Abstract
The biology, feeding ecology and phylogenetic relationships of marine snails in the family Turridae remain poorly understood. Here we report our study on four deep-water species in the genus Gemmula, a major group in this family. The four species G. speciosa (Reeve 1843), G. sogodensis (Olivera 2005), G. kieneri (Doumet 1940) and G. diomedea (Powell 1964) were collected at five different sites in the Philippines, and their pattern of distribution in the sites, their feeding behaviour as well as their phylogenetic relationships with each other and with other members of the subfamily Turrinae were investigated. The radular morphology (of two Gemmula species) and potential prey (for one Gemmula species) were also examined. Actual feeding observations were also conducted for Gemmula speciosa and compared with two turrids from other genera.
All four Gemmula species showed strikingly different patterns of distribution; each species was found to be relatively much more abundant at one site but not at the other sites. Molecular phylogenetic analysis based on 16S sequences correlated with previously reported 12S sequences and revealed that the four species all belong to a well-supported Gemmula clade within the subfamily Turrinae; and that this clade appeared more closely related to the clades Xenuroturris, Turris and Lophiotoma than to the other clades in the subfamily (i.e., Turridrupa, Unedogemmula and Polystira). Morphological analysis of the radula of both G. speciosa and G. sogodensis revealed that the radulae of the two species were similar but differed from the other turrids, Lophiotoma acuta and Unedogemmula bisaya, by the absence of central teeth, consistent with the separation of the Gemmula clade from the Lophiotoma and Unedogemmula clade.
To identify the polychaete group that is targeted as prey by species of Gemmula, analysis of regurgitated food fragments was made; phylogenetic analysis of an mtCOI gene fragment that was PCR-amplified from the regurgitated tissue of one specimen (G. diomedea) indicated close affinity of the prey to the terebellid polychaete Amphitritides. Specimens of Gemmula speciosa, when challenged with the terebellid polychaete Loimia sp., were observed to attack the worm suggesting that Gemmula species feed on terebellid polychaetes. Lophiotoma acuta were also observed to feed on the same species of terebellid but were usually group-feeding in contrast to the solitary feeding of G. speciosa. Unedogemmula bisaya did not feed on the terebellid which also supports the separation of the Gemmula and Unedogemmula clade.
Two lines of proof (i.e. the molecular phylogenetic analysis and the feeding challenge) supporting the toxin homology findings previously reported, provide consistent evidence that Gemmula is a distinct clade of worm-hunting Turrinae that feeds on Terebellidae.
Introduction
Marine snails belonging to the family Turridae (the “turrids”) comprise a large and highly diverse subgroup within the superfamily Conoidea, a group consisting of venomous marine gastropods. In fact, turrids comprise the largest family of deep-sea gastropods (Rex et al. 2000) and account for the vast majority of conoidean biodiversity (Bouchet et al. 2002). However, the biology, ecology, and species-level phylogenetic relationships of these organisms are still poorly known. A number of factors contribute to the difficulty of studying these organisms, such as high morphological similarity among species, poor accessibility of their habitats (50–500 m depth), their small size, and their nocturnal and burrowing behavior. Recent studies have started to shed light on the biochemistry, toxinology and toxin gene expression of this group, revealing its potential as a source of bioactive peptides (López-Vera et al. 2004, Watkins et al. 2006, Heralde et al. 2008). Understanding the ecology, i.e., feeding behavior, and genetic diversity of these organisms will therefore be of interest to marine biologists as well as marine biotechnologists.
As part of an initial effort to characterize this group, this study focuses on the genus Gemmula (the “gem turrids”). The members of this genus largely occur in deeper tropical waters (at depths of 50–500 meters) and comprise a major group in the subfamily Turrinae (Powell 1964). Other genera conventionally included in this subfamily are Turris, Lophiotoma, Unedogemmula, Turridrupa and Polystira (Heralde et al. 2007, Powell 1964, Taylor et al. 1993). Several species of Gemmula have been collected in relatively large numbers in Philippine waters. In this study, we particularly focused on four deep-water Gemmula species, namely, G. speciosa (Reeve 1843) (the “splendid gem turrid”), G. sogodensis (Olivera, 2004), G. diomedea (Powell 1964), and G. kieneri (Doumet 1840) (Figure 1). We investigated their distribution and phylogeny, as information on the pattern of their distribution is scant and the phylogeny of these species has not yet been elucidated. In the superfamily Conoidae, most previous investigations of molecular phylogeny have been carried out on Conus (Duda and Palumbi 1999, Duda et al. 2001, Espiritu et al. 2001, Monje et al. 1999); phylogenetic relationships for the vast biodiversity of turrids (Powell 1964, Taylor et al. 1993, Bouchet et al. 2002, and Puillandre et al. 2008) are still poorly understood. So far, only one study has been carried out (Heralde et al. 2007) which defined clades in the subfamily Turrinae.
Figure 1.
Shells of Gemmula species. Top to bottom, Gemmula speciosa, Gemmula diomedea, Gemmula sogodensis, Gemmula kieneri.
To further discriminate the species, we examined and compared the foregut anatomy, i.e., radula, of three species. Because their habitats are inaccessible, little is also known of their feeding biology. Although turrids in general are known to feed on polychaetes, there are no available data on which species of polychaetes are preyed on by Gemmula (or any turrid) species. We therefore collected new data on feeding behavior and potential prey preferences.
Materials and Methods
Sample Collection
Snails were purchased from local fishermen as by-catch in trawl nets along the mouth of Manila Bay (from Bataan to Cavite and Batangas) and tangle nets in the seas of Cebu and Bohol. Live snails were kept in seawater until they were processed for anatomical or molecular work. The relative distribution and abundance of Gemmula speciosa along the periphery of Manila Bay was initially assessed by monitoring the collections per trawl of selected boats in August 2005 and from October 2005 to January 2006. The abundance in all the sampling sites was monitored from the snails collected by the fishermen from February to May 2006.
