Abstract
The accumulation of the disaccharide trehalose in anhydrobiotic organisms allows them to survive severe environmental stress. A plant cDNA, SlTPS1, encoding a 109-kD protein, was isolated from the resurrection plant Selaginella lepidophylla, which accumulates high levels of trehalose. Protein-sequence comparison showed that SlTPS1 shares high similarity to trehalose-6-phosphate synthase genes from prokaryotes and eukaryotes. SlTPS1 mRNA was constitutively expressed in S. lepidophylla. DNA gel-blot analysis indicated that SlTPS1 is present as a single-copy gene. Transformation of a Saccharomyces cerevisiae tps1Δ mutant disrupted in the ScTPS1 gene with S. lepidophylla SlTPS1 restored growth on fermentable sugars and the synthesis of trehalose at high levels. Moreover, the SlTPS1 gene introduced into the tps1Δ mutant was able to complement both deficiencies: sensitivity to sublethal heat treatment at 39°C and induced thermotolerance at 50°C. The osmosensitive phenotype of the yeast tps1Δ mutant grown in NaCl and sorbitol was also restored by the SlTPS1 gene. Thus, SlTPS1 protein is a functional plant homolog capable of sustaining trehalose biosynthesis and could play a major role in stress tolerance in S. lepidophylla.
An amazing adaptation that allows survival under complete dehydration is present in yeast cells, fungal spores, certain invertebrate species, and resurrection plants that resume their vital functions as soon as they resume contact with water (Clegg, 1965; Gaff, 1971; Thevelein, 1984). These anhydrobiotic organisms also withstand strong vacuum, high doses of ionizing radiation, and extreme temperatures without suffering damage. In addition, many of these organisms accumulate the nonreducing disaccharide trehalose (Weisburd, 1988; Crowe et al., 1992).
Among several well-characterized osmoprotectors (Yancey et al., 1982), trehalose seems to be one of the most efficient at maintaining lipids in a fluid phase in the absence of water, thus avoiding phase separation, leakage, and membrane fusion (Crowe et al., 1984, 1987). Trehalose (α-d-glucopyranosyl-1,1-α-d-glucopyranoside), like other polyols, plays a key role in the structural and functional stabilization of membranes and proteins in the anhydrous state, apparently by means of water replacement of osmolyte molecules (Clegg, 1985) or formation of a glassy state (Burke, 1985).
The biosynthesis of trehalose consists of two enzymatic steps catalyzed by the oligomeric subunits TPS, which synthesizes trehalose-6-P from Glc-6-P and UDP-Glc, and TPP, which forms trehalose (Cabib and Leloir, 1958; Vandercammen et al., 1989; Londesborough and Vuorio, 1993). The genes encoding both enzymes from bacteria and yeast have already been isolated and sequenced (Luyten et al., 1993; Kaasen et al., 1994). Deletion mutants in the trehalose pathway in these microorganisms cause a reduction in osmotolerance (Giaever et al., 1988; Mackenzie et al., 1988) and thermotolerance (Hengge-Aronis et al., 1991; De Virgilio et al., 1994).
The yeast tps1Δ mutant and its alleles are unable to grow in Glc as the sole carbon source. There is strong evidence to suggest that this defect is attributable to the additional role of the TPS1 subunit in regulating the flow of Glc into the cell (Van Aelst et al., 1993; Neves et al., 1995; Thevelein and Hohmann, 1995; Hohmann et al., 1996).
Here we report the isolation and molecular characterization of a full-length cDNA encoding the enzyme TPS (SlTPS1) from the resurrection plant Selaginella lepidophylla, which is one of the organisms that accumulates trehalose at higher levels (Adams et al., 1990; Müller et al., 1995). We show that SlTPS1 cDNA encodes a functional TPS able to restore growth in fermentable sugars of a yeast tps1Δ mutant. Furthermore, SlTPS1 synthesizes high levels of trehalose and complements osmotolerance and thermotolerance deficiencies in a tps1Δ mutant.
MATERIALS AND METHODS
Plant Material
Selaginella lepidophylla (Hook. & Grev. Spring.) plants were collected from arid regions of Morelos state in Mexico. Plants were rehydrated and maintained in controlled conditions (24°C and 16 h of light with an average of 50% humidity) in growth chambers (Conviron, Asheville, NC). Subsequently, S. lepidophylla microphyll fronds were air dried at the indicated times by placing them on 3MM filter paper (Whatman).
Strains
The cDNA bank was plated in the Escherichia coli strain XL-1-Blue MRF′, and the strain SOLR was used to excise the pBluescript from the λ phage, following the manufacturer's instructions (Stratagene). The E. coli DH5α strain was used to subclone and make constructs. The yeast strains were wild type, W303-1A (Mat a leu2-3, 112 ura3-1, trp1-1, his3-11, 15 ade2-1, can1-100, GAL, SUC2) (Thomas and Rothstein, 1989); tps1Δ, YSH290 (W303-1A, ggs1/tps1Δ::TRP1) (Hohmann et al., 1993); tps2Δ, YSH450 (W303-1A, tps2Δ::LEU2) (Neves et al., 1995); and tps1Δtps2Δ, YSH652 (W303-1A, ggs1/tps1Δ::TRP1, tps2Δ::LEU2) (Neves et al., 1995).
Construction of the cDNA Bank of S. lepidophylla
An expression bank was prepared with mRNA isolated from S. lepidophylla microphylls dehydrated for 2.5 h using the ZAP cDNA synthesis kit, the Uni-ZAP XR vector, and the Gigapack II Gold packaging extracts following the manufacturer's instructions (Stratagene). The initial titer of the bank was 2 × 106 plaques of bacteriophage/mL and 1.5 × 1011 plaques of bacteriophage/mL after bank amplification. After 4 × 105 recombinant bacteriophages from the amplified cDNA bank of S. lepidophylla were plated, 13 plaques were obtained that hybridized with a mixture of the five oligonucleotides TPS5′-1, TPS5′-2, TPS5′-3, TPS3′-1, and TPS3′-2 (see Fig. 1A). In a second screening round, only 6 of the initial 13 plaques hybridized with a mixture of the oligonucleotides TPS5′-1 and TPS5′-2. In a third step, pIBT6 was the only clone that hybridized with the oligonucleotide TPS5′-1, corresponding to the most 5′ end of the selected region for isolation of the cDNA. Plaques were converted into plasmid by in vivo excision according to the manufacturer's instructions (Stratagene). Plasmid DNA was digested with EcoRI and XhoI to excise the corresponding insert, transferred to a Hybond N+ nylon membrane (Amersham), and hybridized to oligonucleotides 32P labeled with T4 polynucleotide kinase.
