Abstract
Ultraviolet (UV) light causes DNA damage in skin cells, leading to more than one million cases of non-melanoma skin cancer diagnosed annually in the United States. Although human cells possess a mechanism (Nucleotide Excision Repair, NER) to repair UV-induced DNA damage, mutagenesis still occurs when DNA is replicated prior to repair of these photoproducts. While human cells have all the enzymes necessary to complete an alternate repair pathway, Base Excision Repair (BER), they lack a DNA glycosylase that can initiate BER of dipyrimidine photoproducts. Certain prokaryotes and viruses produce pyrimidine dimer-specific DNA glycosylases (pdgs) that initiate BER of cyclobutane pyrimidine dimers (CPDs), the predominant UV-induced lesions. Such a pdg was identified in the Chlorella virus PBCV-1 and termed Cv-pdg. The Cv-pdg protein was engineered to contain a nuclear localization sequence (NLS) and a membrane permeabilization peptide (TAT). Here, we demonstrate that the Cv-pdg-NLS-TAT protein was delivered to repair-proficient keratinocytes and fibroblasts, and to a human skin model, where it rapidly initiated removal of CPDs. These data suggest a potential strategy for prevention of human skin cancer.
Introduction
Over one million people in the United States are diagnosed with basal cell carcinomas (BCC) and squamous cell carcinomas (SCC) annually, and billions of dollars are spent in associated health care costs (Bickers et al., 2006). Exposure to UV in sunlight results in SCC and BCC by causing DNA damage that, if unrepaired or repaired incorrectly, can lead to mutations in genes such as the ras oncogene or the tumor suppressor p53 (reviewed in Hussein, 2005). The predominant DNA lesions induced by UV are CPDs. Humans possess only one mechanism, the NER pathway, for repairing CPDs. The importance of this single DNA repair system in limiting SCC and BCC formation is best demonstrated by the study of Xeroderma Pigmentosum (XP) patients who have mutations in genes involved in the NER pathway or in a specialized DNA polymerase that catalyzes DNA synthesis past UV-induced DNA lesions. These patients have more than a 1000-fold increased risk of developing skin cancer compared to the general population (reviewed in Kraemer et al., 2007).
In addition to NER, other organisms utilize the BER pathway for repairing UV-induced DNA lesions. Although humans possess a functional BER pathway for many base lesions, they lack the initiating glycosylase to catalyze BER in response to photo-damage of DNA. It is hypothesized that exogenous delivery of BER-initiating repair enzymes to UV-exposed skin cells may promote rapid, accurate repair of CPDs, providing protection against skin cancer. Indeed, the bacteriophage T4 pyrimidine dimer-specific DNA glycosylase (T4-pdg), also called T4 endonuclease V, enhances repair of CPDs and reduces the frequency of UV-induced mutations in cultured mammalian cells when introduced by DNA transfection or by direct protein delivery via encapsulation into a liposomal delivery vehicle (Cafardi and Elmets, 2008; Francis et al., 2000; Kibitel et al., 1991; Kusewitt et al., 1994; Kusewitt et al., 1998; Yarosh et al., 1992).
XP patients topically treated with liposome-encapsulated T4-pdg enzyme for one year exhibited a 68% decrease in new pre-cancerous actinic keratoses (AKs) and a 30% decrease in new cases of BCC compared to patients treated with placebo lotion (Yarosh et al., 2001). Repair-proficient human populations could also potentially benefit from clinical trials exploring the use of pdg treatment for prevention of skin cancer, but such studies have not yet been published. For example, aged individuals who frequently develop AKs, up to 10% of which convert to SCCs (Dodson et al., 1991), and immune suppressed organ transplant patients, who can be up to 250 times more likely than the general public to develop aggressive, metastatic skin cancer (Abgrall et al., 2002) could both benefit from such studies. It is also unknown what impact pdg delivery could have on skin cancer prevention in the general public, in whom accumulated lifetime UV exposure and number of sunburns lead to SCC, BCC, and melanoma (Dennis et al., 2008; Hussein, 2005).
While T4-pdg has been tested for efficacy to prevent skin cancer in XP patients, no homologues of T4-pdg have been studied for this purpose. Cv-pdg, a pdg with 41% sequence identity to T4-pdg, is expressed by a Chlorella virus that infects the eukaryotic Paramecium bursaria chlorella algae (Furuta et al., 1997). Biochemical characterization of the Cv-pdg enzyme showed that, in addition to cleaving cis-syn CPDs similarly to T4-pdg, Cv-pdg also cleaved less prevalent, but mutagenic types of UV-induced DNA lesions including trans-syn-II CPDs (McCullough et al., 1998), FapyAde and FapyGua (Jaruga et al., 2002). Additionally, Cv-pdg is more active over a broader range of salt concentrations and maintains activity after exposure to more extreme temperatures than T4-pdg (McCullough et al., 1998). Due to these potential advantages over T4-pdg for pharmaceutical use, Cv-pdg was selected for the present study. In an effort to enhance the utility of Cv-pdg to remove CPDs in NER-proficient human skin cells following UV exposure, the enzyme was engineered with a nuclear localization sequence (NLS) to enhance targeting to the nucleus. Since unaided absorption of small molecules into the skin is limited to approximately 500 Da (Bos and Meinardi, 2000), the ~18 kDa Cv-pdg-NLS enzyme was also conjugated to a cell membrane permeabilization sequence, TAT (transcriptional transactivator peptide) from human immunodeficiency virus, that is capable of transdermal delivery of conjugated peptides up to approximately 20 kDa (Lopes et al., 2005).
In this study, we examined the delivery of Cv-pdg-NLS-TAT enzyme to cultured repair-proficient keratinocytes and fibroblasts and to the basal keratinocytes of a stratified human skin model. We also examined the ability of Cv-pdg-NLS-TAT to initiate repair of CPDs in these cells.