Specimen Preparation and DNA Extraction
The snails were segregated by putative species and preserved for various uses. The snails were cracked and tissue samples (hepatopancreas and foot) were obtained and preserved in approximately 10 volumes of RNALater. Voucher specimens were kept in 70% ethanol. DNA extraction was performed in fifty mg tissue samples (hepatopancreas or foot) using the Puregene DNA kit (Invitrogen) or the DNeasy tissue kit (Qiagen) and aliquots were prepared..
Gene amplification
The 16S mitochondrial rRNA gene was amplified using the primers 16SL (5′-GTTTACCAAAAACATGGCTTC-3′) and 16SH (5′-CCGGTCTGAACTCAGATC ACGT-3′) with uracil adaptor sequences. A PCR mix containing 20–40 ng genomic DNA, 2μM of each primer, 2μM of dNTP and 2μM of Taq Polymerase was prepared and cycled with the following profile: 95°C 1 min initial denaturation; 40 cycles of 95°C 20 sec denaturation, 55°C 20 sec annealing and 72°C 30 sec extension; and 72°C 5 min final extension. The PCR product was ligated to pNEB206A (USER Friendly Cloning, New England Biolabs) and introduced into E. coli (DH5a) through chemical transformation. Plasmids from transformants with inserts were sequenced through the ABI 377 DNA Sequencer or submitted to the University of Utah Sequencing Facility.
Phylogenetic Analysis
The 16S rDNA sequences of 22 conoidean samples analyzed in the study were aligned with Genbank-derived sequences for coniids and Rhinoclavis aspera, a mesogastropod as root (Table 2). A minimum evolution-based phylogenetic reconstruction was made using MEGA 3.1 (Kumar et al. 2001). Bootstrap values were calculated and putative clades were marked accordingly. The genetic distances were computed using the Kimura-2-parameter model to determine the range of distances of the specimens that belong to a food type cluster.
Table 2.
List of gastropod species utilized for analysis in this study
| Species | Source | 12S Sequence | 16S Sequence |
|---|---|---|---|
| Gemmula sogodensis | Sogod, Cebu, Philippines | EF467337* | GU434132 |
| Gemmula speciosa | Batangas, Philippines | EF467338* | GU434131 |
| Gemmula diomedea | Panglao Is. Bohol, Philippines | EF467334* | GU434133 |
| Gemmula lisajoni | Sogod, Cebu, Philippines | EF467335* | GU434134 |
| Gemmula rosario | Sogod, Cebu, Philippines | EF467336* | GU434135 |
| Gemmula ambara | Marinduque, Philippines | - | GU471196 |
| Lophiotoma acuta | Marinduque, Philippines | EF467339* | GU471195 |
| Lophiotoma albina | Marinduque, Philippines | - | GU471186 |
| Lophiotoma kingae | Cawoy, Olango Island, Philippines | Submitted to GB | GU434137 |
| Lophiotoma olangoensis | Cawoy, Olango Island, Philippines | EF467345* | GU434136 |
| Lophiotoma polytropa | Bohol, Philippines | EF467347* | GU471190 |
| Turris garnonsii | Cawoy, Olango Island, Philippines | EF467352* | GU434139 |
| Turris grandis | Sogod, Cebu, Philippines | EF467353* | GU434138 |
| Turris babylonia | Cawoy, Olango Island, Philippines | EF467351* | GU434140 |
| Turris spectabilis | Cawoy, Olango Island, Philippines | EF467355* | GU471188 |
| Turris normandavidsoni | Sogod, Cebu, Philippines | EF467354* | GU471189 |
| Unedogemmula bisaya | Batangas, Philippines | EF467340* | GU471187 |
| Unedogemmula tayabasensis | Sogod, Cebu, Philippines | EF467348* | GU471185 |
| Clavus unizonalis | Cawoy, Mactan Is, Cebu, Philippines | - | Submitted to GB |
| Drillia regius | Panglao Is. Bohol, Philippines | EF467333* | GU471193 |
| Turridrupa bijubata | Sogod, Cebu, Philippines | Submitted to GB | GU471192 |
| Terebra subulata | Buenavista, Marinduque, Philippines | - | AF174213* |
| Terebra areolata | Buenavista, Marinduque, Philippines | - | GU471184 |
| Terebra crenulata | Buenavista, Marinduque, Philippines | - | GU471194 |
| Conus rolani | Sogod, Cebu, Philippines | - | GU471191 |
| Conus emaciatus | - | - | AF126018* |
| Conus virgo | - | - | AF086616* |
| Conus flavidus | - | - | AF160704* |
| Conus terebra | - | - | AF103815* |
| Conus pulicarius | - | - | AF143992* |
| Conus vexillum | - | - | AF108822* |
| Conus capitaneus | - | - | AF126014* |
| Conus miles | - | - | AF108821* |
| Rhinoclavis aspera | - | - | AF174212* |
| Lophiotoma cingulifera | Cawoy, Olango Island, Philippines | EF467342* | Submitted to GB |
| Lophiotoma cerithiformes | Oahu, Hawaii | EU682298* | EU682307* |
| Unedogemmula panglaoensis | Panglao Is., Bohol, Philippines | EF467346* | Watkins (unpubl) |
| Hastula hectica | Panglao Is. Bohol, Philippines | Submitted to GB | Submitted to GB |
| Terebra guttatta | Cawoy, Olango Island, Philippines | Watkins (unpubl) | Watkins (unpubl) |
| Polystira oxytropis | - | Watkins (unpubl) | Watkins (unpubl) |
| Polystira picta | - | Watkins (unpubl) | Watkins (unpubl) |
| Unedogemmula indica | - | EF467343* | Watkins (unpubl) |
| Unedogemmula leucotropis | - | Watkins (unpubl) | Watkins (unpubl) |
| Turridrupa elongata | Cawoy, Olango Island, Philippines | Watkins (unpubl) | Watkins (unpubl) |
| Lophiotoma jickeli | Cawoy, Olango Island, Philippines | EF467344* | Watkins (unpubl) |
| Gemmula kieneri | Panglao Is. Bohol, Philippines | Submitted to GB | Watkins (unpubl) |
Genbank Derived Sequences
Note: The GU series of accession numbers are sequences obtained in this paper.
