Abstract
Purpose
Despite growing concerns toward maintaining participants’ privacy, individual investigators collecting tissue and other biological specimens for genomic analysis are encouraged to obtain informed consent for broad data sharing. To assess the effect on research enrollment and data sharing decisions of three different consent types (traditional, binary, or tiered) with varying levels of control and choices regarding data sharing.
Methods
A single blind, randomized controlled trial was conducted with 323 eligible adult participants being recruited into one of six genome studies at Baylor College of Medicine in Houston, Texas between January 2008 and August 2009. Participants were randomly assigned to one of three experimental consent documents (traditional, n=110; binary, n=103; tiered, n=110). Debriefing in follow-up visits provided participants a detailed review of all consent types and the chance to change data sharing choices or decline genome study participation.
Results
Before debriefing, 83.9% of participants chose public data release. After debriefing, 53.1% chose public data release, 33.1% chose restricted (controlled access database) release, and 13.7% opted out of data sharing. Only one participant declined genome study participation due to data sharing concerns.
Conclusion
Our findings indicate that most participants are willing to publicly release their genomic data, however, a significant portion prefer restricted release. These results suggest discordance between existing data sharing policies and participants’ judgments and desires.
Keywords: data sharing, genome research, ethical issues, participant perspectives, consent
Introduction
An increasing number of investigators are prospectively collecting and storing biological specimens for genomic analysis. Investigators engaged in this research activity are strongly encouraged to comply with genomic data sharing policies, which have historically called for the rapid public release of all generated DNA data.1–3 Making data publicly accessible is cost efficient and maximizes the scientific utility of genomic information. De-identification, or the removal of all personally identifying information prior to public release, has been the traditional means of protecting the privacy of individuals participating in genomic research. However, it has been shown that individuals can be uniquely identified on the basis of just 30–80 statistically independent single nucleotide polymorphisms (SNPs)4 and it is now even possible to identify an individual from pooled or aggregated DNA data. 5 These findings raise concern about the privacy of research participants and have led to the creation of controlled access, or restricted, scientific databases.6, 7
Some have criticized this shift in data access policy as being overly protective8 and some projects will only enroll participants who agree to full public data release.9 We have argued that all data sharing decisions involve an unavoidable trade-off between protecting privacy and advancing research and since individuals may vary in their judgments about this trade-off, decisions about DNA data release ought to be made by research participants during the informed consent process.10 However, a major policy concern is that giving participants control over decisions about data sharing will lead to excessive anxiety about protecting privacy and a reluctance to share data, negatively impacting research. We conducted a single blind, randomized controlled trial of three different types of consent, each affording varying levels of control over the decision about data sharing, in order to assess their impact on research enrollment into an underlying genomic study and participants’ data sharing preferences.
Materials and Methods
Study Participants and Procedures
Participants were adult (18 years or older) patients (n=205), parents/guardians of pediatric patients (n=103), and family members acting as matched case controls (n=28) who were recruited to one of six ongoing genomic studies (pediatric brain cancer, pediatric brain controls, pediatric autism, adult/pediatric epilepsy, adult/pediatric liver cancer, and adult pancreatic cancer) at Baylor College of Medicine (BCM) in Houston, Texas between January 2008 and August 2009.
Participants eligible for the randomized consent study were English proficient and were enrolled with a waiver of consent obtained from the BCM Institutional Review Board (IRB). Participants considering enrollment in one of the genomic studies were randomized to one of three experimental consent types via a centralized, web-based randomization program using permuted blocks and stratified by genomic study. Genomic study PI’s who could not use the online randomization system were provided with sealed, pre-randomized envelopes each containing the assignment. Informed consent into the genomic study was obtained in a face-to-face setting by the genomic study PI, a research nurse, or a medical resident with one of the three experimental consent documents. The consent process varied slightly depending on the design of the underlying genomic study; however, the overall process did not differ by randomized consent type.
After providing informed consent for the genomic study, participants were debriefed by a designated research coordinator from this consent study. Those who were ineligible or declined participation in the genomic study but had seen or signed one of our experimental consent documents were debriefed by the genomic study PI or research nurse; most refusals were due to general research concerns (e.g., fear of blood draw) or lack of time. One individual reportedly refused participation in the genomic study specifically because of concerns about data sharing and was debriefed by a consent study coordinator. Debriefing took place either in a private hospital room during an inpatient stay or in a waiting or exam room during a follow-up clinic visit. Twenty-seven participants did not return for a follow-up visit and were debriefed by phone or U.S. mail. During the debriefing, participants were given information about the consent study and the randomization process, a detailed review of the data sharing options in each experimental consent document, and an opportunity to change their data sharing choice.