Results
Purified pdg proteins retained activity in vitro after addition of NLS and TAT peptides to enhance cellular delivery
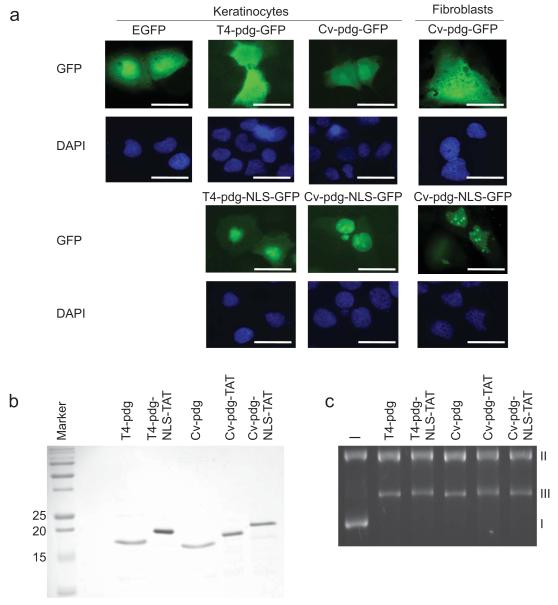
Since T4-pdg lacks a native NLS, the majority of T4-pdg used in previous studies remained cytoplasmic and entered the nucleus only through passive diffusion (Yarosh et al., 1994). To direct T4-pdg and Cv-pdg to the nuclei of human keratinocytes and fibroblasts to initiate repair of UV-induced DNA damage, both pdgs were modified to express a NLS peptide (Pro-Lys-Lys-Arg-Lys-Arg-Arg-Leu) at the carboxy terminus. A green fluorescent protein tag was also added, generating the modified T4-pdg-NLS-GFP and Cv-pdg-NLS-GFP proteins. Following transient transfection of these plasmids into HaCaT keratinocytes and immortalized human skin fibroblasts, localization of these proteins was shown to be predominantly nuclear (Figure 1a). In contrast, expression of the unmodified T4-pdg-GFP and Cv-pdg-GFP proteins demonstrated predominantly cytoplasmic localization. These data provide the first evidence that engineered targeting of these enzymes effectively increased the local concentration in the nucleus.
Figure 1. Purified pdg proteins retain activity in vitro after addition of NLS and TAT peptides to enhance cellular delivery.
a. Human keratinocytes and skin fibroblasts were transfected with expression vectors for GFP, pdg-GFP, or pdg-NLS-GFP and GFP localization was visualized by direct immunofluoresence microscopy. DAPI (blue) indicates nuclei. Scale bar = 10 μm. b. Coomassie blue stained 15% SDS-PAGE gel showing the purity of pdg, pdg-NLS, or pdg-NLS-TAT enzymes after protein expression and purification. c. Plasmid nicking assay on UVC-irradiated DNA showing in vitro activity of the purified pdg enzymes after addition of NLS and TAT peptides. I = covalently closed circular, supercoiled plasmid DNA; II = nicked duplex DNA; III = linear duplex DNA; (-) = plasmid not treated with a pdg.
In order to achieve delivery of pdg-NLS proteins to skin cells without making use of gene transfection, the T4-pdg-NLS and Cv-pdg-NLS genes were further engineered at the carboxy terminus, following the NLS, to encode a peptide known to facilitate transmembrane protein delivery (TAT - Tyr-Gly-Arg-Lys-Lys-Arg-Arg-Gln-Arg-Arg-Arg). Following addition of the NLS and TAT sequences and protein expression, the resulting purified proteins (Figure 1b) were tested for activity using a plasmid nicking assay. The engineered pdg enzymes retained glycosylase/AP lyase activity to cleave CPDs in UVC-irradiated plasmid DNA (Figure 1c). The conversion of form I DNA (covalently closed circular, supercoiled plasmid DNA) to forms II (nicked duplex DNA) and III (linear duplex DNA) indicated active cleavage of CPDs by the pdg enzymes compared to the plasmid not treated with a pdg enzyme (-). Since Cv-pdg has both a broader substrate specificity and greater enzymatic stability, the remaining cellular experiments presented herein tested only the engineered Cv-pdg-NLS-TAT protein.
Delivery of Cv-pdg-NLS-TAT protein to the nuclei of cultured human skin cells and to a stratified human skin model
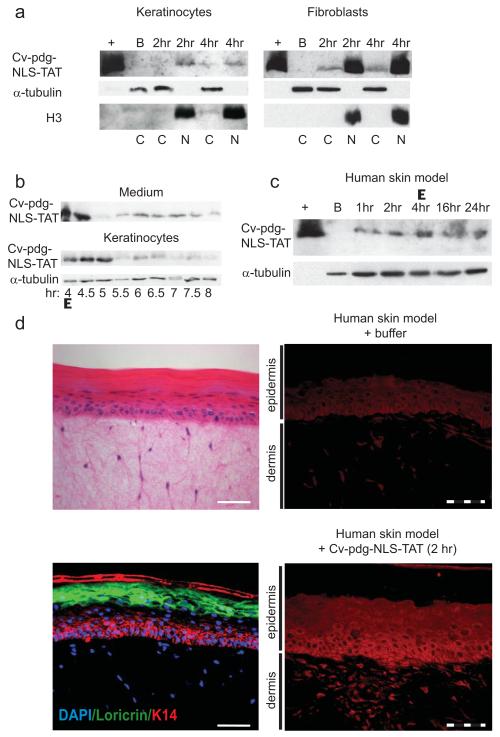
To determine whether the exogenously delivered ~20 kDa Cv-pdg-NLS-TAT protein could traverse skin cell membranes and localize to the nuclei, cells were incubated with 4 μg/ml Cv-pdg-NLS-TAT protein in the medium for 4 hours and harvested for fractionation. Immunoblots of fractionated cell lysates revealed differences between keratinocytes and fibroblasts with regard to enzyme delivery (Figure 2a). Cv-pdg-NLS-TAT was detected in the nuclei of both keratinocytes and fibroblasts by 2 hours, with higher levels in the nuclei of the fibroblasts. Since the mechanism by which TAT mediates transduction of proteins across cellular membranes remains controversial and may change depending on the cargo (Green et al., 2003; Richard et al., 2005), it is unknown whether differences in cell membrane composition or endocytosis capabilities might explain the observed differences between keratinocytes and fibroblasts with regard to uptake of Cv-pdg-NLS-TAT.
