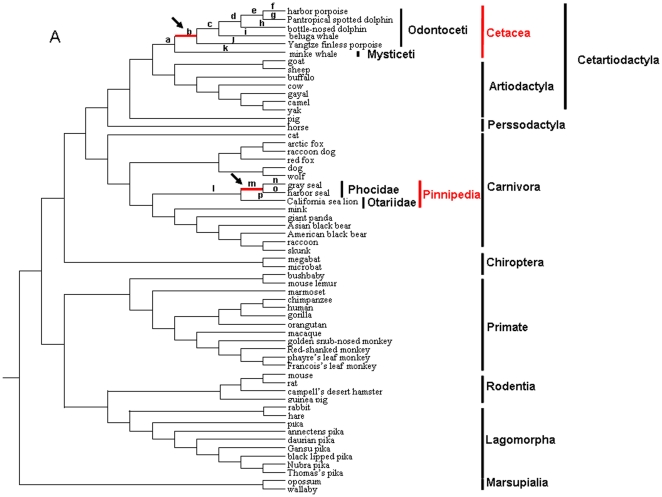
From the leptin gene tree shown in Figure S3, we noticed a long branch leading to the Phocidae family of Pinnipedia, suggesting rapid rates of sequence evolution. This result is consistent with that of Hammond et al. (2005) where a substantially large number of amino acid substitutions in seal leptin sequences were observed [13]. We performed codon-based maximum likelihood analyses for the leptin genes of Cetacea and Pinnipedia (Tables 1 and S2) to determine whether rapid evolution in Phocidae leptin was driven by positive selection, and to examine the selective pattern of Cetacean leptins. Interestingly, the LRT tests based on the branch-site models for all of the branches in Cetacea and Pinnipedia (16 branches in total, a-p as indicated in Figure 1) suggest there was significant evidence of positive selection along both the lineage leading to the common ancestor of the four Odontoceti species in Cetacea (branch b; 2ΔL = 11.046212, p = 0.0009), and that leading to the common ancestor of the two Phocidae species in Pinnipedia (branch m; 2ΔL = 11.165508, p = 0.0008) (Table 1). After performing Bonferroni correction for multiple testing, the LRT tests were still significant in both branches (p<0.003). Therefore, our analyses suggest that positive selection has operated on leptin genes in both Cetacea and Pinnipedia during mammalian evolution.
Table 1. PAML analyses for leptin gene and evidence of positive selection for Odontoceti and Phocidae.
| Models | lnL a | Parameter Estimates | 2ΔL b(P value) | Positively Selected Sites |
| Branch-site models (test 2)c | ||||
| Branch b (ancestral Odontoceti) | ||||
| Null | −2743.730055 | ω 0 = 0.17773 ω 1 = 1 ω 2 = 1 p0 = 0.29440 p1 = 0.10628 p2a = 0.44034 p2b = 0.15897 | ||
| Alternative | −2738.206949 | ω 0 = 0.17871 ω 1 = 1 ω 2 = 999.00000 p0 = 0.72841 p1 = 0.25653 p2a = 0.01114, p2b = 0.00392 | 11.046212 (P = 0.0009) | H46T (0.975) |
| Branch m (ancestral Phocidae) | ||||
| Null | −2731.716272 | ω 0 = 0.15465 ω 1 = 1 ω 2 = 1 p0 = 0 p1 = 0 p2a = 0.75775, p2b = 0.24225 | ||
| Alternative | −2726.133518 | ω 0 = 0.15317 ω 1 = 1 ω 2 = 14.56744 p0 = 0.43153 p1 = 0.14201 p2a = 0.32087, p2b = 0.10559 | 11.165508 (P = 0.0008) | S31C (0.974); Q34P (0.980);L45V (0.972);L49R (0.979);S50T(0.975);T57I (0.996); N72S (0.977); S77A (0.977); E80A (0.976); D85A (0.971);H88R(0.958);S94A (0.970);L104S (0.995);E115R (0.999) |
a lnL is the log-likelihood scores; b likelihood ratio test (LRT) to detect positive selection; c branch b and m are the lineages leading to ancestral Odontoceti and ancestral Phocidae, respectively, in Figure 1.
Figure 1. Phylogenetic trees of leptin gene used for codon-based maximum likelihood analysis in PAML.
Tree topology corresponds to those from Murphy et al. (2001a) and Vogel (2005). Use of the alternative trees (Murphy et al. 2001b, 2007; Springer et al. 2004; Beck et al. 2006) gave essentially the same results. Branches a–p in the tree are used in the branch-site models tests. The thick and red branches (branches b and m) are those with significant evidence of positive selection in PAML analyses.
As summarized in Table 1, the Bayesian approach in PAML predicted one site, site 46 (corresponding to numbers in leptin alignment of Figure S1), as positively selected for branch b with a high Bayesian posterior probability of 0.975. This site is Threonine (T) in all Odontoceti leptin sequences, whereas for Mysticeti leptin, it is Leucine (L). For branch m, there are fourteen sites in total identified to be adaptive with posterior probability larger than 0.95. Interestingly, site 46 was also detected to be under positive selection in Phocidae, but with moderate Bayesian posterior probability of 0.886.
Based on these sequence rate and identification of positively selected sites, we conclude that adaptive amino acid substitutions in the leptin gene occurred more frequently in the Phocidae lineage than in the Odontoceti lineage.