Eligible participants were invited to participate in a structured follow-up interview to assess understanding, comfort in decision making, and to examine preferences and attitudes regarding data sharing. To prevent bias, those who agreed to the interview were not shown the other consent forms or data sharing options until partway through the interview. Analysis of interview responses will be reported elsewhere. All materials and methods for this study were reviewed and approved by the BCM IRB.
Study Instruments
Three experimental consent templates were developed by a review of the informed consent literature and were refined with input from an interdisciplinary panel of experts at BCM and focus group research conducted by two of the authors (AM, AG).11 The experimental consent templates were adapted for each genomic study. All of the consent documents contained specific information about the respective genomic study, including purpose, risks, benefits, compensation, and access to health records. Data sharing was explained in each consent document (see Supplemental Digital Content 1, which contains excerpted text from each consent type on data sharing). Participants were told that personally identifying information (e.g., their name) would never be released. Risks of data sharing were described as small potential breaches in privacy if DNA were traced back to the individual. Participants were cautioned that these risks could increase in the future. It was noted that a researcher’s obligation to protect privacy and confidentiality in restricted databases offers participants an extra layer of protection. Benefits of data sharing were characterized as aiding in the advancement of medical research by speeding up research and allowing other investigators to utilize the data to answer future research questions.
Each experimental consent document offered some combination of the following three data release options: (1) public data release (release of genetic and clinical information into both publicly accessible (opzen access via the internet) and restricted (accessible only to approved researchers) scientific databases), (2) restricted release (release of genetic and clinical information into restricted databases only), and (3) no release (accessible only to the genomic study PI and his or her staff) (Table 1).
Table 1.
Consent Form Data Release Options
| Type of Consent |
Option to participate in study but refuse data sharing |
Data Sharing Options |
|---|---|---|
| Traditional | No |
|
| Binary | Yes |
|
| Tiered | Yes |
|
Those who signed the traditional consent agreed by default to release their genetic and clinical information into both publicly accessible and restricted scientific databases. The binary consent allowed participants to choose between full public data release and no release. Tiered consent presented all three options; participants could choose public data release, restricted release only, or no release.
The primary outcomes were: (a) the rate of refusal and withdrawal within each consent type, and (b) the difference in data sharing choices between the three randomized groups.
Data Analysis
Participant characteristics were described with the use of frequencies for categorical variables and means or medians for continuous variables. Differences between groups were tested with chi-square tests for categorical variables and one-way analysis of variance (ANOVA) for continuous variables. Differences in post-debriefing data release selections were examined with multinomial logistic regression which allows for polytomous instead of dichotomous outcomes, adjusting for potential confounders. Results were presented as odds ratios (OR) with 95% confidence interval (CI) comparing restricted or no data release to public data release. Participants’ age and the time lapse between consent and debrief were treated as continuous variables. All other factors included in the multivariate analysis were categorical variables; this included socio-demographics: gender, race and ethnicity, marital status, religious affiliation, education and income, and participant characteristics, including randomized consent type and consentee relationship (either adults providing consent for themselves or providing parental consent for their child). For all tests, a significance level of p < 0.05 (two-tailed) was used. All analyses were conducted using SPSS 17 or SAS 9.2 (SPSS, Inc., Chicago, IL; SAS Institute Inc., Cary, NC).
Results
Three hundred seventy-eight individuals were approached for recruitment into one of the six genomic studies; 42 were deemed ineligible or chose not to enroll and were removed from the randomization. A total of 349 experimental consent documents were randomized to 336 individual participants (Figure 1). Most of the participants were either consenting adult patients or parents or guardians of pediatric patients. Two of the genomic studies (autism and epilepsy) also enrolled patients’ family members to serve as matched case controls. Parents of pediatric patients who enrolled as matched case controls made two consent choices, one for their child who was the primary subject (i.e., parental consent) and one for themselves as a matched case control (i.e., adult/self consent); these cases were treated as a single participant making two distinct decisions (n=13). All participating members of the same family were randomized to the same experimental consent document (n=18 families comprised of 34 individuals making 47 distinct decisions).
Figure 1.