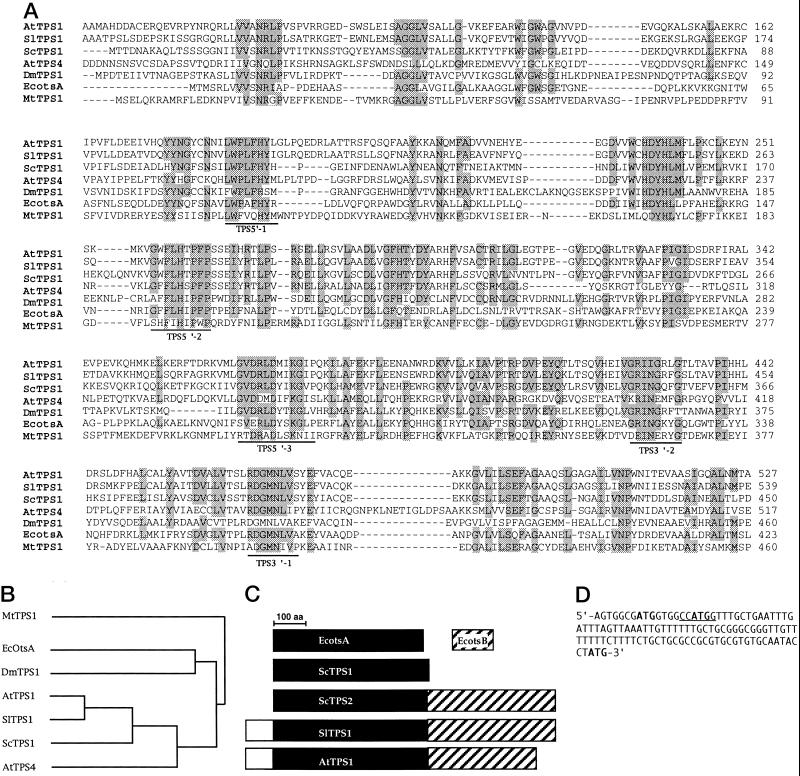
Figure 1.
Comparison of S. lepidophylla SlTPS1 with other TPS protein sequences. A, Alignment of an internal region of plant TPS amino acid sequences with bacteria, yeast, and animal sequences. Gaps that were introduced to optimize the alignment are indicated by dashes; identical residues are shaded. B, Dendrogram of TPS sequences. Amino acid sequences of TPS from (top to bottom) M. thermoautotrophicum (MtTPS1; accession no. AE000931); E. coli (EcOtsA; accession no. X69160); D. melanogaster (DmTPS1; accession no. AC004373); A. thaliana (AtTPS1; accession no. Y08568); S. lepidophylla (SlTPS1; accession no. U96736); S. cerevisiae (ScTPS1; accession no. X68214); and A. thaliana (AtTPS4; accession no. Z97344). DmTPS1 and AtTPS4 expressed sequence tags were found in databases from the respective systematic genome-sequencing programs. C, Diagram of TPS and TPP protein sequences. A comparison of protein size and structure is shown based on sequence-alignment data presented in A and compared with E. coli (EcotsB) and S. cerevisiae (ScTPS2) sequences. Protein size is shown in amino acid (aa) length. D, The 5′ leader sequence of the SlTPS1 gene is shown. Three ATG triplets are present (boldface), but only the one close to the 3′ end is in frame with the open reading frame sequence; thus, it is considered to be the putative initiation codon. The NcoI restriction site is underlined.
The following degenerated oligonucleotides were synthesized for screening of the cDNA bank: TPS5′-1, 5′-YTNTGGCCNBCNTTYCAYTAY-3′; TPS5′-2, 5′-GGNTKBTTYYTNCAYAYNCCNTTYCC-3′; TPS5′-3, 5′-MGNY-TNGAYTAYWBNAARGGNBTNCC-3′; TPS3′-1, 5′-SWN-ACNARRTTCATNCCRTCNCK-3′; and TPS3′-2, 5′-CCRW-ANTKNCCRTTDATNCKNCC-3′ (single-letter abbreviations for wobble nucleotides are B, C, G, or T; K, G or T; M, A or C; N, A or C or G or T; R, A or G; S, C or G; W, A or T; D, A, G, or T; and Y, C or T). These oligonucleotides were designed based on conserved domains of TPS sequences (depicted in Fig. 1A) using the following accession numbers: E. coli, X69160; Schizosaccharomyces pombe, Z29971; Aspergillus niger, U07184; Saccharomyces cerevisiae, X68214; and Kluyveromyces lactis, X72499.
Hybridization of Nucleic Acids
To screen the cDNA bank, the bacteriophage plaques were transferred to a nylon membrane and the filter was hybridized with oligonucleotides and labeled with the 32P isotope by means of T4 polynucleotide kinase using 6× SSC (1× SSC = 0.15 m NaCl and 0.015 m sodium citrate) at 37°C. The filter was washed three times at the same temperature for 10 min each under the following conditions: 6× SSC, 4× SSC, and 2× SSC.
Southern- and northern-blot techniques were used according to standard protocols (Sambrook et al., 1989) with the following modifications. For the genomic Southern-blot analysis, the DNA was fractionated on a 0.8% agarose gel in TBE (0.09 m Tris-borate and 0.002 m EDTA) buffer and transferred to a nylon membrane. The filter was hybridized using SlTPS1 cDNA labeled with 32P isotope as a probe, using 2× SSC at 65°C. The filter was washed three times at the same temperature for 20 min each time under the following conditions: 2× SSC, 1× SSC, and 0.5× SSC. For the northern-blot analysis, a 1.2% agarose gel was used in a Mops-formaldehyde buffer and a Hybond N+ nylon membrane was also used for the transfer. Hybridization conditions were in 50% formamide and 2× SSC at 42°C. The three successive filter washings were performed with 2× SSC, 2× SSC, and 1× SSC at 55°C.
DNA Sequencing
Nested deletions of the insert were created with the enzymes exonuclease III and nuclease S1 from the selected clone to subsequently determine the nucleotide sequence of both strands using Sequenase version 2.0 (United States Biochemical). Protein sequence alignments were analyzed using the CLUSTAL W software program (Thompson et al., 1994).
DNA Manipulation and Constructs
Recombinant DNA techniques such as bacterial transformation, isolation of DNA from plasmid, λ bacteriophage, and labeling of radioactive fragments were carried out according to standard procedures (Sambrook et al., 1989). For expression in yeast, two new shuttle-expression vectors named pSAL4 and pRS6 were constructed. pSAL4 is similar to pSAL1 (Mascorro-Gallardo et al., 1996), but it has the pRS426 backbone (Christianson et al., 1992) with the 2 μ origin of replication and the URA3 marker. The pRS6 vector was constructed using the PMA1 gene promoter from the pRS699 vector (Serrano and Villalba, 1995), a polylinker, and the CYC1 terminator. This cassette was cloned in the pRS423 backbone (Christianson et al., 1992) with the 2 μ origin of replication and the HIS3 marker.
Two out-of-frame ATG triplets in SlTPS1 cDNA leader (see Fig. 1D) were removed by digesting the pIBT6 clone with NcoI and treatment with nuclease S1. The intact leader precludes expression of SlTPS1 in yeast (data not shown). The 3.2-kb SlTPS1 cDNA was subcloned into pSAL4 or pRS6, and the resulting constructions were designated pSTS1 and pRTS1, respectively. The ScTPS1 and ScTPS2 yeast genes were isolated from genomic yeast DNA by PCR using primers ScTPS1-5′ (5′-CCGCTCGAGGGTACTCACATACAGAC-3′), ScTPS1-3′ (5′-ATAGTTTTGCGGCCGCATCGGGTTCATCAG-3′), ScTPS2-5′ (5′-CCGCTCGAGCACTATTTCTGTGCCG-3′), and ScTPS2-3′ (5′-CGGG-GTACCATGGTGGGTTGAGAC-3′). Amplified fragments were cloned into pSAL4 to give plasmids pSTS2 and pSTS3 or into pRS6 to give plasmids pRTS2 and pRTS3. A truncated SlTPS1 (SlTPS1ΔC) was constructed by deleting the DNA sequence coding for the 431-amino acid C terminus. The SlTPS1 region coding for the resting 563-amino acid N terminus was amplified by PCR with primers SlDC400-5′ (5′-CATGCCATGGCTATGCCTCAGCCTTACC-3′) and SlDC400-3′ (5′-CGGGGTACCTCACTTTGACTCCGAGTACTTTGC-3′) with TAG termination codon. A deletion of SlTPS1 (SlTPS1ΔN) comprising the 396-amino acid C terminus was constructed by PCR with primer SlDN600-5′ (5′-CCGCTCGAGCCATGGTGCATATTCCGCCTCAATTGCC-3′) and universal primer (5′-GTAAACGACGGCCAGT-3′). Both SlTPS1ΔC and SlTPS1ΔN were amplified using pIBT6 as a template. The PCR products were subcloned into pSAL4 vector to give plasmid pSTS4 or into pRS6 vector to give plasmid pRTS4. PCR was conducted using the Expand High Fidelity PCR System (Boehringer Mannheim), and reaction conditions were 1 cycle at 94°C for 3 min; 30 cycles at 94°C for 1 min, at 55°C for 1 min, and at 72°C for 2 min; and 1 cycle at 72°C for 10 min.
