Abstract
Background. Epstein-Barr virus (EBV) is a ubiquitous herpesvirus, and Kaposi’s sarcoma–associated herpesvirus (KSHV) has a restricted seroprevalence. Both viruses are associated with malignancies that have an increased frequency in individuals who are coinfected with human immunodeficiency virus type 1 (HIV-1).
Methods. To obtain an overview of humoral immune responses to these viruses, we generated a protein array that displayed 174 EBV and KSHV polypeptides purified from yeast. Antibody responses to EBV and KSHV were examined in plasma from healthy volunteers and patients with B cell lymphoma or with AIDS-related Kaposi’s sarcoma or lymphoma.
Results. In addition to the commonly studied antigens, IgG responses were frequently detected to the tegument proteins KSHV ORF38 and EBV BBRF and BGLF2 and BNRF1 and to the EBV early lytic proteins BRRF1 and BORF2. The EBV vIL-10 protein was particularly well recognized by plasma IgA. The most intense IgG responses to EBV antigens occurred in HIV-1–positive patients. No clear correlation was observed between viral DNA load in plasma and antibody profile.
Conclusions. The protein array provided a sensitive platform for global screening; identified new, frequently recognized viral antigens; and revealed a broader humoral response to EBV compared with KSHV in the same patients.
Epstein-Barr virus (EBV) and Kaposi’s sarcoma–associated herpesvirus (KSHV) are human herpesviruses that differ dramatically in their geographic prevalence. EBV seropositivity in healthy populations is >90% worldwide. KSHV seropositivity is low (0–5%) in the United States, Canada, and Europe; intermediate (7–24%) in Italy and the Mediterranean; and high (23–70%) in sub-Saharan Africa [1–4]. Seroprevalence for KSHV increases in individuals coinfected with human immunodeficiency virus 1 (HIV-1) and, in the United States and Europe, in men who have sex with men [5–7]. Primary infection with EBV can cause infectious mononucleosis, and both EBV and KSHV are associated with human cancers. EBV is associated with nasopharyngeal carcinoma, African Burkitt’s lymphoma, a subset of Hodgkin’s lymphoma, AIDS-related lymphoma, gastric carcinoma, peripheral T cell lymphoma, and posttransplant and other lymphoproliferative diseases in the immunodeficient [8]. KSHV is associated with Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. Immunosuppression, such as that imposed by concurrent HIV-1 infection or transplantation regimens, increases the incidence of some of these virus-associated cancers [8, 9].
The DNA genomes of EBV and KSHV encode an estimated 80 and 86 proteins, respectively. The degree to which these proteins are expressed differs depending on the stage of the viral life cycle. In vivo, the reservoir of EBV latent infection is resting memory B cells that have either no EBV protein expression or transient expression of the EBNA1 protein. Expression of the other latency proteins, EBNA2, EBNA3A, EBNA3B, EBNA3C, EBNA-LP, LMP-1, and LMP2A and LMP2B, has been detected in peripheral blood B cells shortly after primary infection and in B cells in tonsillar tissues [10, 11]. Lytic replication and expression of the EBV lytic proteins takes place in plasma cells and in epithelial cells [12–14]. Less is known about in vivo persistence of KSHV. B cells form a reservoir for latent KSHV infection, and differentiation of these cells also triggers lytic KSHV induction [15, 16]. KSHV endothelial and B cell tumors express the LANA, v-cyclin, and v-FLIP latency proteins, and vIRF3 is also detected as a latency protein in primary-effusion lymphoma cells. In addition, a small percentage of tumor cells express lytic cycle proteins such as the v-IL6 and the viral G-protein coupled receptor (v-GPCR), suggesting that KSHV lytic infection may contribute to the tumorigenic phenotype [17].
Transcriptional profiling arrays have been developed to examine genome-wide expression of EBV and KSHV [18, 19]. However, humoral immune responses to EBV and KSHV proteins have predominantly been measured using virus-infected cell lines or enzyme-linked immunosorbent assays that incorporate just a few selected viral proteins [2–4, 20–22]. Although these assays have proven valuable for diagnostic and epidemiological studies, they do not provide a complete picture of the full repertoire of antibody responses generated by EBV and KSHV infection. High-throughput technologies allow cloning and expression of entire pathogen proteomes [23, 24]. We used this technology to generate proteomic microarrays for EBV and KSHV. These arrays allowed us to compare the global humoral immune responses to EBV vs KSHV in the same patient samples and within the same assay.
METHODS
Generation of EBV Plus KSHV Protein Microarrays
EBV and KSHV open reading frames (ORFs) were cloned and expressed as previously described [25]. Briefly, the open reading frames were polymerase chain reaction (PCR) amplified using primers based on the GenBank sequences V01555, AJ507799, and U756981. The bacterial artificial chromosome plasmids Akata BXI (EBV) and BAC36 (KSHV) [26] were used as templates (gifts from L. Hutt-Fletcher and S-J Gao). PCR products were cloned into the vector pDONR201. ORFs representing different virus strains were amplified directly from clinical samples or from cell lines. Proteins containing extensive repeats that limited expression in yeast (eg, EBNA1 and LANA) were amplified as N-terminal and C-terminal fragments. Escherichia coli bacteria were transformed with the reaction products and DNA prepared for sequence verification. Correct ORF-containing plasmid DNAs were then moved into a yeast destination vector, pEGH-A, a derivative of the yeast glutathione S-transferase (GST) vector, pEGH. Yeast cultures were induced with 2% galactose, and GST fusion proteins were isolated and purified on glutathione beads. One hundred and seventy-four appropriately sized EBV and KSHV proteins (Table S1; available online) were successfully purified based on immunoblot analysis using anti-GST antibody and were printed using a 48-pin contact printer (Bio-Rad) in duplicate on modified glass (Full Moon Biosystems) microscope slides along with controls. Each slide had 784 spots: 348 EBV and KSHV proteins, 112 control proteins (Table S1; available online), and 324 blank spots. The controls were used for orientation and to detect nonspecific interaction with proteins such as the GST fusion partner. Protein arrays were stored at −80°C.
