Abstract
Glutamine synthetase (GS) is the key enzyme in ammonia assimilation and catalyzes the ATP-dependent condensation of NH3 with glutamate to produce glutamine. GS in plants is an octameric enzyme. Recent work from our laboratory suggests that GS activity in plants may be regulated at the level of protein turnover (S.J. Temple, T.J. Knight, P.J. Unkefer, C. Sengupta-Gopalan [1993] Mol Gen Genet 236: 315–325; S.J. Temple, S. Kunjibettu, D. Roche, C. Sengupta-Gopalan [1996] Plant Physiol 112: 1723–1733; S.J. Temple, C. Sengupta-Gopalan [1997] In C.H. Foyer, W.P. Quick, eds, A Molecular Approach to Primary Metabolism in Higher Plants. Taylor & Francis, London, pp 155–177). Oxidative modification of GS has been implicated as the first step in the turnover of GS in bacteria. By incubating soybean (Glycine max) root extract enriched in GS in a metal-catalyzed oxidation system to produce the ·OH radical, we have shown that GS is oxidized and that oxidized GS is inactive and more susceptible to degradation than nonoxidized GS. Histidine and cysteine protect GS from metal-catalyzed inactivation, indicating that oxidation modifies the GS active site and that cysteine and histidine residues are the site of modification. Similarly, ATP and particularly ATP/glutamate give the enzyme the greatest protection against oxidative inactivation. The roots of plants fed ammonium nitrate showed a 3-fold increase in the level of GS polypeptides and activity compared with plants not fed ammonium nitrate but without a corresponding increase in the GS transcript level. This would suggest either translational or posttranslational control of GS levels.
GS (EC 6.3.1.2) is a key enzyme in nitrogen metabolism. It catalyzes the biosynthesis of Gln from Glu, ATP, and ammonium. GS from bacteria consists of 12 identical subunits arranged in two hexamers stacked face to face; the side-to-side interface of a pair of subunits constitutes an active site containing two Mn2+ ions (Yamashita et al., 1989). The two divalent metal ions in the active site are distinguished by their dissociation constants (Villafranca et al., 1985). Saturation of the high-affinity site, n1, in each subunit by Mn2+ or Mg2+ induces a conformational change, converting the enzyme from a catalytically inactive to a catalytically active conformation (Hunt and Ginsburg, 1980). The metal ion at the n1 site also plays a catalytic role in the binding of Glu, whereas the second metal ion, n2, is involved in the binding of ATP (Hunt and Ginsburg, 1980; Liaw et al., 1993).
In bacteria GS has been shown to be regulated by cumulative feedback inhibition, covalent modification, and repression/derepression (Stadtman, 1990). Although normally stable, bacterial GS is turned over when cells are starved for nitrogen (Fulks and Stadtman, 1985), suggesting that the intracellular level of GS in bacterial cells is also regulated by proteolysis. The degradation of GS in Escherichia coli and Klebsiella aerogenes appears to involve two steps: (a) the enzyme is inactivated by oxidative modification of a single His residue per subunit (Levine, 1983a; Rivett and Levine, 1990) and (b) the altered enzyme is then degraded by endogenous proteases that are capable of degrading the oxidized enzyme but exhibit little activity on native GS (Roseman and Levine, 1987; Stadtman and Berlett, 1997).
