Abstract
Evidence is accumulating that small open reading frames (sORF, <100 codons) play key roles in many important biological processes. Yet, they are generally ignored in gene annotation despite they are far more abundant than the genes with more than 100 codons. Here, we demonstrate that popular homolog search and codon-index techniques perform poorly for small genes relative to that for larger genes, while a method dedicated to sORF discovery has a similar level of accuracy as homology search. The result is largely due to the small dataset of experimentally verified sORF available for homology search and for training ab initio techniques. It highlights the urgent need for both experimental and computational studies in order to further advance the accuracy of sORF prediction.
INTRODUCTION
An increasing body of evidence shows that proteins translated from small open reading frames (sORFs; <100 codons) are involved in a variety of important functional classes. These biological functionality includes but not limited to mating pheromones, energy metabolism, proteolipids, chaperonins, stress proteins, transporters, transcriptional regulators, nucleases, ribosomal proteins, thioredoxins, metal ion chelators and transmembrane proteins [1]. For example, tarsal-less (tal) gene, a 33-nucleotide-long ORF, is translated into 11-amino-acid-long peptide and controls gene expression and tissue folding in the Drosophila [2]. Another sORF gene, polished rice (pri), which is of 11–32 amino acids long, controls epidermal differentiation in Drosophila by modifying the transcription factor Shavenbaby [3, 4]. In Bacillus subtilis, the 46-amino-acid-long Sda protein inhibits the onset of sporulation by preventing activation of a transcription factor required for sporulation [5]. In plants, the products of sORF protein coding genes are important components of photosynthetic supracomplex [6]. These identified and characterized examples indicate that sORFs are ubiquitous and play significant roles in various biological processes.
While some sORFs have important function, the majority of sORFs are probably meaningless and arisen by chance. Thus, identifying functional sORFs from huge number of fake sORFs is a daunting task for genome annotation projects. In practice, sORFs are generally eliminated from any genome annotations (i.e., only ORFs with at least 100 codons are annotated) [7] and the functions of proteins encoded by sORFs are largely un-explored. Hence, it is critical to employ effective computational methods to pinpoint potentially genuine sORFs “buried” under piles of meaningless ones [1]. Such an accurate sORF list (narrowed down by computational methods to a practical scale) is highly desirable for investigators to perform follow-up experimental characterization.
The most commonly used computational method for sORF discovery is BLAST (or analog of BLAST), which identifies members of a sORF coding gene family based on protein-sequence homology information [1, 2, 8–11]. The dependence on homologous, known sORFs thus limits the power of BLAST in searching novel sORFs. ab initio methods, on the other hand, goes beyond sequence similarity by finding novel sORFs based on general features common to sORF coding genes. One widely used ab initio sORF-prediction technique is to evaluate whether the pattern of codon usage in a potential sORF is characteristic of genuine genes [7, 12, 13]. Others predicted coding potential of small genes by employing sequence transformation [14], hidden Markov models (HMM) or hybrid HMM-SVM with various sequence features [15–17], and the hexamer composition bias between coding and non-coding sequences [18–20]. In this mini-review, we compiled two datasets of genes with <100 and 100–150 codons for Saccharaomyces cerevisiae from literature and made a comparative assessment of BLAST and two representative ab initio discovery techniques (codonW [12] and sORF finder [20]) that were optimized for sORF prediction in Saccharaomyces cerevisiae. Such an assessment is necessary in order to further improve existing methods.