A second phylogenetic analysis was made using the combined 12S rDNA and 16S rDNA (12S previously reported in Heralde et al. 2007) sequences. The concatenated sequences were aligned using Clustal X. The alignments were refined manually using MacClade 4.08. The process was repeated for some highly variable regions as long as further refinement by eye seemed possible.
Trees were optimized using the individual rRNA gene sequence alignments and the concatenated alignments (presented herein). Final analyses were restricted to model-based maximum likelihood (PAUP4b10) and Bayesian inference (MrBayes 3.1.2) to account for the complexity of sequence evolution. Sequence evolution parameters were optimized by a GTR+I+G model that includes six possible substitution types (GTR), allows some sites to be invariant (I), allows across-site rate heterogeneity (G) and allows unequal base frequencies.
The maximum likelihood optimization used TBR branch swapping with 10 searches, each using a random addition of taxa. The analysis ended when the PAUP default criteria for convergence of the log-likelihood were met.
The Bayesian analysis was run for two million generations with the first 500,000 generations discarded as burn-in trees. Two MCMCMC runs (Metropolis-Coupled Markov-Chain Monte-Carlo), using four chains each, were used to thoroughly explore tree space. Convergence of the likelihoods was judged adequate by monitoring the ASED (Average Standard Error of the Difference) in split frequencies between the two runs and by comparing plots of the tree log-likelihood trees from generation 500,000 to 2 million. By the last generation, average standard error was 0.0039; the plot of likelihoods versus generation had stabilized. Furthermore, the PSRF (Potential Scale Reduction Factor) reached 1.00 for the total tree length and for each model parameter.
Since maximum likelihood and Bayesian analyses converged to the same tree, only the Bayesian results are presented below (ML results are available from PSC).
Species in the phylogenetic analysis included the following: Turrinae: Gemmula speciosa, Gemmula diomedea, Gemmula. kieneri, Gemmula sogodensis, Lophiotoma albina, Lophiotoma acuta, Lophiotoma cerithiformis, Lophiotoma olangoensis, Lophiotoma cingulifera, Lophiotoma kingae, Lophiotoma jickelli, Lophiotoma polytropa, Turris gamonsii, Turris babylonia, Turris spectabilis, Turris normandavidsoni, Turris grandis, Turridrupa elongata, Turridrupa bijubata, Unedogemmula bisaya, Unedogemmula leucotropis, Unedogemmula tayabasensis, Unedogemmula indica, Unedogemmula panglaoensis, Polystira oxytropis, Polystira picta; and Terebridae:Hastula hectica and Terebra guttata,.
Scanning Electron Microscopy (SEM) of the Radula
Live snails were relaxed in 1% cold magnesium chloride (MgCl2) for 2–3 hours and preserved in 95% ethanol; the SEM of their radulae was carried out as described previously (Imperial et al. 2007).
Molecular regurgitate analysis
Six samples of Gemmula diomedea caught by trawling at depths of 231–271 meters in the Panglao 2005 Expedition were relaxed in cold 1% magnesium chloride for at least 2 hours and regurgitated tissues were recovered. Genomic DNA was extracted from the tissues using the DNeasy tissue kit (Qiagen). An aliquot was prepared as template for mtCOI amplification using modified universal primers with USER adaptor sequences (Simison 2000). Subsequent cloning into pNEB206A, transformant screening and plasmid sequencing were as described above. The mtCOI sequence obtained was searched in the Genbank database using BLAST (Basic Local Alignment Search Tool).
Tank feeding preference and competition experiments
The snails used for the feeding experiments were maintained indoor using a 56-liter aquarium containing seawater with salinity maintained at a range between 35–37 ppt. A filtration system and an aerator were in place while the feeding behavior of G. speciosa and other turrids was observed. The snails used in the experiment had been in the tanks for a period of 2–4 weeks with artificial lighting following a 12 hour light-dark cycle. The introduction of live terebellid worms into the tank was done at night.
Results
Species distribution
The four species of Gemmula were obtained from two different biogeographic regions. The first three sites came from Manila Bay in the South China Sea region: site 1 is close to the Bataan peninsula, site 2 is off Corregidor Island and site 3 is off the Batangas coast. At these three sites, Gemmula specimens were obtained as by-catch of commercial fish trawlers operating in these areas. The only larger Gemmula species collected was G. speciosa (Olivera 2005, Powell 1964); no specimens of G. sogodensis or G. diomedea were found at sites 1–3 (Table 1). Specimens were mostly trawled at depths of 50 to 100 meters.
Table 1.
Gemmula species collected at various sites in the period February-May, 2006
| Species | Sampling Sites 1–3: Bataan, Corregidor Is., Batangas, Luzon | Sampling Site 4: Sogod, Cebu | Sampling Site 5: Panglao Is., Bohol | |||
|---|---|---|---|---|---|---|
| Total Number | (%) | Total Number | (%) | Total Number | (%) | |
| G. speciosa | 868 | 87.7% | 1 | 0.2% | 0 | 0.0% |
| G. sogodensis | 0 | 0.0% | 333 | 76.6% | 1 | 0.8% |
| G. diomedea | 0 | 0.0% | 4 | 0.9% | 108 | 81.2% |
| G. kieneri | 0 | 0.0% | 0 | 0.0% | 24 | 18.0% |
| U. bisaya | 122 | 12.3% | 97 | 22.3% | 0 | 0.0% |
G. speciosa was only rarely collected at the southern sites within the Visayan Seas biogeographic region (off Sogod, Cebu and off the island of Panglao in Bohol). The primary method for collection at the latter sites was tangle nets mostly laid at greater depths. At the Sogod, Cebu site, the major Gemmula species collected was G. sogodensis (Olivera 2005). Off Panglao, Bohol, G. diomedea was the dominant Gemmula species; specimens of G. kieneri were also collected. A summary of the number of specimens collected at these sites is shown in Table 1.