Transformation, Complementation, and Stress Assays in Yeast
Yeast was grown at 30°C in minimal medium (0.7% Bacto-yeast nitrogen base without amino acids, pH 6.0, supplemented with 0.002% adenine, 0.002% His, 0.003% Leu, 0.003% Trp, and 0.002% uracil) plus 2% Gal. Transformation was performed as described previously (Elble, 1992), and transformants were selected in medium without uracil for pSAL4 or in medium without His for pRS6. For each construct, at least three independent transformants were chosen to test their ability to restore the growth defect on minimal medium plus 2% Glc. The candidates were streaked on agar plates with minimal medium with 2% Gal, Glc, or Fru. As a control, the same strains were transformed with the pSAL4 or the pRS6 vector alone. After 3 d, growth was visible in positive control and complemented mutants.
For the thermotolerance assays, liquid cultures were grown at 25°C in minimal medium plus 2% Gal up to the mid-log phase (4 × 106 colony-forming units/mL, corresponding to 0.4 A600), then shifted to 39°C for 1 h for thermoinduction, and further incubated at 50°C for different times to evaluate induced thermotolerance. Decimal dilutions were plated in solid YPGal (2% Bacto-peptone, 1% yeast extract, 2% Gal, and 2% agar) and grown at 25°C for 3 d before colony counting. The level of thermotolerance was expressed as the ratio (percentage) of viability after the 50°C treatment and immediately before the 50°C treatment.
For the osmotolerance assays, exponential cultures were first diluted until A600 was 0.1 and then serially diluted 5-fold at each step. Spots were made with 4 μL of each dilution and plated in solid YPGal supplemented with 0.9 m NaCl or 1.6 m sorbitol. Growth was recorded after 3 d for the control plates and after 4 d for the osmotic stress treatments.
Trehalose Determination
Essentially, yeast cells (25–50 mg fresh weight) were collected by vacuum filtration through 0.22- to 0.45-μm membranes (Gelman Sciences, Ann Arbor, MI) and washed several times with ice-cold water to remove external Glc. Yeast cells were scraped from the filter membrane before being frozen in liquid nitrogen and stored at −80°C or immediately transferred to a screw-capped tube containing 1 mL of 0.25 m Na2CO3 and boiled in a water bath for 20 min. After cooling, samples were centrifuged in a microfuge and 0.2 mL of supernatant was mixed with 0.1 mL of 1 m acetic acid to neutralize before the addition of 0.1 mL of buffer T (0.3 m sodium acetate plus 0.03 m CaCl2, pH 5.5). For trehalose quantification, 0.05 mL of samples or trehalose standards and 0.05 mL of Humicola grisea trehalase (or water to determine Glc not derived from trehalose) were incubated for 45 min at 40°C. One milliliter of Tris-HCl buffer, pH 8.0, containing 100 units each of Glc oxidase and peroxidase (Sigma) plus 0.1 mg of o-dianisidine (Sigma) was added. Incubation was for 1 h at 30°C, and the reaction was stopped with 0.5 mL of 56% (v/v) sulfuric acid. The A546 was determined before 1 h. H. grisea trehalase was purified according to the method of Neves et al. (1994).
RESULTS
Cloning and Sequence Analysis of S. lepidophylla SlTPS1
To isolate a gene involved in trehalose synthesis, a comparison of the deduced amino acid sequences of TPS was made from the reported nucleotide sequences from bacteria and yeast (see Methods). Highly conserved regions were selected to synthesize degenerated oligonucleotides (Fig. 1A), which were used to screen a cDNA library from S. lepidophylla (see Methods). The largest clone isolated, pIBT6 (3.2 kb), contains a 110-nucleotide 5′ leader sequence, a 96-nucleotide 3′-untranslated sequence, and an open reading frame coding for a protein of 994 amino acids with a calculated molecular mass of 109 kD; it was designated SlTPS1. The alignment of deduced amino acid sequences of TPS from bacteria, yeast, plants, and animals showed several regions of homology (Fig. 1A). Only the most conserved region in all of the compared TPS sequences is shown. These deduced amino acid sequences were plotted in a dendrogram (Fig. 1B). The most distant sequence from SlTPS1 was the archaebacterium Methanobacterium thermoautotrophicum MtTPS1, which is only 27% identical to S. lepidophylla SlTPS1. The E. coli EcOtsA and the Drosophila melanogaster DmTPS1 sequences shared 35% and 40% identity to SlTPS1, respectively. Plant and yeast TPS sequences were found closely related to each other. SlTPS1 is the most related to the recently reported AtTPS1 sequence from Arabidopsis (Blázquez et al., 1998), and both sequences share an identity of 67%, whereas the S. cerevisiae ScTPS1 is 50% identical to both SlTPS1 and AtTPS1. It is interesting that another Arabidopsis TPS homolog, AtTPS4, is more closely related to ScTPS1 (41% identity) than to SlTPS1 and AtTPS1 (37% identity). AtTPS4 may represent a divergent TPS gene that probably arose by duplication of the ScTPS1 ancestor gene.
An additional feature of plant TPS sequences is the presence of two putative domains (Fig. 1C). Only the N-terminal sequence of SlTPS1, which contains 563 amino acids, has similarity to TPS sequences (Fig. 1, A and B), whereas the remaining 431-amino acid sequence of SlTPS1, which constitutes its C-terminal region, is also present in AtTPS1 and AtTPS4 but is absent in yeast and E. coli TPS sequences (Fig. 1C). This C-terminal region of SlTPS1 shows a certain degree of identity to sequences encoding TPP: 29% to the yeast ScTPS2 subunit and 22% to the E. coli EcOtsB enzyme.
Expression Pattern and Copy Number of SlTPS1
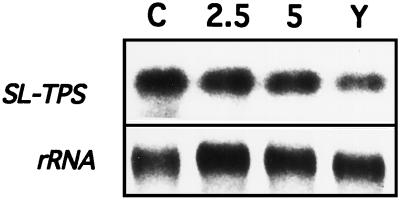
RNA-blot analysis was performed to investigate the expression pattern of the SlTPS1 gene. Poly(A+) RNA was isolated from S. lepidophylla microphylls (lycophyte leaves) that were fully turgid or desiccated for different lengths of time. SlTPS1 mRNA is expressed as a single band corresponding to a 3.2-kb transcript present in fully hydrated and dehydrated S. lepidophylla microphylls at similar levels (Fig. 2). This constitutive expression of the S1TPS1 gene is in agreement with the comparable levels of trehalose in both nonstressed and desiccated S. lepidophylla plants (Adams et al., 1990).