Figure 2. Delivery of Cv-pdg-NLS-TAT protein to the nuclei of cultured human skin cells and to a stratified human skin model.
a. Immunoblot of cytoplasmic (C) and nuclear (N) lysates after incubation with buffer (B) or Cv-pdg-NLS-TAT. Histone H3 and α-tubulin mark nuclei and cytoplasm, respectively. (+) = purified Cv-pdg-NLS-TAT. b. Immunoblot of Cv-pdg-NLS-TAT in concentrated medium or whole keratinocyte lysates. c. Immunoblot of lysates from skin model tissues treated with buffer (B) or Cv-pdg-NLS-TAT. d. Upper left: H&E staining of the skin model; lower left: IIF for expression of K14 (red) and Loricrin (green) with DAPI (blue) for nuclei; right: IIF of Cv-pdg (red) after culturing with buffer (upper) or Cv-pdg-NLS-TAT (lower). Solid bar = 50 μm. Dashed bar = 20 μm.
To determine the persistence of the enzyme in the cells after removal of excess enzyme in the medium, cells were incubated for 4 hours with Cv-pdg-NLS-TAT and then washed (Δ) with phosphate buffered saline (PBS). Fresh medium without enzyme was added and whole cell lysates were examined by immunoblot at various times after enzyme removal. The fibroblasts maintained the protein for up to 44 hours after medium change (48 hours total incubation, data not shown). However, the keratinocytes exhibited decreased levels of Cv-pdg-NLS-TAT protein by 1.5 hours after medium change and immunoblot analyses revealed that the protein was increasingly found in the medium (Figure 2b). Following this observation, to rule out the possibility that the Cv-pdg-NLS-TAT protein was influencing overall cell membrane permeability, keratinocytes and fibroblasts were stained with 0.4% Trypan Blue both immediately and 0.5, 1, and 2 hours after removal of extracellular Cv-pdg-NLS-TAT. The number of cells that stained with Trypan Blue (< 2%) did not change in the presence of Cv-pdg-NLS-TAT in either cell type (data not shown), suggesting that overall changes in membrane permeability were not the reason for the reduction of Cv-pdg-NLS-TAT in the keratinocytes and its appearance in the medium.
The Cv-pdg-NLS-TAT enzyme must be delivered through the stratum corneum of intact skin in order to catalyze DNA repair in basal keratinocytes from which a majority of non-melanoma skin cancers arise. To determine whether the Cv-pdg-NLS-TAT protein could traverse the stratum corneum, the enzyme was applied to the surface of a human skin model (Mattek Epiderm-FT™). This model is composed of dermal fibroblasts and stratified epidermal layers including basal keratinocytes, differentiated keratinocytes, and a stratum corneum. Cv-pdg-NLS-TAT was successfully delivered to the skin model as determined by immunoblot of whole tissue cellular lysates (Figure 2c). To test persistence of Cv-pdg-NLS-TAT in the skin, the enzyme was washed from the skin model surface with PBS after 4 hours of incubation (Δ), and remained detectable by immunoblot up to 20 hours after washing (24 hours total incubation).
To determine whether the Cv-pdg-NLS-TAT protein was delivered to basal keratinocytes in addition to differentiated keratinocytes, skin model samples that were incubated for 2 hours with buffer or Cv-pdg-NLS-TAT were analyzed by indirect immunofluorescence (IIF) using the Cv-pdg antibody. The upper left panel of Figure 2d shows Hematoxylin-Eosin (H&E) staining of the skin model and the lower left panel shows expression of specific epidermal markers, Keratin 14 and Loricrin, to demarcate basal keratinocytes and more differentiated keratinocytes, respectively. The upper right panel shows background fluorescence of the skin model incubated with buffer, while the lower right panel demonstrates that Cv-pdg-NLS-TAT was localized throughout the stratified epidermis, including the basal keratinocytes. While Cv-pdg-NLS-TAT was nuclear in some of the basal keratinocytes and almost all of the dermal fibroblasts, the enzyme surprisingly was primarily cytoplasmic in many basal and differentiated keratinocytes. This observation cannot be explained by loss of the NLS-TAT peptides since the protein size was not altered upon delivery to the skin (comparing sizes of pdgs with and without the NLS and TAT peptides in Figures 2c and 1b). Rather, the observation may be consistent with data from cultured keratinocytes that exhibited less Cv-pdg-NLS-TAT uptake and reduced retention of the enzyme compared to fibroblasts (Figures 2a and 2b).
Cv-pdg-NLS-TAT catalyzed rapid CPD cleavage in UVB-exposed skin cells
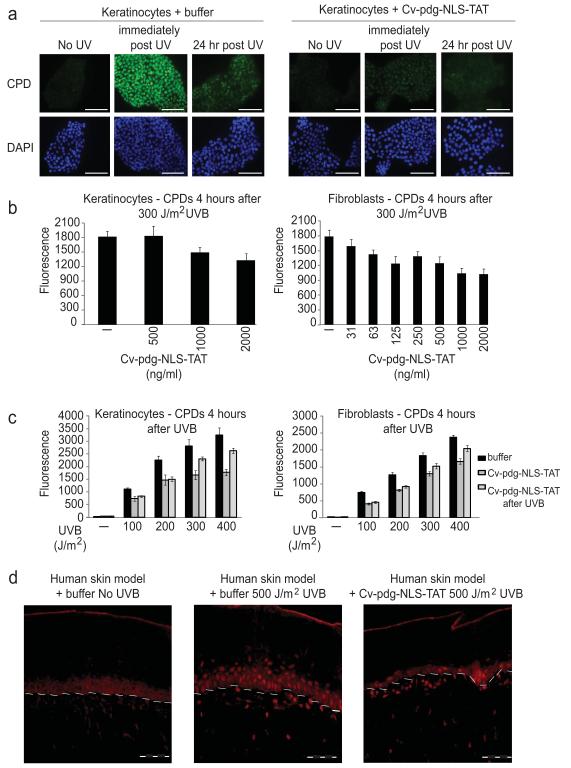
Although the Cv-pdg-NLS-TAT protein retained activity in vitro as purified from Escherichia coli, it was not known whether the enzyme would remain active to cleave CPDs once it had crossed the stratum corneum and the cellular and nuclear membranes of human skin cells. To address this question, cells plated on microscope cover slips were incubated with buffer or 2 μg of Cv-pdg-NLS-TAT per ml of culture medium for 4 hours prior to exposure to 100 J/m2 UVB. After exposure, medium containing Cv-pdg-NLS-TAT or buffer was refreshed so that the cells were incubated in the continual presence of the enzyme. The cells were fixed immediately after UVB exposure or 24 hours later and stained for detection of intact CPDs by IIF microscopy. Due to their normal NER proficiency, the buffer-treated keratinocytes exhibited a decrease in CPD detection by 24 hours (Figure 3a left series of panels). In striking contrast, the CPDs were nearly undetectable by IIF in Cv-pdg-NLS-TAT-treated keratinocytes as rapidly as the cells could be harvested after UVB exposure (Figure 3a right series of panels). Similar results were obtained for the NER-proficient fibroblasts (data not shown).