Patient Samples
Plasma from healthy blood donors and from patients with follicular B cell lymphoma (without HIV-1), AIDS-related lymphoma (ARL), and AIDS Kaposi’s sarcoma was obtained with written informed consent in accordance with the Declaration of Helsinki after approval by the Johns Hopkins institutional review board.
Plasma was separated from peripheral blood collected in standard EDTA or acid citrate dextrose tubes and stored at −80°C.
Antibody Screening
Protein arrays were preincubated in 4 mL Superblock (Pierce) plus 2% bovine serum albumen (BSA) at 4°C overnight. Slides were assembled with 4-well modules and incubated with plasma (300 uL diluted in phosphate buffered saline [PBS] plus 2% BSA) for 1 hour at room temperature. Slides were washed with prewarmed (42°C) PBS Tween-20 (PBST) and incubated with secondary Cy3-labeled antihuman IgG or IgA antibody (1:2000; Sigma and Jackson, respectively) diluted in PBS plus 2% BSA for 1 hour at room temperature. Slides were then washed 2× with prewarmed PBST, 1× with prewarmed distilled water, dried, scanned, and analyzed with GenePix Pro software (Molecular Devices).
Statistical Analysis
The software GenePix Pro 7 was used to obtain the median foreground and background intensity for each spot. The raw intensity of the spot was defined as the ratio of the foreground to background median intensity. There are 324 “blank” spots on each protein chip, and the raw intensity distribution of these spots has a mean value of approximately 1. Assuming that the raw intensity distributions of “blank” spots are the same across all microarrays, we can standardize the protein signals such that
where Z is Z-score of each spot, I is raw intensity of the spot, and m and σ are mean value and standard deviation, respectively, of “blank” spots on the microarray. Each protein has 2 duplicated spots. The identified hits were defined as those proteins with Z-scores for both spots at or above the 5 SD cutoff.
When comparing the appearance frequency for individual antigens for HIV/KS patients vs lymphoma or healthy normals, the enrichment score was reflected by . We adopted binominal probability to calculate the significance between comparisons such that
where p is the probability that the protein was hit for at least k HIV/KS patients, n is the total number of HIV/KS patients, and f1 is the frequency of the protein in the comparison group (either lymphoma or healthy normals). Proteins were selected as a significant biomarker for HIV+/KS patients only if they had at least 50% frequency in HIV+/KS patients, the enrichment score ES was larger than 1.6, and the P value was less than .005.
Plasma DNA Detection
EBV and KSHV DNA in plasma were detected as previously described [27]. Viral DNA levels were determined by real-time, quantitative PCR using K8 primers for KSHV and BamHI-W primers for EBV. Controls were constructed by the addition of viral DNA to serum from healthy donors. Standard curves (with duplicate serial 10-fold dilutions of plasmid DNA that included a target sequence from 105 to 10 copies) were run in parallel with each analysis.
RESULTS
Establishment of the Protein Array Assay
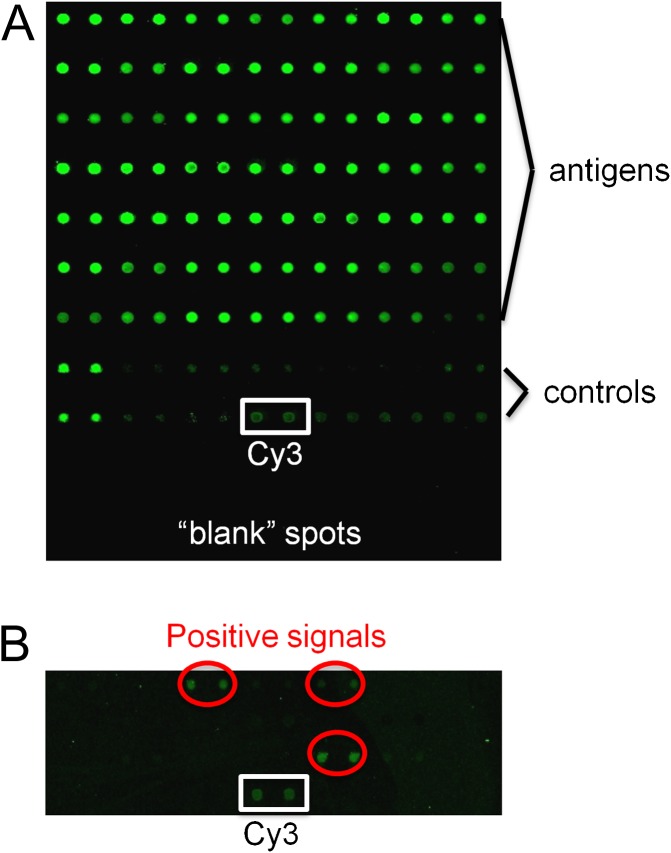
To systematically compare EBV and KSHV antibody responses, we utilized proteomic arrays displaying 174 virus proteins plus controls. The printing quality and the quantity of the immobilized proteins on the chip were monitored using anti-GST antibody followed by Cy3-labeled secondary antibody (Figure 1A). Preliminary analyses with arrays containing the 82 EBV and 92 KSHV ORFs revealed that plasma diluted up to 1:10 000 gave a signal on the arrays that was readily detected with Cy3-tagged antihuman IgG. A positive signal was set as one that was 5 SD or more above background. A section of the array illustrating positive and control signals is shown in Figure 1B.
Figure 1.
Protein detection on the EBV and KSHV protein array. (A) Proteins on the array detected with anti-GST primary antibody and Cy3-tagged secondary antibody. (B) Section of an array incubated with HIV+/KS plasma (1:10 000) and detected with Cy3-tagged antihuman IgG to illustrate positive and Cy3 signals.