Figure 2.
Expression of SlTPS1 mRNA in S. lepidophylla. Poly(A+) RNA (2 μg) was extracted from fully hydrated (lane C) S. lepidophylla microphylls or dehydrated for 2.5 h (lane 2.5), 5 h (lane 5), or 1 year (lane Y). The RNA blot was hybridized with 32P-labeled SlTPS1 cDNA (top row). An rRNA gene fragment was hybridized to the same filter as a loading control (bottom row).
The copy number of the SlTPS1 gene was determined by DNA gel-blot analysis. S. lepidophylla genomic DNA was digested with EcoRI, which cuts internally, and BamHI and XbaI, which do not cut the cDNA. After probing with the full-length SlTPS1 insert, two expected bands were obtained with EcoRI (Fig. 3). Digestion with XbaI corresponded to a single band, but two bands were obtained using BamHI, suggesting either that there are two SlTPS1 genes or that the BamHI site is present in an intron. To test this latter possibility, we used specific oligonucleotides matching the coding 5′ and 3′ ends of SlTPS1 to amplify genomic homologs by PCR. A single fragment was obtained that led to two bands after digestion with BamHI, thus suggesting the possibility of a BamHI site in an intron. Partial nucleotide sequence of these PCR fragments matched exactly the cDNA sequence (data not shown). Therefore, according to these data, SlTPS1 seems to be a single-copy gene.
Figure 3.
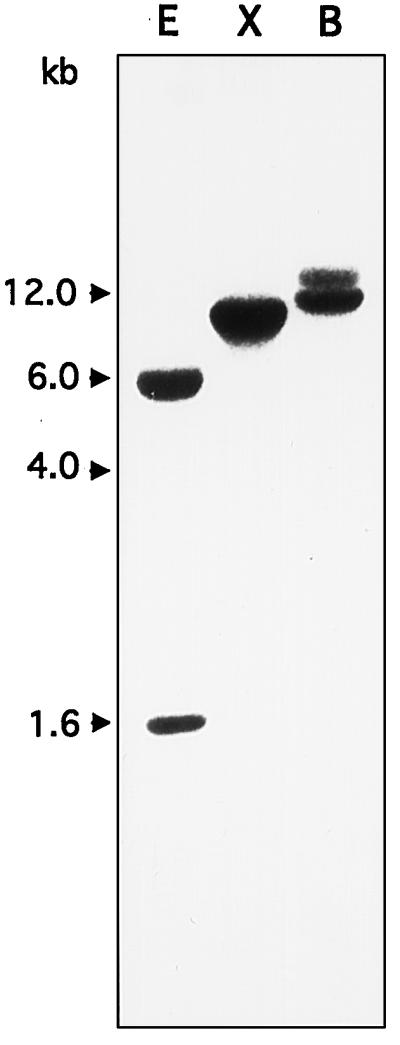
DNA gel-blot analysis of the SlTPS1 gene. Genomic DNA (20 μg) of S. lepidophylla was digested with EcoRI (lane E), XbaI (lane X), or BamHI (lane B). The filter was probed with 32P-labeled SlTPS1 cDNA. The positions of the DNA markers are indicated on the left.
Functional Analysis of SlTPS1 in Yeast
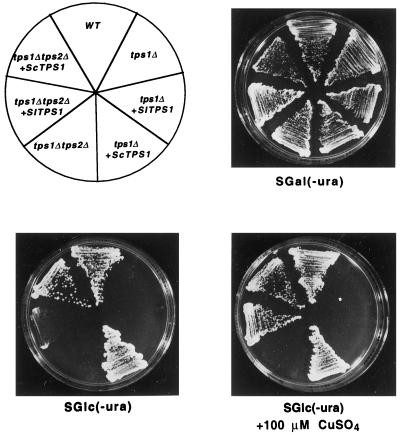
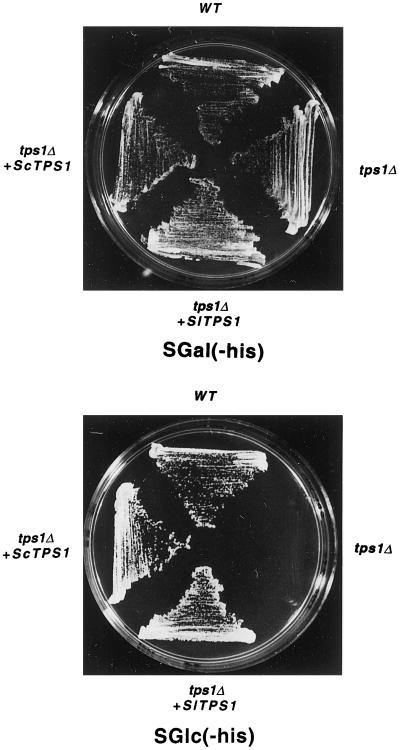
To determine whether the SlTPS1 gene can complement the physiological defects of mutant yeast cells devoid of trehalose biosynthesis genes, the corresponding 3.2-kb cDNA was subcloned into the yeast pSAL4 vector behind a CUP1 promoter that is inducible by copper ions (Mascorro-Gallardo et al., 1996). The resulting plasmid pSTS1 was used to transform the yeast tps1Δ and tps1Δtps2Δ mutants. These mutants are unable to grow with Glc or Fru as the carbon source, apparently because of the role of TPS1 in regulating the flow of Glc in glycolysis (Van Aelst et al., 1993; Thevelein and Hohmann, 1995). After transformation with pSTS1 containing the SlTPS1 gene, the tps1Δtps2Δ mutant was able to grow in Glc in the presence of copper ions (Fig. 4), whereas no growth was observed for the tps1Δ mutant. All strains grew well in Gal, as expected. Transformation of the tps1Δ mutant with plasmid pSTS2, which harbors the homologous ScTPS1 gene, restored growth for both the tps1Δ and tps1Δtps2Δ mutants (Fig. 4). Trehalose levels were measured in the tps1Δ and tps1Δtps2Δ mutants transformed with pSTS1 plasmid (containing SlTPS1) grown in different carbon sources (Table I). Neither the tps1Δ nor the tps1Δtps2Δ mutant transformed with SlTPS1 accumulated trehalose above the level detected for the same mutants transformed with the pSAL4 vector alone. We decided to express SlTPS1 cDNA under the control of a stronger promoter to observe a possible accumulation of trehalose. Thus, the promoter of the PMA1 gene that encodes the H+-ATPase (Serrano and Villalba, 1995) was subcloned in a multicopy vector to give plasmid pRS6 (see Methods). After SlTPS1 was subcloned under the control of the PMA1 promoter (plasmid pRTS1), this construction was used to transform the yeast tps1Δ mutant. It was observed that the tps1Δ mutant transformed with pRTS1 plasmid was able to grow in Glc (Fig. 5).
Figure 4.
Complementation of the tps1Δtps2Δ mutant by SlTPS1 under the control of the CUP1 promoter. Yeast mutants lacking the TPS1 protein (tps1Δ) or both TPS1 and TPS2 (tps1Δtps2Δ) were transformed with pSTS1 (pSAL4 containing SlTPS1), pSTS2 (pSAL4 containing ScTPS1), or pSAL4 vector alone and spread on 2% agar plates in minimal medium without uracil (−ura) supplemented with 2% Gal (SGal), 2% Glc (SGlc), or 2% Glc with copper sulfate (SGlc +100 μm CuSO4). The wild-type control strain W303-1A (WT) was also transformed with pSAL4.
