Abstract
Background
Cross reactivity between peanuts and tree nuts implies that similar IgE epitopes are present in their proteins.
Objective
To determine whether walnut sequences similar to known peanut IgE binding sequences, according to the property distance (PD) scale implemented in the Structural Database of Allergenic Proteins (SDAP), react with IgE from sera of patients with allergy to walnut and/or peanut.
Methods
Patient sera were characterized by Western blotting for IgE-binding to nut protein extracts, and to peptides from walnut and peanut allergens, similar to known peanut epitopes as defined by low PD values, synthesized on membranes. Competitive ELISA was used to show that peanut and predicted walnut epitope sequences compete with purified Ara h 2 for binding to IgE in serum from a cross-reactive patient.
Results
Sequences from the vicilin walnut allergen Jug r 2 which had low PD values to epitopes of the peanut allergen Ara h 2, a 2s-albumin, bound IgE in sera from five patients who reacted to either walnut, peanut or both. A walnut epitope recognized by 6 patients mapped to a surface-exposed region on a model of the N-terminal pro-region of Jug r 2. A predicted walnut epitope competed for IgE binding to Ara h 2 in serum as well as the known IgE epitope from Ara h 2.
Conclusions
Sequences with low PD value (<8.5) to known IgE epitopes could contribute to cross-reactivity between allergens. This further validates the PD scoring method for predicting cross-reactive epitopes in allergens.
Keywords: SDAP, food allergy, peanut allergy, walnut allergy, epitope
Introduction
As clinically significant peanut and tree nut allergies often occur in the same individual, it is important to determine which areas of similar proteins in these nuts are responsible for cross-reactions, rather than separate allergic sensitizations(1, 2). As hypersensitivity to nuts is life threatening, in vitro tests to determine the likelihood of a patient’s reaction to similar allergenic proteins in other food sources are desirable(3–5). Discrete linear IgE binding peptides have been defined for the major peanut allergens Ara h 1, 2 and 3(6–10), and limited data is available for a few tree nut allergens including the walnut (Jug r 1, Jug r 2, Jug r 4)(11), cashew (Ana o 1, Ana o 2)(12) and hazelnut (Cor a 9)(13).
Here we provide experimental evidence that the “PD” (property distance) tool in SDAP((14–16); http://fermi.utmb.edu/SDAP) can accelerate discovery of potentially cross-reactive epitopes in nut allergens, using the data on linear epitopes stored in SDAP and the Immune Epitope Database(17). Previously, we established that the PD scale identified sequences with similar physicochemical properties (PCP) and structure to known IgE epitopes from the major peanut allergens Ara h 1 and Ara h 2 (18), and that PD values correlated with IgE binding to sequences similar to known epitopes of the cedar pollen allergen Jun a 1(19). Starting from a given sequence, the PD tool identifies the most similar areas from all the allergenic proteins stored in SDAP, and outputs a table of the linear sequences with calculated PD scores and indicators for statistical significance. The lower the PD between two peptides, the more similar they are (0 for identical). The experiments presented here show that sequences from nut allergens with low PD values to known IgE epitopes of the major peanut allergen Ara h 2(6, 18, 20–24) are recognized by sera from patients with clinically relevant sensitivity to peanuts and walnuts. Further, a peptide representing a novel Jug r 2 epitope competed with purified Ara h 2 for binding to IgE in serum from a patient allergic to both peanuts and walnuts. Thus the PD tool can identify similar regions, even in allergens with low overall identity, that can contribute to IgE binding and cross-reactivity.
Materials and Methods
Patient sera
Sera from peanut and walnut allergic adults were collected after informed consent at the University of Arkansas for Medical Sciences (Little Rock, AR) and the University of California, Davis, Health Care System in accordance with the rules and regulations of the institutional review boards. While food challenge for research purposes was precluded in some severely allergic patients, all those selected had early childhood onset and recurrent severe systemic allergic reactions to peanut and/or walnut resulting in emergency department visits as children and adults. There is little possibility of such patients “outgrowing” the allergy, indicating the involvement of extremely relevant IgE epitopes. Specific IgE to walnut or peanut was measured by ImmunoCAP (Phadia, Uppsala, Sweden) (Table 1). The atopic control serum was from a patient with clinical grass pollinosis, with a specific IgE of >100 kU/L by ImmunoCap with no history of food allergy. ImmuoCaps against food allergens were therefore not performed.
TABLE 1.