Comparison of Plasma From Healthy Donors, HIV− Lymphoma Patients, and HIV+/KS Patients
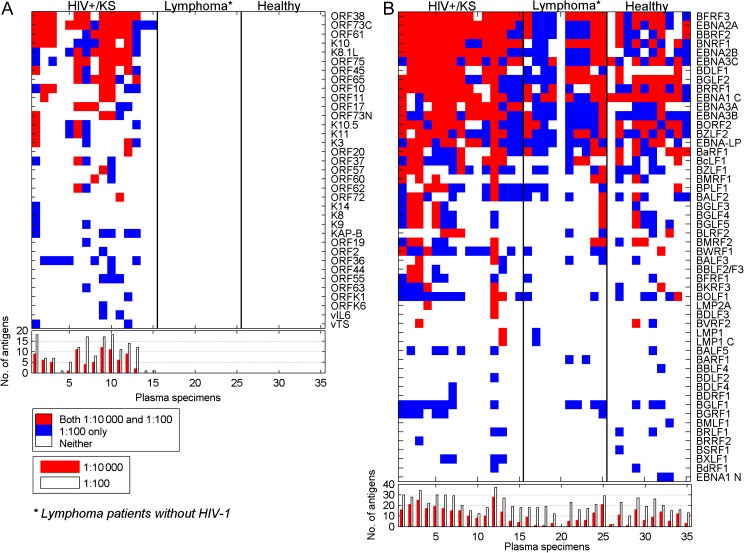
To compare the humoral immune responses in different populations, arrays were incubated with plasma from 10 healthy donors, 10 patients with B cell lymphoma (follicular, marginal zone, mantle cell) known not to be EBV associated, and 15 patients with HIV-1 infection and Kaposi’s sarcoma (HIV+/KS). Plasma was incubated with the protein arrays at dilutions of 1:10 000 and 1:100. Analysis of the KSHV antigens detected on the arrays (Figure 2A) revealed that none of the healthy normal or HIV-negative lymphoma patient plasma specimens that were tested reacted with the array proteins at either the 1:100 or 1:10 000 dilutions. However, positive signals were seen with the plasma from HIV-positive individuals with KS. The antigens most frequently detected at 1:10 000 dilution were ORF73C (10/15; 66% positive) and ORF38 (10/15; 66% positive). ORF38 is a tegument protein that has not been widely examined as an antigen. ORF73 encodes the latency LANA protein that is a gold standard for KSHV serology studies. The central region of LANA contains repeated sequences that limit effective expression of the protein in yeast. The N-terminus and C-terminus of LANA were therefore expressed separately for presentation on the array. The LANA C-terminus was recognized more frequently than the N-terminus (14 vs 4 positive plasma). This is consistent with a peptide mapping study that found more sera reacting to peptides mapping to the LANA C-terminus than to the N-terminus [28]. The median number of KSHV antigens recognized by the LANA-positive HIV+/KS plasma at 1:10 000 serum dilution was 6.
Figure 2.
KSHV and EBV protein recognition by plasma from HIV-1–positive patients with KS and HIV-1–negative lymphoma patients, and healthy volunteers. (A) KSHV protein recognition. (B) EBV protein recognition. Upper panels: Plasma was diluted 1:10 000 or 1:100 and reactivity was detected with Cy3-labeled anti-human IgG. All antigens recognized at 1:10 000 dilution were also positive at 1:100 dilution. The cutoff for a positive signal was 5 SD over background. Lower panel: Number of antigens recognized by each sample at 1:10 000 (red) and 1:100 (white) dilutions.
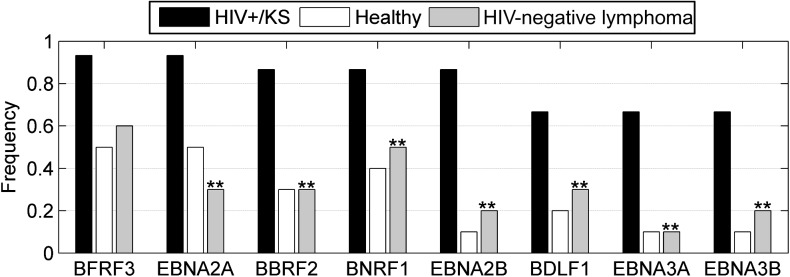
Analysis of the EBV proteins recognized in the same assay by the same plasma samples revealed a different picture. EBV is a ubiquitous virus, and EBV proteins on the array were recognized using plasma from healthy donors, HIV− lymphoma patients, and HIV+/KS patients (Figure 2B). The median number of EBV proteins recognized at 1:100 serum dilution was 17, 18, and 27 for plasma from healthy donors, lymphoma patients, and HIV+/KS patients, respectively. Ninety-one percent of plasma specimens tested (32/35) reacted with the carboxy terminus of EBNA1 and 97% (34/35) reacted with the combination of EBNA3B and EBNA3C at 1:100 or 1:10 000 dilution. The single EBNA3-negative plasma specimen was from a lymphoma patient whose plasma did not recognize any EBV antigens. The difference between the normal and HIV− lymphoma population and the HIV+/KS population lay not in the individual antigens recognized, but rather in the heightened antigenic response to a range of EBV proteins. This is illustrated by the increased median number of antigens detected at 1:10 000 plasma dilution seen with HIV+/KS samples (15) vs the median number with plasma from healthy donors and lymphoma patients (7 and 5, respectively). Antigens with a statistically significant difference in appearance between healthy donors and HIV+/KS patients at a 1:10 000 dilution are shown in Figure 3, which also includes the data for HIV-negative lymphoma patients for comparative purposes. The BFRF3 (p18) capsid antigen has been previously advocated as a serological marker for nasopharyngeal carcinoma diagnosis [29].
Figure 3.
EBV antigens with a statistically significant difference in frequency between healthy and HIV+/KS individuals. Frequency of recognition of EBV antigens at 1:10 000 dilution in plasma from healthy (white) vs HIV+/KS patients (black) who meet criteria of significance (enrichment score > 1.6; P <.005). The antigens for which a significant difference in frequency is also seen between HIV+/KS and HIV-negative lymphoma patients (grey) are indicated by (**).