Of the three species, the largest number of specimens collected was G. speciosa from sites 1–3. Between August 2005 and January 2006, the by-catch was systematically analyzed from site 3 (Table 1). The mean number of G. speciosa collected per trawl was 20±9 or a mean catch rate of 1.3±0.6 per trawl-hour. The specimens ranged in length from 2.0 to 6.7 centimeters (mean: 4.0±1.1 cm.) and collected at night. It must be noted that the fishing gear used effectively captured only those organisms that were either on or close to the surface of the substrate (unlike dredges that go deeper).
Two attempts were made to collect G. speciosa during the day. A dredging trip to the Batangas area was carried out; this was unproductive (only three specimens of G. speciosa were collected). However, between November 2003 and April 2004, there were commercial divers operating in the Batangas area using the hookah method (air compressor/tanks onboard boats are used to supply air to divers). Apparently, it was necessary to go down to a depth of around 50 meters, and then to dig in the sandy-muddy bottom with shovels, followed by sieving the substrate to recover the G. speciosa and other mollusks. The conoidean taxa found to co-occur with G. speciosa using the shoveling/sieving technique included Conus longurionus, Conus mucronatus, U bisaya, T. crispa and sporadically, four to five other small turrid species. In aquarium set-ups and based on the fishers’ knowledge, G. speciosa appear to be nocturnal species, burrowing into the substrate during the day and active at night.
At Sites 1–3 and 4, the major species of Turridae collected in addition to Gemmula was U. bisaya (= L. bisaya (Olivera 2004)). At both localities, fewer specimens of Unedogemmula were collected compared to the dominant Gemmula species (see Table 1). It is notable that although a considerable number of specimens of U. bisaya were collected at the two sites, there was little overlap in the species of Gemmula collected.
Molecular phylogeny
The molecular analysis on selected members of Turrinae (Table 2) for 16S rRNA gene marker has shown congruency in the phylogenetic tree earlier reported for 12S rRNA (Heralde et al. 2007) (Figure 2). Here, the three major genera under the Subfamily Turrinae, namely Gemmula, Turris and Lophiotoma were observed to form distinct clusters, except for the Gemmula 1/Lophiotoma 1 clade. This clade is where the food type/preference analysis is focused. The occurrence of Unedogemmula as a separate clade in the Turrinae (Heralde et al. 2007) is also well-supported as indicated by a high bootstrap value (82%).
Figure 2.
Phylogeny of 31 conoideans based on mitochondrial 16S rRNA gene. Rhinoclavis aspera, a mesogastropod was utilized as outgroup. The bootstrap values indicate separation of turrid and coniid clades. The clustering of worm-hunting coniids are shown: S1 – sedentary polychaete feeders, mainly Terebellidae, S2 – sedentary polychaete feeders, mainly Capitellidae and E6 – errant polychaete feeders, mainly Eunicidae) based on the clade grouping of Duda et al (2001). On the right side columns are indicated, the presence (+) or absence (−) of central teeth (Note: the Turris clade data are published elsewhere); the molecular regurgitate analysis result indicating G. diomedea as a Terebellidae feeder (shown by a “+” sign), and the validation of 16S clustering prediction through a Terebellidae feeding challenge where L. acuta and G. speciosa showed feeding responses to Loimia (indicated by “+” sign) and no feeding response from U. bisaya (−).
The genetic distances calculated for the members of the putative turrid clades were compared to the Conus clades which have similar food type/preference (i.e., among worm-hunting cone species) (Table 3). The working hypothesis applied was that closely related species would assume similar food type/preference; this observation was positively demonstrated in coniids (Duda et al. 2001, Espiritu et al. 2001). We selected three clades of worm-hunting coniids (i.e. S1 – sedentary polychaete feeders, mainly Terebellidae, S2 – sedentary polychaete feeders, mainly Capitellidae and E6 – errant polychaete feeders, mainly Eunicidae) based on the clade grouping of Duda et al (2001) for the genetic distance calculation. We noted the distances in Conus (S1:0.02–0.10, S2:0.05 and E6:0.03–0.05) to be larger than, if not similar to, those calculated for Gemmula (0.01–0.05) (Table 3). The largest distance range occurs among Terebellidae feeders (i.e., the S1 clade), thus in Conus, there are more distantly related species feeding on terebellid worms than in Gemmula.
Table 3.
Comparison of genetic distance between 16S rRNA gene sequences of Gemmula species and selected vermivorous cone clades. Clade S1, sedentary polychaete feeders, mainly Terebellidae; Clade S2, sedentary polychaete feeders, mainly Capitellidae; and Clade E6, errant polychaete feeders, mainly Eunicidae.