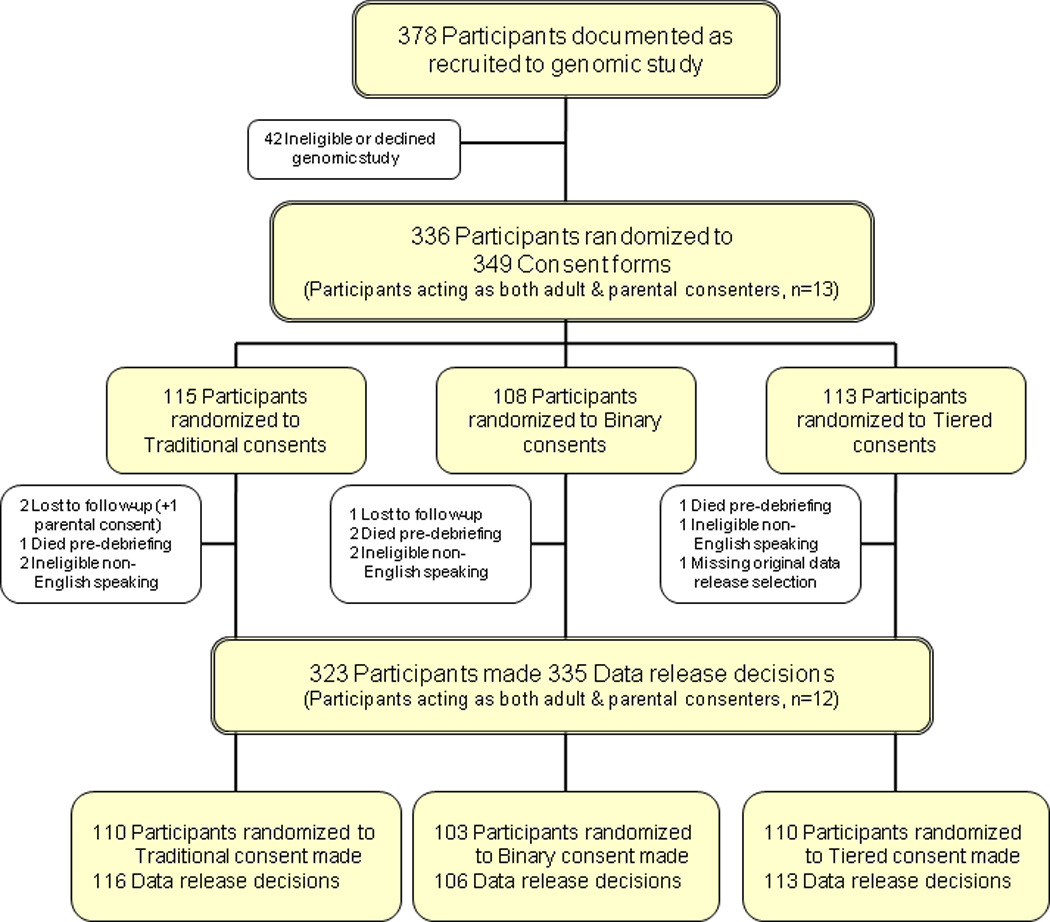
Study Consort Diagram
Thirteen participants were deemed ineligible: five turned out to have limited English proficiency; four died during the course of the study and could not be debriefed, three were lost to follow up (one participant consented on behalf of a child and as a matched case control for a total of four distinct data release decisions lost to follow up), and one did not provide a data release option. The remaining 323 individual participants were enrolled into the consent study, and 335 distinct data sharing decisions were analyzed.
The median age of participants was 48.5 years old (range 18–86). Most participants were female (57.3%) and non-Hispanic white (56.1%). The majority reported being married (63.7%), Christian (81.3%), and roughly two thirds indicated completing at least one year of college (67.8%) (Table 2).
Table 2.