BLAST SEARCH FOR sORF PREDICTION
BLAST search is a commonly used technique to compare a query sequence with the sequences contained in a library of known genes. It tabulates the query sequence as short sequences (seeds), scans the database for matches, and subsequently extends the matches to high-scoring segments [21]. There are two databases of genes: Swiss-Prot and NCBI NR database. Swiss-Prot is a highly-curated, highly-cross-referenced, annotated non-redundant protein sequence database [22], while NR is a more comprehensive and less curated protein sequence database automatically compiled from GenBank CDS translations, PDB, Swiss-Prot, PIR and PRF resources[23]. A BLASTp search of a query sequence will produce an output file containing all homologous hits with sequence identity and an estimated significance based on the expected value (the probability for such a match to occur purely by chance). In this study, we delete those hits that are labeled as “hypothetical”, “putative”, or “unnamed protein product”. Obviously, the number of predicted sORF by BLAST can be controlled by changing the expected value (Evalue) and sequence identity. To investigate the performance of BLAST, we group the hits at different cutoff values of sequence identity and choose the hit based on Evalue after excluding the hits that are above a certain sequence-identity cutoff to the query sequence. The threshold of Evalue is employed as a parameter to control true and false positive rates.
Ab Initio sORF Prediction
CodonW
This method is based on the assumption that the codon usage of well expressed ORF (i.e., with biological functions) is non-random, and a true sORF should possess an optimal codon pattern [24]. Along this assumption, different codon usage indices have been developed for coding potential evaluation [25]. codonW is a program which implemented three popular codon usage indices to analyze the coding potential of a given ORF [12]. We evaluated the performance of each of the three indices, namely, CBI (codon bias index, measuring how biased the codons in an ORF are towards highly “preferred codons” in coding amino acids), Fop (the frequency of optimal codons) and CAI (codon adaption index). The latter two indices measure the degree to which the codon usage of a gene has adapted toward the usage of optimal codons. The indices optimized specifically for S. cerevisiae are chosen for comparative studies.
sORF finder
sORF finder is a more recent program specifically designed for identifying small open reading frames with high coding potential [18, 19]. It is based on the hexamer composition bias between coding sequence (CDS) and non-coding sequences (NCDS). We used the web-based sORF application tool (http://evolver.psc.riken.jp/sORFfinder/cgi-bin/run1.html) for this study, and selected the S. cerevisiae specific training model for CDS and NCDS. The output file assigns scores (the greater the score is, the higher confidence it has) only to the sORFs that the program discovered, while a score of “−100” is assigned arbitrarily to each unidentified sequence. The sORF finder can only apply to predictions with less than 100 codons.
DATASET FOR ASSESSMENT
Assessing the accuracy of sORF computational discovery methods is difficult because there are only a relatively small number of confirmed protein-coding sORFs (i.e., true positive, TP), and even less for the number of confirmed meaningless sORFs (i.e., true negative, TN). Although in some studies intron and/or intergenic regions have been used as an alternative to meaningless sORFs for method evaluation [19], it is not clear whether the meaningless sORFs share the same properties as these regions.
Saccharaomyces cerevisiae (budding yeast) is one of a few extensively studied model organisms where firm evidence for a significant number of genuine sORFs and meaningless sORFs are available. Because the 100-codon boundary is arbitrarily defined in genome sequencing project, annotated ORFs of 100–150 codons inevitably include a fraction of artificial ORFs [7]. We thus assess the computational ORF discovery at both ranges of <100 and 100–150 codons.
We have compiled a curated dataset of 660 positive and 657 negative ORF genes with less than 150 codons, including 311 positive and 167 negative sORF genes with less than 100 amino acids in Saccharaomyces cerevisiae. The positive ORF set is from the annotated ORFs by SGD team [26], including both verified ORF for which experimental evidence exists that a gene product is produced in S. cerevisiae, and uncharacterized ORF that is likely to encode an expressed protein. The negative sORF set comes from the dubious ORF records curate in SGD. A Dubious ORF is one that is unlikely to encode an expressed protein, which meet the criteria including but not exclusively that 1) the ORF is not conserved in other Saccharomyces species, 2) there is no well-controlled, small-scale, published experimental evidence that a gene product is produced, and 3) a phenotype caused by disruption of the ORF can be ascribed to mutation of an overlapping gene. For data quality control, we also made use of the existing large-scale protein expression data [13], protein localization data [27] and gene-deletion data [1]. Specifically, from the positive set, we filtered out the ORFs showing negative status in any of the three large-scale functional genomics datasets. For the negative set, we filtered out the ORFs showing positive status in any of the three large-scale functional genomics datasets.