LPR Gene Evolution in Cetacea and Pinnipedia
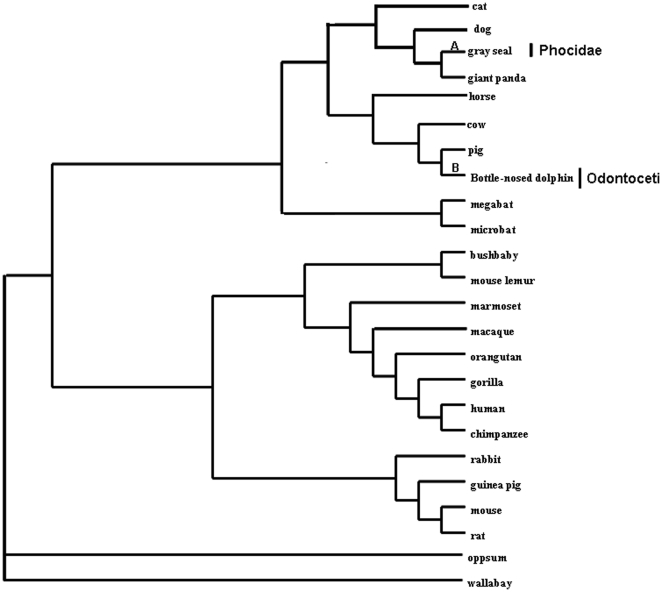
Due to the complex multi-exon structure of the LPR gene (20 exons in mammalian LPR genes), fewer mammalian LPR gene sequences were available from the published data than leptin sequences. For Cetacea and Pinnipedia, only the LPR gene sequences from Halichoerus grypus (gray seal; branch A in Figure 2) and Tursiops truncatus (bottle-nosed dolphin; branch B in Figure 2) are included in the two lineages of Phocidae and Odontoceti, respectively. From the LPR gene tree shown in Figure S4, we found, in contrast to the leptin gene tree, that there were no significant rate changes for the LPR genes of Pinnipedia compared with those of most other mammals.
Figure 2. Phylogenetic trees of leptin receptor gene (LPR) used for codon-based maximum likelihood analysis in PAML.
Tree topology corresponds to those from Murphy et al. (2001a) and Vogel (2005). Use of the alternative trees (Murphy et al. 2001b, 2007; Springer et al. 2004; Beck et al. 2006) gave essentially the same results. Branches A and B in the tree are used in the branch-site models tests.
The same model tests as described in the leptin gene analyses were performed for these 24 LPR genes. As seen in Table 2, none of the LRT tests for the LPR genes were significant. In addition, we conducted analyses based on only cytokine receptor homology domain 2 (CRH2) located in the extracellular region of the LPR genes. Previous structural and mutational studies have shown that the CRH2 domain is the main high-affinity binding site for leptin [30], [31], [32], [33], [34], [35]. However, the LRT tests for the CRH2 domain were not significant either (Table 2). Thus, in contrast to the leptin genes, we did not find evidence for positive selection in these cetacean and pinniped LPR genes. However, it should be noted that fewer LPR genes than leptin genes from mammalian species were available in the present analyses. Therefore, future studies with more LPR genes, especially those from species represented in the leptin dataset but not in the LPR dataset here, are necessary to test for positive selection of the LPR genes in marine mammals.
Table 2. PAML analyses for leptin receptor (LPR) gene.
| Models | lnL a | Parameter Estimates | 2ΔL b(P value) | Positively Selected Sites |
| Branch-site models (test 2)c | ||||
| Dataset 1 (complete mature protein) | ||||
| Branch A (bottle-nosed dolphin) | ||||
| Null | −24857.51845 | ω 0 = 0.15154 ω 1 = 1 ω 2 = 1 p0 = 0.65326 p1 = 0.25041 p2a = 0.06964, p2b = 0.02669 | ||
| Alternative | −24857.32236 | ω 0 = 0.15164 ω 1 = 1 ω 2 = 3.91631 p0 = 0.70244 p1 = 0.26897 p2a = 0.02068, p2b = 0.00792 | 0.39218 (P = 0.5312) | None |
| Branch B (gray seal) | ||||
| Null | −24858.17141 | ω 0 = 0.15231 ω 1 = 1 ω 2 = 1 p0 = 0.72215 p1 = 0.27785 p2a = 0, p2b = 0 | ||
| Alternative | −24858.17141 | ω 0 = 0.15231 ω 1 = 1 ω 2 = 1 p0 = 0.72215 p1 = 0.27785 p2a = 0, p2b = 0 | 0 (P = 1.0000) | None |
| Dataset 2 (CRH2 domain) | ||||
| Branch A (bottle-nosed dolphin) | ||||
| Null | −4027.907504 | ω 0 = 0.09390 ω 1 = 1 ω 2 = 1 p0 = 0.76166 p1 = 0.23834 p2a = 0, p2b = 0 | ||
| Alternative | −4027.907504 | ω 0 = 0.09390 ω 1 = 1 ω 2 = 1 p0 = 0.76166 p1 = 0.23834 p2a = 0, p2b = 0 | 0 (P = 1.0000) | None |
| Branch B (gray seal) | ||||
| Null | −4027.844849 | ω 0 = 0.09346 ω 1 = 1 ω 2 = 1 p0 = 0.72395 p1 = 0.22627 p2a = 0.03793, p2b = 0.01185 | ||
| Alternative | −4027.844849 | ω 0 = 0.15805 ω 1 = 1 ω 2 = 1 p0 = 0.72395 p1 = 0.22627 p2a = 0.03793, p2b = 0.01185 | 0 (P = 1.0000) | None |
a lnL is the log-likelihood scores; b likelihood ratio test (LRT) to detect positive selection; c branch A and B are the lineages leading to bottle-nosed dolphin and gray seal, respectively, in Figure.
Discussion
Physiological function and molecular evolution of the leptin gene in marine mammals have been poorly studied. For the first time, we show evidence of positive selection in the leptin genes of both Cetacea and Pinnipedia, providing valuable insights for understanding the diverse physiological roles it plays in various mammals and also for how such diversity arose during mammalian evolution. Positive selection on the leptin gene detected in Cetacea and Pinnipedia appears to have occurred in ancestral Odontoceti after the Odontoceti-Mysticeti split (about 32 million years ago [36], [37], [38]) and ancestral Phocidae after the Phocidae-Otariidae split (about 23 million years ago [39], [40]), respectively. However, no sign of positive selection was detected in suborder Mysticeti of Cetacea and family Otariidae of Pinnipedia with our limited samples.