Participant Characteristics by Randomized Consent Type
| Randomized Consent Typeab | ||||
|---|---|---|---|---|
| Characteristic | All Participants (n=323) |
Traditional (n=110) |
Binary (n=103) |
Tiered (n=110) |
| Genomic Study | ||||
| Autism | 8 (2.5) | 4 (3.6) | 2 (1.9) | 2 (1.8) |
| Brain Cancer | 76(23.5) | 26(23.6) | 24(23.3) | 26(23.6) |
| Brain Control | 16 (5.0) | 5 (4.6) | 6 (5.8) | 5 (4.6) |
| Epilepsy | 29 (9.0) | 10 (9.1) | 10 (9.7) | 9 (8.2) |
| Liver | 93(28.8) | 30(27.3) | 29(28.2) | 34(30.9) |
| Pancreas | 101(31.2) | 35(31.8) | 32(31.1) | 34(30.9) |
| Age – Yr. | ||||
| Mean(SD) | 48.8(15.4) | 50.6(15.7) | 46.0(15.5) | 49.67(14.7) |
| Median | 48.5 | 49 | 45 | 49 |
| Range | 18–86 | 18–82 | 19–81 | 18–86 |
| Sex | ||||
| Male | 138(42.7) | 51(46.4) | 40(38.8) | 47(42.7) |
| Female | 185(57.3) | 59(53.6) | 63(61.2) | 63(57.3) |
| Ethnicity and Racec | ||||
| Non-Hispanic white | 175(56.1) | 52(48.6) | 58(59.32) | 65(60.7) |
| Non-Hispanic minority | 74(23.7) | 28(26.2) | 21(21.4) | 25(23.4) |
| Hispanic | 63(20.2) | 27(25.2) | 19(19.4) | 17(15.9) |
| Unknown | 11 | 3 | 5 | 3 |
| Marital Status | ||||
| Married | 149(63.7) | 46(64.8) | 49(62.8) | 54(63.5) |
| Divorced, widowed, or separated | 64(27.4) | 17(23.9) | 22(28.2) | 25(29.4) |
| Never married | 21 (8.9) | 8(11.3) | 7 (9.0) | 6 (7.1) |
| Unknown | 89 | 39 | 25 | 25 |
| Religion d | ||||
| Christian | 191(81.3) | 62(84.9) | 60(77.9) | 69(81.2) |
| Other | 44(18.7) | 11(15.1) | 17(22.1) | 16(18.8) |
| Unknown | 88 | 37 | 26 | 25 |
| Education | ||||
| ≤ 11th Grade | 17 (7.3) | 4 (5.6) | 5 (6.6) | 8 (9.4) |
| High School/GED | 58(24.9) | 17(23.6) | 24(31.6) | 17(20.0) |
| 1–3 yrs. of college | 83(35.6) | 31(43.0) | 19(25.0) | 33(38.8) |
| 4+ yrs. of college | 47(20.2) | 11(15.3) | 20(26.3) | 16(18.8) |
| Graduate degree | 28(12.0) | 9(12.5) | 8(10.5) | 11(12.9) |
| Unknown | 90 | 38 | 27 | 25 |
| Annual Household Income | ||||
| <$20,000 | 33(16.2) | 11(17.2) | 12(17.9) | 10(13.7) |
| $20,000–$40,000 | 50(24.5) | 15(23.4) | 18(26.9) | 17(23.3) |
| $40,000–$60,000 | 30(14.7) | 9(14.1) | 7(10.4) | 14(19.2) |
| >$60,000 | 91(44.6) | 29(45.3) | 30(44.8) | 32(43.8) |
| Unknown | 119 | 46 | 36 | 37 |
Not all respondents answered all questions.
No. (%) of participants unless otherwise indicated.
Ethnicity and race were self-reported. Non-Hispanic minority includes: Asian, Native American/Alaskan Native, Pacific Islander and Other.
Christian includes: Catholic, Protestant, Evangelical; Other includes: Jewish, Muslim, Atheist/Agnostic and Other.
Consent Type and Data Sharing Decisions
All eligible participants randomized to traditional consent agreed to participate in the genomic study and by default to public release. Most participants (84.9%) randomized to binary consent chose public data release, while the remaining individuals (15.1%) opted out of data sharing (no release). The majority of participants (66.4%) randomized to tiered consent agreed to public data release, less than a fifth (19.5%) chose restricted release, and the remainder (14.1%) chose no release.
Following the debriefing, participants were given an opportunity to change their data release option; the majority (67.8%) stayed with their original choice. Of those who changed, only three chose an option that was less restrictive then their original choice (i.e., changed from no release to restricted release). Those randomized to tiered consent were less likely to change (21.2%) than those randomized to binary (37.7%) or traditional consent (37.9%) (chi-square test, p=0.01).
A majority of participants (53.1%) chose public data release as their final data sharing decision, a third (33.1%) chose restricted release, and the remaining individuals (13.7%) chose no release (Table 3). Final data sharing decisions and whether this choice differed from their original selection were significantly associated with randomized consent type (final decision chi-square test, p=.02; changing decision chi-square test, p=.01). Those randomized to traditional consent were most likely to choose public data release as their final data sharing decision (62.1%). Only 6% of participants randomized to traditional consent chose not to release their data at all, compared to nearly 20% of those randomized to either binary or tiered consent. Participants randomized to tiered consent were less likely to change their data sharing decision before and after debriefing. 78.8% of those randomized to tiered consent changed in their final data release selection compared to 62.1% randomized to traditional consent and 62.3% randomized to binary consent.