| S1 | S2 | E6 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
| 1. Gemmula speciosa | 0.00 | |||||||||||||
| 2. Gemmula sogodensis | 0.01 | 0.00 | ||||||||||||
| 3. Gemmula diomedea | 0.03 | 0.03 | 0.00 | |||||||||||
| 4. Gemmula lisajoni | 0.04 | 0.05 | 0.04 | 0.00 | ||||||||||
| 5. Gemmula rosario | 0.04 | 0.03 | 0.04 | 0.04 | 0.00 | |||||||||
| 6. Conus emaciatus | 0.16 | 0.16 | 0.17 | 0.17 | 0.17 | 0.00 | ||||||||
| 7. Conus terebra | 0.17 | 0.17 | 0.17 | 0.16 | 0.18 | 0.02 | 0.00 | |||||||
| 8. Conus virgo | 0.16 | 0.17 | 0.17 | 0.17 | 0.18 | 0.04 | 0.03 | 0.00 | ||||||
| 9. Conus flavidus | 0.19 | 0.19 | 0.18 | 0.19 | 0.19 | 0.10 | 0.09 | 0.08 | 0.00 | |||||
| 10. Conus rolani | 0.16 | 0.16 | 0.16 | 0.16 | 0.17 | 0.09 | 0.09 | 0.09 | 0.10 | 0.00 | ||||
| 11. Conus pulicarius | 0.17 | 0.17 | 0.17 | 0.17 | 0.18 | 0.08 | 0.08 | 0.08 | 0.09 | 0.05 | 0.00 | |||
| 12. Conus capitaneus | 0.18 | 0.19 | 0.19 | 0.19 | 0.19 | 0.11 | 0.11 | 0.11 | 0.12 | 0.09 | 0.09 | 0.00 | ||
| 13. Conus miles | 0.17 | 0.19 | 0.18 | 0.18 | 0.19 | 0.10 | 0.10 | 0.10 | 0.12 | 0.08 | 0.09 | 0.04 | 0.00 | |
| 14. Conus vexillum | 0.17 | 0.18 | 0.18 | 0.18 | 0.19 | 0.10 | 0.10 | 0.10 | 0.11 | 0.07 | 0.07 | 0.05 | 0.03 | 0.00 |
The phylogenetic relationship of the four species of Gemmula (G. speciosa, G. sogodensis, G. kieneri and G. diomedea; images of their shells are shown in Figure 1) with each other and with other forms in the subfamily Turrinae was further inferred from the 12S and 16S mitochondrial rRNA gene sequences. Both Bayesian and Maximum Likelihood methods, as described under Experimental Procedures, were used. The phylogenetic tree, shown in Figure 3, groups the species into seven well-supported clades (labelled I to VII) and reveals that the four Gemmula species comprise a distinct well-supported group (the Gemmula clade), separate from other groups in the subfamily Turrinae that were included in the analysis. Furthermore, the analysis suggests that the sister group of the Gemmula species is clade II, i.e., the Xenuroturris clade (Olivera 2002, Powell 1964), and that the four groups Gemmula, Xenuroturris, Turris and Lophiotoma (clades I to IV, respectively, in Figure 3) form a major monophyletic group within the Turrinae, which is strongly supported by the analysis.
Figure 3.
Phylogenetic Tree. Optimal tree for the combined 12S rRNA and 16S rRNA gene sequences for Gemmula and their relatives based on Bayesian inference. (An identical tree was returned by a full maximum likelihood analysis of the sequence data.) Branches are labeled with Bayesian confidence values expressed as percentages. For clarity, some of the outgroup species used in the analysis have been pruned from this figure (see Methods for the full list). Shown are various forms in the subfamily Turrinae, including the four species that are the subject of this article (shells of these species are shown in Figure 1). The seven clades identified by Roman numerals all have 100% support based on both Bayesian and Maximum Likelihood analysis and have the following generic/subgeneric assignments within the subfamily Turrinae: I. Gemmula; II. Xenuroturris (presently a subgenus of Lophiotoma); III. Turris; IV. Lophiotoma s.s.; V. Turridrupa; VI. Unedogemmula; VII. Polystira.
Anatomy and morphology
There were significant morphological variations in the shells of G. speciosa specimens collected (i.e., gemmule shape, inter-gemmule distance, length and diameter ratio, etc). However, when molecular analysis was done to evaluate the specimens with different shell morphotypes, no significant differences could be detected in the rRNA gene sequences of the morphological variants. Thus, the shell morphological variation does not appear to be correlated with any significant genetic (12S and 16S rRNA gene) divergence.
A morphological analysis of the foregut anatomy of G. speciosa revealed a strong similarity to that previously reported for Gemmula deshayesi (Taylor et al. 1993) The radula had type 2 wishbone teeth that were robust, short and curved, sometimes with a knifelike cutting edge on the main limb and a large accessory limb with a formula of 1+0+1+0+1 (following Powell’s system). An analysis of the radular structure of G. sogodensis revealed similar radular morphology. However, both G. speciosa and G. sogodensis differed from the radula of L. acuta and U. bisaya by the presence of central teeth. The relevant radular preparations are shown in Figures 4. The data support the clustering of Gemmula spp. into one group of the phylogenetic tree (Figure 3) and their separation from the Lophiotoma and Unedogemmula clades.
Figure 4.
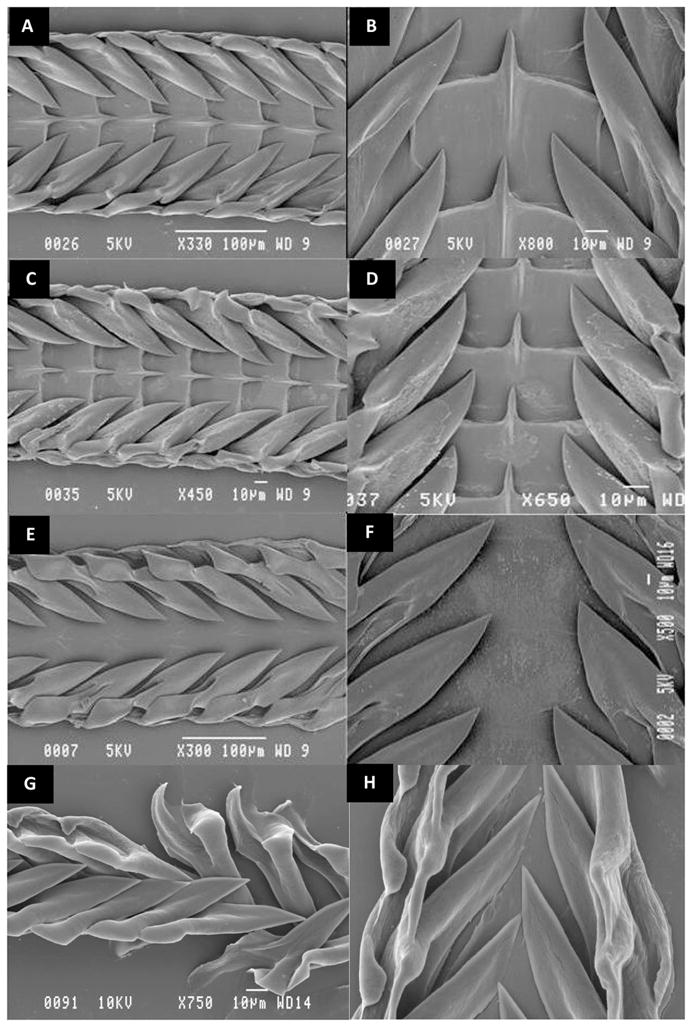
Radular morphology. Scanning electron microscopy images of the radula of Gemmula speciosa (A–B). Gemmula sogodensis (C–D), Unedogemmula bisaya (E–F) and Lophiotoma acuta (G–H).. The central tooth is prominent in both Gemmula species and absent in U. bisaya and L. acuta
Feeding ecology
Little is known regarding the prey preference of any species of Gemmula. A freshly-collected specimen of G. diomedea was observed to regurgitate its gut contents when placed in cold 1% MgCl2 solution (Figure 5); a fragment of mtCOI gene was PCR-amplified from the regurgitate, sequenced, and compared with sequences in GenBank.