In previous physiological assays, leptin in marine mammals has been intensely used as a test case for examining whether leptin plays a significant role in body fat regulation and energy metabolism [41], [42], [43], [44], [45], [46], [47]. Marine mammals provide an interesting study model because they have evolved a fat-based metabolism associated with a blubber layer, the main source of energy stores and essential for body insulation from cold marine waters [48], [49], [50], an adaptation to an aquatic environment and prolonged fasting [43], [51]. All of these studies, however, found no correlation between fat mass and serum leptin levels in seal and sea lion species examined so far. Thus, the primary physiological role of leptin in marine mammals may not be an indicator of body fat and energy reserves. Our study provides indirect evidence in support of this argument: if leptin in marine mammals has contributed to regulation of fat deposits and energy balance, adaptive evolution of leptin should have occurred across all marine mammals, including Odontoceti and Mysticeti (Cetacea), and Phocidae and Otariidae (Pinnipedia). Yet, we reject this hypothesis because this pattern was not seen in our study.
Therefore, the unusual pattern of the leptin gene evolution observed in some lineages of Cetacea and Pinnipeds is intriguing, raising the possibility that new tissue specificity or additional physiological functions of the leptin genes might have arisen in Odontoceti and Phocidae. A number of previous studies have suggested that leptin is expressed in a variety of tissues and blood fluids besides adipose tissues, including placenta, stomach, bone marrow and mammary epithelium [28], [45], [52], [53], [54], [55], [56], and is involved in growth regulation, gastric function, brain development, hematopoiesis, and inflammation [45], [52], [55], [56], [57]. Hammond et al. (2005) found that the leptin in deep-diving seals was expressed in the lung, and proposed a role of leptin in pulmonary surfactant production related to respiratory physiology. A comparison of the diving behaviors between Odontoceti and Mysticeti in Cetacea [58], [59] reveals that Odontoceti generally seeks larger prey at deeper water depths [60], [61], whereas Mysticeti feed on small organisms near the surface of the water and do not hunt their prey or dive to great depths [62]. Therefore, the observed adaptive evolution of leptin in Odontoceti, but not in Mysticeti, might reflect the deep-diving Odontoceti's increased demand for pulmonary or circulatory adaptations to hypoxic conditions. This hypothesis is consistent with the finding that there is adaptive evolution of leptin in deep-diving Phocidae, but not in shallow-diving Otariidae. In the future, it will be interesting to test the expression pattern of leptin in cetacean species and the other marine mammals to determine whether the agent causing the observed positive selection of leptin gene in Odontoceti and Phocidae is indeed the adaptive responses to respiratory physiology of deep-diving marine mammals.
In contrast to other cytokines and their receptors, e.g., growth hormone (GH) and the growth hormone receptor (GHR) [63], [64], and prolactin (PRL) and the prolactin receptor (PRLR) [65], the evolutionary dynamics of leptin and its receptor (LPR) remain unknown. It has been shown that some cytokines and their receptors evolve in a coordinated manner [65], which is consistent with the ligand-receptor co-evolution hypothesis. Therefore, one could expect that the LPR genes in marine mammals would show adaptive evolution similar to that of the leptin gene. However, our study did not detect positive selection in LPR gene of Cetacea and Pinnipedia, unlike its ligand, the leptin genes. However, this finding is not entirely unexpected for a reason. Even though leptin exerts its physiological effects on energy balance via direct binding to the leptin receptor (LPR) [2], [10], [28], novel leptin functions other than energy balance in Cetacea and Pinnipedia may be independent of the physical interaction between leptin and its receptor. The discrepancy in evolutionary patterns of leptin and LPR observed here may have resulted from the multi-functionality of leptin and leptin receptor, supporting the view that the biological roles of leptin vary from species to species [18]. The major novelty of our study is to provide clues about the evolutionary scenarios of leptin/receptor signaling system in marine mammals at the molecular level. In future analyses, the inclusion of more LPR sequences from marine mammals and other mammals would make more rigorous tests of the co-evolution of leptin and its receptor genes possible.
In summary, our study of leptin not only shows that positive selection has promoted rapid divergence of Phocidae leptin, but also demonstrates adaptive evolution of Odontoceti leptin. These findings increase the current knowledge of evolution and function of leptin in mammals, supporting the possibility that new tissue specificity or more crucial physiologic functions of leptin genes may have been developed in both Odontoceti and Phocidae. Furthermore, our study revealed several potentially adaptive amino acid changes, providing a foundation for further experimental investigations (e.g. site-directed mutagenesis) to elucidate functional implications of these substitutions in marine mammals. In the future, evolutionary analyses of leptin from a wider range of mammals with varying life histories, including more marine mammals, and functional assays of adaptive amino acid changes, should contribute to better understanding of the ecology and the selective agent(s) acting on the evolution of leptin genes.
Materials and Methods
Amplification and Sequencing of Cetacea leptin genes
The leptin gene sequences from four cetacean species, i.e., Delphinapterus leucas (Beluga whale; family Monodontidae, suborder Odontoceti), Stenella attenuata (Pantropical spotted dolphin; family Delphinidae; suborder Odontoceti), Neophocaena phocaenoides asiaeorientalis (Yangtze finless porpoise; family Phocoenidae, suborder Odontoceti), and Balaenoptera acutorostrata (Minke whale; family Balaenopteridae; suborder Mysticeti), were determined in this study. We obtained the D. leucas specimen from Harbin Polarland, which was imported from the Okhotsk Sea in Russia, the S. attenuata specimen from Dayawan Bay in Shenzhen, China, the N. p. asiaeorientalis specimen from Dongting Lake in Hunan Province of China, and the B. acutorostrata specimen from the Zhanjiang Coast in GuangDong Province, China. Among these four specimens, the first was obtained from biopsy, while the other three specimens were obtained from stranded animals. For each sample, total genomic DNA was isolated from blood or frozen tissues using a standard proteinase K, phenol/chloroform extraction method [66].
The leptin gene has an approximate length of 4.5 kb, spanning two coding exons and an intron, and was amplified by long PCR using the external primer pair Ceta-F (5′-GACAACCACAAGCAGAAAGCAAATCT-3′ and Ceta-R (5′-GCCTTTGGAA GAAGAGCGGCTTAGAG-3′). PCR amplification was done in a 25-µl reaction mixture containing 5 pM of each primer, 100 µM of each dNTP, 2.5 µl 10ΧLA PCR Buffer, 1.25 units of Takara LA Taq® (Takara Biotechnology Co., Ltd), and 100 ng genomic DNA. The PCR amplification reaction was performed with 34 cycles of 10 sec at 97°C, 7 min at 66°C, with an initial step of 1 min at 94°C and a final step of 10 min at 72°C.