Table 3.
Pre- and Post-Debriefing Data Release Selections by Randomized Consent Type, Genomic Study and Consentee Relationship
| Pre-Debriefing | Post-Debriefing | ||||||
|---|---|---|---|---|---|---|---|
|
Public Release |
Restricted Release |
No Release |
Public Release |
Restricted Release |
No Release |
P Value |
|
| Randomized Consent Type (%) | |||||||
| Traditional (n=116) | 100.0 | 0.0 | 0.0 | 62.1 | 31.9 | 6.0 | |
| Binary (n=106) | 84.9 | 0.0 | 15.1 | 50.9 | 30.2 | 18.9 | .02 |
| Tiered (n=113) | 66.4 | 19.5 | 14.2 | 46.0 | 37.2 | 16.8 | |
| Genomic Study (%) | |||||||
| Autism (n=12) | 75.0 | 0.0 | 25.0 | 58.3 | 16.7 | 25.0 | |
| Brain Cancer (n=76) | 75.0 | 9.2 | 15.8 | 43.4 | 36.8 | 19.7 | |
| Brain Control (n=16) | 56.3 | 18.8 | 25.0 | 31.3 | 43.8 | 25.0 | .04 |
| Epilepsy (n=37) | 78.4 | 8.1 | 13.5 | 43.2 | 35.1 | 21.6 | |
| Liver (n=93) | 94.6 | 2.2 | 3.2 | 61.2 | 28.0 | 10.8 | |
| Pancreas (n=101) | 88.1 | 6.9 | 5.0 | 59.4 | 34.7 | 5.9 | |
| Consentee Relationshipa(%) | |||||||
| Adult/Self Consentee (n=221) | 89.1 | 5.0 | 5.9 | 61.1 | 29.0 | 9.9 | <.001 |
| Parental Consentee (n=113) | 73.5 | 9.7 | 16.8 | 38.1 | 40.7 | 21.2 | |
Missing data release decision for 1 developmentally disabled adult participant (not parental or adult consentee).
Other Factors Influencing Data Sharing Decisions
Hispanic participants were significantly less likely to choose public data release compared to non-Hispanic white participants (restricted release OR, 2.94; CI, 1.16–7.43; no release OR, 3.94; CI, 1.05–1.76). Unmarried participants, including those who were divorced, widowed, separated, or never married, were more likely to choose restricted data release (OR, 2.40; CI 1.05–5.44). When choosing between restricted and public data release, participants with some college or a college degree were also more likely to choose restricted data release (some college, OR 3.52; CI 1.02–12.14; college graduate, OR 4.67; CI 1.35–16.12) (Table 4).
Table 4.
Multinomial Logistic Regression Analysis of Factors Associated with Participants’ Final Data Release Selection
| Final Data Release Selection Compared to Public Releaseab | ||||
|---|---|---|---|---|
|
Restricted Release (n=111) |
No Release (n=46) |
|||
| OR | (95% CI) | OR | (95% CI) | |
| Randomized Consent Type | ||||
| Traditional | 1.00 | 1.00 | ||
| Binary | 1.13 | (0.50 – 2.54) | 3.14 | (0.82 – 12.03) |
| Tiered | 1.48 | (0.68 – 3.24) | 3.84 | (1.01 – 14.54) |
| Consentee Relationship | ||||
| Adult/Self Consentee | 1.00 | 1.00 | ||
| Parental Consentee | 3.56 | (1.57 – 8.08) | 4.78 | (1.46 – 15.64) |
| Consent-Debrief Time Lapse | 1.00 | (1.00 – 1.01) | 1.00 | (0.99 – 1.01) |
| Age | 1.03 | (1.00 – 1.06) | 1.02 | (0.98 – 1.06) |
| Gender | ||||
| Male | 1.00 | 1.00 | ||
| Female | 1.11 | (0.54 – 2.25) | 1.54 | (0.50 – 4.70) |
| Race/Ethnicityc | ||||
| Non-Hispanic white | 1.00 | 1.00 | ||
| Non-Hispanic minority | 1.73 | (0.74 – 4.05) | 2.08 | (0.61 – 7.09) |
| Hispanic | 2.94 | (1.16 – 7.43) | 3.94 | (1.05 – 14.76) |
| Marital Statusd | ||||
| Married | 1.00 | 1.00 | ||
| Unmarried | 2.40 | (1.05 – 5.44) | 3.07 | (0.98 – 9.65) |
| Religious Affiliatione | ||||
| Christian | 1.00 | 1.00 | ||
| Other | 1.24 | (0.53 – 2.89) | 1.97 | (0.60 – 6.53) |
| Education Level | ||||
| <11th Grade | 3.14 | (0.49 – 20.12) | 1.57 | (0.13 – 19.55) |
| HS Graduate/GED | 1.49 | (0.38 – 5.83) | 1.41 | (0.24 – 8.42) |
| Some College | 3.52 | (1.02 – 12.14) | 1.32 | (0.25 – 6.81) |
| College Graduate | 4.67 | (1.35 – 16.12) | 1.41 | (0.27 – 7.36) |
| Graduate School | 1.00 | 1.00 | ||