Two subtypes of EBV, type 1 and type 2 (also called A and B), were originally defined on the basis of differences in the EBNA2 gene [30]. In this set, 10/35 sera showed discordant responses to the 2 EBNA2 proteins with either a different sensitivity of detection (1:100 vs 1:10 000) or recognition of only 1 of the 2 EBNA2 proteins. It is likely that these individuals are infected with only 1 of the 2 EBV subtypes. Note that 9/10 discordant sera had preferential recognition of type 1 EBNA2, which is consistent with the greater prevalence of type 1 EBV in clinical samples from the United States. Positivity to both proteins would represent recognition of common epitopes or reflect concurrent infection with both virus subtypes.
Comparison of Plasma From HIV-Positive Patients With KSHV-Associated Versus Nonviral-Associated Cancer
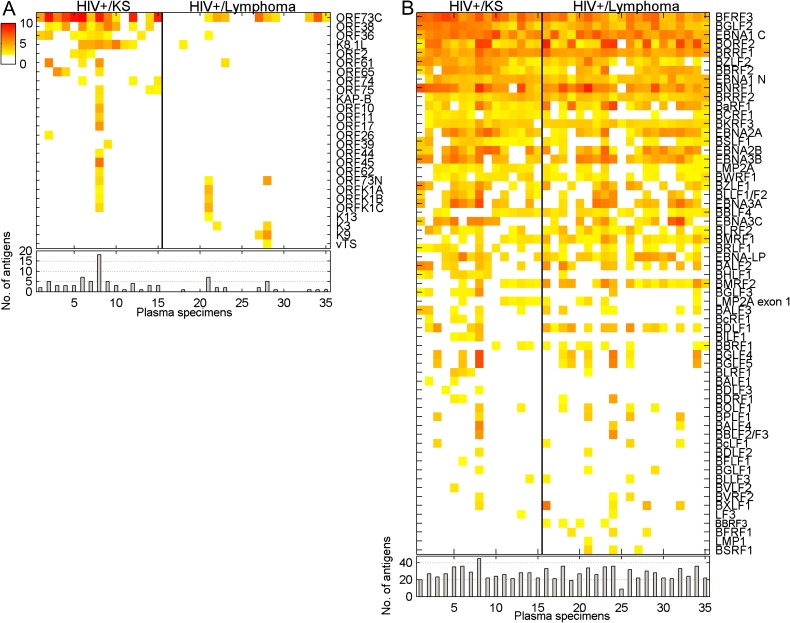
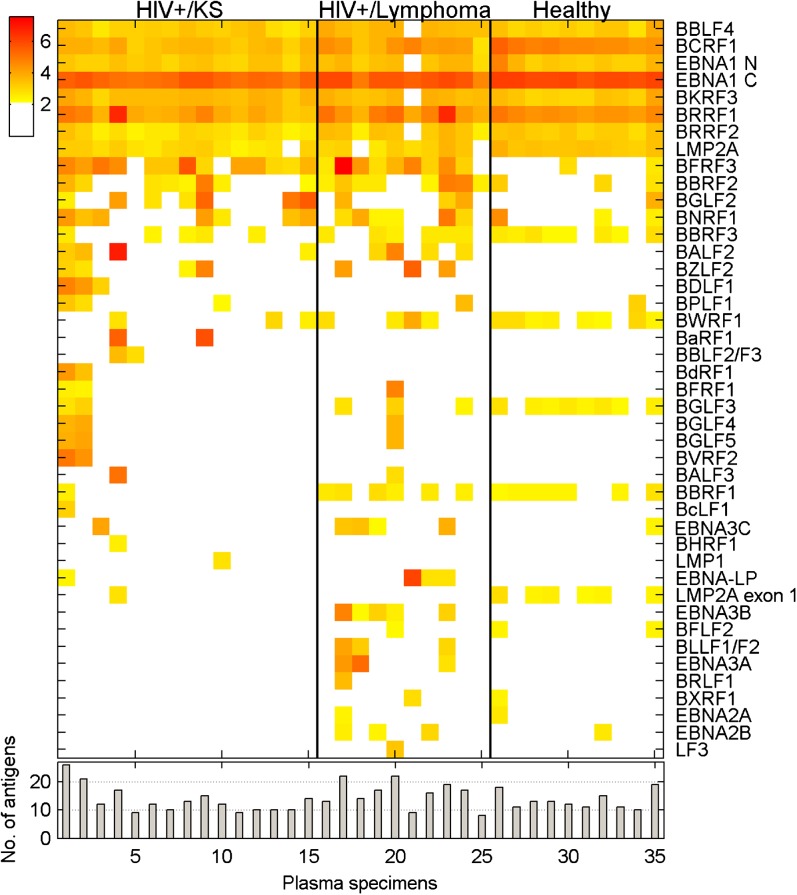
We next compared plasma IgG responses to KSHV in 2 populations, both of which were HIV-1–positive and had a diagnosis of cancer, but where 1 group had KS, a KSHV-associated disease, and the other had lymphoma. Plasma samples were evaluated at a 1:10 000 dilution. Nine of the 20 HIV-positive lymphoma patients recognized ORF73C at the 1:10 000 dilution vs 13 of the 15 HIV-positive KS patients (Figure 4A). The average signal intensity for ORF73C was higher in the HIV-positive KS population than in the HIV-positive lymphoma population. There was also an indication of increased lytic KSHV replication in the KS patients. Sixty-six percent (10/15) of the HIV-positive KS plasma specimens had antibody responses to KSHV ORF38, a myristylated tegument protein, while 0/9 LANA-positive, HIV-positive lymphoma patients had titers to ORF38 at the 1:10,000 dilution. Similarly, 53% (8/15) of the HIV-positive KS serum had antibody responses to KSHV ORF36, the conserved protein kinase that is a homolog of HSV-1 UL13, while only 1/9 (11%) of LANA-positive HIV-positive lymphoma patients had titers to ORF36. The most commonly used antigen to test for antibodies to lytic KSHV infection is the envelope glycoprotein K8.1 [4, 31, 32]; 8/15 (53%) of HIV-positive KS sera recognized K8.1.