CHARACTERISTICS OF THE PATIENT SERA.
| Subject Number | Age of onset (yrs) | Reaction to Peanut | Reaction to Walnut | ImmunoCAP IgE to Peanut (KU/L) | ImmunoCAP IgE to Walnut (KU/L) | Peanut Proteins detected on WB | Walnut Proteins detected on WB |
|---|---|---|---|---|---|---|---|
| 1. | 1 | Yes, anphx | No | 21.5 | 0.24 | Ara h 1, Ara h 2, Ara h 3, Ara h 6 | Jug r 2, Jug r 4 other |
| 2. | 2 | Yes, multiple ER visits | Yes, anphx | 1.8 | 2.21 | Ara h 1, Ara h 2, Ara h 3, | Jug r 1, Jug r 2, Jug r 4 |
| 3. | 1 | Yes, anphx | Yes, throat swelling, angioedema | >100 | 8.7 | Ara h 1, Ara h 2, Ara h 3 | Jug r 1, Jug r 2, Jug r 4 |
| 4. | <2 | Yes, anphx | No | 80 | 0.01 | Ara h 2 | none |
| 5. | < 4 | Yes, anphx | No | 23.1 | 0.12 | Ara h 1, Ara h 2, Ara h 6 | none |
| 6. | 2 | No | Yes, anphx | 0.94 | 10.2 | Ara h 1, Ara h 2, Ara h 3, Ara h 6 | Jug r 1 Jug r 2 |
yrs=years; KU/L=Kilounits/liter; anphx=anaphylaxis; WB=western blot
IgE Immunoblotting
Extracts from defatted peanut or walnut flours were subjected to sodium dodecyl polyacrylamide gel electrophoresis (SDS-PAGE) on 4–20% Novex Tris-HCl precast gels with Magic Mark (MM), Molecular Weight Marker (Invitrogen Corp., Carlsbad, California), followed by transfer to PVDF membranes and incubated overnight at 4 C with patient sera (1:10 dilution in PBST, phosphate buffered saline + 0.5% Tween 20), washed with PBST, incubated with anti-human IgE horseradish peroxidase (HRP)-labeled secondary antibody (Sigma Chemical Company, St. Louis, MO); diluted 1:10,000 in 2% nonfat dried milk dissolved in PBST) for 30’; washed with PBS; incubated with ECL-Plus substrate (Amersham Bioscience Corp., Piscataway, NJ); and visualized using a CCD camera (Fuji Photo Film Co., Ltd., Duluth, GA).
PD scores were calculated using the automatic tool in SDAP, as previously described(14, 15, 18, 19), starting from the tabulated epitopes of Ara h 1 and Ara h 2. Generally, peptides with a recognizable similarity in their PCPs have PD values below 10, while those for unrelated peptides average to 15 or more. In Table 2, sequences are listed in order of decreasing similarity and increasing PD to the starting Ara h 2 epitope.
Table 2. A search performed by PD tool of SDAP using two known Ara h 2 epitopes.
A weak (line 1) and a strong/immunodominant (line 4) Ara h 2 epitope were used to identify related sequences in 2 other peanut allergens (Ara h 6 and 7) and the walnut allergens Jug n 2 and Jug r 2.
| Spot number Fig. 3 | Allergen name | PD value# | Start * | Sequence | End* |
|---|---|---|---|---|---|
| 1 | Ara h 2 | 0.00 | 24 | QWELQGDR | 31 |
| 2 | Jug n 2 | 8.03 | 351 | SFEDQGRR | 358 |
| 3 | Jug r 2 | 8.21 | 463 | SYEGQGRR | 470 |
| 4 | Ara h 2 | 0.00 | 27 | DRRCQSQLER | 36 |
| 5 | Ara h 7 | 5.23 | 42 | DDQCQRQLQR | 51 |
| 6 | Jug r 2 | 5.87 | 140 | QRQCQQRCER | 149 |
| 7 | Ara h 7 | 7.08 | 96 | EQRCCNELNR | 105 |
| 8 | Jug r 2 | 8.00 | 76 | YEQCQQQCER | 85 |
| 9 | Jug r 2 | 8.12 | 49 | DQRSQEERER | 58 |
| 10 | Jug r 2 | 8.22 | 101 | QRRQQEERER | 110 |
| 11 | Ara h 6 | 8.48 | 94 | DRQMVQHFKR | 103 |
| 12 | Jug r 2 | 8.65 | 532 | QNNIINQLER | 541 |
Start and End refer to the start and end positions of the amino acids in the sequence of the indicated protein;
PD value=property distance score
Probing of SPOTs membranes with sera from allergic patients
A membrane with derivatized peptides (Sigma-Genosys, St. Louis, MO) was wet with methanol, washed with Tris-buffered saline (TBS, 8.0 g of NaCl, 0.2 g of KCl, and 6.1 g of Tris-base in 1 liter of water, pH 7.0), blocked overnight with membrane blocking solution (MBS, No. SU-07-250; Sigma-Genosys), in TBST (TBS + 0.5% Tween-20), pH 7.0 and incubated overnight with an optimized dilution (1:10–1:120) of a patient sera. The membrane was washed, incubated 30 min with goat anti-human IgE/HRP conjugate (Sigma-Genosys) diluted 1:10,000 in MBS for 30 minutes, washed, exposed to ECL-Plus substrate, and spot density determined with a FUJIFILM Luminescent Image Analyzer LAS-1000plus (Fuji Photo Film Co., Ltd., Duluth, GA). The membrane was stripped (1% SDS in TBS, 0.5% 2-mercaptoethanol, pH 7.0), re-blocked, checked for complete stripping by incubating with the secondary antibody, then restripped before probing with a new patient sera. Negative control SPOTS were peptides designed to have a high PD value (low similarity) to 3 different Jun a 1 IgE-epitopes(19).