Table I.
Trehalose content of the transformed tps1Δ and tps1Δtps2Δ mutants with pSAL4-derived vectors
| Carbon Source | Strain | Trehalose | Trehalose Content |
|---|---|---|---|
| μmol g−1 yeast wet wt | % | ||
| Gal | Wild type (pSAL4) | 71.70 ± 25.18 | 100 |
| tps1Δtps2Δ (pSAL4) | 5.25 ± 6.77 | 7 | |
| tps1Δtps2Δ (SlTPS1) | 6.32 ± 6.23 | 9 | |
| tps1Δtps2Δ (SlTPS1ΔC) | 2.13 ± 0.52 | 3 | |
| tps1Δtps2Δ (ScTPS1) | 19.27 ± 14.93 | 27 | |
| tps1Δ (pSAL4) | 4.80 ± 4.90 | 7 | |
| tps1Δ (S1TPS1) | 2.83 ± 1.65 | 4 | |
| tps1Δ (ScTPS1) | 48.43 ± 21.16 | 67 | |
| Glc | Wild type (pSAL4) | 50.90 ± 9.02 | 100 |
| tps1Δtps2Δ (SlTPS1) | 5.42 ± 4.58 | 11 | |
| tps1Δtps2Δ (ScTPS1) | 18.50 ± 12.93 | 36 | |
| tps1Δ (ScTPS1) | 36.48 ± 10.69 | 72 | |
| Fru | Wild type (pSAL4) | 81.77 ± 31.03 | 100 |
| tps1Δtps2Δ (SlTPS1) | 6.95 ± 5.01 | 7 | |
| tps1Δtps2Δ (ScTPS1) | 21.13 ± 13.04 | 26 | |
| tps1Δ (ScTPS1) | 54.40 ± 24.92 | 66 |
Three independent transformants of wild-type yeast, the tps1Δ mutant, or the tps1Δtps2Δ mutant transformed with pSAL4 vector alone or harboring the SlTPS1, SlTPS1ΔC, or ScTPS1 genes were grown in minimal medium with the indicated carbon source plus 100 μm CuSO4. Trehalose content was determined in the stationary phase (7.0 A600). Values are means ± sd.
Figure 5.
Complementation of the tps1Δ mutant by SlTPS1 under the control of the PMA1 promoter. The yeast mutant lacking TPS1 protein (tps1Δ) was transformed with pRTS1 (pRS6 containing SlTPS1), pRTS2 (pRS6 containing ScTPS1), or pRS6 vector alone and spread on 2% agar plates in minimal medium without His (−his) supplemented with 2% Gal (SGal) or 2% Glc (SGlc). The wild-type control strain W303-1A (WT) was also transformed with pRS6.
The trehalose content was determined for tps1Δ mutant strains transformed with the pRTS1 (containing SlTPS1) and pRTS2 (harboring ScTPS1) plasmids. Trehalose was detected in the tps1Δ mutant complemented with SlTPS1 grown in different carbon sources (Table II). The trehalose levels were higher when the tps1Δ mutant was complemented with the S. cerevisiae ScTPS1 gene than when SlTPS1 was used. These results indicate that heterologous S. lepidophylla SlTPS1 is not completely fulfilling the function of the yeast ScTPS1 protein, considering the sequence divergence and the fact that the SlTPS1 polypeptide is larger than ScTPS1. Therefore, we constructed a deletion mutant of SlTPS1, named SlTPS1ΔC, to remove the sequence encoding its C-terminal domain of 431 amino acids, which lacks similarity to ScTPS1 or EcOtsA (Fig. 1C). Plasmid pRTS4 containing SlTPS1ΔC was used to transform the tps1Δ and tps1Δtps2Δ mutants. Growth in Glc was complemented in both mutants by the C-terminal deletion of SlTPS1. When SlTPS1ΔC was cloned under control of the CUP1 promoter (plasmid pSTS4), only partial complementation was observed in the tps1Δtps2Δ mutant: Growth in Glc was slow compared with that in the same mutant transformed with the full-length SlTPS1 gene in pSAL4 (data not shown). Trehalose levels in the tps1Δ mutant transformed with pRTS4 were 2.5 times lower than the levels with the full-length SlTPS1 gene but significantly higher than in the tps1Δ mutant transformed with vector alone (Table II). These results show that the C-terminal region of SlTPS1 is required for full activity.
Table II.
Trehalose content of the transformed tps1Δ mutant with pRS6-derived vectors
| Carbon Source | Strain | Trehalosea | Trehalose Content |
|---|---|---|---|
| μmol g−1 yeast wet wt | % | ||
| Gal | Wild type (pRS6) | 16.45 ± 4.59 | 100 |
| tps1Δ (pRS6) | 1.45 ± 0.07 | 9 | |
| tps1Δ (SlTPS1) | 16.55 ± 0.35 | 100 | |
| tps1Δ (SlTPS1ΔC) | 6.59 ± 0.72 | 40 | |
| tps1Δ (ScTPS1) | 26.20 ± 2.97 | 159 | |
| Glc | Wild type (pRS6) | 29.45 ± 0.64 | 100 |
| tps1Δ (SlTPS1) | 18.95 ± 5.87 | 64 | |
| tps1Δ (ScTPS1) | 27.85 ± 7.42 | 95 | |
| Fru | Wild type (pRS6) | 29.00 ± 7.07 | 100 |
| tps1Δ (SlTPS1) | 20.40 ± 3.39 | 70 | |
| tps1Δ (ScTPS1) | 34.05 ± 4.59 | 117 |
Three independent transformants of wild-type yeast or the tps1Δ mutant transformed with pRS6 vector alone or harboring the SlTPS1, SlTPS1ΔC, or ScTPS1 genes were grown in minimal medium with the indicated carbon source.
Trehalose content was determined in the stationary phase (7.0 A600). Values are means ± sd.
Given that the C-terminal region of SlTPS1 shares relative similarity to TPPs (Fig. 1C), we tested the ability of full-length SlTPS1 and its C-terminal domain (SlTPS1ΔN) to complement the defect to grow at 37.5°C associated with the yeast tps2Δ and tps1Δtps2Δ mutants. pRTS1 (containing SlTPS1) and pRTS5 (harboring SlTPS1ΔN) were used to transform these mutants. It is known that the tps2Δ mutant grows normally in Glc but is unable to grow continuously at 37.5°C (De Virgilio et al., 1993). The tps1Δtps2Δ mutant has both growth defects. SlTPS1 was not able to restore the growth of the tps2Δ and tps1Δtps2Δ mutants at 37.5°C. Transformation with pRTS3, which harbors the homologous ScTPS2 gene, complemented both mutants (data not shown).