Figure 5.

Prey Determination. A freshly collected G. diomedea with regurgitated prey tissue. The samples were collected in the location code CP2340; 231–271 meters deep, by trawl in Bohol Sea on a sandy/mud bottom.
A high sequence similarity between the regurgitate COI and the COI sequence of the tube-dwelling polychaete Amphitritides harpa (Hutchings and Glasby 1988) was found (Figure 6). A. harpa belongs to the family Terebellidae, a sedentary clade of polychaetes. Given the significant homology in the toxin sequences from the three Gemmula species previously reported (Heralde et al. 2007), it seems a reasonable working hypothesis that the prey of Gemmula species are sedentary polychaetes belonging to the family Terebellidae.
Figure 6.
Molecular Regurgitate Analysis. BLAST hit for COI gene from regurgitated tissue (Query sequence) indicating the generic identity of the polychaete to be Amphitritides sp. (Subject sequence).
This hypothesis was experimentally evaluated by challenging the species that could be maintained successfully in an aquarium, G. speciosa, with a terebellid polychaete. The species of Terebellidae most accessible was a Loimia species. The addition of a terebellid to the tank containing turrids elicited activities such as movement of siphon and the active hunt for the prey for many of the turrids including the G. speciosa.
In the first Gemmula-Loimia interaction observed, the snail detected and located the worm ~5 minutes after the latter was dropped into the aquarium. The worm moved toward one end of the worm and pinned it down using its muscular and flexible foot. It rolled its foot into a barrel-like form that can be easily mistaken as a mouth. The snail tried to fully engulf the worm with its foot but was unsuccessful. A smaller Loimia sp. was dropped into the aquarium. The same “foot-folding” behavior was exhibited by the snail. The rostrum was observed to have expanded as it extended towards it captured prey. The muscular foot was observed to not only pin down the worm but also helped to bring it near the snail’s mouth. The snail remained motionless after ingesting ~50% of the worm’s body length. The entire worm was consumed after 2 hours.
L. acuta attacked the terebellid in groups. However, in some feeding events, it was also capable of feeding alone. In both cases, the L. acuta extended its proboscis and quickly stabbed the worm. It then attached itself and remained motionless for an extended period of time. A closer look at the snail’s mouth shows that it takes in a small portion of the worm tissue and is not limited to just sucking. It is most likely that digestion is already taking place even in the mouth. After leaving a single worm being fed on by a L. acuta overnight, the latter was able to eat up the anterior portion of the worm including the tentacles. When L. acuta are group feeding, the prey is completely consumed.
U. bisaya was not observed to react to the addition of Loimia sp. into the tank. In most feeding experiments, it remained submerged in the substrate. The closest interaction between U. bisaya and Loimia sp. documented was when the snail crawled on top of the worm being fed on by an L. acuta (http://msiconusproject.multiply.com/video).
To test the specificity of the snails’ prey, other worms (< 2 cm.), e.g., blood worms (Glycera sp.) and fireworms, were also introduced in the tank. Several trials yielded no result as the turrids showed no activity after introduction of the worms both during day and night time. The snails remained partially burrowed under the substrate and the worms remained untouched.
Discussion
The four species of Gemmula investigated in this study occur in relatively deep water (>50 meters). What is interesting is the striking difference in the pattern of their abundance across the collection sites. Each species appeared to exhibit a similar distribution pattern (Table 1), being much more abundant at one site and scarce at the other sites, but the site where a species was most abundant was different for each species. The other turrid species, U. bisaya, showed another distribution pattern, being abundant at two sites (Sites 1–3 and 4) but not at the third site (Site 5, which is geographically close to Site 4). The field distribution of gastropods is usually associated with their larval life cycles (i.e. planktonic or non-planktonic) (Jablonski and Lutz 1983). These larval life cycles are determined from the type of protoconch that each species possess. The Gemmula species have polygyrate protoconch (Olivera 2005) and thus have planktonic larval life cycle (Jablonski and Lutz 1983), while Unedogemmula species have paucispiral protoconch and have short planktonic life (Olivera 2004). Surprisingly, the observed field distribution of Gemmula and Unedogemmula runs in contrast with the expected pattern of larval distribution. This result warrants further exploration to explain the ecological factors that govern this distribution pattern.
The differences in distribution pattern among the four species could also be seen when comparing their abundance. From the collection by commercial fishing vessels near Manila Bay, the number of specimens of G. speciosa collected was greater than the other three (G. sogodensis, G. diomedea and G. kieneri); the latter two species were entirely absent from site 1-3 (see Table I). In sites 4 and 5 respectively, G. sogodensis and G. diomedea were the predominant species found, with only a minor amount of overlap. The fourth species, G. kieneri, was only collected at site 5. Again, the ecological factors that could explain these interspecies differences in the spatial pattern of abundance remain to be investigated.
