Abstract
(See the editorial commentary by Cunningham and Booth, on pages 1645–7.)
Background. Herpes simplex virus type 1 (HSV-1) infects >70% of the United States population. We identified a 3-megabase region on human chromosome 21 containing 6 candidate genes associated with herpes simplex labialis (HSL, “cold sores”).
Methods. We conducted single nucleotide polymorphism (SNP) scans of the chromosome 21 region to define which of 6 possible candidate genes were associated with cold sore frequency. We obtained the annual HSL frequency for 355 HSV-1 seropositive individuals and determined the individual genotypes by SNPlex for linkage analysis and parental transmission disequilibrium testing (ParenTDT).
Results. Two-point linkage analysis showed positive linkage between cold sore frequency and 2 SNPs within the C21orf91 region, 1 of which is nonsynonymous. ParenTDT analysis revealed a strong association between another C21orf91 SNP, predicted to lie in the 3′ untranslated region, and frequent HSL (P = .0047). C21orf 91 is a predicted open reading frame of unknown function that encodes a cytosolic protein.
Conclusions. We evaluated candidate genes in the cold sore susceptibility region using fine mapping with 45 SNP markers. 2 complementary techniques identified C21orf91 as a gene of interest for susceptibility to HSL. We propose that C21orf91 be designated the Cold Sore Susceptibility Gene-1 (CSSG1).
Herpes simplex labialis (HSL) is a common and ubiquitous infection of the skin caused by reactivation of infection with herpes simplex virus type 1 (HSV-1). The frequency of these outbreaks is extremely variable, ranging, in some individuals, from rare episodes every 5–10 years to monthly or more frequent outbreaks among a small proportion of participants [1]. Most persons, 70%–80%, are infected with (ie, seropositive for) HSV-1, and about two-thirds of these infected individuals have experienced at least 1 recognizable HSL outbreak [2].
Researchers believe 3 components account for reactivation of HSV-induced diseases. The first component is the virus itself, including strain, infecting inoculum, and the burden of latent virus within the ganglion [3, 4]. The second component is exposure to environmental factors including fever, wind, sunburn, and surgical manipulation of the ganglion [5].
The third component of susceptibility to HSL is host genetics. Several older studies have linked human leukocyte antigen (HLA) types to HSL susceptibility, although these studies were limited by their inability to reliably separate HSV-1 and herpes simplex virus type 2 (HSV-2) seropositivity among the participants [6–9]. The HLA-A1 allele frequency is increased in patients with frequent genital herpes outbreaks, whereas the HLA-B27 allele appears to protect against outbreaks [10]. Ocular HSV-1 infection is associated with an increased allele frequency of the HLA-B5 and Aw30 alleles [9]. Somewhat surprisingly, a recent family-linkage study showed no significant association between HLA alleles and the expression of HSL [2]. Lundberg et al identified a locus on mouse chromosome 6 near the gene for the tumor necrosis factor receptor [11]. This murine HSV susceptibility region is influenced by the sex of the animal and appears to be inherited in an autosomal dominant fashion. Rare primary immunodeficiencies of human toll-like receptor 3 and UNC-93B may confer susceptibility to herpes encephalitis, at least in children (reviewed in [12]). Changes in toll-like receptor 2 levels increased the number of genital outbreaks and the rate of genital shedding among 128 participants with HSV-2 infection [13].
In this study, we have studied HSL frequency as a phenotype of HSV-1 disease and used transmission disequilibrium testing to determine its association with genotypes in a cohort of related individuals. Transmission disequilibrium testing measures the frequency of transmission of an allele from heterozygous parents to affected offspring compared with frequency expected by random assortment. An allele that is associated with a phenotype will be transmitted to the affected offspring at a higher frequency. The parental transmission disequilibrium test (ParenTDT) is more powerful than the conventional transmission disequilibrium test (TDT) because it also incorporates phenotypes of parents, not just those of the offspring [14]. This method allows not only the detection of associations within families (as the TDT does), but also the detection of associations between families (as a case–control study would).
We previously identified a region of chromosome 21 that is significantly linked to HSL disease. This region was defined using a genome-wide, family-based linkage study for human genes that may be linked to frequent HSL [2]. This approach has been successful in several previous studies [15–18]. The smallest HSL susceptibility region we identified includes 6 known or putative genes mapping from chromosomal position 15.7 to 18.6 Mb. In the current study, we refined our analysis of this region using additional SNP genotyping. The association of individual SNP genotypes with HSL frequency was evaluated using linkage analysis and transmission disequilibrium testing. Our results provide evidence that a novel candidate gene, C21orf91, plays a role in the susceptibility to frequent orolabial herpes outbreaks.
MATERIALS AND METHODS
Participants
The study families comprise 618 people, from 43 three-generation families, whose DNA was used among that of 61 large families collected by the Centre d'Etude du Polymorphisme Humain (CEPH, http://www.cephb.fr/) to map the human genome [19, 20] and were recently resampled as the Utah Genetic Reference Project (UGRP) [16, 17]. All participants were ≥18 years of age at the time of sampling. All individuals provided informed consent approved by the University of Utah Institutional Review Board (IRB #6090-96) before participating in the UGRP.