COMPARATIVE ASSESSMENT
We assess various methods by using Receiver Operating Characteristic (ROC) curve. A ROC curve shows the True Positive Rate (TPR, fraction of correctly predicted positive ORF in the total number of positive ORFs) versus the False Positive Rate (FPR, fraction of incorrectly predicted positive ORF in the total number of negative ORFs). The area under the ROC curve (AUC) is a measure of discrimination accuracy. All the ROC analysis is performed by the ROCR package in R programming environment [28]. The overall assessment of performance is shown in Table 1. As the number of sORF with 1–99 codons is huge in any sequenced eukaryotic genome, it is highly desirable that prediction methods can reach high true positive rate (TPR) at a low threshold of false negative rate (FPR).
Table 1.
The Performance of sORF Prediction Methods on the Dataset with 1–99 Codons and 100–150 Codons, Separately
| ORF with 1–99 Codons | ORF with 100–150 Codons | |||||
|---|---|---|---|---|---|---|
| TPR (at 5% FPR) |
TPR (at 20% FPR) |
Overall AUC Value |
TPR (at 5% FPR) |
TPR (at 20% FPR) |
Overall AUC Value |
|
| sORF finder | 0.531 | 0.650 | 0.771 | N/A | N/A | N/A |
| CBI | 0.248 | 0.354 | 0.639 | 0.444 | 0.645 | 0.787 |
| CAI | 0.257 | 0.405 | 0.662 | 0.473 | 0.699 | 0.824 |
| Fop | 0.244 | 0.389 | 0.660 | 0.433 | 0.705 | 0.815 |
| BLAST (30%, NR) | 0.251 | 0.412 | 0.630 | 0.650 | 0.731 | 0.824 |
| BLAST (30%, SP) | 0.187 | 0.428 | 0.629 | 0.570 | 0.768 | 0.858 |
| BLAST (50%, NR) | 0.492 | 0.572 | 0.704 | 0.903 | 0.931 | 0.949 |
| BLAST (50%, SP) | 0.360 | 0.489 | 0.694 | 0.785 | 0.845 | 0.920 |
| BLAST (70%, NR) | 0.418 | 0.595 | 0.716 | 0.914 | 0.946 | 0.959 |
| BLAST (70%, SP) | 0.270 | 0.531 | 0.704 | 0.800 | 0.854 | 0.918 |
| BLAST (90%, NR) | 0.286 | 0.630 | 0.709 | 0.880 | 0.951 | 0.952 |