| Annual Household Income | ||||
| <$20,000 | 0.46 | (0.14 – 1.53) | 0.43 | (0.08 – 2.48) |
| $20,000–$40,000 | 0.52 | (0.19 – 1.45) | 0.21 | (0.04 – 1.19) |
| $40,000–$60,000 | 0.43 | (0.15 – 1.24) | 1.48 | (0.38 – 5.71) |
| >$60,000 | 1.00 | 1.00 | ||
Public data release (n=178) is reference group.
Table represents total number of final data release selections (n=335); characteristics of participants acting as adult/self and parental consenters (n=12) are used twice.
Non-Hispanic minority includes: Asian, Native American/Alaskan Native, Pacific Islander, and Other.
Unmarried includes: Never married, divorced, widowed and separated.
Christian includes: Catholic, Protestant and Evangelical; Other includes: Jewish, Muslim, Atheist/Agnostic and Other.
Genomic study was also found to be significantly associated with final data release selection (Table 3). Participants from studies conducting pediatric research (autism, brain cancer, and brain control) were more restrictive in their final data release choices than individuals from studies targeting mostly adult populations (liver and pancreatic cancers) (chi-square test, p=.04). To determine if these differences could be categorized based on consentee relationship, parental consent decisions (n=113) were compared with adult/self consent decisions (n=221). Consentee relationship was significantly associated with one’s final data release selection (chi-square test, p<.001). After controlling for other variables, consentee relationship remained a significant predictor; participants providing parental consent were significantly less likely to chose public data release than adults consenting for themselves (restricted release OR, 3.56; CI, 1.57 – 8.08; no release OR, 4.78; CI, 1.46 – 15.64) (Table 4). Those participants who made decisions both for themselves (adult/self consent) and on behalf of their child (parental consent) (n=12) made the same data sharing choice for themselves as for their child.
Another possible explanation for the difference between genomic studies could be the amount of time that lapsed between obtaining informed consent and debriefing the study participant. Most of the participants from the autism, brain cancer, and brain control studies were debriefed immediately after the informed consent process, while some individuals from the pancreatic cancer and liver cancer studies were not debriefed until months later (at a subsequent post-operative visit). However, when we controlled for other factors, time lapse between consent and debrief was not found to be a significant predictor of one’s final data release selection (restricted release OR, 1.00; CI, 1.00–1.01; no release OR, 1.00; CI, 0.99–1.01) (Table 4).
Refusal and Withdrawal Rates
All of the genomic studies reported high enrollment rates (autism, 85.7%; brain cancer, 80.9%; brain control, 61.5%; epilepsy, 85.7%; liver cancer, 97%; pancreatic cancer, 98.1%). Variations in genomic study enrollment rates were due to individual recruitment methods and the populations under study and were not reflective of issues with the consent process or study or data sharing concerns. Only 20 individuals overall declined participation. Of those, four were randomized to traditional consent, six to binary, three to tiered, and seven were not randomized to a consent type prior to declining. Most who declined participation in the genomic study reported that they did so because of general research-related concerns (e.g., blood draw, no time). Only one participant, randomized to binary consent, specified apprehension about data sharing.