Phenotyping of the Utah CEPH/UGRP Participants
Prior exposure to HSV-1 was determined by performing glycoprotein G–based HSV-1 type-specific enzyme-linked immunosorbent assay (ELISA) on serum available from 480 individuals from 43 families (HerpeSelect test, Focus Diagnostics). Sera was considered positive for anti–HSV-1 antibodies if the ELISA index was ≥1.1, negative if ≤0.9, and equivocal if 0.9–1.1.
Information about HSL triggers, lifetime episodes, prodromal symptoms and annual HSL frequency was collected via a standardized questionnaire, as described previously [2]. Annual HSL frequency in the 355 (74%) HSV-1 seropositive individuals was used to classify participants as:
“frequently affected” if they experienced ≥2 HSL episodes annually (N = 97)
“unknown” if they experienced 0.1–1.9 episodes annually (N = 162)
“unaffected” if they had never experienced any HSL episodes during their lifetime but were still HSV-1 seropositive (N = 96)
Genotyping
We used the SNPBrowser software package from Applied Biosystems to select SNPs for genotyping. We selected SNPs to include all predicted nonsynonymous SNPs in coding regions, SNPs in 5′ and 3′ untranslated regions (UTRs) with ≥10% heterozygosity in the white population, and tagging SNPs to improve coverage between genes in the region. We obtained SNPlex (Applied Biosystems) genotyping primers for 67 tagging or nonsynonymous SNPs across the 3 Mb chromosome 21 candidate region. All 618 individuals in the UGRP were genotyped by SNPlex. We checked genotypes in the 43 family pedigrees for misinheritance and Hardy–Weinberg equilibrium. We calculated Hardy–Weinberg equilibrium values using the method of Wigginton et al as implemented in Haploview [21].
Linkage Analysis
The 43 pedigrees had 97 frequently affected, 96 unaffected, and 162 unknown HSV-1 seropositive individuals (total 355). The 43 pedigrees include 4 additional families that were phenotyped and genotyped after our original linkage findings were reported [2]. To provide a direct comparison with these earlier data, linkage analysis was also performed using the subset of 39 pedigrees from that previous report, referred to hereafter as the “original families.”
We tested both autosomal dominant and autosomal recessive modes of inheritance [2]. We performed pairwise 2-point genetic linkage analysis using the MLINK subroutine of the Fastlink (v4.1p) program to compute a logarithm of odds (LOD) score [22–26]. LOD scores measure the likelihood of observing linkage purely by chance between each SNP allele and the frequently affected versus unaffected phenotypes.
Parental Transmission Disequilibrium Analysis
ParenTDTs for associations between the disease phenotype and markers from unrelated families. We performed the ParenTDT on sets of completely unrelated trios (2 parents and 1 frequently affected child). We studied 25 trios that fit these criteria. We performed ParenTDT analysis using the method of Purcell et al as implemented in the program Haploview (http://www.broadinstitute.org/science/software) [27]. Linkage analysis was also performed on all the phenotyped individuals within the 25 families from which the TDT trios were selected (ParenTDT families).
Haplotype Analysis in the C21orf91 Gene Region
SNP genotypes along the chromosome are not necessarily independent of each other because combinations or blocks of SNPs are inherited together (a haplotype) along each chromosome from each parent. An individual’s genotype is the sum of these 2 haplotypes. To determine whether particular haplotypes were associated with HSL, susceptibility haplotypes were predicted using the PLINK 1.07 software developed at the Broad Institute (http://pngu.mgh.harvard.edu/purcell/plink/) [28]. Among the 355 HSV-1 seropositive participants, 302 participants had complete genotype data for the 6 SNPs across the C21orf91 region yielding 604 haplotypes. Of the 302 participants, 94 (31%) were frequently affected individuals, 93 (31%) were unaffected individuals, and the remaining 115 individuals were of the unknown or intermediate phenotype. Individuals were scored for the presence or absence of each of the 5 major haplotypes (frequency ≥4%).
Expression and Visualization of C21orf91 Protein
We determined the localization of the C21orf91 protein within human cells by confocal microscopy of HEK293 cells transfected with the C21orf91 gene expressed as a fusion protein with the red fluorescent protein, mCherry. Full-length human C21orf91 (Accession # BC015468) was cloned by polymerase chain reaction (PCR) from complementary DNA and ligated into pEFBOS-C-term-mCherry using PCR-generated XhoI and BamHI restriction sites (generously provided by E. Latz, University of Massachusetts Medical School). We generated the K115, N115, D136, and E136 variants of C21orf91 by site-directed mutagenesis using Quickchange reagents (Stratagene). For confocal imaging, we plated 5 × 105 HEK293T cells in collagen-coated 35 mm glass-bottom microwell plates (MatTek Corporation). The following day, we transfected cells with 1μg C21orf91-mCherry plasmids, encoding the 4 different variants, using Genejuice (Novagen) according to manufacturer’s instructions. At 24 hours posttransfection, we fixed cells with 4% paraformaldehyde for 20 minutes and stained them with Alexa 488–conjugated cholera toxin subunit B (Invitrogen) according to manufacturer’s protocol. To visualize cell nuclei, we added Hoechst 34580 (Invitrogen) to cells 30 minutes prior to imaging at a final concentration of 4μg/mL. We captured images via a Leica SP2 AOBS confocal laser-scanning microscope. We acquired multicolor images by sequential scanning with only 1 laser active per scan to avoid cross-excitation.