In plants GS is an octamer and has a native molecular mass of approximately 320 to 380 kD (Stewart et al., 1980). Conservation in the amino acid sequence in the active site of GS across kingdoms suggests that plant GS is mechanistically similar to bacterial GS (Shatters and Kahn, 1989; Sanangelantoni et al., 1990). There are, however, differences in the ATP-binding site within the active site between the GS in plants and that in bacterial GS (Kim and Rhee, 1988). It is generally believed that GS activity in plants is regulated at the transcriptional level, and most of the research on GS regulation has focused on this aspect (Hirel et al., 1987; Bennett et al., 1989; Forde et al., 1989; Walker and Coruzzi, 1989; Edwards et al., 1990; Cock et al., 1991, 1992; Miao et al., 1991; Roche et al., 1993; Sukanya et al., 1994; Temple et al., 1995). Very little is known about the regulation of plant GS at the level of translation, the assembly of holoenzyme, and enzyme turnover. However, it has been shown that GS in plants is not regulated by the adenylylation/deadenylylation cascade utilized by many Gram-negative bacteria (Tate and Meister, 1971). Recent work from our laboratory suggests that, aside from transcriptional regulation, GS activity in plants might be regulated at the level of enzyme assembly or turnover (Temple et al., 1993, 1996; Temple and Sengupta-Gopalan, 1997). To our knowledge, there have been no reports of how GS in plants is turned over; therefore, in this study we have made the initial step in understanding the mechanism of turnover of GS in plants by determining whether regulation by oxidative modification has a role.
The first step in protein oxidation requires the production of oxygen radicals. This process is mediated by several enzymatic and nonenzymatic systems. In plants oxygen radicals are generated during normal physiological processes such as photosynthetic electron transport, mitochondrial respiration, and nitrogen fixation (Allen, 1995; Dalton, 1995). Some enzymatic redox systems can also generate reactive oxygen species (Levine et al., 1981; Stadtman and Oliver, 1991; Harding et al., 1997; del Rio et al., 1998). The production of reactive oxygen species increases during physiological disorders that result from environmental stresses such as temperature changes, drought stress, and herbicide toxicity (Iturbe-Ormaetxe et al., 1998), from exposure to high radiance (Landgraf et al., 1997) or high levels of ozone (Pell et al., 1997), from defense against pathogens (Low and Merida, 1996), and during plant senescence (del Rio et al., 1998). The injury caused to plant tissues during environmental stresses is a result of an imbalance between the production of oxygen radicals and antioxidant defense responses (Foyer et al., 1994).
Nonenzymatic oxidase systems, including the ascorbate/metal/oxygen system (Levine et al., 1983b) and the mercaptan-mediated MCO system, in the presence of transition metal ions such as Fe3+ or Cu2+ (Rhee et al., 1990; Netto and Stadtman, 1996), are capable of generating ·OH radicals in vitro. In these systems a series of reactions take place in which the Fe3+ ion is reduced to Fe2+ with ascorbate or DTT as the reductants. The Fe2+ replaces Mn2+ at the n2 site of the GS. The hydrogen peroxide generated during the reduction of Fe3+ interacts with the Fe2+-GS complex and the Fe2+-peroxide complex and then dissociates into two reactive species, the ·OH radical and Fe-O (the ferryl ion) (Liaw et al., 1993; Netto and Stadtman, 1996). Both are extremely reactive and attack the side chains of amino acid residues proximal to the n2 metal-binding site (Liaw et al., 1993).
We demonstrate that, like the GS from E. coli, GS from soybean (Glycine max) roots can be oxidized in vitro with the MCO system, and the oxidized form is susceptible to degradation with proteases present in the plant root extract. We also present data suggesting that turnover of GS in vivo is mediated via an oxidation step and that in plants grown with exogenous nitrogen the GS is less susceptible to oxidative modification and proteolytic turnover.
MATERIALS AND METHODS
Plant Material
Soybean (Glycine max L. cv Williams) roots were obtained from 4-d-old seedlings germinated aseptically in aluminum trays. Roots were also obtained from 18- to 21-d-old plants that were planted in hydroponic culture vessels (Magenta, Chicago, IL) with vermiculite and grown in nutrient solution without nitrogen for 15 to 17 d; at this stage the nutrient solution was supplemented for 3 to 4 d before harvesting with one of the following: 10 mm KCl, 10 mm ammonium nitrate, or 0.1% hydrogen peroxide. Root tissues were frozen in liquid nitrogen and stored at −80°C until use.