Figure 4.
KSHV and EBV protein recognition by plasma from HIV-1–positive patients with KS vs AIDS-related lymphoma. (A) KSHV protein recognition. (B) EBV protein recognition. Upper panels: Heat maps comparing antigen recognition at 1:10 000 dilution by IgG from a set of HIV-1–positive patients with KS and HIV-1–positive patients with a diagnosis of lymphoma. Yellow: 5 SD cutoff; red: highest intensity signal. Lower panels: Number of antigens recognized by each sample.
In contrast to the antibody responses to KSHV proteins, the profiles of antibody responses to EBV proteins in these plasma samples were similar in the KS and lymphoma-carrying HIV-positive populations (Figure 4B). There was no significant difference in either the intensity of the positive signals or the number of antigens being recognized at a 1:10 000 serum dilution. Thus, the presence of KSHV-associated disease did not appear to have any impact on humoral responses to EBV antigens in the setting of concurrent HIV-1 infection. Again, the broader antigenic response to EBV compared with KSHV is striking. The W1 exon of EBV BamHI-W encodes 22 amino acids that form part of the EBNA-LP protein [33, 34]. The protein encoded by the entire EBV BamHI-W ORF (BWRF1) and the EBNA-LP protein were both printed on the array. We noted that 23/24 (95%) of the specimens that recognized EBNA-LP also recognized the protein encoded by BWRF1, implying that the common 22 amino acids contribute significantly to antibody recognition of EBNA-LP.
Comparison of Virus Load in Plasma Versus Humoral Immune Responses
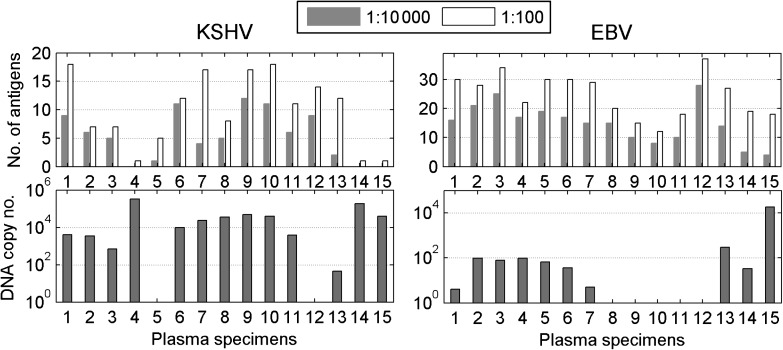
EBV and KSHV DNA are detected infrequently in serum or plasma from healthy individuals, but there is an increased prevalence in cancer patients and in HIV-1–infected individuals [35, 36]. A consistent correlation between EBV or KSHV DNA levels in serum or plasma and antibody titers to viral proteins has not been apparent [37]. To see if the more global antibody responses detected on the arrays would provide any additional insight, we examined KSHV and EBV DNA levels in the plasma from HIV-positive KS patients (see Figure 2A). The plasma copy number for KSHV and EBV DNA was measured by real-time quantitative PCR using previously published primers [27]. A comparison of DNA copy number with antibody responses did not reveal any obvious correlation for either KSHV or EBV (Figure 5). However, the 2 samples with the highest KSHV DNA copy number (>180 000 copies) did not recognize either the K8.1 or ORF38 lytic proteins, whereas 61% (8/13) of the samples with <50 000 copies recognized both of these antigens. Two samples are insufficient to allow any conclusions to be drawn, but the observation does raise the possibility that inadequate antibody production may have contributed to the robust KSHV replication detected in these individuals.
Figure 5.
Comparison of KHSV and EBV plasma DNA loads with IgG responses in HIV-1–infected patients with KS. Plasma KSHV and EBV DNA copy numbers as determined by real-time PCR are compared with the frequency of IgG responses in the same samples determined at 1:10 000 and 1:100 dilutions.
IgA Recognition of EBV Antigens
IgA responses to EBV EBNA1 and viral capsid antigens have long been used as a diagnostic tool for nasopharyngeal carcinoma [38]. In addition to nasopharyngeal carcinoma, IgA responses to VCA and to EBNA1 are frequently elevated in lymphoma patients and in individuals who are HIV-1 positive [39]. IgA responses to EBV proteins were examined at a 1:1000 dilution for 15 of the HIV-positive KS patients, 10 healthy individuals, and 10 HIV-positive lymphoma patients. Seven EBV antigens were consistently recognized by IgA across these patient groups, namely, EBNA1, the helicase BBLF4, the viral IL10 BCRF1, the uracil DNA glycoslyase BKRF3, the early protein BRRF1, the tegument protein BRRF2 [40], and the latency membrane protein LMP2A (Figure 6). The most intense IgA response detected was to EBNA1, which appeared to be recognized even more readily by IgA than IgG. All 25 normal and HIV-positive KS plasma tested recognized EBNA1 at a 1:1000 dilution, whereas in 4 of these samples there was no recognition of EBNA-1 by IgG even at a 1:100 dilution. However, the IgA response to the functionally equivalent protein in KSHV, LANA, was very different. Of 14 HIV-positive KS plasma samples that had IgG responses to LANA at a 1:10 000 dilution, only 2 had IgA responses at a 1:1000 dilution (data not shown). In 1 previous study, IgA antibody to KSHV LANA was detected in saliva but not in serum [41].
Figure 6.
IgA responses to EBV proteins. Plasma specimens from HIV-1–positive KS patients, AIDS-related lymphoma patients, and healthy individuals were incubated on the protein arrays at a 1:1000 dilution (upper panel). Heat map showing responses detected with Cy3-labeled anti-human IgA that were above the 5 SD cutoff (lower panel). Number of antigens recognized by each sample.