RESULTS
We used SNPlex to identify the genotypes of DNA from 618 UGRP participants in the chromosome 21 HSL candidate region [2]. Of the 68 SNPlex primer sets, 8 primer sets (12%) failed to give genotypes. Genotypes for 14 primer sets (21%) were homozygous for all individuals and were thus uninformative. Genotypes for 1 SNP (rs1389072) produced 6 Mendelian errors. This SNP was excluded from further analysis, resulting in a final set of 45 SNPs that could be analyzed within the region of interest.
Linkage Analysis
Original Families Data Set
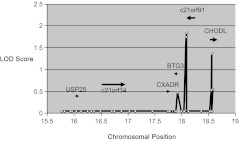
We generated LOD scores in the original families across the chromosome 21 susceptibility region for nonsynonymous, 3′-UTR, and 5′-UTR SNPs in candidate genes. We found that 2 genes emerged as candidates for HSL susceptibility: an open reading frame at C21orf91 and a region encoding the Chondrolectin (CHODL) gene as indicated by positive LOD scores for individual SNPs. For the 45 SNPs with identified genotypes, we generated 2-point LOD scores, in which θ refers to the recombination frequency within the population being studied, using the Fastlink program. The LOD scores in the nonrecombinant region ranged from -4.46 at SNP rs2251818 to a maximum of 2.00 at SNP rs1047978 under the dominant model. All reported LOD scores are at θ = 0, except as otherwise noted. SNP rs1047978 is predicted to alter the amino acid 136 from aspartic acid (D) to glutamic acid (E) in C21orf91 (D136E). The dominant model analysis gave 3 additional LOD scores >1.0 in the region, indicating linkage: 1.96 at SNP rs243588 (approximately 6.7 kb 3′ of C21orf91) and, within the Chondrolectin gene (CHODL), 1.68 at SNP rs7279148 and 1.37 at SNP rs1051526 (data not shown). Nonparametric analysis also supports linkage of this region with the cold-sore phenotype (P = .005).
The recessive inheritance analysis using the fine-mapping SNP genotypes gave maximum LODs of 2.75 at SNP rs7279148 in CHODL and 1.75 (θ = 0.05) at SNP rs1047978 (D136E) in C21orf91. These data seemed to favor either C21orf91 under the dominant model or CHODL under the recessive model as the best candidate genes in the region that affect HSL frequency.
43-Family Expanded Data Set
We extended linkage analysis to DNA from 43 multigenerational families. LOD scores for SNPs in the chromosome 21 susceptibility region confirmed the association of C21orf91 candidate gene with HSL susceptibility (Figure 1). The 2-point linkage analysis on the complete set of 43 fully phenotyped families in the region showed the 2 highest LOD scores (1.85 and 1.75) under the dominant model, again at SNPs rs1047978 (D136E) and rs243588 in and near C21orf91. The only other LOD score >1.0 was 1.37 at SNP rs1051526 within CHODL. Under the recessive model, the maximum LOD score was 1.11 (θ = 0.10) at SNP rs7279148 in CHODL.
Figure 1.
Single nucleotide polymorphism (SNP) linkage plot. Logarithm of odds (LOD) scores are shown for each SNP tested within the human chromosome 21 region of interest. White circles denote the chromosomal locations (bp x 106) of the tested SNPs. LOD scores, shown on the y-axis, were computed with the Fastlink application using a dominant mode of inheritance of the cold sore phenotype (frequently affected vs unaffected) in the expanded data set of 43 families. The labeled black bars show the approximate gene locations with arrows indicating the direction of transcription.
Parental Transmission Disequilibrium Testing
To determine the transmission of specific genotypes and susceptibility phenotypes from parent to child, a subset of 25 trios (2 parents and 1 frequently affected child) from the UGRP families were selected for ParenTDT analysis. For families with shared grandparents, we included only 1 family (whichever had the largest number of affected individuals) in the analysis to ensure that each family trio was unrelated to the others. To ensure that the above linkage results were not due solely to the contribution of the removed families, 2-point linkage analysis was also performed on all individuals from these 25 distinct families. This generated LOD scores similar to the scores obtained at the same SNPs with the larger, 43-family data set (data not shown).