Protein Extraction
All procedures were carried out at 4°C. Roots were ground in liquid nitrogen with 15% (w/w) insoluble polyvinylpolypyrrolidone and homogenized with 2 (old tissues) or 5 (young tissues) volumes of extraction buffer (50 mm Tris-Cl, pH 8.0, 5 mm EDTA, 5% [v/v] ethylene glycol, 20% [v/v] glycerol, 1 mm magnesium acetate, 1 mm DTT, and a mixture of protease inhibitors: 50 μg/mL antipain, 1 μg/mL cystatin, 10 μg/mL chymostatin, 2 μg/mL leupeptin, and 1 mm PMSF). The homogenate was centrifuged for 15 min at 20,000g.
For in vitro oxidation experiments, ammonium sulfate was added to the 4-d-old root extracts to a 30% to 70% saturation level. The protein precipitate was resuspended in 50 mm imidazole buffer, pH 7.4, and desalted in Sephadex G-25 spin-out columns against 50 mm imidazole, pH 7.4. For in vivo GS oxidation analysis, root extracts were desalted in Sephadex G-25 columns against a buffer containing 10 mm Tris-Cl, pH 8.0, 1 mm EDTA, 5% (v/v) ethylene glycol, 20% (v/v) glycerol, 1 mm magnesium acetate, 1 mm DTT, and a mixture of protease inhibitors: 50 μg/mL antipain, 1 μg/mL cystatin, 10 μg/mL chymostatin, 2 μg/mL leupeptin, and 1 mm PMSF.
Assay of Enzyme Activity and Protein Determination
Protein concentration was measured by the Bradford (1976) assay, using BSA as a standard. GS activity was measured spectrophotometrically at 500 nm by the transferase assay reported by Ferguson and Sims (1971). Transferase units were calculated from a standard curve of γ-glutamyl hydroxamate. One unit of transferase activity is equivalent to 1 μmol γ-glutamyl hydroxamate min−1 produced at 30°C. GS activity data presented are the averages of at least three independent experiments.
Oxidative Inactivation of GS
Inactivation of root GS was carried out by three different MCO systems, all modifications of the mercaptan-mediated MCO system described by Rhee et al. (1990). Samples of desalted 4-d-old root extract (30%–70% ammonium sulfate fraction) equivalent to 25 μg of protein were incubated in 50 μL of solution containing 50 mm imidazole, pH 7.4, different concentrations of FeCl3 (up to 1 mm), and 5 mm DTT, 10 mm GSH, or 20 mm ascorbate at 4°C. Incubation was performed with and without 1 mm EDTA. Samples were assayed for transferase activity after incubation. Results are presented as the percentages of activity compared with the control (no addition). Data are the averages of at least three independent experiments.
Analysis of the Stability of the GS Protein
Samples (30%–70% ammonium sulfate fraction) of desalted root extract from 4-d-old soybean plants were incubated in a MCO reaction containing 50 mm imidazole, pH 7.4, 0.125 mm FeCl3, 5 mm DTT, and 500 μg/mL root protein for 2 h at 4°C. The root extract for this experiment did not include any of the protease inhibitors. Incubation was performed with and without 1 mm EDTA. Samples were then incubated at 30°C to observe the stability of the GS activity and protein for the next 22 h. Aliquots were taken at several times for estimation of GS activity and electrophoretic analysis. In one set of experiments, an oxidized sample after 2 h of incubation at 4°C in the MCO system was passed through a Sephadex G-25 spin-out column equilibrated with 50 mm imidazole, pH 7.4, 5 mm DTT, and 1 mm EDTA to remove Fe3+ ions. This sample was then incubated at 30°C for the next 22 h, and the stability of the GS protein was monitored by one-dimensional SDS-PAGE and then by western blotting. This experiment was repeated several times, and the result of a typical experiment is presented here.