Figure 3. Cv-pdg-NLS-TAT catalyzed rapid CPD cleavage in UVB-exposed skin cells.
a. IIF to detect CPDs (green) in UVB-exposed keratinocytes pre-incubated with buffer or Cv-pdg-NLS-TAT. DAPI (blue) indicates nuclei. Scale bar = 50 μm. b. Graphical representation of CPD IIF intensity 4 hours after skin cells pre-incubated with buffer (−) or Cv-pdg-NLS-TAT were exposed to 300 J/m2 UVB. c. Same as b. except with increasing UVB doses (- = no UVB). Error bars = standard deviation of mean nuclear pixel intensities of 8 wells imaged. d. IIF to detect CPDs (red) in the skin model pre-treated with buffer or Cv-pdg-NLS-TAT and allowed 4 hour recovery after exposure to no UVB or 500 J/m2 UVB. White dashed line = dermal-epidermal junction. Dashed bar = 20 μm.
To further characterize the ability of Cv-pdg-NLS-TAT to initiate repair of CPDs in UVB-exposed skin cells, cells were pre-incubated for 4 hours with buffer or varying concentrations of Cv-pdg-NLS-TAT, exposed to 300 J/m2 UVB, allowed to recover for 4 hours in the continual presence of buffer or enzyme, and then fixed and analyzed by IIF to detect CPD presence using an automated imager. As shown in the left graph of Figure 3b, keratinocytes treated with 1 μg or 2 μg of Cv-pdg-NLS-TAT per ml of medium exhibited reduced CPD levels compared to cells treated with buffer alone. The detection of CPDs was reduced in fibroblasts treated with as little as 63 ng Cv-pdg-NLS-TAT per ml of medium compared to cells incubated with only buffer (right graph of Figure 3b), consistent with the data showing increased cellular uptake of Cv-pdg-NLS-TAT in fibroblasts (Figure 2a). For comparison, it is noted that a concentration of 1 μg of T4-pdg/ml was used in several previously published studies making use of that enzyme (Hacker et al., 2010; Yarosh et al., 2001).
To determine whether the UVB dose impacted the ability of CV-pdg-NLS-TAT to initiate CPD repair, cells were either incubated with buffer before and after exposure to increasing UVB doses (black bars Figure 3c), pre-incubated for 4 hours with 1 μg/ml Cv-pdg-NLS-TAT followed by addition of fresh 1 μg/ml Cv-pdg-NLS-TAT immediately after UVB exposure (dark grey bars Figure 3c), or incubated with buffer for 4 hours prior to UVB exposure followed by addition of 1 μg/ml Cv-pdg-NLS-TAT immediately after UVB exposure (light grey bars Figure 3c). Cells were fixed 4 hours after UVB exposure and analyzed by automated microscopy for IIF indicating CPD presence. Incubation with CV-pdg-NLS-TAT reduced the detectable CPDs in both keratinocytes and fibroblasts exposed to up to 400 J/m2 UVB compared to cells incubated with buffer alone. At the 300 and 400 J/m2 UVB exposures, Cv-pdg-NLS-TAT incubation pre- and post-exposure led to fewer detectable CPDs compared to cells that were given Cv-pdg-NLS-TAT only after UVB exposure. The difference between these two conditions was more subtle in the fibroblasts than in the keratinocytes, again possibly correlating with the more efficient enzyme uptake and retention observed in the fibroblasts (Figure 2a).
Next, experiments were conducted to determine whether Cv-pdg-NLS-TAT was capable of cleaving CPDs in the UVB-exposed skin model. The tissues were incubated for 4 hours with buffer or 3 μg Cv-pdg-NLS-TAT. After washing the enzyme from the skin model surface with PBS (Δ), the tissues were exposed to no UVB or 500 J/m2 UVB (a higher dose than for cultured cells because UVB must penetrate through the layers of the skin model) followed by a 4 hour recovery. IIF microscopy using the CPD antibody revealed reduced CPD fluorescence in the UVB-exposed cells of the skin model that had been pre-treated with Cv-pdg-NLS-TAT compared with those incubated with buffer (Figure 3d). Although Cv-pdg-NLS-TAT appeared to localize in the cytoplasm of many basal and differentiated keratinocytes (Figure 2d lower right panel), the reduced CPD levels presented in Figure 3d demonstrate that nuclear levels of the enzyme were sufficient to induce rapid cleavage of CPDs following UVB exposure in the majority of skin cells.
Discussion
The aim of this study was to demonstrate the feasibility of delivering an active DNA repair enzyme, Cv-pdg, to NER-proficient human keratinocytes and fibroblasts to initiate BER of UV-induced DNA damage. The data demonstrate that by engineering Cv-pdg to be expressed as a fusion with NLS and TAT peptides, such a modified enzyme can be delivered to the nuclei of cultured human keratinocytes and fibroblasts, as well as across the stratum corneum to the basal keratinocytes of a stratified human skin model. This Cv-pdg-NLS-TAT enzyme significantly increased the rate of initiation of CPD repair in NER-proficient skin cells following UVB exposure.