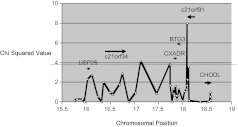
ParenTDT trio analysis confirmed the association of C21orf91 with HSL frequency (Figure 2). This analysis revealed a strong association between the frequently affected phenotype and the C allele of SNP rs1062202 (χ2 = 8.0, P = .0047), predicted to lie in the 3′ UTR of C21orf91 (Figure 2, Table 1). Another SNP, rs10446073, which was approximately 1 kb upstream of the start site of transcription of C21orf91, also showed a moderate association (χ2 = 3.57, P = .06) with the cold-sore phenotype. Notably, the SNPs that showed positive linkage in CHODL, rs7279148 and rs1051526, were not significantly associated (χ2 = 0.04, P = .84 and χ2 = 0.52, P = .47, respectively) with the cold-sore phenotype by ParenTDT analysis. These results suggest that C21orf91 is the best candidate gene affecting the cold sore phenotype in this region. The locations of the SNPs within and adjacent to C21orf91 are displayed in Figure 3.
Figure 2.
Parent transmission disequilibrium testing in the chromosome 21 region. Single nucleotide polymorphism (SNP) genotypes were tested for association with the cold sore phenotype (frequently affected vs unaffected) among 25 family trios (50 parents + 25 offspring) from the Utah Genetic Reference project. White circles denote the chromosomal locations (bp × 106) of the tested SNPs plotted against the computed χ2. The black bars show the approximate gene locations. Arrows indicate the direction of transcription of the gene. The dashed line across the figure represents P ≤ .05 (χ2 ≈ 3.8).
Table 1.
Parental Transmission Disequilibrium Testing Results of Single Nucleotide Polymorphisms (SnPs)s Within and Adjacent to C21orf91.
| SNP | Location | Change | χ2 | P valuea |
| rs10446073 | 5′ of geneb | G > T | 3.57 | .06 |
| rs2824499 | Intron 2 | C > T | 2.25 | .13 |
| rs2824495 | Exon 3 | C > G: N115K | 2.25 | .13 |
| rs1047978c | Exon 3 | C > G: D136E | 1.69 | .19 |
| rs1062202 | 3′ UTR | T > C | 8.00 | .005 |
| rs243588c | 3′ of gened | A > G | 1.28 | .26 |
Parental transmission disequilibrium P values across the C21orf91 gene are shown. Computed χ2 values across the chromosome 21 candidate region are shown in Figure 2.
Abbreviations: SNP, single nucleotide polymorphism; UTR, untranslated region.
Derived from parental transmission disequilibrium testing.
Located approximately 1.1 kb upstream of the start site of transcription—potential promoter region.
SNPs with positive LOD scores by linkage analysis.
Located approximately 6.7 kb downstream of the 3′ end of the transcript.
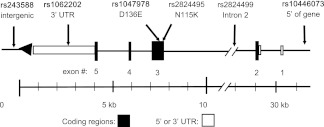
Figure 3.
Map of the SNPs genotyped and tested within and adjacent to C21orf91.The C21orf91 gene spans 30 300 bp of chromosome 21. It is transcribed from the minus strand (right to left in this figure). The locations of the 6 single nucleotide polymorphisms (SNPs) presented in Table 1 are indicated by arrows. Nonsynonymous (amino acid–changing) SNPs are shown (D136E and N115K). The SNPs labeled in boldface type were associated with the cold-sore phenotype by linkage or transmission disequilibrium testing. The distance along the chromosome in kb is shown.
Linkage Disequilibrium
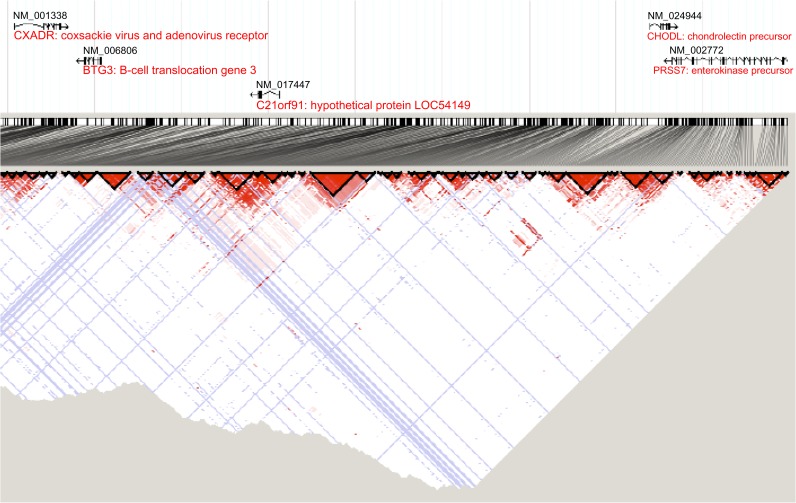
Linkage disequilibrium (LD) within the herpes gene candidate region was examined using data obtained from the HapMap CEU database (http://hapmap.ncbi.nlm.nih.gov). This data includes SNPs in the region with frequency >1% in the population. A plot was generated in the Haploview computer application (Figure 4). This data indicates that significant LD is not present between C21orf91 and any of the other genes in the chromosome 21 candidate region.
Figure 4.
Linkage disequilibrium within the cold sore susceptibility region. This plot showing linkage disequilibrium (LD) was generated in the Haploview computer application using data obtained from the HapMap CEU database (http://hapmap.ncbi.nlm.nih.gov). Areas of high LD are shown in red. Blue indicates areas without informative data. The relative locations of the genes are indicated above the plot. This data indicates that significant LD is not present between C21orf91 and the adjacent genes CHODL, BTG3, and CXADR.