A comparison of the IgG and IgA responses in the plasma from healthy donors also revealed differential responses to certain EBV proteins. All 10 normal sera had IgA responses at a 1:1000 dilution to the viral IL10 BCRF1, the tegument protein BRRF2, and the latency membrane protein LMP2A. In contrast, even at a 1:100 dilution, IgG responses to BCRF1, BRRF2, or LMP2A were not detected in these plasma samples. The IgA response to vIL10 is particularly interesting because vIL10 is an immune modulator that can have an impact on localized immune escape of EBV-infected cells.
DISCUSSION
Testing for EBV and KSHV has relied on assays such as immunofluorescence staining and Western blotting of infected cells. Assays using individual viral proteins have also been described. However, in the absence of a global comparison of protein immunogenicity, there has been inadequate information on which to base the choice of antigens to incorporate into these assays. The EBV latency proteins EBNA1, EBNA2, and EBNA3C and the KSHV LANA latency and K8.1 lytic proteins have been widely used as antigens, and these antigens were also frequently detected on the arrays. Our protein array screens also identified the EBV tegument proteins BBRF2, BGLF2, and BNRF1 (FGARAT); the small capsid protein BFRF3; the early protein BRRF1; and the KSHV ORF38 tegument protein as lytic viral proteins that are particularly well recognized by IgG. The screens also identified BBLF4, BCRF1, BKRF3, and BBRF1 as EBV lytic proteins that were well recognized by IgA. It should be noted that the EBV and KSHV proteins for our array were expressed in yeast which do not have the same machinery as mammalian cells for complex glycosylation. It is therefore likely that our assays underestimate the immunological response to glycosylated viral antigens such as the EBV envelope glycoprotein gp350/220 and KSHV K8.1.
Globally, the array analyses revealed a stark contrast between antibody responses to EBV and KSHV in the same patients. Multiple EBV proteins were consistently recognized by plasma IgG at the dilutions examined, whereas only the LANA (ORF73), ORF38, and K8.1 proteins of KSHV were detected with any consistency, even in patients with KS. This difference carried over to serum IgA responses where 2 latency EBV proteins and 5 lytic proteins were detected by the majority of the sera, while the only KSHV antigen detected by IgA was LANA, and that was seen only rarely. These observed differences may reflect the biology of in vivo persistence. Both viruses latently infect B cells, and both viruses are secreted into saliva [42, 43]. However, EBV can be routinely detected in circulating latently infected B cells of healthy seropositive individuals, whereas detection of KSHV in blood is rare [44–46]. The preferential ability to detect serum IgA responses against EBV vs KSHV on the protein arrays may also be related to in vivo infection of mucosal epithelial cells by EBV because the most frequently observed antigens were latency proteins or lytic replicative proteins known to be expressed in epithelial cells [47–50]. KSHV infects endothelial cells as well as B cells, but the contribution of endothelial cells to in vivo viral persistence is not well understood.
The protein array described here provided a highly sensitive platform. Plasma with EBNA1 titers between 1:80 and 1:320 in conventional anticomplement immunofluorescence assays had titers of 1:10,000 or greater in the array assay (data not shown). The assay showed specificity. IgG responses to KSHV proteins were detected in sera from HIV-positive individuals with a diagnosis of KS or lymphoma but were not detected in a panel of HIV-negative lymphoma patients or healthy normals. This result, along with comparisons between EBV and KSHV responses in the HIV-positive samples, indicates that cross-reactivity between homologous proteins encoded by EBV and KSHV was not a confounding factor. This array provides a new platform for investigating the contribution of humoral immune responses in patients with EBV- and KSHV-associated pathologies.
Notes
Acknowledgments.
We thank Lindsey Hutt-Fletcher and S-J Gao for the bacmids used to generate the EBV and KSHV ORF libraries.
Financial support.
This work was supported by Public Health Service grants from the National Cancer Institute of the National Institutes of Health (R37 CA42245 and R01 CA30356 to S. D. H, R21 CA138163 to S. D. H. and H. Z., RC2 CA148402 to G. S. H., and Cancer Center Core Grant P30CA006973 to William Nelson).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Bagni R, Whitby D. Kaposi's sarcoma-associated herpesvirus transmission and primary infection. Curr Opin HIV AIDS. 2009;4:22–6. doi: 10.1097/COH.0b013e32831add5a. [DOI] [PubMed] [Google Scholar]
- 2.Dollard SC, Butler LM, Jones AM, et al. Substantial regional differences in human herpesvirus 8 seroprevalence in sub-Saharan Africa: insights on the origin of the “Kaposi's sarcoma belt.”. Int J Cancer. 2010;127:2395–401. doi: 10.1002/ijc.25235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao SJ, Kingsley L, Li M, et al. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat Med. 1996;2:925–8. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 4.Pfeiffer RM, Wheeler WA, Mbisa G, et al. Geographic heterogeneity of prevalence of the human herpesvirus 8 in sub-Saharan Africa: clues about etiology. Ann Epidemiol. 2010;20:958–63. doi: 10.1016/j.annepidem.2010.07.098. [DOI] [PubMed] [Google Scholar]