Electrophoretic Analysis of GS Polypeptides and Holoenzyme
Protein aliquots were taken at different times during the incubation in the MCO system. Samples were either boiled in 2% SDS or brought to 20% glycerol and placed in a −80°C freezer for electrophoretic analysis in denaturing and native conditions, respectively. One-dimensional PAGE was performed in a Mini-Protean II electrophoretic apparatus (Bio-Rad). Analysis of total protein patterns was performed by SDS-PAGE (Laemmli, 1970) using gradient PAGE gels from 7% to 15% in which a standard acrylamide:Bis-acrylamide solution was used. Gels were run at a constant 100 V. Proteins were visualized by silver staining (Morrissey, 1981).
For analysis of the GS protein the concentration of the crosslinker used was decreased to 1% of the total acrylamide solution for both native and SDS-PAGE to increase the separation of the GS1 isoforms in the gels. SDS-PAGE was run in 12% gels at constant voltage. Native gel electrophoresis was run at 4°C using 7.5% gels for 4 h at a constant 100 V.
GS was also analyzed by two-dimensional PAGE (O'Farrell, 1975) with the modifications previously described (Temple et al., 1996), except that 10 mm DTT replaced 2-mercaptoethanol in all cases. Protein samples were boiled in 2% SDS, 1 μg/μL urea was added to the sample, and the samples were allowed to dissolve at 37°C for 30 min before loading. Volumes equivalent to 0.035 transferase unit (transferase activity) were loaded in each tube gel, and the tube gels were extruded from the tubes and equilibrated for 30 min in 62.5 mm Tris-HCl, pH 6.8, 2% SDS, 10 mm DTT, and 10% glycerol before being subjected to SDS-PAGE. After electrophoresis, gels were electroblotted onto nitrocellulose, and the GS immunoreactive bands or spots were detected with antibodies raised against nodule GS (Cullimore et al., 1983). Native GS bands were detected with antibodies against the native enzyme (Lara et al., 1984). The blots were scanned, and the GS immunoreactive bands were quantified by using Intelligent Quantifier software (Bio-Image, Ann Arbor, MI). Experiments were repeated at least three times, and the results presented here represent a typical experiment. For the analysis of in vivo oxidation of GS, western blots from four different experiments were quantified. To compare between experiments, band intensities were standardized against the total GS intensity in the control samples (plants fed KCl).
Protection of GS from Oxidative Inactivation
Amino acid and GS substrates were analyzed for their role in preventing the oxidative inactivation of GS. Protection experiments were performed by the addition of amino acids, ATP, magnesium acetate, or MnCl2 at a concentration of 5 mm to desalted 4-d-old ammonium sulfate-precipitated root extracts before incubation in 100 μL of a MCO reaction (125 μmol FeCl3, 5 mm DTT, 5 mm protectant, and 50 μg of root protein in 50 mm imidazole, pH 7.4) with or without 1 mm EDTA. GS-transferase activity was assayed after 40 min of incubation at 4°C. At this time control samples with no protectants were inactivated by 40% to 60%. Inactivation of GS was calculated as the difference in GS activity between the control sample, in which 1 mm EDTA was included in the MCO reaction, and the samples incubated in the MCO system containing either the amino acids or the substrates indicated in Table II. Values are presented as the percentages of the GS activity lost relative to the control sample, in which no amino acid or GS substrate was added. Values of approximately 100% mean no protection, values lower than 100% mean some protection, and values higher than 100% mean an enhancement in the inactivation of GS.
Table II.