Haplotypes
Among the 302 participants whose haplotypes were identified, 5 major haplotypes had a frequency >4%. Haplotype 1 (H1, N = 176/604, 29%) is the evolutionarily conserved allele (GCCCTA) within this region, as defined by the University of California at Santa Cruz Genome browser (http://genome.ucsc.edu/, GRCh36/hg18 assembly) [29]. Haplotype 2 (H2, N = 190/604, 32%) is defined by presence of the D136E amino acid changing SNP and the 3′ Tag SNP. Haplotype 3 (H3, N = 64/604, 11%) is defined by the 3′ Tag SNP alone. Haplotype 4 (H4, N = 131/604, 22%) includes the intron 2 SNP, the N115K amino acid changing SNP, and the 3′ UTR SNP. Haplotype 5 (H5, N = 26/604, 4%) includes the 3′ UTR SNP and the 5′-SNP found 1 kb upstream of the start site of transcription. Overall, H2 appeared to be more common in the unaffected participants, and H4 and H5 were more common in the frequently affected participants. These results are summarized in Table 2. The haplotype frequency data suggests that the H2 haplotype may be protective while the H4 and H5 haplotypes may be susceptible, all compared with the (ancestral) H1 haplotype.
Table 2.
Definition of the C21orf91 Haplotypes and Their Possible Effects on Cold Sore Frequency. PLINK Identified 5 Major Haplotypes Among 6 SNPs Spanning the C21orf91 Gene Region.
| Haplotypea | SNP genotypesb (5′ – 3′) | Haplotype frequency in unaffected (N = 186) | Haplotype frequency in frequently affected (N = 188) | Mean cold sore frequencyc (N = 604) | Interpretationd |
| H1 | GCCCTA | 26% | 27% | 1.49 | Neutral |
| H2 | GCCGTG | 36% | 27% | 1.30 | Protected |
| H3 | GCCCTG | 11% | 10% | 1.16 | Neutral |
| H4 | GTGCCA | 21% | 29% | 1.79 | Susceptible |
| H5 | TCCCCA | 0% | 5% | 2.03 | Susceptible |
The frequency of each haplotype among the participants who are “unaffected” (no cold sores) vs “frequently affected” (≥2 cold sores per y) are shown. N refers to the number of haplotypes observed in each group (2 chromosomes, therefore 2 haplotypes per person). Mean annual cold sore frequencies are reported among the 302 herpes simplex virus type 1 seropositive individuals genotyped from the 43 Utah Genetic Reference Project families. Statistical comparison of the mean cold sore frequencies could not be employed because of the relatedness of the participants.
Abbreviations: SNP, single nucleotide polymorphism; H1, haplotype 1; H2, haplotype 2; H3, haplotype 3; H4, haplotype 4; H5, haplotype 5.
Definition of haplotypes: H1 = Ancestral genotype. H2 = D136E (rs1047978) + 3'-Tag SNP (rs243588). H3 = 3'-Tag SNP only (rs243588). H4 = Intron 2 (rs2824499) + N115K (rs2824495) + 3'-UTR (rs1062202). H5 = 5′ of gene (rs10446073) + 3'-UTR (rs1062202).
The following SNPs (5′ – 3′) were used to derive these haplotypes: rs10446073, rs2824499, rs2824495, rs1047978, rs1062202, rs243588. See also Figure 3.
All herpes simplex virus type 1-302 seropositive individuals contributed their reported cold sore frequency twice—once for each haplotype—for calculation of these estimated frequencies.
As defined by the haplotype frequency differences between the unaffected and frequently affected groups.
Cold-Sore Frequencies Among the C21orf91 Haplotypes
The mean cold-sore frequency found for H1 was 1.49, for H2 was 1.30, for H3 was 1.16, for H4 was 1.79, and for H5 was 2.03 cold sores annually (Table 2). Because of the relatedness of offspring within sibships (ie, nonindependence of samples), statistical testing was not employed. Although the data are preliminary, the H2 and H3 haplotypes may be associated with lower cold sore frequencies while H4 and H5 haplotypes may be associated with higher cold sore frequencies, all compared to the (ancestral) H1 haplotype.