Competitive Inhibition ELISA
Purified native Ara h 2 (25) (50 ul/well,1 ng/ul in 0.1 M NaHCO3 (pH 9.6)) was added to a 96 well plate, allowed to bind for 75 minutes at 37°C, washed three times with PBST, blocked with 2% dry milk in PBST for one hour at 37°C and washed. Diluted peptides (EZBiolab, Carmel, IN) (80, 40, 15, 3, 0.6, 0.12, 0.024, 0.0048, and 0 ng/μl) were mixed with equal volumes of serum (diluted 1:10) and the mixtures rotated at 4 °C for 2.5 hours before adding to the Ara h 2-bound plates for 1 hour at 37°C. Plates were washed, incubated with a 1:1000 dilution of HRP-conjugated goat anti-human IgE (Sigma Chemical Company, St. Louis, MO), for one hour at 37°C, washed, and incubated with the HRP substrate 3, 3', 5, 5'-tetramethylbenzidine (KPL, Gaithersburg, MD)and fluorescence determined in a Tecan Sunrise microtitre plate reader (Tecan US, San Jose, CA).
3D-models and surface exposure analysis
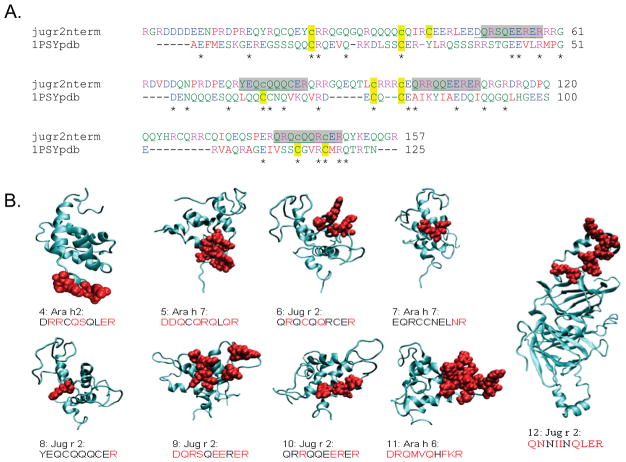
Epitopes were mapped on the crystal structure of a portion of Ara h 2 (as a fusion protein with maltose binding protein(26), PDB structure 3OB4 ) or on models produced with MPACK followed by FANTOM minimization(27–29), available in the Structural Database of Allergenic Proteins (SDAP). The Jug r 2 N-terminal pro-region was modeled on the crystal structure of the ricin Ric c 3 protein (PDB file 1PSY) using the alignment shown in Figure 3a. Four disulfides were specified, based homology to the template, between residues 20–90, 33–74,78–138, and 94–142.The final FANTOM energy of the model was −220kcal/mole. Surface exposure of residues in the selected sequences on the models was determined with GETAREA(30), using default water radius (1.4Å) and a cutoff for exposure as 30% of total area of the side chain.
Figure 3. 3D-characterization of peptides (Table 2, 4–12) tested in this study.
a) Alignment of the Jug r 2 N-terminal region with the modeling template, 1PSY-pdb (Ric c 3). The residues are colored to show amino acid properties. Cys residues involved in disulfide bonding are highlighted in yellow, IgE binding sequences identified in this study (6, 8–10 in Table 2) are highlighted in gray. b) Mapping of peptides on a crystal structure of Ara h 2 and models, from SDAP or specifically prepared for this study. Structures are shown in ribbon format, with the potential epitopes shown space filling in red. Side chains with >30% surface exposure according to GETAREA are highlighted in red in the text below each.