Amplified PCR products were purified and sequenced in both directions with an ABI PRISM™ 3700 DNA sequencer (PE Biosystems, USA). The full-length 167- amino-acid coding region consisted of a 21-amino-acid signal peptide and a 146- amino-acid mature peptide obtained by designing additional internal sequencing primers Ceta-R1 (5′-GGGACACCGGACCGTTATG-3′), Ceta-R2 (5′-CGCCCAGG CTCTCCAAGGT-3′), Ceta-F1 (5′-AGTGGAGGGCAGGGTGGTT-3′), and Ceta-F2 (5′-CTTCATCCCTGGGCTCACC-3′). Sequence data were assembled using the Lasergene SeqMan program (DNAStar, Madison, Wisc.) and visually checked for accuracy. The sequences were deposited in GenBank under accession numbers HQ689399-HQ689402.
Database Searches for leptin and LPR genes
Besides the four cetacean leptin sequences, we also collected 55 publicly available leptin coding sequences for analysis, including 24 from the EMSEMBLE Genome Database (http://www.ensembl.org/) and 31 from published data (http://www.ncbi. nlm.nih.gov/) (Table S1).
In addition to leptin genes, the coding sequences of leptin receptor, LPR, genes consisting of a 22- amino-acid signal peptide and a 1140-amino-acid mature peptide were obtained from the same 24 mammalian species with available genome sequences (http://www.ensembl.org/) as those in the leptin dataset except for the pika Ochotona princeps, whose LPR gene is unavailable (Table S1). It has been reported that human and mouse LPR have multiple alternatively spliced forms [35], [67], [68], [69], [70], [71], [72], [73], and that the full-length form is essential for leptin signaling [69], [74]. Thus, in those cases where multiple splice variants of LPR were available for a species, only the full-length form from that species was used for analysis. In addition, the publicly available LPR gene from Halichoerus grypus (gray seal) (GenBank Accession No. HM448474) was also included.
Phylogenetic Reconstructions
Nucleotide sequences from the leptin coding region and LPR were separately aligned using CLUSTAL X program [75]. A 501-bp alignment of 59 leptin sequences and a 3519-bp alignment of 24 LPR sequences were used for phylogenetic analyses. The amino acid alignments of their mature peptides are shown in Figure S1 and S2, respectively.
Phylogenetic reconstructions were performed using MEGA4 (Kumar et al. 2008) for neighbor-joining (NJ) analyses, PAUP*4.0b8 [76] for maximum- parsimony (MP) analyses, and MrBayes 3.1.2 [77] for Bayesian inference. In NJ analysis, Kimura's 2-parameter nucleotide model with pairwise deletion option for gaps was used. In MP analysis, a heuristic search strategy was employed with the TBR branch swapping algorithm, random addition of taxa and 1000 replicates per search. The reliability of the tree topologies was evaluated using bootstrap support (BS; [78]) with 1000 replicates for NJ and MP analyses. In Bayesian analyses, the best-fit models of sequence evolution were selected using the Akaike Information Criterion (AIC; [79], [80]) with jModeltest [81]. The chosen models were used in the priors of Bayesian inference. Four Metropolis-coupled Markov chain Monte Carlo (MCMC) analyses were run for 2×106 generations, sampling trees every 100 generations. The average standard deviation of split frequencies was below 0.001 when the run ended. The first 25% were discarded as the burn-in. A 50% majority-rule consensus of post burn-in trees was constructed to summarize posterior probabilities (PP) for each branch. Opossum and wallaby sequences were used as outgroups in all analyses.
Molecular Evolutionary Analysis
The nonsynonymous to synonymous rate ratio ω (dN/dS) provided an indication of the change of selective pressures. A dN/dS ratio = 1, <1, and >1 are indicative of neutral evolution, purifying selection, and positive selection at the protein involved, respectively. We applied the codon substitution models implemented in the CODEML program in the PAML package [82]. Only gene regions coding for the mature leptin (438 bp in alignment) and LPR peptide (3453 bp in alignment) were analyzed. All models correct the transition/transversion rate and codon usage biases (F3×4). The ambiguous sites were removed in PAML analysis of leptin gene (clean data = yes). Different starting ω values were also used to avoid the local optima on the likelihood surface [83].
The branch-site models accommodating ω ratios to vary both among lineages of interest and amino acid sites were considered here [84], [85]. We used branch-site Model A for stringent testing (test 2) and identification of sites under positive selection along the lineages of interest. Significant differences between models were evaluated by calculating twice the log-likelihood difference following a χ2 distribution, with the number of degrees of freedom equal to the difference in the numbers of free parameters between models. The presence of sites with ω>1 is suggested when the positive-selection model (Model A) fits the data significantly better than the corresponding null model (M1a). We used the conservative Bayes Empirical Bayes (BEB) approach, which assigns a prior to the model parameters and integrates over their uncertainties, to calculate the posterior probabilities of a specific codon site and to identify those most likely to be under positive selection [85], [86]. In addition, the Bonferroni correction [87] was applied for multiple testing in the analysis.
In all PAML analyses, the tree topology ((((Cetartiodactyla, Perissodactyla), Carnivora), Chiroptera), ((Rodentia, Lagomorpha), Primates), Marsupialia) (Figure 1 and Figure 2), based on an analysis of nearly 10,000 bp of 18 genes from 64 mammalian species [88], [89] was used. In fact, to avoid a possible effect of tree topology used on the results, we also conducted the analyses using alternative tree topologies among the published mammalian phylogenies [90], [91], [92], [93] and different tree topologies at family level, and obtained the same results.
Supporting Information
The mature protein alignment of 59 leptin sequences used in this study. The positions of four α-helices (helices A-D) and a distorted helix E in the CD loop are indicated. The species in Cetacea and Pinnipedia are crossed.
(TIF)
The mature protein alignment of 24 leptin receptor (LPR) sequences used in this study. The positions of extracellular, transmembrane, and intracellular regions, as well as the binding regions for leptin (cytokine receptor homology; CRH2) are indicated. The species in Cetacea and Pinnipedia are crossed.