To our knowledge, TAT-mediated delivery of an active DNA repair enzyme across the barrier of a human skin model has previously not been reported, though other studies show TAT-mediated delivery of other enzymes and peptides to mouse and porcine skin (Lopes et al., 2005; Nakashima-Kamimura et al., 2008; Song et al., 2008). While the current study made use of a cultured model of human skin, the stratum corneum and thickness of the epidermal layers are highly similar to those found in vivo and it is thought that the use of such models represents a valid way to initially test epidermal drug delivery (Zghoul et al., 2001). Our data demonstrate that Cv-pdg-NLS-TAT delivery to a skin model is feasible, thus advancing a method for delivering therapeutic proteins across the skin barrier without the need for encapsulation into a delivery vehicle.
The data suggest differences between keratinocytes and skin fibroblasts with regard to the efficiency of TAT-mediated enzyme uptake, warranting further study. It is possible that the Cv-pdg-NLS-TAT sequence or delivery conditions can be further optimized to enhance uptake and retention of the enzyme, particularly in basal keratinocytes. Since these keratinocytes divide to replenish the upper cell layers and have been shown to be able to proliferate in the presence of potentially mutagenic CPDs (Courdavault et al., 2005), these cells are perhaps the most critical targets for Cv-pdg-NLS-TAT activity toward skin cancer prevention. Further, the persistence of CPDs in a particular subset of UV-irradiated basal keratinocytes is thought to be a key component of cellular transformation during skin carcinogenesis (Mitchell et al., 2001).
While normal human skin cells are capable of repairing UV-induced DNA damage through the NER pathway, the kinetics of CPD repair are relatively slow (Mouret et al., 2008). Interestingly, the most frequently formed CPDs, thymine-thymine dimers, are especially poorly recognized by NER machinery. In the Mouret et al. study, approximately 40% and 50% of these lesions remained in the DNA of cultured primary human keratinocytes and fibroblasts, respectively, 48 hours after exposure to 200 J/m2 UVB. In skin biopsies exposed to 1000 J/m2 UVB, the kinetics of repair of thymine-thymine dimers were even slower with ~60% of lesions remaining in the genome after 48 hours. It was shown that enhancing the rate of CPD repair using the CPD-photolyase enzyme (Jans et al., 2005) or T4-pdg (Bito et al., 1995; Yarosh et al., 1992) reduced the incidence of non-melanoma skin tumors in NER-proficient mice. In the current study, Cv-pdg-NLS-TAT, a novel DNA repair enzyme with broader substrate specificity than the previously studied T4-pdg (Jaruga et al., 2002; McCullough et al., 1998), was delivered to NER-proficient human skin cells and rapidly initiated CPD repair.
Cv-pdg-NLS-TAT may be a useful agent for prevention of skin cancer in NER-deficient XP patients, as well as specific NER-proficient populations, such as aged individuals and immunosuppressed organ transplant patients. Increased age correlates with a decreased rate of CPD repair following UV exposure, likely contributing to the exponentially increasing incidence of skin cancer associated with advancing age (Goukassian et al., 2000). Immune suppressed organ transplant patients frequently develop aggressive, metastatic SCCs (Abgrall et al., 2002). Regular sunscreen use decreases the skin cancer incidence in transplant patients, suggesting that reducing UV-induced DNA damage could lead to skin cancer prevention in this population (Ulrich et al., 2009). In conjunction with sunscreen use and sun protective behaviour, Cv-pdg-NLS-TAT treatment could be recommended to patients taking immunosuppressive drugs and aged patients to increase the cellular capacity to initiate CPD repair to prevent skin cancer.
Cv-pdg-NLS-TAT may also be useful for the prevention of SCC, BCC, and even melanoma in the general public. The total number of sunburns experienced throughout life and, specifically, DNA lesions in the form of CPDs play a role in melanoma etiology (Dennis et al., 2008; Mitchell et al., 2007; Wang et al., 2009). It should be noted that treatment with T4-pdg (lacking a NLS) prior to neonatal UV exposure of a melanoma-prone mouse model yielded no effect on the incidence or age of onset of melanoma compared to placebo treated controls, even though the enzyme initiates rapid repair of CPDs in the skin of the treated mice (Hacker et al., 2010). However, Cv-pdg-NLS-TAT could be more useful for melanoma prevention than T4-pdg due to its broader substrate specificity and the inclusion of the NLS to improve nuclear concentration. Cv-pdg-NLS-TAT will need to be tested for safety before efficacy trials can be conducted for prevention of skin cancer in humans. While humans treated with T4-pdg showed no adverse effects or changes in skin histology (Yarosh et al., 1996), Cv-pdg and T4-pdg share only 41% homology and it is unknown whether Cv-pdg protein or the NLS or TAT peptides will produce an allergic response or other negative effects in humans.
In conclusion, we showed that delivery of Cv-pdg-NLS-TAT to human skin cells leads to rapid initiation of CPD repair following exposure to UVB. Future studies will investigate the subsequent enzymatic steps and the biological consequences following initiation of CPD repair by Cv-pdg-NLS-TAT in repair-proficient human skin cells.
Materials and Methods
Cell and skin model culture, and UVB exposure
Spontaneously immortalized, NER-proficient human HaCaT keratinocytes (Boukamp et al., 1988; Takasawa et al., 2005) and an E6/E7 immortalized human skin fibroblast line (Iordanov et al., 2000) from a healthy donor were grown in high glucose Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone, Thermo Fisher Scientific, South Logan, UT) and 1% antibiotics/antimycotics (Invitrogen, Carlsbad, CA). The Epi-derm FT™ human skin model (Mattek, Ashland, MA) was cultured as suggested by the manufacturer for 48 hours prior to experiments. For UVB experiments, cells (in sufficient PBS to minimally cover the culture plate surface) or tissues were irradiated under a F15T8 UVB bulb with peak irradiance at 302 nm wavelength in a Blak-Ray XX-15M UV Bench Lamp (Ultraviolet Products Inc., Upland, CA). The exposure time (seconds) required to obtain the UVB doses (J/m2) used throughout this study was calculated by measuring μW/cm2 using an IL1400A photometer (International Light Technologies, Peabody, MA) calibrated annually by the manufacturer. The samples were exposed for 15 seconds for each 100 J/m2 UVB at a distance of 14 cm from the light source.