Results
IgE binding to peanut and walnut proteins
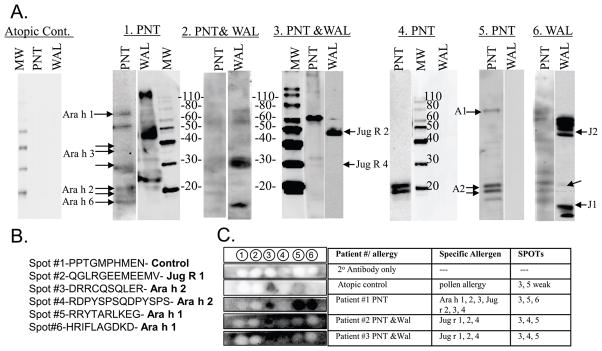
Six clinically well-characterized patients (P1–6) were chosen based on their reactivity to peanuts and/or walnuts and their IgE binding profiles to peanut and/or walnut proteins (Table 1 and Figure 1A). IgE reactivity was not strictly reflective of clinical sensitivity. The IgE in the sera of P1, allergic to peanut, and P6, allergic to walnut, bound to both peanut and walnut proteins, suggesting that the two sources might contain similar epitopes.
Figure 1. Immunoblots of IgE binding by peanut and or walnut allergic individuals.
A) Western blots: Patients are depicted by numbers (1–6) above each blot and the clinical allergy to peanut (PNT) or walnut (WAL). The Molecular Weight Marker (MW) is shown. Ara h 1, 2, Jug r 1, 2 and 4 are indicated as A1, A2 and J1–J4, respectively. Negative control serum is Atopic Control. B) Sequence of membrane-bound, synthetic peptides in C. C) From left to right, column 1 shows IgE binding to spots of a “no serum control” (row 1) and 4 sera to the six membrane-bound synthetic peptides (row 2–5), specific allergens recognized in western blot (column 2), and peptide spots recognized by each serum (column 3).
Selected sera all bound a major Ara h 2 epitope (spot #3, Figure 1C) when tested for binding to previously characterized linear IgE epitopes of Ara h 1, Ara h 2 and Jug r 1 on SPOT strips (Figure 1B and 1C); none recognized a negative control sequence (spot 1) or the Jug r 1 epitope. This clinical relevance of the Ara h 2 epitope, spot 3, (peptide 4 in Table 2) is undetermined, but it seems to be quite cross reactive.
PD searches from previously defined IgE epitopes of peanut allergens revealed similar areas in walnut allergens
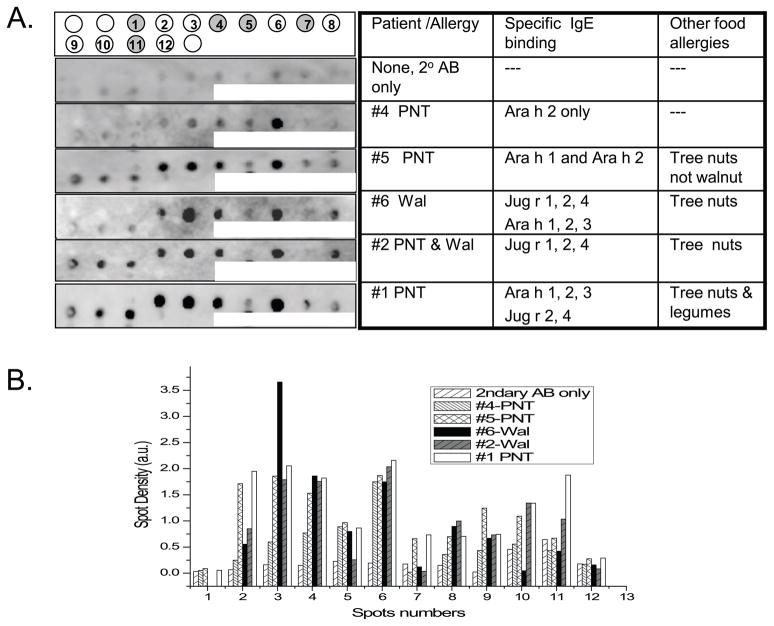
Sequences with low PD value (high similarity) to minor and major (peptides 1 and 4 respectively of Table 2) epitopes of Ara h 2 were identified in other peanut 2S albumins (Ara h 6 and Ara h 7) and in the walnut vicilin, Jug r 2. Peptides were synthesized on a SPOTs membrane (Figure 2A) and incubated with P1–6 sera. The predicted peptides 2 and 3 in Table 2, from walnut vicilins, exhibit higher IgE binding by densitometry for all patients (Figure 2B) than the original Ara h 2 epitope (QWELQGDR; spot 1) even for the peanut-not-walnut-allergic patient sera. Spot 6, a Jug r 2 peptide similar to the major peanut IgE epitope (spot 4) was recognized by all patient sera. P1 and P2 sera recognized additional sequences from Jug r 2 (spots 8, 9, 10). Spots 5 and 7, from peanut proteins, were primarily recognized by patients with peanut allergy. Sequence 12 in Table 2, with the highest PD value to the starting epitope, did not react with any of the sera.
Figure 2. Immunoblot of IgE binding to membrane-bound known and predicted synthetic peptide epitopes of peanut and walnut allergens.