The phylogeny of Turrinae was reconstructed based on the 16S rRNA gene sequence and congruency was observed with 12S rDNA-based tree reported by Heralde et al (2007). The agreement of results from two independent gene markers provides strong support for the phylogenetic relationship of members of Turrinae under study. The Gemmula species in particular have shown a consistent clustering where three major groups emerge: the Gemmula speciosa group, the Gemmula diomedea group and the Gemmula lisajoni group. The close association reflected between G. diomedea group and G. speciosa group more than the G. lisajoni group, indicates a sharing of biological characteristics (like food type/preference). We have shown, by molecular regurgitate analysis, the diet of Gemmula diomedea to be a tube-dwelling polychaete belonging to the Terebellid group. Similar attempts in G. speciosa have been unsuccessful. We utilized the molecular approach of inferring food type from the phylogenetic relatedness based on the 16S rRNA gene sequence (Duda et al. 2001, Espiritu et al. 2001) and validated this prediction with a Terebellidae feeding challenge.
In the combined 12S and 16S sequence analysis, the four Gemmula species (G. speciosa, G. sogodensis, G. kieneri and G. diomedea) are clearly closely related phylogenetically (Figure 3). The morphological similarity of their shells is shown in Figure 1. These species form a distinct well-supported Gemmula clade in the tree relative to the other groups in the subfamily Turrinae. This result further warrants the Gemmula clade as likely Terebellid worm feeders analogous to the worm-hunting coniids, the S1 – sedentary polychaete feeders, feeding mainly on Terebellidae ( Duda et al. 2001).
The phylogenetic results also have implications on the current understanding of the phylogeny of the group. If the phylogenetic scheme revealed by our analysis is confirmed by a more extensive analysis, it would seem justified to separate Xenuroturris (i.e., clade II, consisting of what is currently recognized as Lophiotoma species) from Lophiotoma (clade IV) at the generic level. It must be noted that in this scheme, the form Lophiotoma albina, which is generally not included in Xenuroturris (but in Lophiotoma s.s.) based on shell morphology, is grouped with the Xenuroturris (clade II). This is consistent with the previous observation by Olivera (2002) that L. albina was very closely related to species assigned previously to Xenuroturris (Powell 1964). The outgroup taxa used were in the family Terebridae (Hastula hectica and Terebra guttata). Our results further indicate that the subfamily Turrinae is a monophyletic assemblage; the closer kinship of Gemmula, Xenuroturris, Turris, and Lophiotoma to each other than to the other turrine groups is strongly supported by this analysis. All of the taxa shown are in the Pacific, except for the two Polystira species which appear to be outliers with respect to the other branches shown within the subfamily. This result also points out the value of combined 12S and 16S rDNA gene markers in determining a reasonably sound phylogenetic inference for the members of Turrinae.
The analysis of the radular structure shows a clear distinction between G. speciosa and G. diomedea, on the one hand, versus L. acuta and U. bisaya, on the other hand. The radula is an important structure for prey hunting of venomous gastropods. In coniids, the hypodermic type of radula is found in fish-hunting cones (Olivera 1997) while a typical hypodermic type radula, enrolled, with a wide opening at the base is found in the terebrid, Hastula hectica (Imperial et al. 2007); thus radular structure may define the likely prey to a particular snail. The emergence of the central tooth to the Gemmula radula suggests a prey-specificity that could be validated by the polychaete feeding challenge.
The recent molecular analysis of Heralde et al. (2007) demonstrated that contrary to the phylogenetic scheme proposed by Powell (1964), Unedogemmula is not a subgenus of Gemmula and in fact, within the subfamily Turrinae, it is one of the more distant groups from Gemmula. The radular differences documented here provide additional support for the generic separation of Gemmula from Unedogemmula. This conclusion is further strengthened by the phylogenetic analysis carried out in this study (Figure 3). The data shown provide the basis for a comparison of molecular phylogeny with the sequences of toxins (described in studies presented elsewhere (Heralde et al. 2008). Both the present data and the toxinological data are consistent with the four species of Gemmula defining a distinct clade within the subfamily Turrinae. It should be noted, however, that the taxonomic status of the genus Gemmula needs reevaluation. Recent unpublished data (C. Meyer, personal communication) suggest that Atlantic and Eastern Pacific forms of Gemmula do not belong in the same clade as the Indo-Pacific species in Figures 1 and 3. This would create a problematic taxonomic situation because the type species of Gemmula is Pleurotoma gemmata (= G. hindsiana) from the Eastern Pacific.
Finally, we have provided the first data suggesting what species (or group) of polychaetes the Gemmula species might target as prey. A regurgitate from G. diomedea collected in the field was analyzed by the barcode (COI gene) sequence; the match with the polychaete, A. harpa, provided the first evidence for Gemmula being predators of sedentary polychaetes in the family Terebellidae. We have tested this hypothesis more directly: a Gemmula species that could be maintained in an aquarium for extended periods was challenged with the Terebellid species most readily collected alive, a species in the genus Loimia. When Loimia was presented to Gemmula, this triggered an attack; the worm was engulfed and ingested by the Gemmula. The molecular analysis combined with the aquarium challenge experiment is consistent with members of Terebellidae being major prey of this clade of Gemmula species. The feeding experiment on L. acuta expanded the group of turrids to prey on terebellids. This response of L. acuta was consistent with the prediction of the 16S-based clustering of turrids with similar prey type/preference. The non-responsive behavior of U. bisaya towards Loimia sp. indicated that it may not be its prey preference. This also supports the generic separation of Gemmula from Unedogemmula. Correlating this result with radular data, the absence of central teeth does not appear to support prey type/preference as observed in L, acuta.