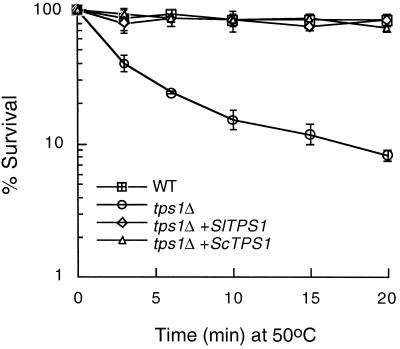
Trehalose is required for the acquisition of thermotolerance in yeast and E. coli (Hengge-Aronis et al., 1991; De Virgilio et al., 1994). The yeast tps1Δ, tps2Δ, and tps1Δtps2Δ deletion mutants are deficient in both induced and noninduced thermotolerance. The phenotypes are restored by complementation with the corresponding homologous gene. To determine whether the SlTPS1 gene can also complement these deficient stress responses, the yeast tps1Δ mutant was transformed with plasmid pRTS1 (harboring SlTPS1) or pRTS2 (containing ScTPS1). We assayed the ability to survive both a sublethal heat shock of 39°C for 1 h (thermoinduction) and a lethal heat shock of 50°C for 20 min after an acclimation treatment of 39°C for 1 h (induced thermotolerance). The viability of the tps1Δ mutant after thermoinduction decreased to 30%, but almost complete viability was recovered after transformation with the SlTPS1 or the ScTPS1 gene (Table III). After lethal heat shock, S. lepidophylla SlTPS1 restored the induced thermotolerance as effectively as yeast ScTPS1 in tps1Δ mutant cells compared with the level in the wild-type cells (Fig. 6). Both SlTPS1- and ScTPS1-transformed cells displayed a 10-fold higher induced thermotolerance than the tps1Δ mutant cells transformed with the pRS6 vector alone after 20 min at 50°C.
Table III.
Viability of tps1Δ mutant cells transformed with SlTPS1 gene after thermoinduction
| Strain | Surviving Cells |
|---|---|
| % | |
| Wild type (pRS6) | 127.00 ± 19.52 |
| tps1Δ (pRS6) | 29.67 ± 6.66 |
| tps1Δ (SlTPS1) | 80.00 ± 21.16 |
| tps1Δ (ScTPS1) | 96.00 ± 2.65 |
Thermoinduction was performed by incubating yeast cells in liquid SGal (-His) medium up to approximately 0.4 A600 at 25°C, and then shifted for 1 h at 39°C. Aliquots for colony counting were taken just before and after thermoinduction. Data represent the mean of three independent transformants and their sd.
Figure 6.
Complementation of thermotolerance deficiency of the tps1Δ deletion mutant transformed with the SlTPS1 gene. Transformed strains were grown in liquid culture and assayed for induced thermotolerance (see Methods). Values shown for each construct are the averages of three independent transformants. WT, Wild-type cells transformed with the pRS6 vector alone; tps1Δ, mutant transformed with the pRS6 vector alone; tps1Δ+SlTPS1, mutant transformed with plasmid pRTS1; tps1Δ+ScTPS1, mutant transformed with plasmid pRTS2. Error bars denote ±sd.
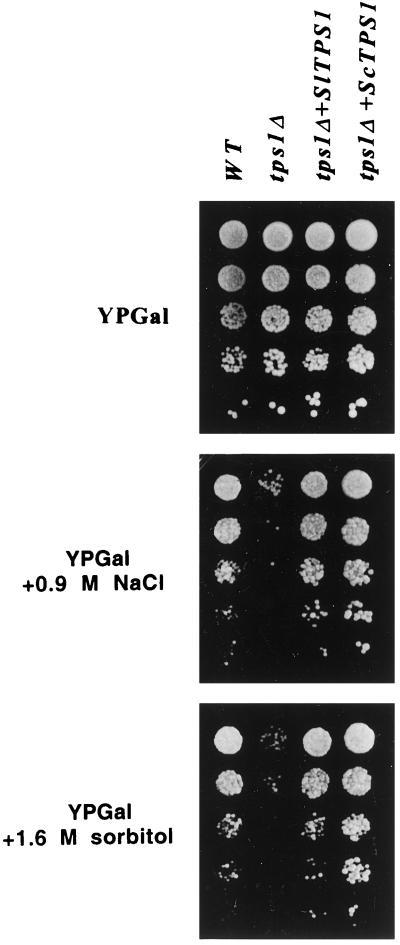
Finally, it has been shown that the tps1Δ mutant is osmosensitive (Hounsa et al., 1998). To determine whether SlTPS1 could restore the osmotolerance defect of the tps1Δ mutant, this strain was transformed with the plasmids pRTS1 and pRTS2 and growth was evaluated under osmotic stress conditions. The tps1Δ mutant transformed either with the SlTPS1 or the ScTPS1 gene was able to grow in medium containing 1.6 m sorbitol or 0.9 m NaCl (Fig. 7). As a positive control, we used the wild-type strain, which also grew normally. In contrast, the tps1Δ mutant transformed with the vector alone was unable to grow in high-osmoticum conditions. All of these results suggest that the levels of trehalose accumulated in the tps1Δ mutant transformed with SlTPS1 are sufficient to restore growth under osmotic and heat stress.
Figure 7.
SlTPS1 restores the osmotolerance defect of a yeast tps1Δ deletion mutant. Osmotolerance assay of a tps1Δ deletion mutant transformed with the SlTPS1 gene. Yeast mutants lacking the protein TPS1 (tps1Δ) were transformed with pRTS1 (containing SlTPS1), pRTS2 (containing ScTPS1), or pRS6 vector alone and spotted on 2% agar plates in rich YPGal medium supplemented with 0.9 m NaCl or 1.6 m sorbitol. The wild-type control strain W303-1A (WT) was also transformed with pRS6.
DISCUSSION
Trehalose is one of the most efficient osmoprotectors and thermoprotectors in nature (Colaço et al., 1992; Crowe et al., 1992). Although much experimental work concerning trehalose has been done in bacteria and yeast, little information has been reported from animals or plants. Here we report the molecular and functional characterization of a full-length cDNA encoding a novel TPS (SlTPS1) from the resurrection plant S. lepidophylla. This plant is known to accumulate trehalose at levels comparable to yeast and other fungi and at levels much higher than those found in other plants (Adams et al., 1990; Müller et al., 1995). It is likely that a major factor determining the anhydrobiotic ability of resurrection plants is their capacity to accumulate high levels of osmoprotectors such as trehalose. Until a few years ago, it was thought that only resurrection plants had the capacity to synthesize trehalose (Müller et al., 1995). Recently, the presence of trehalose was reported in tobacco, potato (Goddijn et al., 1997), and rice (Garcia et al., 1997), but at levels about 3000-fold less than in S. lepidophylla (Adams et al., 1990). Although the actual level of trehalose in Arabidopsis has not yet been reported, presumably it is not significant because AtTPS1 mRNA is expressed at very low levels (Blázquez et al., 1998). Additionally, it is possible that the low levels of trehalose in most higher plants could be the result of very active trehalase. Nevertheless, when Goddijn et al. (1997) incubated tobacco and potato plants in the presence of the trehalase inhibitor validamycin A, trehalose did not reach concentrations comparable to those in a resurrection plant.
Another possibility is that substitutions in the amino acid sequence that occurred during evolution in different plant TPS have an important influence on trehalose synthesis among plant species.
In this work, we present evidence that the expression of the SlTPS1 gene in the yeast tps1Δ mutant under control of the strong PMA1 promoter leads to an accumulation of trehalose in the stationary phase equivalent to 64% to 100% of the levels reached by wild-type yeast, depending on the carbon source. Similarly, Blázquez et al. (1998) used the PGK1 promoter to express AtTPS1, which resulted in 25% of the trehalose found in the wild-type yeast grown in Gal. These results might be attributable to either a difference between PGK1 and PMA1 promoter strength or an increased capacity of SlTPS1 to synthesize trehalose. Therefore, it remains to be shown if the difference in the trehalose content between S. lepidophylla and nonresurrection plants is the result of transcriptional control and/or balance between trehalase and TPS enzyme activities.