Expression vectors and enzyme purification
For cell transfection to demonstrate nuclear localization of pdg enzymes following addition of an NLS, the T4-pdg, T4-pdg-NLS, Cv-pdg, or Cv-pdg-NLS genes were subcloned into the pEGFP-N3 vector (Clontech, Mountain View, CA). For pdg enzyme purification, the above genes were subcloned into the pET-22b(+) vector with a carboxy-terminal 6 histidine tag (Novagen, EMD chemicals, Inc., Gibbstown, NJ). Standard purification protocols using Ni-NTA agarose beads were used to purify these enzymes to apparent homogeneity. The Supplemental Text details methods for pdg protein purification.
Transfection and visualization
HaCaT keratinocytes and immortalized human fibroblasts were cultured to 60% confluence on microscope cover slips (Fisher Scientific, Houston, TX) in a 6-well tissue culture dish, transiently transfected with the appropriate pEGFP-N3 or pdg-pEGFP-N3 vector with Lipofectin Reagent (Invitrogen), and grown for 16 hours. Cells were fixed in 4% formalin solution for 30 minutes at room temperature (RT) and washed 3 times in PBS. Cover slips were mounted on slides using ProLong Gold antifade reagent with DAPI (Invitrogen). Cells were visualized for direct GFP fluorescence with a Leica DM IRB microscope under a 63X magnification objective lens.
In vitro pdg activity assay
To determine if the pdg-NLS and pdg-NLS-TAT proteins were active to cleave DNA containing CPDs, pBR322 (Fermentas, Glen Bernie, MD) plasmid DNA was irradiated with UVC to induce more than 25 CPDs per plasmid using a Spectroline ENF-280C lamp (Spectronics Corporation, Westbury, NY) at 254 nm wavelength (peak intensity of 1290 μW/cm2 at 15.25 cm) and incubated with each purified pdg protein at 37°C for 1 hour in reaction buffer [25 mM Tris-HCl (pH 8.0), 20 mM NaCl, 1 mM EDTA, 100 μg/ml BSA]. Supercoiled, nicked, and linearized DNA products were separated by electrophoresis through a 1% agarose gel.
Preparation of Cv-pdg-NLS-TAT for delivery to human skin cells
Purified Cv-pdg-NLS-TAT protein in elution buffer containing 400 mM imidazole (see Supplemental Text for protein purification methods) was concentrated by centrifugation using an Amicon device with a low-binding Ultracel membrane that has a 3 kDa molecular weight cut off (Fisher Scientific, Houston, TX). PBS was added and the centrifugation was repeated 5 times to dilute the imidazole concentration to less than 20 mM. A control buffer for cell culture experiments was prepared by treating elution buffer alone with 5 additions of PBS. The amount of Cv-pdg-NLS-TAT protein delivered in these studies was as follows: 4 μg/ml culture medium for cells harvested for immunoblots to show protein delivery (Figures 2a and 2b); 2 μg/ml culture medium for cells grown on cover slips for IIF microscopy of CPDs (Figure 3a); the concentrations indicated in Figures 3b and 3c for automated IIF microscopy of CPDs; and a total of 3 μg topically delivered in 100 μl of PBS to each human skin model sample (Figures 2c, 2d, and 3d).
Primary antibodies used for immunoblot and IIF
Primary antibodies included Cv-pdg (produced in rabbits against the full length non-denatured protein), α-tubulin (Sigma-Aldrich, Saint Louis, MO), histone H3 (Active Motif, Carlsbad, CA), anti-cytokeratin 14 (Fitzgerald, Concord, MA), anti-loricrin (Covance, Emeryville, CA), and CPD (Cosmo Bio Co., LTD, Tokyo, Japan using a protocol as described in (Schul et al., 2002)). Detailed methods describing preparation of samples as well as immunoblot analyses of Cv-pdg-NLS-TAT delivery, IIF microscopy for proteins and for CPDs, and histology of the skin model can be found in the Supplemental Text.
Automated imaging for detection of CPDs
Cells were cultured until ~70% confluence in 24-well plates (Corning, Corning, NY) and incubated for 4 hours with buffer or Cv-pdg-NLS-TAT. Following UVB exposure, cells from each 24-well sample were trypsinized and plated into 4 wells of a non-siliconized type I collagen-coated 96-well view plate (Greiner Bio-One, Monroe, NC). The replating step was necessary due to the UV-protective shadowing effect of the wells in a 96-well plate. The detection of CPDs by IIF was as detailed in (Schul et al., 2002). The Alexa Fluor 647-conjugated secondary goat anti-mouse IgG antibody (Invitrogen) was used. Total nuclear DNA was stained using 5 μg/ml of 4′, 6-diamidino-2-phenylindole (DAPI, KPL, Gaithersburg, MD). Cells were imaged with 405 nm and 647 nm lasers using a 10X objective on an OPERA LX reader (Perkin-Elmer, Waltham, MA) at the Oregon Translation Research and Drug Development Institute (Portland, OR). Images were analyzed using a script developed under the Acapella software environment (Perkin-Elmer). Briefly, the nuclei of individual cells were identified based on the DAPI fluorescence (read at 405 nm), then the areas in the correlating 647 nm (CPD antibody stain) image were mapped and the pixel intensities recorded on a cell-by-cell basis. More than 5000 cells were analyzed for each sample and the mean nuclear 647 nm fluorescence intensity was graphically reported.
Supplementary Material
Acknowledgements
This research was funded in part by NIH R01 ES04091 and Restoration Genetics, Inc. STTR grant R41 CA114923. Dr. Johnson’s salary was supported by the Ruth L. Kirschstein National Research Service Training Grant Award ES007060-28 from Oregon State University’s Department of Environmental and Molecular Toxicology and some research supplies were provided by a Dermatology Foundation Research Grant. We acknowledge Dr. Yih-Tai Chen at OTRADI for his expertise with automated microscopy. Dr. Mihail Iordanov provided the E6/E7 immortalized skin fibroblasts. We thank Lauriel Earley, James Lagowski and Drs. Anuradha Kumari and Irina G. Minko for insightful discussions concerning these investigations and critically reading the manuscript.