Expression of C21orf91 in a Human Cell Line: Localization to the Cytosol
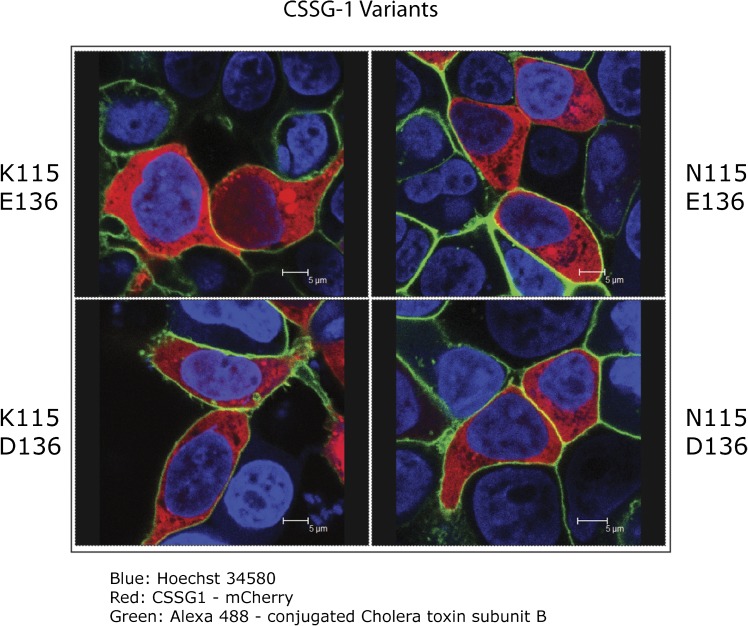
The candidate protein encoded by the ancestral C21orf91 gene is likely cytosolic as it lacks a signal sequence for secretion and lacks a consensus transmembrane domain. We confirmed that the protein indeed localized to the cytosol using confocal microscopy (Figure 5). Of the 3 most common haplotypes, 2 contain a different nonsynonymous amino acid change within C21orf91. We used in vitro transfection experiments to determine the localization of C21orf91 variants. A C21orf91 construct was cloned and expressed as a fusion protein with an mCherry fluorescent tag at the COOH terminus. We transfected variants encoding proteins with either lysine (K) or asparagine (N) at position 115 and with either D or E at position 136 in HEK293FT cells and used confocal microscopy to visualize the expression of the proteins (Figure 5). C21orf91 protein was expressed mainly in the cytoplasm, we observed no apparent difference in the expression of the variants. These results suggest that the amino acid alterations do not alter protein stability or subcellular localization of the protein.
Figure 5.
Expression of C21orf91 in human embryonic kidney (HEK293) cells. HEK293FT cells expressing C21ORF91 with a C-terminal mCherry fluorescent tag (red) were imaged by confocal microscopy. Nuclei were stained with Hoechst 34580 (blue). Plasma membranes were stained with cholera toxin B subunit (green). HEK293FT cells were transfected with complementary DNAs encoding C21ORF91 proteins with either lysine (K) or asparagine (N) at position 115 and either aspartic acid (D) or glutamic acid (E) at position 136.
DISCUSSION
Using 2 complementary techniques, transmission disequilibrium and SNP linkage, we show that SNP variations adjacent to and within the human gene C21orf91 yield the best fine-mapping linkage and TDT association results for susceptibility or resistance to labial herpes outbreaks. None of the 39 SNPs in the other 5 genes investigated appeared to be related to the cold-sore phenotype by both methods.
These results are somewhat surprising because the coxsackie and adenovirus receptor gene (CXADR), a well-known viral receptor, also lies within the chromosome 21 region studied. SNPs within the other genes in the chromosome 21 region, USP25, C21orf34, CXADR, and BTG3, were not linked with the cold-sore frequency phenotype nor were they significantly associated by ParenTDT. Although 2 SNPs within CHODL had LOD scores >1, the scores were not confirmed by ParenTDT.
For all SNPs with identified genotypes in the region, the total LOD score was lower in the full set of 43 families compared with that of the original families (data not shown). An examination of the LOD scores at each SNP by individual family (data not shown) revealed that this decrease was because 28% of the added families showed positive linkage to the chromosome 21 region, 44% were linkage neutral, and 28% were negative or unlinked. The additional families provided some support for the original locus but also demonstrated that cold-sore frequency in some of the families was not attributable to SNPs in susceptibility genes within the chromosome 21q region. It is possible, however, that genes outside the 21q region regulate the function or expression of 21q genes in these families.
Within the nonrecombinant region of chromosome 21 under investigation, a maximum LOD of 2.26 for short tandem repeat (STR) marker abmc65 (D21S409) was found under the dominant model within the original families [2]. This marker maps the region between C21orf91 and CHODL. Therefore, in this study the positive SNP LOD scores in and near these genes correlate well with the previous findings. The D21S409 marker is a 10-allele system, whereas all the SNPs genotyped here are 2-allele systems. This fact likely accounts for the higher STR marker LOD scores compared with the SNP LOD scores observed in this region.
The highest 2-point LOD scores were consistently found for SNP rs1047987 under the dominant model tested with the original families, the expanded 43-family data set, and the 25-family ParenTDT data set. This SNP encodes a nonsynonymous change of residue 136 from aspartic acid to glutamic acid (D136E). Positive LOD scores were also observed with the SNP rs243588 in all 3 data sets. This SNP lies 3′ (downstream) from the end of the predicted transcript and shows some linkage disequilibrium (r2 = 0.71) with the D136E SNP. SNP rs243588 is found in both haplotypes that may protect against frequent cold sores, H2 and H3 (see Table 2). An additional C21orf91 SNP, rs1062202, was identified by ParenTDT analysis. This 3′-UTR SNP was found in the haplotypes that may increase susceptibility to frequent cold sores, H4 and H5. SNPs in 3′ UTR of other genes have been shown to alter messenger RNA (mRNA) stability or protein translation. The specific effects of these C21orf91 genotype changes on mRNA and protein expression in different tissues are unknown at this time.