- 5.Martin JN, Ganem DE, Osmond DH, Page-Shafer KA, Macrae D, Kedes DH. Sexual transmission and the natural history of human herpesvirus 8 infection. N Engl J Med. 1998;338:948–54. doi: 10.1056/NEJM199804023381403. [DOI] [PubMed] [Google Scholar]
- 6.Martro E, Esteve A, Schulz TF, et al. Risk factors for human herpesvirus 8 infection and AIDS-associated Kaposi's sarcoma among men who have sex with men in a European multicentre study. Int J Cancer. 2007;120:1129–35. doi: 10.1002/ijc.22281. [DOI] [PubMed] [Google Scholar]
- 7.O'Brien TR, Kedes D, Ganem D, et al. Evidence for concurrent epidemics of human herpesvirus 8 and human immunodeficiency virus type 1 in US homosexual men: rates, risk factors, and relationship to Kaposi's sarcoma. J Infect Dis. 1999;180:1010–7. doi: 10.1086/315039. [DOI] [PubMed] [Google Scholar]
- 8.Ambinder RF, Cesarman E. Clinical and pathological aspects of EBV and KSHV infection. In: Arvin A, Campadelli-Fiume G, Moore PS, editors. Human herpesviruses biology, therapy, and immunoprophylaxis. New York: Cambridge University Press; 2007. pp. 885–914. [PubMed] [Google Scholar]
- 9.Nguyen ML, Farrell KJ, Gunthel CJ. Non-AIDS-defining malignancies in patients with HIV in the HAART era. Curr Infect Dis Rep. 2010;12:46–55. doi: 10.1007/s11908-009-0075-6. [DOI] [PubMed] [Google Scholar]
- 10.Thorley-Lawson DA, Duca KA, Shapiro M. Epstein-Barr virus: a paradigm for persistent infection - for real and in virtual reality. Trends Immunol. 2008;29:195–201. doi: 10.1016/j.it.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Tierney RJ, Steven N, Young LS, Rickinson AB. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J Virol. 1994;68:7374–85. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borza CM, Hutt-Fletcher LM. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat Med. 2002;8:594–9. doi: 10.1038/nm0602-594. [DOI] [PubMed] [Google Scholar]
- 13.Hadinoto V, Shapiro M, Sun CC, Thorley-Lawson DA. The dynamics of EBV shedding implicate a central role for epithelial cells in amplifying viral output. PLoS Pathog. 2009;5:e1000496. doi: 10.1371/journal.ppat.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laichalk LL, Thorley-Lawson DA. Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo. J Virol. 2005;79:1296–307. doi: 10.1128/JVI.79.2.1296-1307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassman LM, Ellison TJ, Kedes DH. KSHV infects a subset of human tonsillar B cells, driving proliferation and plasmablast differentiation. J Clin Invest. 2011;121:752–68. doi: 10.1172/JCI44185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu F, Feng J, Harada JN, Chanda SK, Kenney SC, Sun R. B cell terminal differentiation factor XBP-1 induces reactivation of Kaposi's sarcoma-associated herpesvirus. FEBS Lett. 2007;581:3485–8. doi: 10.1016/j.febslet.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 17.Mesri EA, Cesarman E, Boshoff C. Kaposi's sarcoma and its associated herpesvirus. Nat Rev Cancer. 2010;10:707–19. doi: 10.1038/nrc2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dittmer DP, Hilscher CJ, Gulley ML, Yang EV, Chen M, Glaser R. Multiple pathways for Epstein-Barr virus episome loss from nasopharyngeal carcinoma. Int J Cancer. 2008;123:2105–2. doi: 10.1002/ijc.23685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fakhari FD, Dittmer DP. Charting latency transcripts in Kaposi's sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J Virol. 2002;76:6213–3. doi: 10.1128/JVI.76.12.6213-6223.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng WM, Chan KH, Chen HL, et al. Assessing the risk of nasopharyngeal carcinoma on the basis of EBV antibody spectrum. Int J Cancer. 2002;97:489–92. doi: 10.1002/ijc.1641. [DOI] [PubMed] [Google Scholar]
- 21.Fachiroh J, Schouten T, Hariwiyanto B, et al. Molecular diversity of Epstein-Barr virus IgG and IgA antibody responses in nasopharyngeal carcinoma: a comparison of Indonesian, Chinese, and European subjects. J Infect Dis. 2004;190:53–62. doi: 10.1086/421245. [DOI] [PubMed] [Google Scholar]
- 22.Ng WT, Yau TK, Yung RW, et al. Screening for family members of patients with nasopharyngeal carcinoma. Int J Cancer. 2005;113:998–1001. doi: 10.1002/ijc.20672. [DOI] [PubMed] [Google Scholar]
- 23.Davies DH, Molina DM, Wrammert J, et al. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics. 2007;7:1678–86. doi: 10.1002/pmic.200600926. [DOI] [PubMed] [Google Scholar]
- 24.Zhu H, Hu S, Jona G, et al. Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray. Proc Natl Acad Sci U S A. 2006;103:4011–6. doi: 10.1073/pnas.0510921103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu J, Liao G, Shan L, et al. Protein array identification of substrates of the Epstein-Barr virus protein kinase BGLF4. J Virol. 2009;83:5219–31. doi: 10.1128/JVI.02378-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou FC, Zhang YJ, Deng JH, et al. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J Virol. 2002;76:6185–96. doi: 10.1128/JVI.76.12.6185-6196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin L, Lee JY, Kaplan LD, et al. Effects of chemotherapy in AIDS-associated non-Hodgkin's lymphoma on Kaposi's sarcoma herpesvirus DNA in blood. J Clin Oncol. 2009;27:2496–502. doi: 10.1200/JCO.2008.20.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen SJ, Sarid R, Chang Y, Moore PS. Evaluation of the latency-associated nuclear antigen (ORF73) of Kaposi's sarcoma-associated herpesvirus by peptide mapping and bacterially expressed recombinant western blot assay. J Infect Dis. 2000;182:306–10. doi: 10.1086/315689. [DOI] [PubMed] [Google Scholar]