Abbreviations
- UV
ultraviolet light
- NER
nucleotide excision repair
- BER
base excision repair
- XP
Xeroderma Pigmentosum
- Pdg
pyrimidine dimer glycosylase
- CPD
cyclobutane pyrimidine dimer
- Cv
Chlorella virus
- NLS
nuclear localization sequence
- TAT
transcriptional transactivator peptide
- IIF
indirect immunofluorescence
- SCC
squamous cell carcinoma
- BCC
basal cell carcinoma
Footnotes
Delivery of DNA repair enzymes to skin cells
Conflict of Interest: OHSU and Drs. McCullough and Lloyd have a financial interest in Restoration Genetics, Inc., a company that may have commercial interest in the results of this research and technology. This potential conflict of interest has been reviewed and managed by OHSU and the Integrity Program Oversight Council.
References
- Abgrall S, Orbach D, Bonhomme-Faivre L, et al. Tumors in organ transplant recipients may give clues to their control by immunity. Anticancer Res. 2002;22:3597–3604. [PubMed] [Google Scholar]
- Bickers DR, Lim HW, Margolis D, et al. The burden of skin diseases: 2004 a joint project of the American Academy of Dermatology Association and the Society for Investigative Dermatology. J Am Acad Dermatol. 2006;55:490–500. doi: 10.1016/j.jaad.2006.05.048. [DOI] [PubMed] [Google Scholar]
- Bito T, Ueda M, Nagano T, et al. Reduction of ultraviolet-induced skin cancer in mice by topical application of DNA excision repair enzymes. Photodermatol Photoimmunol Photomed. 1995;11:9–13. doi: 10.1111/j.1600-0781.1995.tb00130.x. [DOI] [PubMed] [Google Scholar]
- Bos JD, Meinardi MM. The 500 Dalton rule for the skin penetration of chemical compounds and drugs. Exp Dermatol. 2000;9:165–169. doi: 10.1034/j.1600-0625.2000.009003165.x. [DOI] [PubMed] [Google Scholar]
- Boukamp P, Petrussevska RT, Breitkreutz D, et al. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafardi JA, Elmets CA. T4 endonuclease V: review and application to dermatology. Expert Opin Biol Ther. 2008;8:829–838. doi: 10.1517/14712598.8.6.829. [DOI] [PubMed] [Google Scholar]
- Courdavault S, Baudouin C, Charveron M, et al. Repair of the three main types of bipyrimidine DNA photoproducts in human keratinocytes exposed to UVB and UVA radiations. DNA Repair (Amst) 2005;4:836–844. doi: 10.1016/j.dnarep.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Dennis LK, Vanbeek MJ, Beane Freeman LE, et al. Sunburns and risk of cutaneous melanoma: does age matter? A comprehensive meta-analysis. Ann Epidemiol. 2008;18:614–627. doi: 10.1016/j.annepidem.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson JM, DeSpain J, Hewett JE, et al. Malignant potential of actinic keratoses and the controversy over treatment. A patient-oriented perspective. Arch Dermatol. 1991;127:1029–1031. [PubMed] [Google Scholar]
- Francis MA, Bagga P, Athwal R, et al. Partial complementation of the DNA repair defects in cells from xeroderma pigmentosum groups A, C, D and F but not G by the denV gene from bacteriophage T4. Photochem Photobiol. 2000;72:365–373. doi: 10.1562/0031-8655(2000)072<0365:pcotdr>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Furuta M, Schrader JO, Schrader HS, et al. Chlorella virus PBCV-1 encodes a homolog of the bacteriophage T4 UV damage repair gene denV. Appl Environ Microbiol. 1997;63:1551–1556. doi: 10.1128/aem.63.4.1551-1556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goukassian D, Gad F, Yaar M, et al. Mechanisms and implications of the age-associated decrease in DNA repair capacity. FASEB J. 2000;14:1325–1334. doi: 10.1096/fj.14.10.1325. [DOI] [PubMed] [Google Scholar]
- Green I, Christison R, Voyce CJ, et al. Protein transduction domains: are they delivering? Trends Pharmacol Sci. 2003;24:213–215. doi: 10.1016/S0165-6147(03)00076-2. [DOI] [PubMed] [Google Scholar]
- Hacker E, Muller HK, Hayward N, et al. Enhancement of DNA repair using topical T4 endonuclease V does not inhibit melanoma formation in Cdk4(R24C/R24C)/Tyr-Nras(Q61K) mice following neonatal UVR. Pigment Cell Melanoma Res. 2010;23:121–128. doi: 10.1111/j.1755-148X.2009.00643.x. [DOI] [PubMed] [Google Scholar]
- Hussein MR. Ultraviolet radiation and skin cancer: molecular mechanisms. J Cutan Pathol. 2005;32:191–205. doi: 10.1111/j.0303-6987.2005.00281.x. [DOI] [PubMed] [Google Scholar]
- Iordanov MS, Wong J, Newton DL, et al. Differential requirement for the stress-activated protein kinase/c-Jun NH(2)-terminal kinase in RNAdamage-induced apoptosis in primary and in immortalized fibroblasts. Mol Cell Biol Res Commun. 2000;4:122–128. doi: 10.1006/mcbr.2000.0266. [DOI] [PubMed] [Google Scholar]
- Jans J, Schul W, Sert YG, et al. Powerful skin cancer protection by a CPD-photolyase transgene. Curr Biol. 2005;15:105–115. doi: 10.1016/j.cub.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Jaruga P, Jabil R, McCullough AK, et al. Chlorella virus pyrimidine dimer glycosylase excises ultraviolet radiation- and hydroxyl radical-induced products 4,6-diamino-5-formamidopyrimidine and 2,6-diamino-4-hydroxy-5-formamidopyrimidine from DNA. Photochem Photobiol. 2002;75:85–91. doi: 10.1562/0031-8655(2002)075<0085:cvpdge>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Kibitel JT, Yee V, Yarosh DB. Enhancement of ultraviolet-DNA repair in denV gene transfectants and T4 endonuclease V-liposome recipients. Photochem Photobiol. 1991;54:753–760. doi: 10.1111/j.1751-1097.1991.tb02086.x. [DOI] [PubMed] [Google Scholar]