| BLAST (90%, SP) | 0.174 | 0.508 | 0.690 | 0.800 | 0.860 | 0.914 |
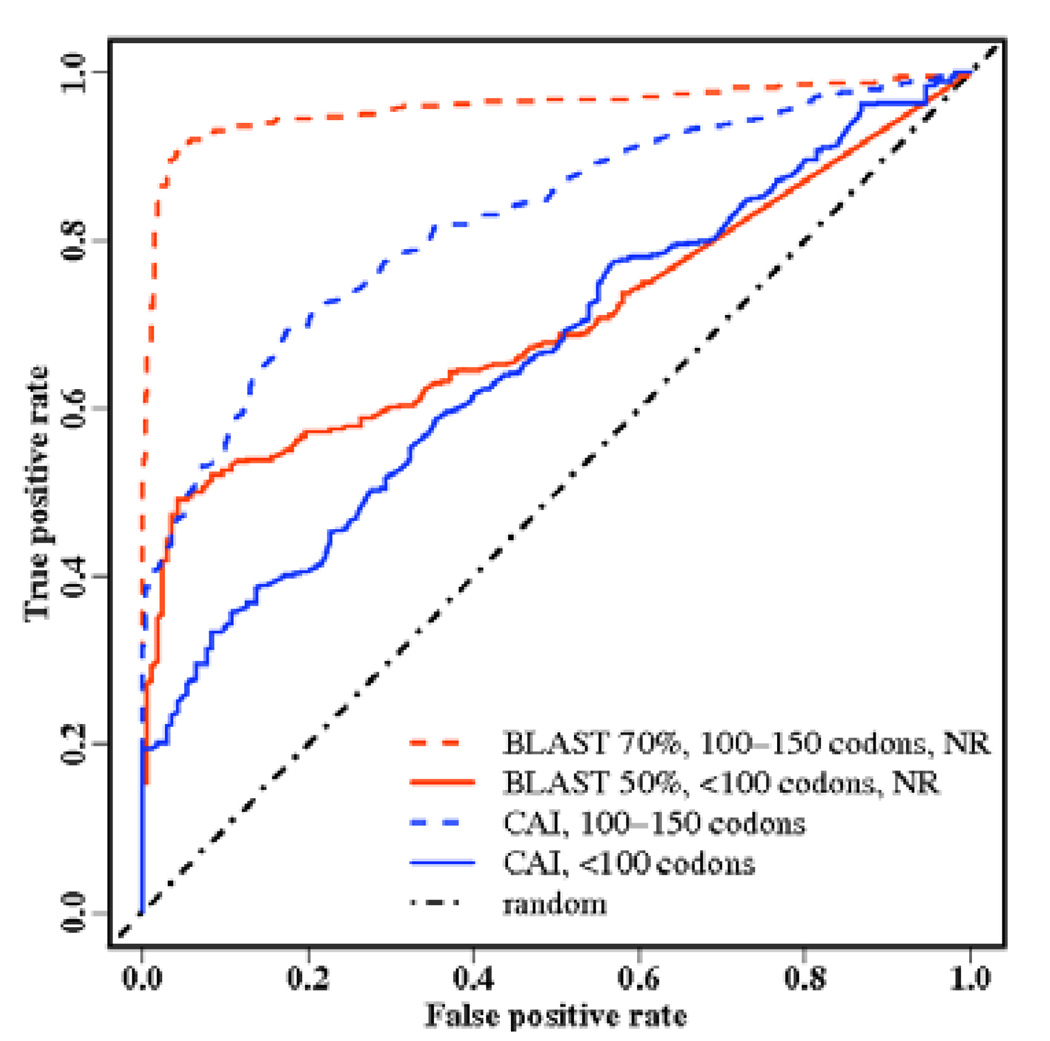
To evaluate BLAST, we exclude homologous hits if it is higher than a given sequence identity to examine its ability to detect genes in the absence of highly homologous sequences. The expected value (Evalue) is changed to obtain the ROC curve. Swiss-Prot is a highly curated database while NR is automatically compiled from multiple resources. However, at low FPR threshold (e.g., 5%), the usage of more comprehensive NR database has improved TPR over that obtained from the Swiss-Prot (Table 1). Table 1 further shows that the AUC value for ≥100 codons is above 0.9 even after excluding homologous hits with 50% or more sequence identity to the query sequence, while is only around 0.7 for <100 codons even if only >90% homologous hits are excluded. At the 5% false positive rate, the highest true positive rate is 91% for ≥100 codons for BLAST with 70% sequence identity cutoff based on the NR database, while the corresponding value is only 49% for <100 codons for BLAST with 50% sequence identity cutoff based on the NCBI NR database. Thus, as shown in Fig. (1), BLAST is a powerful search technique for searching homologous genes with >100 codons but not for <100 codons, i.e., sORFs.
Fig. (1).
Significant drop in performance is observed for the popular homolog-search technique BLAST (in Red) and ab initio predictor codonW (in Blue). The receiver operating curves are shown for the best performed BLAST search in NR database for sORFs (excluding 50% or more homologous sequences) and for ORFs with 100–150 codons (excluding 70% or more homologous sequence) along with the performance of codonW for sORFs and ORFs with 100–150 codons, respectively.