Hence, two lines of proof were provided to demonstrate Gemmula as a distinct clade of worm-hunting Turrinae feeding on Terebellidae. The molecular phylogenetic analyses (i.e., the 16S-based clustering of snails with similar food type and combined 12S and 16S rRNA gene sequences that define the Gemmula clade) and the polychaete feeding challenge provide consistent evidence of this distinction. The radular anatomy did not support prey preference in contrast with the 16S clustering data. The radular anatomy’s non-correlation with prey preference could be validated in the genus Turris, where the central teeth presence/absence is diverse within a specific 16S cluster. Furthermore, the 16S clustering further validated the expanded spectrum of Terebellidae feeding turrids which includes L. acuta. Meanwhile, the field distribution study demonstrates an unusual pattern that requires further investigation.
Acknowledgments
This work was supported in part by grant GM46877 (to B.M.O.).
Footnotes
There are no financial or professional conflicts of interest in this paper.
References
- Bouchet P, Lozouet P, Maestrati P, Heros V. Assessing the magnitude of species richness in tropical marine environments: exceptionally high numbers of molluscs at a New Caledonia site. Biol J Linnaean Soc. 2002;75:421–436. [Google Scholar]
- Duda TF, Jr, Palumbi SR. Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc Nat Acad Sci USA. 1999;96:6820–3. doi: 10.1073/pnas.96.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda TF, Jr, Kohn AJ, Palumbi SR. Origins of diverse feeding ecologies within Conus, a genus of venomous marine gastropods. Biol J Linnaean Soc. 2001;73:391–409. [Google Scholar]
- Espiritu JD, Watkins M, Monje VD, Cartier GE, Cruz LJ, Olivera BM. Venomous cone snails: molecular phylogeny and the generation of toxin diversity. Toxicon. 2001;39:1899–1916. doi: 10.1016/s0041-0101(01)00175-1. [DOI] [PubMed] [Google Scholar]
- Heralde FMIII, Imperial J, Bandyopadhyay PK, Olivera BM, Concepcion GP, Santos AD. A rapidly diverging superfamily of peptide toxins in venomous. Gemmula species Toxicon. 2008;51:890–897. doi: 10.1016/j.toxicon.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heralde FM, III, Watkins M, Ownby J-P, Bandyopadhyay PK, Santos AD, Concepcion GP, Olivera BM. Molecular phylogeny of some Indo-Pacific genera in the family Turrinae H. Adams and A. Adams. Nautilus. 2007;121:131–138. [Google Scholar]
- Hutchings PA, Glasby CJ. The Amphitritinae (Polychaeta: Terebellidae) from Australia. Records of the Australian Museum. 1988;40:1–60. [Google Scholar]
- Imperial J, Kantor Y, Watkins M, Heralde FMIII, Stevenson BJ, Chen P, Ownby P-J, Bouchet P, Olivera BM. The venomous auger snail Hastula (Impages) hectica (Linnaeus, 1758): molecular phylogeny, foregut anatomy and comparative toxinology. J Exp Zoo. 2007;6(Part B):744–756. doi: 10.1002/jez.b.21195. [DOI] [PubMed] [Google Scholar]
- Jablonski D, Lutz R. Larval ecology of marine benthic invertebrates: paleobiological implications. Biol Rev. 1983;58:21–89. [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- López-Vera E, Heimer de la Cotera EP, Maillo M, Riesgo-Escovar JR, Olivera BM, Aguilar MB. A novel structural class of toxins: the methionine-rich peptides from the venoms of turrid marine snails (Mollusca, Conoidea) Toxicon. 2004;43:365–374. doi: 10.1016/j.toxicon.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Monje VD, Ward R, Olivera BM, Cruz LJ. 16S mitochondrial ribosomal RNA gene sequences: a comparison of seven Conus species. Phil J Sci. 1999;128:225–237. [Google Scholar]
- Olivera BM. The gastropod genus Xenuroturris (Iredale, 1929) evaluated and a new turrid Lophiotoma olangoensis, described from the central Philippines. Sci Diliman. 2002;14:39–49. [Google Scholar]
- Olivera BM. Larger forms of Lophiotoma: four new species described in the Philippines and three from elsewhere in the Indo-Pacific. Sci Diliman. 2004;16:1–28. [Google Scholar]
- Olivera BM. Evaluation of Philippine Gemmula: I. Forms related to G. speciosa and G. kieneri. Sci Diliman. 2005;17:1–14. [Google Scholar]
- Olivera BM. Conus venom peptides, receptor and ion channel targets, and drug design: 50 million years of neuropharmacology. Essay E.E. Just Lecture, 1996. Molec Bio Cell. 1997;8:2101–2109. doi: 10.1091/mbc.8.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AWB. The family Turridae in the Indo-Pacific: Part 1 - The subfamily Turrinae. Indo-Pacific Mollusca. 1964;1:227–340. [Google Scholar]
- Puillandre N, Samadi S, Boisselier M-C, Sysoev AV, Kantor YI, Cruaud C, Couloux A, Bouchet P. Starting to unravel the toxoglossan knot: molecular phylogeny of the “turrids” (Neogastropoda: Conoidea) Mol Phylogen Evol. 2008;47:1122–1134. doi: 10.1016/j.ympev.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Rex MA, Stuart CT, Coyne G. Latitudinal gradients of species richness in the deep-sea benthos of the North Atlantic. Proc Nat Acad Sci USA. 2000;97:4082–4085. doi: 10.1073/pnas.050589497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simison WB. PhD Dissertation. UC-Berkeley: 2000. Evolution and phylogeography of new world gastropod faunas; p. 202. [Google Scholar]
- Taylor JD, Kantor YI, Sysoev AV. Foregut anatomy, feeding mechanisms, relationships and classification of the Conoidea (=Toxoglossa) (Gastropoda) Bull Nat His Mus London (Zool) 1993;59:125–170. [Google Scholar]
- Watkins M, Hillyard DR, Olivera BM. Genes expressed in a turrid venom duct: divergence and similarity to conotoxins. J Mol Evol. 2006;62:247–56. doi: 10.1007/s00239-005-0010-x. [DOI] [PubMed] [Google Scholar]