Because of the limited number of SNPs available for genotyping, it is likely that additional SNPs are present within the main candidate gene, C21orf91. Deletions, frame-shifts, and insertions may also be present, genetic features not detectable in this study. The possibility that the SNPs from C21orf91 might be in linkage disequilibrium (LD) with neighbouring genes was addressed and no significant LD was detected. Because the LD data does not include rare variants (SNP frequency <1%), it is unlikely but still possible that there are rare variants of larger effect in the neighbouring genes that the C21orf91 SNPs could be tagging. Another limitation is the size and familial structure of the population being studied. The conclusions stated here are based on ParenTDT testing from 75 individuals plus linkage on 355 individuals. Despite the limited number of SNPs and individuals analyzed, positive linkage signals and a significant association (P < .005) by ParenTDT analysis were seen with SNPs mapping in or near C21orf91. Although these results do not definitively rule out a role for other genes in this region, they support a potential role for C21orf91 in modulation of the cold-sore phenotype.
The results of this study indicate that the C21orf91 gene may play a role in the frequency of labial herpes outbreaks. This gene has an open reading frame of 293 amino acids, a 3′ UTR of about 4400 nucleotides, and encodes a protein of unknown function. The C21orf91 protein is the sole member of the “Early Undifferentiated Retina and Lens” proteins [30]. It is expressed in chick retinal precursor cells as well as in the anterior epithelial cells of the lens during early development. We demonstrated that the protein was localized to the cytoplasm. Additional studies are needed to determine how C21orf91 affects HSV-1 pathogenesis.
In summary, this study reports a new association between a previously obscure human gene, C21orf91, and frequent cold sores. The authors propose that C21orf91 be designated the Cold Sore Susceptibility Gene 1 (CSSG1). Although these findings await confirmation in a larger, unrelated population, they could have important implications for the development of new drugs that affect determinants of the cold-sore phenotype.
Notes
Acknowledgments.
The authors wish to acknowledge Focus Technologies for the generous contribution of HSV-1 type-specific serology kits. Thanks to Brith E. Otterud and Tami Leppert for performing extensive linkage analyses. We would like to extend our sincere thanks to all family members who participated in the Utah Genetic Reference Project. Thanks also to Andreas P. Peiffer, MD, PhD, UGRP Medical Director, and Melissa M. Dixon, UGRP Study Coordinator.
Financial support.
This work was supported by Public Health Services (R01-AI64349 and PO1-AI083215 to R. W. F.); the Cold Sore Research Foundation (CSRF-04); National Center for Research Resources (M01-RR00064 to the Huntsman General Clinical Research Center at the University of Utah); the W. M. Keck Foundation (gift); andthe George S. and Delores Doré Eccles Foundation (gift). The work is referable to United States patent #7449294 B2, issued 11 November 2008.
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Spruance S, Kriesel J. Treatment of herpes simplex labialis. Herpes. 2002;9:6–11. [PubMed] [Google Scholar]
- 2.Hobbs MR, Jones BB, Otterud BE, Leppert M. Kriesel JD. Identification of a herpes simplex labialis susceptibility region on human chromosome 21. J Infect Dis. 2008;197:340–6. doi: 10.1086/525540. [DOI] [PubMed] [Google Scholar]
- 3.Hoshino Y, Pesnicak L, Cohen J, Straus S. Rates of reactivation of latent herpes simplex virus from mouse trigeminal ganglia ex vivo correlate directly with the viral load and inversely with the number of infiltrating CD8+T cells. J Virol. 2007;81:8157–64. doi: 10.1128/JVI.00474-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawtell N. The probability of in vivo reactivation of herpes simplex virus type 1 increases with the number of latently infected neurons in the ganglia. J Virol. 1998;72:6888–92. doi: 10.1128/jvi.72.8.6888-6892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spruance SL, Kriesel JD, Evans TG, McKeough MB. Susceptibility to herpes labialis following multiple experimental exposures to ultraviolet radiation. Antiviral-Res. 1995;28:57–67. doi: 10.1016/0166-3542(95)00038-n. [DOI] [PubMed] [Google Scholar]
- 6.Seppanen M, Lokki ML, Timonen T, et al. Complement C4 deficiency and HLA homozygosity in patients with frequent intraoral herpes simplex virus type 1 infections. Clin Infect Dis. 2001;33:1604–7. doi: 10.1086/323462. [DOI] [PubMed] [Google Scholar]
- 7.Jabbar AA, al-Samarai AM, al-Amar NS. HLA antigens associated with susceptibility to herpes simplex virus infection. Dis-Markers. 1991;9:281–7. [PubMed] [Google Scholar]