Discussion
When given a choice about genomic data sharing, just over half of the participants in this study chose public data release. Genomic data generated during the course of research have traditionally been treated primarily as a community resource and made widely available through publicly accessible scientific databases. Some human DNA data are still available to the general public (e.g., http://hapmap.ncbi.nlm.nih.gov/, http://www.1000genomes.org/page.php), but the vast majority of data are now only available to approved researchers through controlled access databases (e.g., http://www.ncbi.nlm.nih.gov/gap). This policy shift was prompted by evidence of the potential vulnerability of de-identified DNA data4, 5 and related concerns about participant privacy.10, 12 Privacy risks have been carefully considered,13 but until now, there has been little empirical data on stakeholder perspectives to help inform these policy decisions.
Studies have shown that participants are apprehensive about potential privacy invasions when participating in genomic research.11, 14, 15 This is the first study to examine in a randomized fashion how these concerns impact research enrollment and actual data sharing decisions. Our findings indicate that, despite privacy concerns, the majority of research participants are “information altruists”16 with regards to the public release of their genomic data.
Another important observation is that parents are less inclined to consent to the public release of their child’s DNA data. Still, the majority is willing to share the data with the broader scientific community via controlled access databases. Additional research on pediatric participants’ attitudes toward data sharing will be important for future policy development.
Our finding that white participants are less restrictive in their data sharing choice than their minority counterparts is consistent with other studies that have found that minorities are less likely to participate in research and are more distrustful of study investigators.17 Educational programs that aim to increase minority participation in research should specifically address concerns about genomic data sharing.
This study has several limitations. All participants were recruited within a clinical setting. They may have formed a trusting relationship with study investigators, which could have influenced their willingness to participate and their data release choice. Additionally, all of the genomic studies were conducted at BCM in Houston, Texas. These findings may not be generalizable to other participant populations or geographic regions. Although all of the genomic studies used the same consent language, the consenting process varied across studies with regards to the consent process facilitator, length of time spent with the participants, and the timing of the consenting process (i.e., whether consent was obtained at a pre-operative visit, just prior to surgery, or during the post-operative period). These are some of the factors that may help explain the variations in data sharing choices among participants from individual studies.
A primary outcome was to determine if offering greater control over DNA data release during the informed consent process affected the rate of participation in or withdrawal from the genomic research. However, the common practice of participant recruitment into research studies may not have allowed us to accurately assess these parameters. Participants eligible for genomic studies were often approached informally by the study PI, a clinical research nurse, or a surgical resident and only those who expressed an interest were formally recruited. All of the genomic studies reported high enrollment rates, but these data may not capture individuals who were approached but not formally recruited. This informal screening typically occurred prior to randomization or exposure to the assigned consent.
Conclusion
There are great scientific benefits to the public availability of genomic data as expressed by Francis Collins: “free and open access to genome data has had a profoundly positive effect on progress”.18 However, data sharing policies must balance the scientific benefits with ethical obligations to participants. This study raises the important question of whether existing policies achieve an appropriate balance or whether they are overly restrictive. In this study, more than half of the participants consented to the public release of their DNA data, and nobody declined enrollment when participation was conditioned on public data release. This suggests that mandating full public data release would maximize data availability. However, it would not be consistent with the preferences of the 47% of study participants who chose a more restrictive data sharing option.
Providing options through tiered consent respects participants’ preferences without significantly impeding research. Those who were randomized to tiered consent were less likely to change their consent post-debrief, which suggests that offering options maximizes participant autonomy by allowing participants to make decisions consistent with their preferences. The primary purpose of some large scale genomic studies (e.g., 1000 Genomes Project) is to create a community resource. For those studies, where unrestricted data sharing is an essential component of the research, traditional consent may be most appropriate. However, in studies where data sharing is desired but not required, tiered consent can provide a mechanism to respect individuals’ preferences without imposing an excessive burden on researchers. Participants in this study were generally accepting of broad but controlled data sharing; other groups may be less willing to share their data. Respecting the preferences of individuals within these groups will go a long way towards securing the public’s trust and will maximize the diversity of participation in genomic research.
Additional research is needed to assess the costs and benefits of providing participants with control over data release through tiered consent; to determine if data sharing decisions, attitudes, and preferences differ among disease, geographic, ethnic, and socioeconomic populations; and to better understand any discrepancy between participants’ stated preferences, reported concerns, and actual decisions.