Transformation of the yeast tps1Δ mutant with the SlTPS1 gene under the control of a moderate promoter such as CUP1 did not restore the ability of the wild-type phenotype to grow in Glc, whereas the growth of the tps1Δtps2Δ mutant was complemented. Nevertheless, trehalose levels in both the tps1Δ and tps1Δtps2Δ mutants transformed with SlTPS1 were negligible, thus suggesting that restoration of growth in Glc of the tps1Δtps2Δ mutant by SlTPS1 is independent of the presence of trehalose. This observation is interesting because SlTPS1 expressed in a moderate promoter, such as CUP1, allows separate analyses of both TPS functions, i.e. trehalose synthesis capacity and regulation of Glc influx in glycolysis (Van Aelst et al., 1993; Thevelein and Hohmann, 1995).
Moreover, the fact that tps1Δ was not complemented by the SlTPS1 gene expressed under the control of the CUP1 promoter led us to suggest that maybe SlTPS1 was somehow inhibited by ScTPS2. One possible explanation for these results is a negative interaction or sequestration of SlTPS1 by ScTPS2. In yeast, ScTPS1, ScTPS2, ScTPS3, and ScTSL1 interact with each other to form the holoenzyme complex (Reinders et al., 1997). Thus, given the sequence similarity between yeast and plant TPS1, ScTPS2 and SlTPS1 may interact with each other, resulting in a structural constraint of SlTPS1 enzyme activity.
The SlTPS1 cDNA encodes a 109-kD protein that shares strong similarity to TPS sequences and has a C-terminal extension with some similarity to TPP. We tested whether SlTPS1 had TPP activity by transforming the yeast tps2Δ and tps1Δtps2Δ mutants with the full-length SlTPS1 gene or just its C-terminal region under the control of the strong PMA1 promoter. However, the lack of complementation for growth at 37.5°C suggested that SlTPS1 does not have TPP activity. It has been shown that all TPP enzymes from bacteria to plants have two short and well-conserved regions of homology (Vogel et al., 1998). These sequences are absent in SlTPS1 and AtTPS1, providing further evidence for the absence of TPP activity in SlTPS1.
In yeast, it is well established that trehalose is involved in acquired thermotolerance and tolerance to continuous growth at sublethal temperatures (De Virgilio et al., 1994; Elliot et al., 1996). It has been shown that the tps1Δ mutant is deficient in induced thermotolerance, and this phenotype can be restored after complementation with the homologous ScTPS1 gene (De Virgilio et al., 1994). Here we show that SlTPS1 is able to restore the yeast capacity for both thermoinduction and induced thermotolerance. Another aspect of SlTPS1 that we addressed was its capacity to confer osmotolerance. A recent work (Hounsa et al., 1998) analyzed the osmosensitive phenotype tps1Δ mutant under moderate and severe osmotic stress, revealing the strong correlation between the presence of trehalose in yeast and survival under osmotic stress conditions. In the present study, we showed that both SlTPS1 and ScTPS1 are able to complement and restore growth of yeast cells under osmotic stress. These data suggest that trehalose may play a similar role in S. lepidophylla as a stress protectant.
A few TPS homologs have been cloned from bacteria, fungi, and higher eukaryotes (Fig. 1). It is possible that TPS1 is present in most organisms, although not necessarily involved in the synthesis of significant amounts of trehalose. That plant or animal TPS might have a role similar to that of yeast ScTPS1 as a controller of the influx of sugar into glycolysis is an intriguing possibility. The fact that both SlTPS1 and AtTPS1 are able to complement the growth defect on fermentable sugars of the yeast tps1Δ mutant suggests this possibility and opens a new perspective on the regulation of glycolysis in plants and animals that should be explored.
ACKNOWLEDGMENTS
We thank Paul Gaytán and Eugenio López for oligonucleotide synthesis and Mario Trejo, Elena Arriaga, Guadalupe Ochoa, Martine De Jonge, and Willy Verheyden for technical assistance. We acknowledge Dr. G.J. Ruijter (Wageningen Agricultural University, The Netherlands) for providing purified A. niger hexokinase. J.O.M.-G. thanks Consejo Nacional de Ciencia y Tecnología, Mexico, and B.B. thanks Conselho Nacional de Desenvolvimento Cientifico e Tecnológico, Brazil, for Ph.D. fellowships.
Abbreviations:
- TPP
trehalose-6-P phosphatase
- TPS
trehalose-6-P synthase
Footnotes
This work was supported by grant no. IN202795 to G.I. and J.N.-S. from Dirección General de Asuntos del Personal Académico-Universidad Nacional Autónoma de México, Mexico; by grants to J.M.T. from the Fund for Scientific Research-Flanders and the Research Fund of the Katholieke Universiteit Leuven, Concerted Research Actions, Belgium; and by grant no. 938032MX to R.G. from the European Economic Community, Belgium.
LITERATURE CITED
- Adams RP, Kendall E, Kartha KK. Comparison of free sugars in growing and desiccated plants of Selaginella lepidophylla. Biochem Syst Ecol. 1990;18:107–110. [Google Scholar]
- Blázquez MA, Santos E, Flores C-L, Martínez-Zapater JM, Salinas J, Gancedo C. Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J. 1998;13:685–689. doi: 10.1046/j.1365-313x.1998.00063.x. [DOI] [PubMed] [Google Scholar]
- Burke MJ (1985) The glassy state and survival of anhydrous biological systems. In AC Leopold, ed, Membranes, Metabolism and Dry Organisms. Cornell University Press, Ithaca, NY, pp 358–363
- Cabib E, Leloir LF. The biosynthesis of trehalose phosphate. J Biol Chem. 1958;231:259–275. [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Clegg JS. The origin of trehalose and its significance during the formation of encysted embryos of Artemia salina. Comp Biochem Physiol. 1965;14:135–143. doi: 10.1016/0010-406x(65)90014-9. [DOI] [PubMed] [Google Scholar]
- Clegg JS (1985) The physical properties and metabolic status of Artemia cysts at low water contents: “the water replacement hypothesis.” In AC Leopold, ed, Membranes, Metabolism and Dry Organisms. Cornell University Press, Ithaca, NY, pp 169–187
- Colaço C, Sen S, Thangavelu M, Pinder S, Roser B. Extraordinary stability of enzymes dried in trehalose: simplified molecular biology. Biotechnology. 1992;10:1007–1011. doi: 10.1038/nbt0992-1007. [DOI] [PubMed] [Google Scholar]
- Crowe JH, Crowe LM, Carpenter JF, Aurell Wistrom C. Stabilization of dry phospholipid bilayers and proteins by sugars. Biochem J. 1987;242:1–10. doi: 10.1042/bj2420001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe JH, Crowe LM, Chapman D. Preservation of membranes in anhydrobiotic organisms: the role of trehalose. Science. 1984;223:701–703. doi: 10.1126/science.223.4637.701. [DOI] [PubMed] [Google Scholar]
- Crowe JH, Hoekstra FA, Crowe LM. Anhydrobiosis. Annu Rev Physiol. 1992;54:579–599. doi: 10.1146/annurev.ph.54.030192.003051. [DOI] [PubMed] [Google Scholar]