(TIF)
NJ trees based on leptin alignment (501 nt; 59 sequences). MP and Bayesian analyses (AIC model: GTR+G; Base frequencies: A = 0.2405; C = 0.3097; G = 0.2365 and T = 0.2133; Transition/transversion ratio: R[AC] = 0.8717; R[AG] = 4.7477; R[AT] = 0.2437; R[CG] = 0.9188; R[CT] = 3.5498; R [GT] = 1. 0000; gamma shape = 0.9150) produced similar tree topologies to those of NJ analyses with similar nodal supports. The bootstrap supports shown on the nodes are calculated from NJ/MP/Bayesian analyses. Those not shown on the nodes are poorly supported by all the three analyses.
(TIF)
NJ trees based on LPR alignment (3519 nt; 24 sequences). MP and Bayesian analyses (AIC model: TVM+G; Base frequencies: A = 0.2723; C = 0.2327; G = 0.2286 and T = 0.2665; Transition/transversion ratio: R[AC] = 1.5455; R[AG] = 5.1963; R[AT] = 0.5967; R[CG] = 1.1684; R[CT] = 5.1963; R [GT] = 1.00 00; gamma shape = 0.8000) produced similar tree topologies to those of NJ analyses with similar nodal supports. The bootstrap supports shown on the nodes are calculated from NJ/MP/Bayesian analyses. Those not shown on the nodes are poorly supported by all the three analyses.
(TIF)
List of taxonomic samples and sequences used in this study
(DOC)
Acknowledgments
We thank Professor Oliver A Ryder, David A Liberles and Peng Shi for valuable comments. We thank Dr. Pen-tao Luan and Dr. Xiao-ping Wang for helping data analyses.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the State Key Basic Research and Development Plan (2007CB411600) and National Natural Science Foundation of China (U0836603). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, et al. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 2.Campfield LA, Smith FJ, Guisez Y, Devos R, Burn P. Recombinant mouse OB protein: evidence for a peripheral signal linking adiposity and central neural networks. Science. 1995;269:546–549. doi: 10.1126/science.7624778. [DOI] [PubMed] [Google Scholar]
- 3.Considine RV, Caro JF. Leptin and the regulation of body weight. Int J Biochem Cell Biol. 1997;29:1255–1272. doi: 10.1016/s1357-2725(97)00050-2. [DOI] [PubMed] [Google Scholar]
- 4.Friedman JM, Halaas JL. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 5.Houseknecht KL, Baile CA, Matteri RL, Spurlock ME. The biology of leptin: a review. J Anim Sci. 1998;76:1405–1420. doi: 10.2527/1998.7651405x. [DOI] [PubMed] [Google Scholar]
- 6.Auwerx J, Staels B. Leptin. Lancet. 1998;351:737–742. doi: 10.1016/S0140-6736(97)06348-4. [DOI] [PubMed] [Google Scholar]
- 7.Jiang H, Ren F, Sun J, He L, Li W, et al. Molecular cloning and gene expression analysis of the leptin receptor in the Chinese mitten crab Eriocheir sinensis. PLoS One. 2010;5:e11175. doi: 10.1371/journal.pone.0011175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennedy GC. The role of depot fat in the hypothalamic control of food intake in the rat. Proc R Soc Lond B Biol Sci. 1953;140:578–596. doi: 10.1098/rspb.1953.0009. [DOI] [PubMed] [Google Scholar]
- 9.Friedman JM, Leibel RL, Bahary N, Siegel DA, Truett G. Genetic analysis of complex disorders. Molecular mapping of obesity genes in mice and humans. Ann N Y Acad Sci. 1991;630:100–115. doi: 10.1111/j.1749-6632.1991.tb19579.x. [DOI] [PubMed] [Google Scholar]
- 10.Pelleymounter MA, Cullen MJ, Baker MB, Hecht R, Winters D, et al. Effects of the obese gene product on body weight regulation in ob/ob mice. Science. 1995;269:540–543. doi: 10.1126/science.7624776. [DOI] [PubMed] [Google Scholar]
- 11.Montague CT, Farooqi IS, Whitehead JP, Soos MA, Rau H, et al. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature. 1997;387:903–908. doi: 10.1038/43185. [DOI] [PubMed] [Google Scholar]
- 12.Strobel A, Issad T, Camoin L, Ozata M, Strosberg AD. A leptin missense mutation associated with hypogonadism and morbid obesity. Nat Genet. 1998;18:213–215. doi: 10.1038/ng0398-213. [DOI] [PubMed] [Google Scholar]
- 13.Hammond JA, Bennett KA, Walton MJ, Hall AJ. Molecular cloning and expression of leptin in gray and harbor seal blubber, bone marrow, and lung and its potential role in marine mammal respiratory physiology. Am J Physiol Regul Integr Comp Physiol. 2005;289:R545–R553. doi: 10.1152/ajpregu.00203.2004. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Zhao XQ, Guo SC, Li HG, Qi DL, et al. Leptin cDNA cloning and its mRNA expression in plateau pikas (Ochotona curzoniae) from different altitudes on Qinghai-Tibet Plateau. Biochem Biophys Res Commun. 2006;345:1405–1413. doi: 10.1016/j.bbrc.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 15.Yang J, Wang ZL, Zhao XQ, Wang de P, Qi de L, et al. Natural selection and adaptive evolution of leptin in the ochotona family driven by the cold environmental stress. PLoS One. 2008;3:e1472. doi: 10.1371/journal.pone.0001472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benner SA, Trabesinger N, Schreiber D. Post-genomic science: converting primary structure into physiological function. Adv Enzyme Regul. 1998;38:155–180. doi: 10.1016/s0065-2571(97)00019-8. [DOI] [PubMed] [Google Scholar]
- 17.Benner SA, Chamberlin SG, Liberles DA, Govindarajan S, Knecht L. Functional inferences from reconstructed evolutionary biology involving rectified databases--an evolutionarily grounded approach to functional genomics. Res Microbiol. 2000;151:97–106. doi: 10.1016/s0923-2508(00)00123-6. [DOI] [PubMed] [Google Scholar]