- 29.Fachiroh J, Stevens SJ, Haryana SM, Middeldorp JM. Combination of Epstein-Barr virus scaffold (BdRF1/VCA-p40) and small capsid protein (BFRF3/VCA-p18) into a single molecule for improved serodiagnosis of acute and malignant EBV-driven disease. J Virol Methods. 2010;169:79–86. doi: 10.1016/j.jviromet.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Dambaugh T, Hennessy K, Chamnankit L, Kieff E. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc Natl Acad Sci U S A. 1984;81:7632–6. doi: 10.1073/pnas.81.23.7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corchero JL, Mar EC, Spira TJ, Pellett PE, Inoue N. Comparison of serologic assays for detection of antibodies against human herpesvirus 8. Clin Diagn Lab Immunol. 2001;8:913–21. doi: 10.1128/CDLI.8.5.913-921.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu L, Wang R, Sweat A, Goldstein E, Horvat R, Chandran B. Comparison of human sera reactivities in immunoblots with recombinant human herpesvirus (HHV)-8 proteins associated with the latent (ORF73) and lytic (ORFs 65, K8.1A, and K8.1B) replicative cycles and in immunofluorescence assays with HHV-8-infected BCBL-1 cells. Virology. 1999;256:381–92. doi: 10.1006/viro.1999.9674. [DOI] [PubMed] [Google Scholar]
- 33.Sauter M, Boos H, Hirsch F, Mueller-Lantzsch N. Characterization of a latent protein encoded by the large internal repeats and the BamHI Y fragment of the Epstein-Barr virus (EBV) genome. Virology. 1988;166:586–90. doi: 10.1016/0042-6822(88)90530-2. [DOI] [PubMed] [Google Scholar]
- 34.Wang F, Petti L, Braun D, Seung S, Kieff E. A bicistronic Epstein-Barr virus mRNA encodes two nuclear proteins in latently infected, growth-transformed lymphocytes. J Virol. 1987;61:945–54. doi: 10.1128/jvi.61.4.945-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin JC, Wang WY, Chen KY, et al. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med. 2004;350:2461–70. doi: 10.1056/NEJMoa032260. [DOI] [PubMed] [Google Scholar]
- 36.Lo YM, Chan LY, Lo KW, et al. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 1999;59:1188–91. [PubMed] [Google Scholar]
- 37.Gartner BC, Kortmann K, Schafer M, et al. No correlation in Epstein-Barr virus reactivation between serological parameters and viral load. J Clin Microbiol. 2000;38:2458. doi: 10.1128/jcm.38.6.2458-2458.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsang RK, Vlantis AC, Ho RW, Tam JS, To KF, van Hasselt CA. Sensitivity and specificity of Epstein-Barr virus IGA titer in the diagnosis of nasopharyngeal carcinoma: a three-year institutional review. Head Neck. 2004;26:598–602. doi: 10.1002/hed.20022. [DOI] [PubMed] [Google Scholar]
- 39.Stevens SJ, Smits PH, Verkuijlen SA, et al. Aberrant Epstein-Barr virus persistence in HIV carriers is characterized by anti-Epstein-Barr virus IgA and high cellular viral loads with restricted transcription. AIDS. 2007;21:2141–9. doi: 10.1097/QAD.0b013e3282eeeba0. [DOI] [PubMed] [Google Scholar]
- 40.Johannsen E, Luftig M, Chase MR, et al. Proteins of purified Epstein-Barr virus. Proc Natl Acad Sci U S A. 2004;101:16286–91. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mbopi-Keou FX, Legoff J, Piketty C, et al. Salivary production of IgA and IgG to human herpes virus 8 latent and lytic antigens by patients in whom Kaposi's sarcoma has regressed. AIDS. 2004;18:338–40. doi: 10.1097/00002030-200401230-00030. [DOI] [PubMed] [Google Scholar]
- 42.Niederman JC, Miller G, Pearson HA, Pagano JS, Dowaliby JM. Infectious mononucleosis. Epstein-Barr-virus shedding in saliva and the oropharynx. N Engl J Med. 1976;294:1355–9. doi: 10.1056/NEJM197606172942501. [DOI] [PubMed] [Google Scholar]
- 43.Pauk J, Huang ML, Brodie SJ, et al. Mucosal shedding of human herpesvirus 8 in men. N Engl J Med. 2000;343:1369–77. doi: 10.1056/NEJM200011093431904. [DOI] [PubMed] [Google Scholar]
- 44.Blackbourn DJ, Ambroziak J, Lennette E, Adams M, Ramachandran B, Levy JA. Infectious human herpesvirus 8 in a healthy North American blood donor. Lancet. 1997;349:609–11. doi: 10.1016/S0140-6736(96)10004-0. [DOI] [PubMed] [Google Scholar]
- 45.Pellett PE, Wright DJ, Engels EA, et al. Multicenter comparison of serologic assays and estimation of human herpesvirus 8 seroprevalence among US blood donors. Transfusion. 2003;43:1260–8. doi: 10.1046/j.1537-2995.2003.00490.x. [DOI] [PubMed] [Google Scholar]
- 46.Qu L, Jenkins F, Triulzi DJ. Human herpesvirus 8 genomes and seroprevalence in United States blood donors. Transfusion. 2010;50:1050–6. doi: 10.1111/j.1537-2995.2009.02559.x. [DOI] [PubMed] [Google Scholar]
- 47.Brooks L, Yao QY, Rickinson AB, Young LS. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J Virol. 1992;66:2689–97. doi: 10.1128/jvi.66.5.2689-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Busson P, McCoy R, Sadler R, Gilligan K, Tursz T, Raab-Traub N. Consistent transcription of the Epstein-Barr virus LMP2 gene in nasopharyngeal carcinoma. J Virol. 1992;66:3257–62. doi: 10.1128/jvi.66.5.3257-3262.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heussinger N, Buttner M, Ott G, et al. Expression of the Epstein-Barr virus (EBV)-encoded latent membrane protein 2A (LMP2A) in EBV-associated nasopharyngeal carcinoma. J Pathol. 2004;203:696–9. doi: 10.1002/path.1569. [DOI] [PubMed] [Google Scholar]
- 50.Oh ST, Seo JS, Moon UY, et al. A naturally derived gastric cancer cell line shows latency I Epstein-Barr virus infection closely resembling EBV-associated gastric cancer. Virology. 2004;320:330–6. doi: 10.1016/j.virol.2003.12.005. [DOI] [PubMed] [Google Scholar]