Effect of amino acids and substrates on the protection of GS inactivation by MCO
| Addition | GS Inactivation |
|---|---|
| % of control | |
| Control | 100% |
| Arg | 92.5 ± 1.9 |
| Cys | 45.6 ± 2.9 |
| Glu | 95.4 ± 1.5 |
| Gln | 69.6 ± 3.9 |
| GSH | 104.0 ± 2.8 |
| His | 55.7 ± 1.7 |
| Leu | 93.0 ± 3.3 |
| Lys | 108.2 ± 2.2 |
| Phe | 81.6 ± 1.1 |
| Pro | 87.5 ± 0.6 |
| Trp | 91.5 ± 1.6 |
| Tyr | 85.9 ± 5.2 |
| ATP | 53.6 ± 1.2 |
| Glu/ATP | 38.1 ± 2.0 |
| MgCl2 | 54.2 ± 3.7 |
| MnCl2 | 27.7 ± 4.3 |
A 4-d-old root extract (desalted ammoniun sulfate fraction) was incubated in 50 mm imidazol (pH 7.4) with 5 mm amino acids and GS substrates before the addition of a mixture of FeCl3 and DTT with or without EDTA for 40 min of incubation at 4°C for partial inactivation (Fig. 1). Samples were assayed for transferase activity following incubation in the MCO system. Inactivation was calculated as the difference between each sample and its respective control in which 1 mm EDTA was added. Values are the percentages of inactivation compared with the nonprotected sample. Values below 100% = protection from inactivation; values close to 100% = no protection. Data are the averages ± se of at least three independent experiments (n ≥ 3).
RNA Purification and Northern-Blot Analysis
Total RNA was purified by the LiCl-precipitation procedure (De Vries et al., 1982). RNA (20 μg) was fractionated on a 1.4% agarose/formaldehyde gel and blotted onto nitrocellulose. Probes were prepared from inserts isolated from clones: pGS100, coding for the cytoplasmic “housekeeping” isoform of alfalfa GS1 (Tischer et al., 1986), the 3′-untranslated region from the pGSGmD gene for GS1 from soybean (Roche et al., 1993), and a DNA clone for soybean 28S rRNA (a gift from Dr. F. Ausubel, Harvard University, Boston, MA). Standard hybridization conditions, including 50% formamide (Sambrook et al., 1989), were used. The filter was washed five times with 2× SSC and 0.5% SDS for 20 min at 42°C and exposed. The same filter was used for reprobing after the DNA probe was stripped by three washes in a boiling solution of 0.5% SDS. Exposed films were scanned and RNA-hybridization signals were quantified using Intelligent Quantifier software. Intensities were standardized for loading against the intensity of the 28S rRNA hybridization signal.
RESULTS
Inactivation of Soybean Root GS1 by the Iron/Ascorbate, Iron/GSH, and Iron/DTT Oxidation System
Our first objective was to demonstrate that GS enzyme from soybean roots is inactivated in vitro by active oxygen species. We used three different MCO systems with ascorbate, GSH, or DTT as the reductant and varying concentrations of FeCl3, and we checked for GS activity (Table I). Incubation of desalted ammonium sulfate-precipitated extracts of soybean roots was for 2 h at 4°C. Fe3+ alone at different concentrations had no effect on GS activity. The reductants by themselves appeared to promote enzyme activity to a small extent (20%–25%). DTT with 0.125 mm FeCl3 produced the highest level of inactivation (90%), whereas ascorbate and GSH required a higher concentration of FeCl3 (0.5–1 mm) for maximal inactivation (75% and 64%, respectively). Furthermore, of the three reductants used, DTT exerted the maximum inactivation (90%) after 2 h of incubation. Ascorbate and GSH, although less effective than DTT in inactivating GS, showed the maximum inactivation after 1 h of incubation (data not shown).
Table I.