Supplementary Material
Acknowledgments
This work was supported by grant NIH R01 HG004333 (A.L. McGuire, S.G. Hilsenbeck, P.A. Kelly); The Greenwall Foundation Faculty Scholars Program in Bioethics (A.L. McGuire); DLDCC P30CA125123 (S.G. Hilsenbeck); NINDS R01 NS 29709 (J.L. Noebels); NCI R01 CA109467,R33 CA97874 (C.C. Lau). We thank Laura Beskow, Wylie Burke, Aravinda Chakravarti, Mildred Cho, Rebecca Fisher, Gail Geller, Laura Rodriguez, Louise Strong, and Richard Gibbs for their expert advice throughout this project. We thank Claudette Campbell, Sally E. Hodges, Liz Hinojosa, Morgan Lasala, Melissa Pagaoa, Suzanne Wheeler, and Tiffany Zgabay-Hunsucker for their valuable assistance and research coordination.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.The Wellcome Trust (Bermuda, February 25, 1996) Summary of Principles Agreed at the International Strategy Meeting on Human Genome Sequencing, 1996. [Accessed February 3, 2011]; Available at: http://www.wellcome.ac.uk/About-us/Policy/Policy-and-position-statements/WTD002751.htm.
- 2.The Wellcome Trust (Fort Lauderdale, January 14 – 15, 2003) Sharing Data from Large-Scale Biological Research Projects: A System of Tripartite Responsibility, 2003. [Accessed February 3, 2011]; Available at: http://www.genome.gov/Pages/Research/WellcomeReport0303.pdf.
- 3.National Human Genome Research Institute Reaffirmation and Extension of NHGRI Rapid Release Policies: Large Scale Sequencing and Other Community Resource Projects, February 2003. [Accessed February 3, 2011]; Available at: http://www.genome.gov/10506537.
- 4.Lin Z, Owen AB, Altman RB. Genomic research and human subject privacy. Science. 2004;205:183. doi: 10.1126/science.1095019. [DOI] [PubMed] [Google Scholar]
- 5.Homer N, Szelinger S, Redman M, et al. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 2008;4(8):e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Institutes of Health Policy for Sharing of Data Obtained in NIH Supported or Conducted Genome-Wide Association Studies, 2007. [Accessed February 3, 2011]; Available at: http://grants.nih.gov/grants/guide/notice-files/NOT-OD-07-088.html.
- 7.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39(10):1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert N. Researchers criticize genetic data restrictions. Nature News, September 4, 2008. [Accessed February 3, 2011]; Available at: http://www.nature.com/news/2008/080904/full/news.2008.1083.html. [Google Scholar]
- 9.Lunshof JE, Chadwick R, Vorhaus DB, Church GM. From genetic privacy to open consent. Nat Rev Genet. 2008;9(5):406–411. doi: 10.1038/nrg2360. [DOI] [PubMed] [Google Scholar]
- 10.McGuire AL, Gibbs RA. No longer de-identified. Science. 2006;312:370–371. doi: 10.1126/science.1125339. [DOI] [PubMed] [Google Scholar]
- 11.McGuire AL, Hamilton JA, Lunstroth R, McCullough LB, Goldman A. DNA data sharing: research participants’ perspectives. Genet Med. 2008;10:46–53. doi: 10.1097/GIM.0b013e31815f1e00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowrance WW, Collins FS. Identifiability in genomic research. Science. 2007;317:600–602. doi: 10.1126/science.1147699. [DOI] [PubMed] [Google Scholar]
- 13.National Human Genome Research Institute (NHGRI) Workshop on Privacy, Confidentiality, and Identifiability in Genomic Research, 2006. [Accessed February 3, 2011]; Available at: http://www.genome.gov/19519198.
- 14.Hull SC, Sharp RR, Botkin JR, et al. Patients’ views on identifiability of samples and informed consent for genetic research. Am J Bioeth. 2008;8(10):62–70. doi: 10.1080/15265160802478404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaufman DJ, Murphy-Bollinger J, Scott J, Hudson KL. Public opinion about the importance of privacy in biobank research. Am J Hum Genet. 2009;85(5):643–654. doi: 10.1016/j.ajhg.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohane IS, Altman RB. Health-information altruists – a potentially critical resource. N Engl J Med. 2005;353(19):2074–2077. doi: 10.1056/NEJMsb051220. [DOI] [PubMed] [Google Scholar]
- 17.Corbie-Smith G, Thomas SB, Williams MV, Moody-Ayers S. Attitudes and beliefs of African Americans toward participation in medical research. J Gen Intern Med. 1999;14:537–546. doi: 10.1046/j.1525-1497.1999.07048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F. Has the revolution arrived? Nature. 2010;464:674–675. doi: 10.1038/464674a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