(A) From left to right, the first column shows the IgE binding profile of a control and 5 patient sera to the 12 membrane-bound synthetic peptides. Grey spots at the top indicate peanut peptides. The specific allergens recognized in the blots are indicated in the third column. (B), Densitometric scan of spots in panel A (indicated on x-axis) and reported in arbitrary units (a.u., y-axis).
Structural Characterization of the novel IgE epitopes
To further characterize peptides 5–12 from Table 2, the sequences were displayed on models of Ara h 6 and 7 and the mature and pro-regions of Jug r 2 (Figure 3; models were taken from SDAP except for the pro-region of Jug r 2, which was prepared for this study). The starting epitope (peptide 4) has a high degree of surface exposure on the recently published crystal structure of Ara h 2 (26). Four of the sequences (peptides 6 and 8–10) recognized by patient IgE were in the Jug r 2 N-terminal pro-region, which contains several cysteine and glutamine rich repeats, as does the template used for modeling, Ric c 3 (Figure 3a). This model suggests an α-helical structure, more similar to Ara h 2 than to the mature region of Jug r 2 (a 2 cupin domain vicilin that should have a primarily β-sheet structure). The sequences all have a fair degree of predicted surface exposure, except 7 and 8, which also had relatively lower IgE binding than the other peptides (Fig. 2b,c). The models illustrate that many of the protein areas for the sequences are predicted to be on the surface exposed; absolute solvent exposure may be quite different in the nut or nut extract due to alternate sidechain orientations, glucosylation and/or protein oligomerization.
Selected peptides can compete for binding of IgE with whole Ara h 2
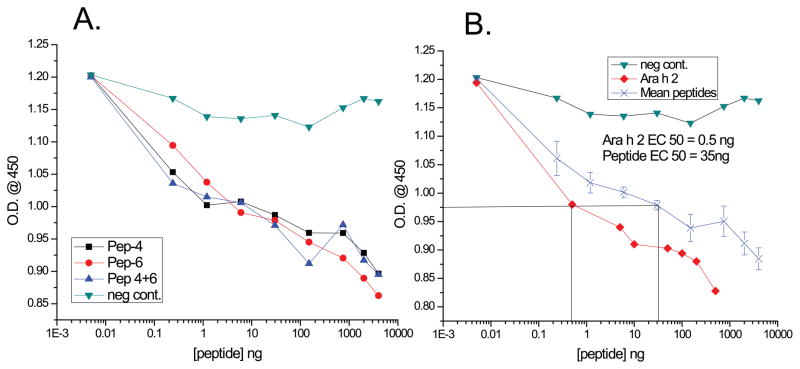
To further explore the potential role of one of the newly discovered walnut epitopes in cross reactivity, peptide 4, the known Ara h 2 epitope, and the new walnut epitope, peptide 6 (Table 2), were synthesized and tested for their ability to inhibit binding of IgE from a peanut allergic patient (P1) to purified Ara h 2. Both the known Ara h 2 IgE epitope and the newly discovered walnut one competed in a similar fashion with Ara h 2 for binding to IgE in patient sera, as measured by ELISA (Figure 4). Combining the peptides did not add to their effect or decrease the concentration of peptide needed to compete with Ara h 2, suggesting that these peptides compete for IgE binding with the same epitope of Ara h 2. A negative control peptide (with a high PD to cedar pollen IgE epitopes (spot 1, Figure 1B) did not significantly inhibit IgE binding to Ara h 2. The mean values for 50% inhibition of IgE binding to intact Ara h 2 was 0.5 ng/ml and for the peptides 32–35 ng/ml, which was ~65–70 fold lower than the ability of free Ara h 2 to compete (4B). This concentration difference would be expected, as the peptides target only one of the many identified epitopes on Ara h 2.
Figure 4. Competitive inhibition ELISA of IgE binding to purified Ara h 2 with synthetic peptides.
A and B: indicated concentrations (x-axis) of peptides: a known Ara h 2 epitope (DRRCQSQLER, ●), a walnut peptide with low PD to the Ara h 2 one (QRQCQQRECER, ■), a combination of the two peptides(▲), intact Ara h 2 (◆) and a negative control (▼) were used to compete with IgE binding to intact Ara h 2. (B) Mean of 3 peptide assays from panel A is also shown (X). The optical density at 450 nm is indicated on the y-axis.