- De Virgilio C, Bürckert N, Bell W, Jenö P, Boller T, Weimken A. Disruption of TPS2, the gene encoding the 100-kDa subunit of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiae, causes accumulation of trehalose6-phosphate and loss of trehalose-6-phosphate phosphatase activity. Eur J Biochem. 1993;212:315–323. doi: 10.1111/j.1432-1033.1993.tb17664.x. [DOI] [PubMed] [Google Scholar]
- De Virgilio C, Hottiger T, Domínguez J, Boller T, Wiemken A. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. I. Genetic evidence that trehalose is a thermoprotectant. Eur J Biochem. 1994;219:179–186. doi: 10.1111/j.1432-1033.1994.tb19928.x. [DOI] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. BioTechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Elliot B, Haltiwanger RS, Futcher B. Synergy between trehalose and Hsp104 for thermotolerance in Saccharomyces cerevisiae. Genetics. 1996;144:923–933. doi: 10.1093/genetics/144.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaff DF. Desiccation-tolerant flowering plants in southern Africa. Science. 1971;174:1033–1034. doi: 10.1126/science.174.4013.1033. [DOI] [PubMed] [Google Scholar]
- Garcia AB, de Almeida Engler J, Iyer S, Gerats T, Van Montagu M, Caplan AB. Effects of osmoprotectants upon NaCl stress in rice. Plant Physiol. 1997;115:159–169. doi: 10.1104/pp.115.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever HM, Styrvold OB, Kaasen I, Strom AR. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J Bacteriol. 1988;170:2841–2849. doi: 10.1128/jb.170.6.2841-2849.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddijn OJM, Verwoerd TC, Voogd E, Krutwagen RWHH, de Graaf PTHM, Poels J, van Dun K, Ponstein AS, Damm B, Pen J. Inhibition of trehalase activity enhances trehalose accumulation in transgenic plants. Plant Physiol. 1997;113:181–190. doi: 10.1104/pp.113.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R, Klein W, Lange R, Rimmele M, Boos W. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J Bacteriol. 1991;173:7918–7924. doi: 10.1128/jb.173.24.7918-7924.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S, Bell W, Neves MJ, Valckx D, Thevelein JM. Evidence for trehalose-6-phosphate-dependent and -independent mechanisms in the control of sugar influx into yeast glycolysis. Mol Microbiol. 1996;20:981–991. doi: 10.1111/j.1365-2958.1996.tb02539.x. [DOI] [PubMed] [Google Scholar]
- Hohmann S, Neves MJ, de Koning W, Alijo R, Ramos J, Thevelein JM. The growth and signalling defects of the ggs1 (fdp1/byp1) deletion mutant on glucose are suppressed by a deletion of the gene encoding hexokinase PII. Curr Genet. 1993;23:281–289. doi: 10.1007/BF00310888. [DOI] [PubMed] [Google Scholar]
- Hounsa C-G, Brandt EV, Thevelein J, Hohmann S, Prior BA. Role of trehalose in survival of Saccharomyces cerevisiae under osmotic stress. Microbiology. 1998;144:671–680. doi: 10.1099/00221287-144-3-671. [DOI] [PubMed] [Google Scholar]
- Kaasen I, McDougall J, Strom AR. Analysis of the otsBA operon for osmoregulatory trehalose synthesis in Escherichia coli and homology of the OtsA and OtsB proteins to the test trehalose-6-phosphate synthase/phosphatase complex. Gene. 1994;145:9–15. doi: 10.1016/0378-1119(94)90316-6. [DOI] [PubMed] [Google Scholar]
- Londesborough J, Vuorio OE. Purification of trehalose synthase from baker's yeast: its temperature-dependent activation by fructose 6-phosphate and inhibition by phosphate. Eur J Biochem. 1993;216:841–848. doi: 10.1111/j.1432-1033.1993.tb18206.x. [DOI] [PubMed] [Google Scholar]
- Luyten K, de Koning W, Tesseur I, Ruiz MC, Ramos J, Cobbaert P, Thevelein JM, Hohmann S. Disruption of the Kluyveromyces GGS1 gene causes inability to grow on glucose and fructose and is suppressed by mutations that reduce sugar uptake. Eur J Biochem. 1993;217:701–713. doi: 10.1111/j.1432-1033.1993.tb18296.x. [DOI] [PubMed] [Google Scholar]
- Mackenzie KF, Singh KK, Brown AD. Water stress plating hypersensitivity of yeasts: protective role of trehalose in Saccharomyces cerevisiae. J Gen Microbiol. 1988;134:1661–1666. doi: 10.1099/00221287-134-6-1661. [DOI] [PubMed] [Google Scholar]
- Mascorro-Gallardo JO, Covarrubias AA, Gaxiola R. Construction of a CUP1 promoter-based vector to modulate gene expression in Saccharomyces cerevisiae. Gene. 1996;172:169–170. doi: 10.1016/0378-1119(96)00059-5. [DOI] [PubMed] [Google Scholar]
- Müller J, Boller T, Wiemken A. Trehalose and trehalase in plants: recent developments. Plant Sci. 1995;112:1–9. [Google Scholar]
- Neves MJ, Hohmann S, Bell W, Dumortier F, Luyten K, Ramos J, Cobbaert P, de Koning W, Kaneva Z, Thevelein JM. Control of glucose influx into glycolysis and pleiotropic effects studied in different isogenic sets of Saccharomyces cerevisiae mutants in trehalose biosynthesis. Curr Genet. 1995;27:110–122. doi: 10.1007/BF00313424. [DOI] [PubMed] [Google Scholar]
- Neves MJ, Terenzi HF, Leone FA, Jorge JA. Quantification of trehalose in biological samples with a conidial trehalase from the thermophilic fungus Humicola grisea var. thermoidea. World J Microbiol Biotechnol. 1994;10:17–19. doi: 10.1007/BF00357555. [DOI] [PubMed] [Google Scholar]
- Reinders A, Bürckert N, Hohmann S, Thevelein JM, Boller T, Wiemken A, De Virgilio C. Structural analysis of the subunits of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiae and their function during heat shock. Mol Microbiol. 1997;24:687–695. doi: 10.1046/j.1365-2958.1997.3861749.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Serrano R, Villalba JM. Expression and localization of plant membrane proteins in Saccharomyces. Methods Cell Biol. 1995;50:481–496. doi: 10.1016/s0091-679x(08)61052-3. [DOI] [PubMed] [Google Scholar]
- Thevelein JM. Regulation of trehalose mobilization in fungi. Microbiol Rev. 1984;48:42–59. doi: 10.1128/mr.48.1.42-59.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM, Hohmann S. Trehalose synthase: guard to the gate of glycolysis in yeast? Trends Biochem Sci. 1995;20:3–10. doi: 10.1016/s0968-0004(00)88938-0. [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein RJ. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aelst L, Hohmann S, Bulaya B, de Koning W, Sierkstra L, Neves MJ, Luyten K, Alijo R, Ramos J, Coccetti P and others. Molecular cloning of a gene involved in glucose sensing in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1993;8:927–943. doi: 10.1111/j.1365-2958.1993.tb01638.x. [DOI] [PubMed] [Google Scholar]
- Vandercammen A, François J, Hers H-G. Characterization of trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase of Saccharomyces cerevisiae. Eur J Biochem. 1989;182:613–620. doi: 10.1111/j.1432-1033.1989.tb14870.x. [DOI] [PubMed] [Google Scholar]
- Vogel G, Aeschbacher RA, Müller J, Boller T, Wiemken A. Trehalose-6-phosphate phosphatase from Arabidopsis thaliana: identification by functional complementation of the yeast tps2 mutant. Plant J. 1998;13:673–683. doi: 10.1046/j.1365-313x.1998.00064.x. [DOI] [PubMed] [Google Scholar]
- Weisburd S. Death-defying dehydration. Sci News. 1988;133:107–110. [Google Scholar]
- Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: evolution of osmolyte systems. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]