In vitro inactivation of soybean root GS by different MCO systems
| Addition | Remaining GS Activity |
|---|---|
| % of control with EDTA | |
| Control (buffer only) | 100.0 |
| 1 mm FeCl3 | 99.63 ± 0.62 |
| 1 mm EDTA | 99.98 ± 0.17 |
| 1 mm FeCl3/1 mm EDTA | 101.89 ± 0.28 |
| 5 mm DTT | 120.34 ± 4.14 |
| 10 mm GSH | 125.53 ± 4.00 |
| 20 mm ascorbate | 122.01 ± 3.15 |
| 5 mm DTT/0.125 mm FeCl3 | 10.10 ± 0.39 |
| 5 mm DTT/0.125 mm FeCl3/1 mm EDTA | 100.59 ± 0.19 |
| 10 mm GSH/0.5 mm FeCl3 | 24.54 ± 0.12 |
| 10 mm GSH/0.5 mm FeCl3/1 mm EDTA | 98.90 ± 0.02 |
| 20 mm ascorbate/0.5 mm FeCl3 | 52.22 ± 0.62 |
| 20 mm ascorbate/1 mm FeCl3 | 36.45 ± 0.51 |
| 20 mm ascorbate/0.5 mm FeCl3/1 mm EDTA | 106.98 ± 0.69 |
A 4-d-old root extract (desalted ammoniun sulfate fraction) was incubated in 50 mm imidazol (pH 7.4) with FeCl3 (0.25–1 mm) and either DTT, GSH, or ascorbate in the presence or absence of 1 mm EDTA for 2 h at 4°C, and samples were assayed for transferase activity. Values are the averages ± se of at least three independent experiments and were calculated as the percentages of the remaining activity compared with the control with no addition (n ≥ 3).
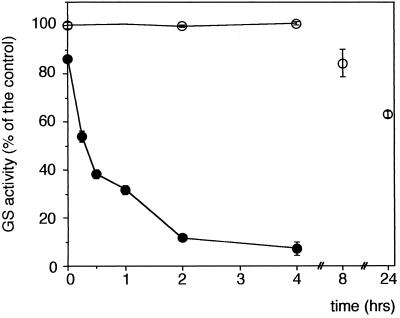
To demonstrate the involvement of the Fe3+-catalyzed oxidation reaction in the oxidation of GS, the desalted ammonium sulfate-precipitated root extracts were incubated with DTT and 0.125 mm FeCl3 in the presence or absence of 1 mm EDTA for different times at 4°C. Inactivation of GS in the absence of EDTA was observed within 30 min, and by 2 h there was a 90% loss in GS activity (Fig. 1). In contrast, there was no loss of GS activity in the presence of EDTA during the first 4 h of incubation at 4°C and only a 40% loss of GS activity after 24 h of incubation at 30°C.
Figure 1.
In vitro inactivation of soybean root GS by a MCO system. A 4-d-old root extract was incubated with an FeCl3 solution (0.125 mm FeCl3 and 50 mm imidazol, pH 7.4) and 5 mm DTT in the presence (○) or absence (•) of 1 mm EDTA for the times indicated. The incubation was at 4°C for the first 2 h and then at 30°C for the next 22 h. At each time the samples were assayed for transferase activity. Values are averages ± se from at least three independent experiments (n = 3–6).