Discussion
The main result of this paper is that similar IgE epitopes on allergenic proteins, defined by the PD scale of SDAP, could account for some of the cross-reactivity between peanuts and tree nuts. This indicates that the PD tool can be useful for predicting cross-reactive, previously unidentified epitopes based on their similarity to known IgE epitopes. Sequences from peanut and walnut allergens that had a low PD value to previously identified peanut epitopes (Table 2) bound IgE from patient sera (Figure 2), and competed for binding of IgE to Ara h 2 in ELISA (Figure 4). Clinically irrelevant IgE binding to peanut epitopes was demonstrated using sera from a walnut allergic-only patient, and conversely, binding of IgE from peanut-only allergic patients to the similar epitopes in walnut. Our study did not look at the total number of IgE epitopes a particular serum could react with, nor the affinity/avidity of the interaction, all of which may be important in actually leading to clinical reactivity. Also, some of the proteins, from which IgE binding peptides were derived, were not recognized by serum IgE in western blots, it is important to note that concentrations of individual peptides synthesized on membranes are much higher than concentration of proteins in western blots, which might allow better detection of IgE binding.
We note that Ara h 2, and Jug r 2 have low overall sequence identity (approximately 13%), although our modeling (Figure 3) suggests that these proteins may have similar 3D-structural elements. That peptides stemming from them should be able to compete for binding to an intact allergen suggests that the 50% overall identity threshold for predicting cross-reactive allergens (31) may be set too high. Other experimental results, such as the ability of between Can f 4 and a related bovine lipocalin-like protein that are only 36% identical to compete for IgE binding(32), and repeated findings that individual amino acid changes can reduce IgE binding, also indicates that allergenicity may lie in discrete regions. Further, proteins may be closely related to one another at the level of physicochemical properties (PCPs), while having a relatively low % identity to one another. The PD tool in SDAP was thus designed to recognize areas of allergens that are similar in their PCPs to known IgE epitopes, thus predicting sequences that are truly cross-reactive in vitro. While our analysis was based only on linear sequence comparisons, we anticipate that 3D- analysis will become important as more experimental structures of allergenic proteins are determined. Our results indicate that IgE reactivity to Jug r 2 and Ara h 2 may explain some cases of peanut/walnut cross-reactivity, which we are investigating further.
The major difficulty one faces in predicting antibody epitopes is achieving statistical significance when comparing short segments of sequence. The PD scale implemented in SDAP depends both on the ability to compare the PCPs of amino acids, as well as the ability to limit the search for similar sequences to known allergenic proteins(33). These results and our current reported findings suggest that we should not limit the search for similar epitopes to proteins within the same Pfam (i.e., proteins with similar predicted overall structure and high sequence identity)(33), and that we should take into account proteins that are genetically encoded, even if they are not present in the mature region. IgE epitopes have been reported for the pro-region of Ara h 1.(22), and proregion polypeptides have been found in a seed protein body(34), indicating they may not be completely degraded after cleavage. The repetitive sequence of the pro-region in Jug r 2 contains several regions that we have shown here may bind the same pool of IgE antibodies in the sera of patients allergic to peanut, specifically the peanut 2S albumins/conglutinins.
These results extend previous observations that many similar epitope sequences may contribute to the extreme reactions caused by nuts(18). The novel epitopes identified in this study will be screened with additional sera from allergic patients to further clarify their relevance to clinically important cross-reactions. This finding also has implications for screening for allergenicity of genetically modified plant products.
Acknowledgments
Declaration of all sources of funding: This research was supported by funds from the Environmental Protection Agency (EPA RE-83406601-0 to CHS), the NIH (R01 A1064913) and the U.S. Department of Agriculture, Agricultural Research Service.
PD scale and web server: Ovidiu Ivanciuc, Werner Braun
Advice on Immunoblotting: Terumi Midoro Horiuti
Abbreviations
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- ELISA
enzyme-linked immunosorbent assay
- IgE
immunoglobulin E
- PD
property distance
- PCP
physicochemical properties
- sIgE
specific immunoglobulin E
- SDAP
Structural Database of Allergenic Proteins
Footnotes
Conflict of interest statement:
None of the authors have any conflict of interests to disclose regarding this manuscript.
Author Contributions: Drs. Maleki, Schein and Teuber conceived of the idea, wrote the manuscript and funded the project, Ms. Cheng performed Western blot and SPOTS membrane IgE binding studies and assisted in data interpretation; Dr. Ruan, performed and interpreted competitive inhibition ELISA assays; Structural modeling: Dr. Chen; IgE binding properties of patient sera: Dr. Comstock.