- 18.Benner SA, Gaucher EA. Evolution, language and analogy in functional genomics. Trends Genet. 2001;17:414–418. doi: 10.1016/s0168-9525(01)02320-4. [DOI] [PubMed] [Google Scholar]
- 19.Benner SA, Caraco MD, Thomson JM, Gaucher EA. Planetary biology--paleontological, geological, and molecular histories of life. Science. 2002;296:864–868. doi: 10.1126/science.1069863. [DOI] [PubMed] [Google Scholar]
- 20.Liberles DA. Evaluation of methods for determination of a reconstructed history of gene sequence evolution. Mol Biol Evol. 2001;18:2040–2047. doi: 10.1093/oxfordjournals.molbev.a003745. [DOI] [PubMed] [Google Scholar]
- 21.Liberles DA, Schreiber DR, Govindarajan S, Chamberlin SG, Benner SA. The adaptive evolution database (TAED). Genome Biol. 2001;2:RESEARCH0028. doi: 10.1186/gb-2001-2-8-research0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siltberg J, Liberles DA. A simple covarion-based approach to analyse nucleotide substitution rates. Journal of Evolutionary Biology. 2002;15:588–594. [Google Scholar]
- 23.Gaucher EA, Miyamoto MM, Benner SA. Evolutionary, structural and biochemical evidence for a new interaction site of the leptin obesity protein. Genetics. 2003;163:1549–1553. doi: 10.1093/genetics/163.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berglund AC, Wallner B, Elofsson A, Liberles DA. Tertiary windowing to detect positive diversifying selection. J Mol Evol. 2005;60:499–504. doi: 10.1007/s00239-004-0223-4. [DOI] [PubMed] [Google Scholar]
- 25.Hoelzel, R A, editors. Oxford: Blackwell Publishing; 2002. Marine mammal biology: an evolutionary approach. [Google Scholar]
- 26.McClellan DA, Palfreyman EJ, Smith MJ, Moss JL, Christensen RG, et al. Physicochemical evolution and molecular adaptation of the cetacean and artiodactyl cytochrome b proteins. Mol Biol Evol. 2005;22:437–455. doi: 10.1093/molbev/msi028. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Yuan L, Rossiter SJ, Zuo X, Ru B, et al. Adaptive evolution of 5′HoxD genes in the origin and diversification of the cetacean flipper. Mol Biol Evol. 2009;26:613–622. doi: 10.1093/molbev/msn282. [DOI] [PubMed] [Google Scholar]
- 28.Yu L, Jin W, Wang JX, Zhang X, Chen MM, et al. Characterization of TRPC2, an essential genetic component of VNS chemoreception, provides insights into the evolution of pheromonal olfaction in secondary-adapted marine mammals. Mol Biol Evol. 2010;27:1467–1477. doi: 10.1093/molbev/msq027. [DOI] [PubMed] [Google Scholar]
- 29.Halaas JL, Gajiwala KS, Maffei M, Cohen SL, Chait BT, et al. Weight-reducing effects of the plasma protein encoded by the obese gene. Science. 1995;269:543–546. doi: 10.1126/science.7624777. [DOI] [PubMed] [Google Scholar]
- 30.Fong TM, Huang RR, Tota MR, Mao C, Smith T, et al. Localization of leptin binding domain in the leptin receptor. Mol Pharmacol. 1998;53:234–240. doi: 10.1124/mol.53.2.234. [DOI] [PubMed] [Google Scholar]
- 31.Sandowski Y, Raver N, Gussakovsky EE, Shochat S, Dym O, et al. Subcloning, expression, purification, and characterization of recombinant human leptin-binding domain. J Biol Chem. 2002;277:46304–46309. doi: 10.1074/jbc.M207556200. [DOI] [PubMed] [Google Scholar]
- 32.Zabeau L, Defeau D, Van der Heyden J, Iserentant H, Vandekerckhove J, et al. Functional analysis of leptin receptor activation using a Janus kinase/signal transducer and activator of transcription complementation assay. Mol Endocrinol. 2004;18:150–161. doi: 10.1210/me.2003-0078. [DOI] [PubMed] [Google Scholar]
- 33.Iserentant H, Peelman F, Defeau D, Vandekerckhove J, Zabeau L, et al. Mapping of the interface between leptin and the leptin receptor CRH2 domain. J Cell Sci. 2005;118:2519–2527. doi: 10.1242/jcs.02386. [DOI] [PubMed] [Google Scholar]
- 34.Peelman F, Iserentant H, De Smet AS, Vandekerckhove J, Zabeau L, et al. Mapping of binding site III in the leptin receptor and modeling of a hexameric leptin.leptin receptor complex. J Biol Chem. 2006;281:15496–15504. doi: 10.1074/jbc.M512622200. [DOI] [PubMed] [Google Scholar]
- 35.Peelman F, Van Beneden K, Zabeau L, Iserentant H, Ulrichts P, et al. Mapping of the leptin binding sites and design of a leptin antagonist. J Biol Chem. 2004;279:41038–41046. doi: 10.1074/jbc.M404962200. [DOI] [PubMed] [Google Scholar]
- 36.Nikaido M, Matsuno F, Hamilton H, Brownell RL, Jr, Cao Y, et al. Retroposon analysis of major cetacean lineages: the monophyly of toothed whales and the paraphyly of river dolphins. Proc Natl Acad Sci U S A. 2001;98:7384–7389. doi: 10.1073/pnas.121139198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hassanin A, Douzery EJ. Molecular and morphological phylogenies of ruminantia and the alternative position of the moschidae. Syst Biol. 2003;52:206–228. doi: 10.1080/10635150390192726. [DOI] [PubMed] [Google Scholar]
- 38.Gatesy J. Whales and even-toed ungulates (Cetartiodactyla). In: Hedges S.B., Kumar S., editors. In the Timetree of Life Pp: 511-515. Oxford University Press; 2009. [Google Scholar]
- 39.Bininda-Emonds OR, Gittleman JL, Purvis A. Building large trees by combining phylogenetic information: a complete phylogeny of the extant Carnivora (Mammalia). Biol Rev Camb Philos Soc. 1999;74:143–175. doi: 10.1017/s0006323199005307. [DOI] [PubMed] [Google Scholar]
- 40.Higdon JW, Bininda-Emonds OR, Beck RM, Ferguson SH. Phylogeny and divergence of the pinnipeds (Carnivora: Mammalia) assessed using a multigene dataset. BMC Evol Biol. 2007;7:216. doi: 10.1186/1471-2148-7-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rea LD, TR Nagy. Changes in serum leptin levels during fasting and food limitation in Steller sea lions (Eumetopias jubatus). Proc Comp Nutr Soc. 2000;3:171–175. [Google Scholar]