- Kraemer KH, Patronas NJ, Schiffmann R, et al. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship. Neuroscience. 2007;145:1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusewitt DF, Budge CL, Ley RD. Enhanced pyrimidine dimer repair in cultured murine epithelial cells transfected with the denV gene of bacteriophage T4. J Invest Dermatol. 1994;102:485–489. doi: 10.1111/1523-1747.ep12373084. [DOI] [PubMed] [Google Scholar]
- Kusewitt DF, Dyble J, Sherburn TE, et al. Altered UV resistance and UV mutational spectrum in repair-proficient murine fibroblasts expressing endonuclease V. Mutat Res. 1998;407:157–168. doi: 10.1016/s0921-8777(98)00004-4. [DOI] [PubMed] [Google Scholar]
- Lopes LB, Brophy CM, Furnish E, et al. Comparative study of the skin penetration of protein transduction domains and a conjugated peptide. Pharm Res. 2005;22:750–757. doi: 10.1007/s11095-005-2591-x. [DOI] [PubMed] [Google Scholar]
- McCullough AK, Romberg MT, Nyaga S, et al. Characterization of a novel cis-syn and trans-syn-II pyrimidine dimer glycosylase/AP lyase from a eukaryotic algal virus, Paramecium bursaria chlorella virus-1. J Biol Chem. 1998;273:13136–13142. doi: 10.1074/jbc.273.21.13136. [DOI] [PubMed] [Google Scholar]
- Mitchell D, Paniker L, Sanchez G, et al. The etiology of sunlight-induced melanoma in Xiphophorus hybrid fish. Mol Carcinog. 2007;46:679–684. doi: 10.1002/mc.20341. [DOI] [PubMed] [Google Scholar]
- Mitchell DL, Volkmer B, Breitbart EW, et al. Identification of a non-dividing subpopulation of mouse and human epidermal cells exhibiting high levels of persistent ultraviolet photodamage. J Invest Dermatol. 2001;117:590–595. doi: 10.1046/j.0022-202x.2001.01442.x. [DOI] [PubMed] [Google Scholar]
- Mouret S, Charveron M, Favier A, et al. Differential repair of UVB-induced cyclobutane pyrimidine dimers in cultured human skin cells and whole human skin. DNA Repair (Amst) 2008;7:704–712. doi: 10.1016/j.dnarep.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Nakashima-Kamimura N, Nishimaki K, Mori T, et al. Prevention of chemotherapy-induced alopecia by the anti-death FNK protein. Life Sci. 2008;82:218–225. doi: 10.1016/j.lfs.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Richard JP, Melikov K, Brooks H, et al. Cellular uptake of unconjugated TAT peptide involves clathrin-dependent endocytosis and heparan sulfate receptors. J Biol Chem. 2005;280:15300–15306. doi: 10.1074/jbc.M401604200. [DOI] [PubMed] [Google Scholar]
- Schul W, Jans J, Rijksen YM, et al. Enhanced repair of cyclobutane pyrimidine dimers and improved UV resistance in photolyase transgenic mice. Embo J. 2002;21:4719–4729. doi: 10.1093/emboj/cdf456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HY, Lee JA, Ju SM, et al. Topical transduction of superoxide dismutase mediated by HIV-1 Tat protein transduction domain ameliorates 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced inflammation in mice. Biochem Pharmacol. 2008;75:1348–1357. doi: 10.1016/j.bcp.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Takasawa R, Nakamura H, Mori T, et al. Differential apoptotic pathways in human keratinocyte HaCaT cells exposed to UVB and UVC. Apoptosis. 2005;10:1121–1130. doi: 10.1007/s10495-005-0901-8. [DOI] [PubMed] [Google Scholar]
- Ulrich C, Jurgensen JS, Degen A, et al. Prevention of non-melanoma skin cancer in organ transplant patients by regular use of a sunscreen: a 24 months, prospective, case-control study. Br J Dermatol. 2009;161(Suppl 3):78–84. doi: 10.1111/j.1365-2133.2009.09453.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Digiovanna JJ, Stern JB, et al. Evidence of ultraviolet type mutations in xeroderma pigmentosum melanomas. Proc Natl Acad Sci U S A. 2009;106:6279–6284. doi: 10.1073/pnas.0812401106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarosh D, Alas LG, Yee V, et al. Pyrimidine dimer removal enhanced by DNA repair liposomes reduces the incidence of UV skin cancer in mice. Cancer Res. 1992;52:4227–4231. [PubMed] [Google Scholar]
- Yarosh D, Bucana C, Cox P, et al. Localization of liposomes containing a DNA repair enzyme in murine skin. J Invest Dermatol. 1994;103:461–468. doi: 10.1111/1523-1747.ep12395551. [DOI] [PubMed] [Google Scholar]
- Yarosh D, Klein J, Kibitel J, et al. Enzyme therapy of xeroderma pigmentosum: safety and efficacy testing of T4N5 liposome lotion containing a prokaryotic DNA repair enzyme. Photodermatol Photoimmunol Photomed. 1996;12:122–130. doi: 10.1111/j.1600-0781.1996.tb00188.x. [DOI] [PubMed] [Google Scholar]
- Yarosh D, Klein J, O’Connor A, et al. Effect of topically applied T4 endonuclease V in liposomes on skin cancer in xeroderma pigmentosum: a randomised study. Xeroderma Pigmentosum Study Group. Lancet. 2001;357:926–929. doi: 10.1016/s0140-6736(00)04214-8. [DOI] [PubMed] [Google Scholar]
- Zghoul N, Fuchs R, Lehr CM, et al. Reconstructed skin equivalents for assessing percutaneous drug absorption from pharmaceutical formulations. ALTEX. 2001;18:103–106. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.