References
- 1.de Leon MP, Drew AC, Glaspole IN, Suphioglu C, O'Hehir RE, Rolland JM. IgE cross-reactivity between the major peanut allergen Ara h 2 and tree nut allergens. Mol Immunol. 2007;44(4):463–71. doi: 10.1016/j.molimm.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 2.de Leon MP, Glaspole IN, Drew AC, Rolland JM, O'Hehir RE, Suphioglu C. Immunological analysis of allergenic cross-reactivity between peanut and tree nuts. Clin Exp Allergy. 2003;33(9):1273–80. doi: 10.1046/j.1365-2222.2003.01761.x. [DOI] [PubMed] [Google Scholar]
- 3.Schein CH, Ivanciuc O, Braun W. Bioinformatics approaches to classifying allergens and predicting cross-reactivity. Immunology and Allergy Clinics of North America. 2007;27(1):1. doi: 10.1016/j.iac.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teuber S, Beyer K, Comstock S, Wallowitz M. The big eight foods: clinical and epidemiological overview. In: Malecki S, editor. Food Allergy. Washington DC: ASM Press; 2006. pp. 49–79. [Google Scholar]
- 5.Teuber SS, Beyer K. Peanut, tree nut and seed allergies. Current Opinion in Allergy and Clinical Immunology. 2004;4(3):201–203. doi: 10.1097/00130832-200406000-00011. [DOI] [PubMed] [Google Scholar]
- 6.Burks AW, Shin D, Cockrell G, Stanley JS, Helm RM, Bannon GA. Mapping and mutational analysis of the IgE-binding epitopes on Ara h 1, a legume vicilin protein and a major allergen in peanut hypersensitivity. Eur J Biochem. 1997;245(2):334–339. doi: 10.1111/j.1432-1033.1997.t01-1-00334.x. [DOI] [PubMed] [Google Scholar]
- 7.Rabjohn P, Burks AW, Sampson HA, Bannon GA. Mutational analysis of the IgE-binding epitopes of the peanut allergen, Ara h 3: a member of the glycinin family of seed-storage proteins. Journal of Allergy and Clinical Immunology. 1999;103(1):S101–S101. [Google Scholar]
- 8.Shin DS, Compadre CM, Maleki SJ, Kopper RA, Sampson H, Huang SK, et al. Biochemical and structural analysis of the IgE binding sites on ara h1, an abundant and highly allergenic peanut protein. J Biol Chem. 1998;273(22):13753–9. doi: 10.1074/jbc.273.22.13753. [DOI] [PubMed] [Google Scholar]
- 9.Stanley JS, King N, Burks AW, Huang SK, Sampson H, Cockrell G, et al. Identification and mutational analysis of the immunodominant IgE binding epitopes of the major peanut allergen Ara h 2. Arch Biochem Biophys. 1997;342(2):244–53. doi: 10.1006/abbi.1997.9998. [DOI] [PubMed] [Google Scholar]
- 10.Barre A, Borges JP, Culerrier R, Rouge P. Homology modelling of the major peanut allergen Ara h 2 and surface mapping of IgE-binding epitopes. Immunol Lett. 2005;100(2):153–8. doi: 10.1016/j.imlet.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 11.Robotham JM, Teuber SS, Sathe SK, Roux KH. Linear IgE epitope mapping of the English walnut (Juglans regia) major food allergen, Jug r 1. J Allergy Clin Immunol. 2002;109(1):143–9. doi: 10.1067/mai.2002.120558. [DOI] [PubMed] [Google Scholar]
- 12.Xia L, Willison LN, Porter L, Robotham JM, Teuber SS, Sathe SK, et al. Mapping of a conformational epitope on the cashew allergen Ana o 2: a discontinuous large subunit epitope dependent upon homologous or heterologous small subunit association. Mol Immunol. 2010;47(9):1808–16. doi: 10.1016/j.molimm.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 13.Robotham JM, Hoffman GG, Teuber SS, Beyer K, Sampson HA, Sathe SK, et al. Linear IgE-epitope mapping and comparative structural homology modeling of hazelnut and English walnut 11S globulins. Mol Immunol. 2009;46(15):2975–84. doi: 10.1016/j.molimm.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 14.Ivanciuc O, Schein CH, Braun W. Data mining of sequences and 3D structures of allergenic proteins. Bioinformatics. 2002;18(10):1358–1364. doi: 10.1093/bioinformatics/18.10.1358. [DOI] [PubMed] [Google Scholar]
- 15.Ivanciuc O, Schein CH, Braun W. SDAP: Database and computational tools for allergenic proteins. Nucleic Acids Res. 2003;31(1):359–362. doi: 10.1093/nar/gkg010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schein CH, Ivanciuc O, Braun W. Structural Database of Allergenic Proteins (SDAP) In: Maleki SJ, Burks AW, Helm RM, editors. Food Allergy. Washington, D.C: ASM Press; 2006. pp. 257–283. [Google Scholar]