- 42.Gurun GD, Noren J, Ramirez A, Ramirez RM, Ortiz CL. Leptin does not correlate with fat mass in northern elephant seal pups(Abstract). FASEB J. 2001;15:A414. [Google Scholar]
- 43.Ortiz RM, Wade CE, Ortiz CL. Effects of prolonged fasting on plasma cortisol and TH in postweaned northern elephant seal pups. Am J Physiol Regul Integr Comp Physiol. 2001;280:R790–795. doi: 10.1152/ajpregu.2001.280.3.R790. [DOI] [PubMed] [Google Scholar]
- 44.Ortiz RM, Houser DS, Wade CE, Ortiz CL. Hormonal changes associated with the transition between nursing and natural fasting in northern elephant seals (Mirounga angustirostris). Gen Comp Endocrinol. 2003;130:78–83. doi: 10.1016/s0016-6480(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 45.Arnould JP, Morris MJ, Rawlins DR, Boyd IL. Variation in plasma leptin levels in response to fasting in Antarctic fur seals (Arctocephalus gazella). J Comp Physiol B. 2002;172:27–34. doi: 10.1007/s003600100224. [DOI] [PubMed] [Google Scholar]
- 46.Guilherme C, Bianchini A, Martinez PE, Robaldo RB, Colares EP. Serum leptin concentration during the terrestrial phase of the Southern elephant seal Mirounga leonina (Carnivora: Phocidae). Gen Comp Endocrinol. 2004;139:137–142. doi: 10.1016/j.ygcen.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Ortiz RM, Noren DP, Litz B, Ortiz CL. A new perspective on adiposity in a naturally obese mammal. Am J Physiol Endocrinol Metab. 2001;281:E1347–1351. doi: 10.1152/ajpendo.2001.281.6.E1347. [DOI] [PubMed] [Google Scholar]
- 48.Ortiz CL, CostaD, Boeuf BJL. Water and energy flux in elephant seal pups fasting under natural conditions. Physiol Zool. 1978;51:166–178. [Google Scholar]
- 49.Young RA. Fat, energy and mammalian survival. Amer Zool. 1976;16:699–710. [Google Scholar]
- 50.Whittow GC. Thermoregulatory adaptations in marine mammals: interacting effects of exercise and body mass: a review. Mar Mamm Sci. 1987;3:220–241. [Google Scholar]
- 51.Worthy GAJ, L DM. Energetics of fasting and subsequent growth in weaned harp seal pups, Phoca groenlandica. Can J Zool. 1983;61:447–456. [Google Scholar]
- 52.Masuzaki H, Ogawa Y, Sagawa N, Hosoda K, Matsumoto T, et al. Nonadipose tissue production of leptin: leptin as a novel placenta-derived hormone in humans. Nat Med. 1997;3:1029–1033. doi: 10.1038/nm0997-1029. [DOI] [PubMed] [Google Scholar]
- 53.Casabiell X, Piñeiro V, Tomé MA, Peinó R, Diéguez C, et al. Presence of leptin in colostrum and/or breast milk from lactating mothers: a potential role in the regulation of neonatal food intake. J Clin Endocrinol Metab. 1997;82:4270–4273. doi: 10.1210/jcem.82.12.4590. [DOI] [PubMed] [Google Scholar]
- 54.Señarís R, Garcia-Caballero T, Casabiell X, Gallego R, Castro R, et al. Casanueva Synthesis of Leptin in Human Placenta. Endocrinology. 1997;138:4501–4504. doi: 10.1210/endo.138.10.5573. [DOI] [PubMed] [Google Scholar]
- 55.Bado A, Levasseur S, Attoub S, Kermorgant S, Laigneau JP, et al. The stomach is a source of leptin. Nature. 1998;394:790–793. doi: 10.1038/29547. [DOI] [PubMed] [Google Scholar]
- 56.Laharrague P, Larrouy D, Fontanilles AM, Truel N, Campfield A, et al. High expression of leptin by human bone marrow adipocytes in primary culture. FASEB J. 1998;12:747–752. doi: 10.1096/fasebj.12.9.747. [DOI] [PubMed] [Google Scholar]
- 57.Torday JS, Sun H, Wang L, Torres E, Sunday ME, et al. Leptin mediates the parathyroid hormone-related protein paracrine stimulation of fetal lung maturation. Am J Physiol Lung Cell Mol Physiol. 2002;282:L405–410. doi: 10.1152/ajplung.2002.282.3.L405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perrin WF, Würsig B, Thewissen H. San Diego, CA: Academic Press; 2002. Encyclopedia of Marine Mammals. [Google Scholar]
- 59.Berta A, Sumich JL, Kovacs KM. San Diego, CA: Elsevier; 2006. Marine mammals: evolutionary biology. 2nd ed. [Google Scholar]
- 60.Heide-Jørgensen MP, Dietz R. Some characteristics of narwhal, Monodon monoceros, diving behaviour in Baffin Bay. Can J Zool. 1995;73:2120–2132. [Google Scholar]
- 61.Martin AR, Smith TG. Strategy and capability of wild belugas, Delphinapterus leucas, during deep, benthic diving. Can J Zool. 1999;77:1783–1793. [Google Scholar]
- 62.Schreer JF, Kovacs KM. Allometry of diving capacity in air-breathing vertebrates. Can J Zool. 1997;75:339–358. [Google Scholar]
- 63.de Vos AM, Ultsch M, Kossiakoff AA. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 64.Liu JC, Makova KD, Adkins RM, Gibson S, Li WH. Episodic evolution of growth hormone in primates and emergence of the species specificity of human growth hormone receptor. Mol Biol Evol. 2001;18:945–953. doi: 10.1093/oxfordjournals.molbev.a003895. [DOI] [PubMed] [Google Scholar]
- 65.Li Y, Wallis M, Zhang YP. Episodic evolution of prolactin receptor gene in mammals: coevolution with its ligand. J Mol Endocrinol. 2005;35:411–419. doi: 10.1677/jme.1.01798. [DOI] [PubMed] [Google Scholar]
- 66.Sambrook J, Russell WD. New York: Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning: A Laboratory Manual, 3rd ed. [Google Scholar]
- 67.Tartaglia LA, Dembski M, Weng X, Deng N, Culpepper J, et al. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 68.Tartaglia LA. The leptin receptor. J Biol Chem. 1997;272:6093–6096. doi: 10.1074/jbc.272.10.6093. [DOI] [PubMed] [Google Scholar]