Similar performance drops between <100 codon and ≥100 codons is observed for codonW Fig. (1). The highest AUC value is 0.82 for ≥100 codons by the CAI index but only 0.66 for <100 codons by either the CAI or the Fop index. At the 5% false positive rate, the highest true positive rate is 47% for ≥100 codons but only 26% for <100 codons. This suggests that codonW is significantly less effective in ab initio prediction of sORF than in predicting larger genes.
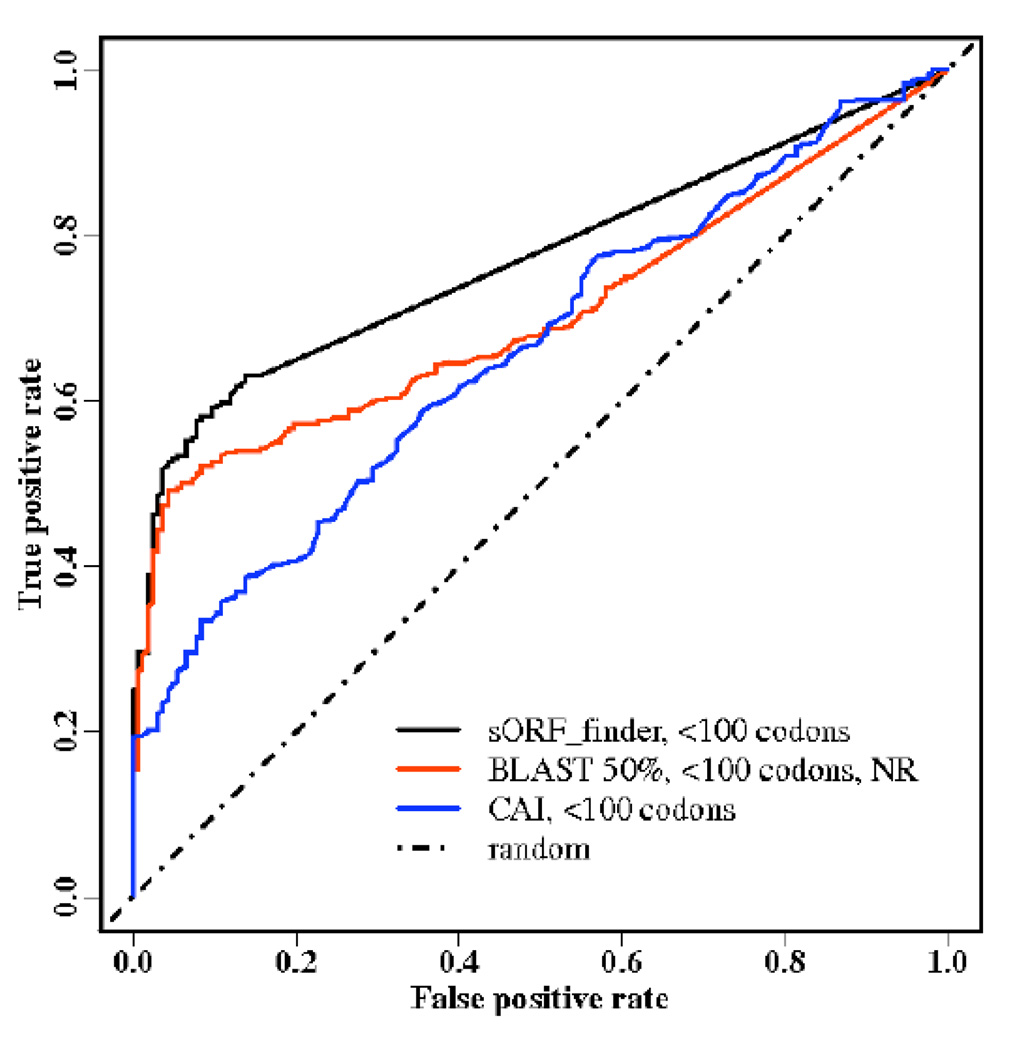
For sORF, the best BLAST performance (at 50% sequence-identity cutoff) is compared to the best codonW performance with CAI index and the sORF finder, the method dedicated to sORF. As shown in Fig. (2), sORF finder has the best performance among the three methods. The true positive rate for sORF finder is 53% at the 5% false positive rate, compared to 50% by BLAST. This small improvement from sORF finder may be due to the fact that the method was specifically trained for detecting small ORF and some of positive and negative examples are in the training set. This ~50% true positive rate at 5% false positive rate, however, is far from satisfactory. It should be emphasized that the direct comparison of BLAST and sORF finder is not suitable because one is a homolog-based technique while the other is an ab initio approach. The purpose here is to demonstrate the remaining challenges facing the detection of small open reading frames regardless of the techniques used.