- 17.Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, et al. The immune epitope database 2.0. Nucleic Acids Res. 2010;38(Database issue):D854–62. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schein CH, Ivanciuc O, Braun W. Common physical-chemical properties correlate with similar structure of the IgE epitopes of peanut allergens. Journal of Agricultural and Food Chemistry. 2005;53(22):8752–8759. doi: 10.1021/jf051148a. [DOI] [PubMed] [Google Scholar]
- 19.Ivanciuc O, Midoro-Horiuti T, Schein CH, Xie L, Hillman GR, Goldblum RM, et al. The property distance index PD predicts peptides that cross-react with IgE antibodies. Mol Immunol. 2009;46(5):873–83. doi: 10.1016/j.molimm.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabjohn P, Helm EM, Stanley JS, West CM, Sampson HA, Burks AW, et al. Molecular cloning and epitope analysis of the peanut allergen Ara h 3. J Clin Invest. 1999;103(4):535–542. doi: 10.1172/JCI5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shreffler WG, Beyer K, Chu TH, Burks AW, Sampson HA. Microarray immunoassay: association of clinical history, in vitro IgE function, and heterogeneity of allergenic peanut epitopes. J Allergy Clin Immunol. 2004;113(4):776–782. doi: 10.1016/j.jaci.2003.12.588. [DOI] [PubMed] [Google Scholar]
- 22.Wichers HJ, De Beijer T, Savelkoul HF, Van Amerongen A. The major peanut allergen Ara h 1 and its cleaved-off N-terminal peptide; possible implications for peanut allergen detection. J Agr Food Chem. 2004;52(15):4903–4907. doi: 10.1021/jf049697o. [DOI] [PubMed] [Google Scholar]
- 23.Beardslee TA, Zeece MG, Sarath G, Markwell JP. Soybean glycinin G1 acidic chain shares IgE epitopes with peanut allergen Ara h 3. Int Arch Allergy Immunol. 2000;123:299–307. doi: 10.1159/000053642. [DOI] [PubMed] [Google Scholar]
- 24.Rabjohn P, West C, Connaughton C, Sampson H, Helm R, Burks A, et al. Modification of peanut allergen Ara h 3: effects on IgE binding and T cell stimulation. Int Arch Allergy Immunol. 2002;128:15–23. doi: 10.1159/000057999. [DOI] [PubMed] [Google Scholar]
- 25.Maleki SJ, Viquez O, Jacks T, Dodo H, Champagne ET, Chung SY, et al. The major peanut allergen, Ara h 2, functions as a trypsin inhibitor, and roasting enhances this function. J Allergy Clin Immunol. 2003;112(1):190–5. doi: 10.1067/mai.2003.1551. [DOI] [PubMed] [Google Scholar]
- 26.Mueller GA, Gosavi RA, Pomés A, Wünschmann S, Moon AF, London RE, et al. Ara h 2: crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity. Allergy. 2011 doi: 10.1111/j.1398-9995.2010.02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanciuc O, Oezguen N, Mathura V, Schein CH, Xu Y, Braun W. Using property based sequence motifs and 3D modeling to determine structure and functional regions in CASP5 targets. Curr Med Chem. 2004;11(5):583–593. doi: 10.2174/0929867043455819. [DOI] [PubMed] [Google Scholar]
- 28.Soman KV, Midoro-Horiuti T, Ferreon JC, Goldblum RM, Brooks EG, Kurosky A, et al. Homology modeling and characterization of IgE epitopes of mountain cedar allergen Jun a 3. Biophys J. 2000;79(3):1601–1609. doi: 10.1016/S0006-3495(00)76410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oezguen N, Zhou B, Negi SS, Ivanciuc O, Schein CH, Labesse G, et al. Comprehensive 3D-modeling of allergenic proteins and amino acid composition of potential conformational IgE epitopes. Mol Immunol. 2008;45(14):3740–7. doi: 10.1016/j.molimm.2008.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comp Chem. 1998;19(3):319–333. [Google Scholar]
- 31.Aalberse RC, Stadler BM. In silico predictability of allergenicity: From amino acid sequence via 3-D structure to allergenicity. Molecular Nutrition & Food Research. 2006;50(7):625–627. doi: 10.1002/mnfr.200500270. [DOI] [PubMed] [Google Scholar]
- 32.Mattsson L, Lundgren T, Olsson P, Sundberg M, Lidholm J. Molecular and immunological characterization of Can f 4: a dog dander allergen cross-reactive with a 23 kDa odorant-binding protein in cow dander. Clin Exp Allergy. 2010 doi: 10.1111/j.1365-2222.2010.03533.x. [DOI] [PubMed] [Google Scholar]
- 33.Ivanciuc O, Garcia T, Torres M, Schein CH, Braun W. Characteristic motifs for families of allergenic proteins. Molecular Immunology. 2009;46(4):559–568. doi: 10.1016/j.molimm.2008.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcus J, Goulter K, Manners J. Peptide Fragments From Plant Vicilins Expressed in Escherichia Coli Exhibit Antimicrobial Activity In Vitro. Plant Mol Biol Rep. 2008;26:75–87. [Google Scholar]