- 8.Malo A, Kampgen E, Wank R. Recurrent herpes simplex virus-induced erythema multiforme: Different HLA-DQB1 alleles associate with severe mucous membrane versus skin attacks. Scand J Immunol. 1998;47:408–11. doi: 10.1046/j.1365-3083.1998.00357.x. [DOI] [PubMed] [Google Scholar]
- 9.Elner VM, Dutt S, Pavilack MA, Sugar A, Foster CS, Elner SG. Intercellular adhesion molecule-1 (ICAM-1) and HLA-DR antigens in herpes keratitis. Ophthalmology. 1992;99:1400–7. doi: 10.1016/s0161-6420(92)31801-9. [DOI] [PubMed] [Google Scholar]
- 10.Lekstrom-Himes JA, Hohman P, Warren T, et al. Association of major histocompatibility complex determinants with the development of symptomatic and asymptomatic genital herpes simplex virus type 2 infections. J Infect Dis. 1999;179:1077–85. doi: 10.1086/314729. [DOI] [PubMed] [Google Scholar]
- 11.Lundberg P, Welander P, Openshaw H, et al. A locus on mouse chromosome 6 that determines resistance to herpes simplex virus also influences reactivation, while an unlinked locus augments resistance of female mice. J Virol. 2003;77:11661–73. doi: 10.1128/JVI.77.21.11661-11673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sancho-Shimizu V, Zhang S, Abel L, et al. Genetic susceptibility to HSV-1 encephalitis in mice and humans. Curr Opin Allergy Clin Immunol. 2007;7:495–505. doi: 10.1097/ACI.0b013e3282f151d2. [DOI] [PubMed] [Google Scholar]
- 13.Bochud PY, Magaret AS, Koelle DM, Aderem A, Wald A. Polymorphisms in TLR2 are associated with increased viral shedding and lesional rate in patients with genital herpes simplex virus Type 2 infection. J Infect Dis. 2007;196:505–9. doi: 10.1086/519693. [DOI] [PubMed] [Google Scholar]
- 14.Purcell S, Sham P, Daly M. Parental phenotypes in family-based association analysis. Am J Hum Genet. 2005;76:249–59. doi: 10.1086/427886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall MA, Norman PJ, Thiel B, et al. Quantitative trait loci on chromosomes 1, 2, 3, 4, 8, 9, 11, 12, and 18 control variation in levels of T and B lymphocyte subpopulations. Am J Hum Genet. 2002;70:1172–82. doi: 10.1086/340090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drayna D, Coon H, Kim UK, et al. Genetic analysis of a complex trait in the Utah Genetic Reference Project: A major locus for PTC taste ability on chromosome 7q and a secondary locus on chromosome 16p. Hum Genet. 2003;112:567–72. doi: 10.1007/s00439-003-0911-y. [DOI] [PubMed] [Google Scholar]
- 17.Malhotra A, Peiffer AP, Ryujin DT, et al. Further evidence for the role of genes on chromosome 2 and chromosome 5 in the inheritance of pulmonary function. Am J Respir Crit Care Med. 2003;168:556–61. doi: 10.1164/rccm.200303-410OC. [DOI] [PubMed] [Google Scholar]
- 18.Sandovici I, Leppert M, Hawk PR, Suarez A, Linares Y, Sapienza C. Familial aggregation of abnormal methylation of parental alleles at the IGF2/H19 and IGF2R differentially methylated regions. Hum Mol Genet. 2003;12:1569–78. doi: 10.1093/hmg/ddg167. [DOI] [PubMed] [Google Scholar]
- 19.Dausset J, Cann H, Cohen D, Lathrop M, Lalouel J, White R. Centre d'etude du polymorphisme humain (CEPH): Collaborative genetic mapping of the human genome. Genomics. 1990;6:575–7. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- 20.White R, Leppert M, Bishop D, et al. Construction of linkage maps with DNA markers for human chromosomes. Nature. 1985;313:101–5. doi: 10.1038/313101a0. [DOI] [PubMed] [Google Scholar]
- 21.Wigginton J, Cutler D, Abecasis G. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–93. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cottingham RW, Jr, Idury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53:252–63. [PMC free article] [PubMed] [Google Scholar]
- 23.Lathrop GM, Lalouel JM. Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet. 1984;36:460–5. [PMC free article] [PubMed] [Google Scholar]
- 24.Lathrop GM, Lalouel JM, Julier C, Ott J. Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci U S A. 1984;81:3443–6. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lathrop GM, Lalouel JM, White RL. Construction of human linkage maps: Likelihood calculations for multilocus linkage analysis. Genet Epidemiol. 1986;3:39–52. doi: 10.1002/gepi.1370030105. [DOI] [PubMed] [Google Scholar]
- 26.Schaffer AA, Gupta SK, Shriram K, Cottingham RW., Jr Avoiding recomputation in linkage analysis. Hum Hered. 1994;44:225–37. doi: 10.1159/000154222. [DOI] [PubMed] [Google Scholar]
- 27.Barrett J, Fry B, Maller J, Daly MPI. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent W, Sugnet C, Furey T, Roskin K, Pringle T, Zahler A. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Godbout R, Andison R, Katyal S, Bisgrove D. Isolation of a novel cDNA enriched in the undifferentiated chick retina and lens. Dev Dyn. 2003;227:409–15. doi: 10.1002/dvdy.10310. [DOI] [PubMed] [Google Scholar]