Fig. (2).
As in Fig.1 but for sORF only with an additional method called sORF finder (in Black).
DISCUSSION AND OUTLOOK
Recent literatures have shown that proteins translated from small open reading frames are involved in a variety of important functional classes. Because the number of sORF candidates far exceeds the number of ORF with more than 100 codons from any eukaryotic genome sequencing project, it is important to develop accurate computational methods to identity genuine sORFs. The most commonly used computational method for sORF discovery is BLAST. However, BLAST relies on known ORFs deposited in a database and hence is unable to discover novel sORFs. Here we showed that both BLAST and ab initio methods based on codon patterns have significant performance drop when applied to small ORFs. This is largely, in part, due to relative small library for verified small genes. The development of an ab initio method dedicated to small ORFs is still at its infancy and the initial accuracy is low.
Two ab initio techniques (codonW and sORF finder) described here are single-feature based methods. Condon index techniques have been used by others in sORF discoveries, and sORF finder is specifically designed for sORF prediction. They were selected as representative here because they were also optimized for Saccharaomyces cerevisiae. There are many other sophisticated gene prediction programs available for genes with 100 or more codons (For recent reviews, see e.g. [29–32]). They often integrate multiple features including nucleotide composition, transcriptional signals (e.g., promoter motifs), splicing signals (e.g., intron splice sites), translational signals (e.g., translational start/stop sites), and un-translated signals (e.g., polyadenylation tails). For example, GeneScan [33] is a probabilistic model of the gene structure built on promoter, splice and translation signals plus additional features of gene and its surrounding regions. The program was optimized for vertebrate, Arabidopsis, and Maize only. GlimmerHMM [34] incorporates features from introns, intergenic regions, and four types of exons into a Generalized Hidden Markov Model. It was optimized for Arabidopsis thaliana, Coccidioides species, Cryptococcus neoformans, and Brugia malayi, C. elegans, Danio rerio (zebrafish), and human. Glimmer3.0 introduces a sophisticated Markov models called interpolated context models (ICM), which can capture dependencies among adjacent nucleotides with a typical window size of 12 [35]. It was optimized for microbial DNA, especially the genomes of bacteria, archaea, and viruses. These complex ab initio gene prediction programs are designed to evaluate the coding likelihood of long ORFs (>100 codons) with rich features. A direct application of them to sORF predictions is not appropriate due to the following two considerations. First, they are not specifically trained to discriminate true sORFs from false sORFs. Hence their performance in sORF prediction does not reflect their genuine capability. Second, as pointed out by previous work [7, 19, 36, 37], the gain of decreased false-positive prediction of genes from integration of multiple features is often associated with increased false-negative gene prediction, which is less desirable for sORF discovery due to the very large pool of meaningless ORFs in any sequenced eukaryotic genomes. Thus, our current assessment is limited to two ab initio methods that focus on single feature of sORFs, and we further point out the need for reorienting those sophisticated programs for sORF discovery.