- 69.Lee GH, Proenca R, Montez JM, Carroll KM, Darvishzadeh JG, et al. Abnormal splicing of the leptin receptor in diabetic mice. Nature. 1996;379:632–635. doi: 10.1038/379632a0. [DOI] [PubMed] [Google Scholar]
- 70.Cioffi JA, Shafer AW, Zupancic TJ, Smith-Gbur J, Mikhail A, et al. Novel B219/OB receptor isoforms: possible role of leptin in hematopoiesis and reproduction. Nat Med. 1996;2:585–589. doi: 10.1038/nm0596-585. [DOI] [PubMed] [Google Scholar]
- 71.Mercer JG, Hoggard N, Williams LM, Lawrence CB, Hannah LT, et al. Localization of leptin receptor mRNA and the long form splice variant (Ob-Rb) in mouse hypothalamus and adjacent brain regions by in situ hybridization. FEBS Lett. 1996;387:113–116. doi: 10.1016/0014-5793(96)00473-5. [DOI] [PubMed] [Google Scholar]
- 72.Wang MY, Zhou YT, Newgard CB, Unger RH. A novel leptin receptor isoform in rat. FEBS Lett. 1996;392:87–90. doi: 10.1016/0014-5793(96)00790-9. [DOI] [PubMed] [Google Scholar]
- 73.Kapitonov VV, Jurka J. The long terminal repeat of an endogenous retrovirus induces alternative splicing and encodes an additional carboxy-terminal sequence in the human leptin receptor. J Mol Evol. 1999;48:248–251. doi: 10.1007/pl00013153. [DOI] [PubMed] [Google Scholar]
- 74.Huang L, Wang Z, Li C. Modulation of circulating leptin levels by its soluble receptor. J Biol Chem. 2001;276:6343–6349. doi: 10.1074/jbc.M009795200. [DOI] [PubMed] [Google Scholar]
- 75.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Swofford DL. Sinauer Associates; 2001. PAUP*: phylogenetic analysis using parsimony (* and other methods). Version 4.0b8 Sunderland (MA) [Google Scholar]
- 77.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 78.Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 79.Akaike H. "A new look at the statistical model identification". IEEE Transactions on Automatic Control. 1974;19:716–723. [Google Scholar]
- 80.Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 81.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 82.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 83.Suzuki Y, Nei M. Reliabilities of parsimony-based and likelihood-based methods for detecting positive selection at single amino acid sites. Mol Biol Evol. 2001;18:2179–2185. doi: 10.1093/oxfordjournals.molbev.a003764. [DOI] [PubMed] [Google Scholar]
- 84.Gillespie JH. Oxford: Oxford University Press Golding GB, Dean AM; 1991. The causes of molecular evolution. [Google Scholar]
- 85.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 86.Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bonferroni CE. Teoria statistica delle classi e calcolo delle probabilità. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze. 1936;8:3–62. [Google Scholar]
- 88.Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, et al. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- 89.Vogel P. The current molecular phylogeny of Eutherian mammals challenges previous interpretations of placental evolution. Placenta. 2005;26:591–596. doi: 10.1016/j.placenta.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 90.Murphy WJ, Eizirik E, O'Brien SJ, Madsen O, Scally M, et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- 91.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Springer MS, Stanhope MJ, Madsen O, de Jong WW. Molecules consolidate the placental mammal tree. Trends Ecol Evol. 2004;19:430–438. doi: 10.1016/j.tree.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 93.Beck RM, Bininda-Emonds OR, Cardillo M, Liu FG, Purvis A. A higher-level MRP supertree of placental mammals. BMC Evol Biol. 2006;6:93. doi: 10.1186/1471-2148-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The mature protein alignment of 59 leptin sequences used in this study. The positions of four α-helices (helices A-D) and a distorted helix E in the CD loop are indicated. The species in Cetacea and Pinnipedia are crossed.
(TIF)
The mature protein alignment of 24 leptin receptor (LPR) sequences used in this study. The positions of extracellular, transmembrane, and intracellular regions, as well as the binding regions for leptin (cytokine receptor homology; CRH2) are indicated. The species in Cetacea and Pinnipedia are crossed.
(TIF)
NJ trees based on leptin alignment (501 nt; 59 sequences). MP and Bayesian analyses (AIC model: GTR+G; Base frequencies: A = 0.2405; C = 0.3097; G = 0.2365 and T = 0.2133; Transition/transversion ratio: R[AC] = 0.8717; R[AG] = 4.7477; R[AT] = 0.2437; R[CG] = 0.9188; R[CT] = 3.5498; R [GT] = 1. 0000; gamma shape = 0.9150) produced similar tree topologies to those of NJ analyses with similar nodal supports. The bootstrap supports shown on the nodes are calculated from NJ/MP/Bayesian analyses. Those not shown on the nodes are poorly supported by all the three analyses.
(TIF)
NJ trees based on LPR alignment (3519 nt; 24 sequences). MP and Bayesian analyses (AIC model: TVM+G; Base frequencies: A = 0.2723; C = 0.2327; G = 0.2286 and T = 0.2665; Transition/transversion ratio: R[AC] = 1.5455; R[AG] = 5.1963; R[AT] = 0.5967; R[CG] = 1.1684; R[CT] = 5.1963; R [GT] = 1.00 00; gamma shape = 0.8000) produced similar tree topologies to those of NJ analyses with similar nodal supports. The bootstrap supports shown on the nodes are calculated from NJ/MP/Bayesian analyses. Those not shown on the nodes are poorly supported by all the three analyses.
(TIF)
List of taxonomic samples and sequences used in this study
(DOC)