One limitation of this assessment is the relative small number of sORF genes available. We obtained 311 positive and 167 negative sORF (<100 codons) from SGD curation, and there exists possibility that some of these negative sORF genes may be translated into functional proteins. To overcome such limitation, some studies have either used ORFs within intergenic/intron regions or randomly generated fragments from such regions as a negative set in method development [19–20]. However, to what extent that genuine negative sORFs share the same properties as intron and/or intergenic regions is still an interesting open question. Thus, the availability of large set of experimentally verified sORFs (both positive and negative genes) is crucial for a thorough assessment of prediction techniques in this field. Previous experimental studies indicate that signals from tiling microarray might be useful for finding novel transcribed regions including sORFs [38]. However, the accuracy of the tiling array results is uncertain owing to concerns about high background noises due to cross-hybridization [39], RNA-Seq, built on the massively parallel next-generation DNA sequencing technologies, provides an unprecedented ability to screen novel transcribed regions including sORFs without being confounded by background noises [40]. The novel transcribed regions, once verified to be genuine protein coding sORFs (or dismissed as non protein-coding sORFs), will undoubtedly provide a rich resource for developing and evaluating sORF prediction techniques.
In summary, we expect that the accuracy of sORF prediction will be likely improved when more computational biologists armed with modern machine learning techniques are interested in this problem and when existing state-of-the-art techniques including those combining homology with ab initio approaches (such as TWINSCAN [41] and CONTRAST [42]) are retooled specifically for sORF discovery. Meanwhile, a significantly larger experimentally validated set of genuine vs. false sORFs is needed urgently in order to better train and evaluate the computational methods.
ACKNOWLEDGEMENT
National Institutes of Health grants R01GM068530 (Y.Z.). Roswell Park Cancer Institute research startup fund (S.L.).
REFERENCES
- 1.Kastenmayer JP, Ni L, Chu A, Kitchen LE, Au WC, Yang H, Carter CD, Wheeler D, Davis RW, Boeke JD, Snyder MA, Basrai MA. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 2006;16(3):365–373. doi: 10.1101/gr.4355406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 2007;5(5):e106. doi: 10.1371/journal.pbio.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat. Cell Biol. 2007;9(6):660–665. doi: 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- 4.Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329(5989):336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- 5.Burkholder WF, Kurtser I, Grossman AD. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell. 2001;104(2):269–279. doi: 10.1016/s0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 6.Shi LX, Schroder WP. The low molecular mass subunits of the photosynthetic supracomplex, photosystem II. Biochim. Biophys. Acta. 2004;1608(2–3):75–96. doi: 10.1016/j.bbabio.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Basrai MA, Hieter P, Boeke JD. Small open reading frames: beautiful needles in the haystack. Genome Res. 1997;7(8):768–771. doi: 10.1101/gr.7.8.768. [DOI] [PubMed] [Google Scholar]
- 8.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423(6937):241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 9.Hemm MR, Paul BJ, Schneider TD, Storz G, Rudd KE. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol. Microbiol. 2008;70(6):1487–1501. doi: 10.1111/j.1365-2958.2008.06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Windsor AJ, Mitchell-Olds T. Comparative genomics as a tool for gene discovery. Curr. Opin. Biotechnol. 2006;17(2):161–167. doi: 10.1016/j.copbio.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Olsen AN, Mundy J, Skriver K. Peptomics, identification of novel cationic Arabidopsis peptides with conserved sequence motifs. In Silico Biol. 2002;2(4):441–451. [PubMed] [Google Scholar]
- 12.Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 14.Guo FB, Zhang CT. ZCURVE_V: a new self-training system for recognizing protein-coding genes in viral and phage genomes. Bmc Bioinformatics. 2006:7–9. doi: 10.1186/1471-2105-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saeys Y, Rouze P, Van de Peer Y. In search of the small ones: improved prediction of short exons in vertebrates, plants, fungi and protists. Bioinformatics. 2007;23(4):414–420. doi: 10.1093/bioinformatics/btl639. [DOI] [PubMed] [Google Scholar]
- 16.Besemer J, Borodovsky M. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 1999;27(19):3911–3920. doi: 10.1093/nar/27.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27(23):4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanada K, Akiyama K, Sakurai T, Toyoda T, Shinozaki K, Shiu SH. sORF finder: a program package to identify small open reading frames with high coding potential. Bioinformatics. 26(3):399–400. doi: 10.1093/bioinformatics/btp688. [DOI] [PubMed] [Google Scholar]
- 19.Hanada K, Zhang X, Borevitz JO, Li WH, Shiu SH. A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res. 2007;17(5):632–640. doi: 10.1101/gr.5836207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanada K, Akiyama K, Sakurai T, Toyoda T, Shinozaki K, Shiu SH. sORF finder: a program package to identify small open reading frames with high coding potential. Bioinformatics. 2010;26(3):399–400. doi: 10.1093/bioinformatics/btp688. [DOI] [PubMed] [Google Scholar]
- 21.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Re.s. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38(Database issue):D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2005;33(Database issue):D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li WH. Models of nearly neutral mutations with particular implications for nonrandom usage of synonymous codons. J. Mol. Evol. 1987;24(4):337–345. doi: 10.1007/BF02134132. [DOI] [PubMed] [Google Scholar]
- 25.Sharp PM, Matassi G. Codon usage and genome evolution. Curr. Opin. Genet. Dev. 1994;4(6):851–860. doi: 10.1016/0959-437x(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 26.Engel SR, Balakrishnan R, Binkley G, Christie KR, Costanzo MC, Dwight SS, Fisk DG, Hirschman JE, Hitz BC, Hong EL, Krieger CJ, Livstone MS, Miyasato SR, Nash R, Oughtred R, Park J, Skrzypek MS, Weng S, Wong ED, Dolinski K, Botstein D, Cherry JM. Saccharomyces Genome Database provides mutant phenotype data. Nucleic Acids Res. 2010;38(Database issue):D433–D436. doi: 10.1093/nar/gkp917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 28.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21(20):3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 29.Do JH, Choi DK. Computational approaches to gene prediction. J. Microbiol. 2006;44(2):137–144. [PubMed] [Google Scholar]
- 30.Sleator RD. An overview of the current status of eukaryote gene prediction strategies. Gene. 2010;461(1–2):1–4. doi: 10.1016/j.gene.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 31.Guigo R, Flicek P, Abril JF, Reymond A, Lagarde J, Denoeud F, Antonarakis S, Ashburner M, Bajic VB, Birney E, Castelo R, Eyras E, Ucla C, Gingeras TR, Harrow J, Hubbard T, Lewis SE, Reese MG. EGASP: the human ENCODE genome annotation assessment project. Genome Biol. 2006;7 Suppl 1:S2.1–S2.31. doi: 10.1186/gb-2006-7-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brent MR, Guigo R. Recent advances in gene structure prediction. Curr. Opinion Struct. Biol. 2004;14(3):264–272. doi: 10.1016/j.sbi.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 33.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268(1):78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 34.Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20(16):2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 35.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23(6):673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Li S, Zhang Y, Zheng H, Xu Z, Ye J, Yu J, Wong GK. Vertebrate gene predictions and the problem of large genes. Nat. Rev. Genet. 2003;4(9):741–749. doi: 10.1038/nrg1160. [DOI] [PubMed] [Google Scholar]
- 37.Claverie JM. Computational methods for the identification of genes in vertebrate genomic sequences. Hum. Mol. Genet. 1997;6(10):1735–1744. doi: 10.1093/hmg/6.10.1735. [DOI] [PubMed] [Google Scholar]
- 38.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308(5725):1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 39.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most "Dark Matter" Transcripts Are Associated With Known Genes. Plos Biol. 2010;8(5):e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genetics. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flicek P, Keibler E, Hu P, Korf I, Brent MR. Leveraging the mouse genome for gene prediction in human: From whole-genome shotgun reads to a global synteny map. Genome Res. 2003;13(1):46–54. doi: 10.1101/gr.830003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gross SS, Do CB, Sirota M, Batzoglou S. CONTRAST: a discriminative, phylogeny-free approach to multiple informant de novo gene prediction. Genome Biol. 2007;8(12):R269. doi: 10.1186/gb-2007-8-12-r269. [DOI] [PMC free article] [PubMed] [Google Scholar]