Characterization of the Oxidatively Modified GS
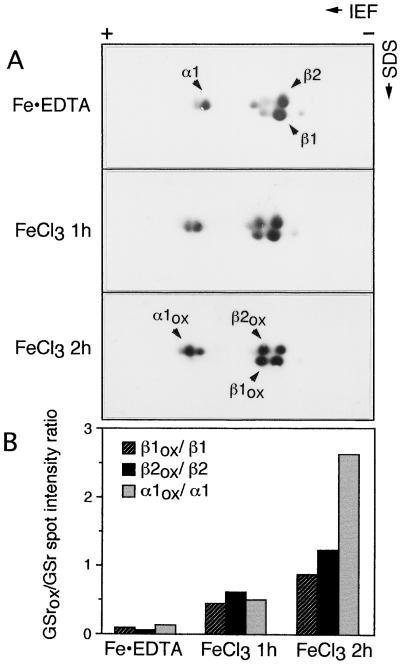
Aguirre and Hansberg (1986) showed that oxidized GS subunits from Neurospora crassa resolved on two-dimensional gels showed a more acidic pI compared with nonoxidized GS. To determine whether the oxidized form of GS from roots of soybean exhibit the same phenomenon, an ammonium sulfate-precipitated extract of 4-d-old soybean roots (after desalting) was incubated with DTT/FeCl3 (1 and 2 h at 4°C) in the presence or absence of EDTA, and samples were subjected to two-dimensional gel electrophoresis and then analyzed by immunoblot (Fig. 2). The GS polypeptides in the EDTA-treated samples resolved into three major subunits: β1, β2, and α1. The samples treated with the MCO system in the absence of EDTA showed an additional set of three GS polypeptides with slightly more acidic IEF values than β1, β2, and α1. The β1ox, β2ox, and α1ox polypeptides represent the oxidized forms of β1, β2, and α1, respectively. These polypeptides were also seen in the samples incubated in the MCO system in the presence of EDTA but at a significantly lower level than in samples that did not contain EDTA. The spots were all quantified using Intelligent Quantifier software, and the ratios of the intensity of the oxidized to the corresponding nonoxidized forms were calculated (Fig. 2B). The ratio for each form showed an increase in level as the time of incubation with the MCO system was increased from 1 to 2 h (Fig. 2). The ratio for the α1ox:α1 forms showed a dramatic increase after 2 h of incubation. The data suggest that the oxidized form of GS subunits can be differentiated from the nonoxidized form on two-dimensional gels and that the oxidized forms show an increase with time of incubation in the MCO system.
Figure 2.
Two-dimensional PAGE profile of soybean GS polypeptides from inactive (oxidized) and EDTA-protected GS enzyme. A, Root extract was incubated in the MCO system with or without EDTA for 1 and 2 h, and the samples were subjected to two-dimensional gel electrophoresis followed by immunodetection with anti-GS antibodies. The native GS polypeptides are labeled as β1, β2, and α1, and their modified versions are labeled as β1ox, β2ox, and α1ox, respectively. B, The spots were all quantified using Intelligent Quantifier software and the ratios of the intensity of the oxidized to the corresponding nonoxidized forms were calculated and plotted on a graph. The figure represents typical results.
Fate of Oxidized GS Polypeptides when Incubated with Root Extract
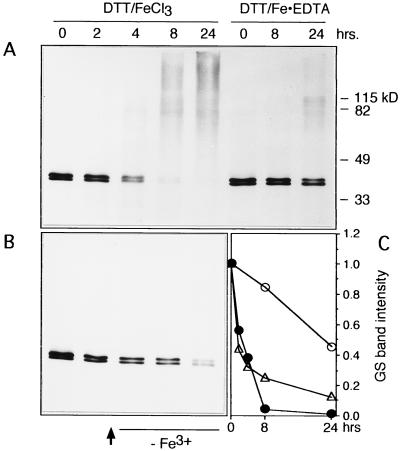
The GS from bacterial systems is known to go through a two-step turnover mechanism that includes an oxidation step followed by the degradation of the oxidized GS by endogenous proteases (Roseman and Levine, 1987). To determine whether the oxidized form of GS from the roots of soybean is more susceptible to endogenous proteases present in the roots, the ammonium sulfate-precipitated root extracts after desalting were incubated in the DTT/FeCl3 system in the presence or absence of EDTA for different times, and the extracts were subjected to SDS-PAGE analysis (Fig. 3A). The intensity of immunostained bands as measured by Intelligent Quantifier software was plotted (Fig. 3C). The two immunoreactive bands showed only a 15% decrease after 8 h of incubation and a 50% decrease after 24 h in samples incubated in the presence of EDTA, whereas in the absence of EDTA the decrease was almost 95% after 8 h. Concomitantly with the loss of GS polypeptides, the oxidized samples showed high-molecular-mass immunoreactive bands. Higher-molecular-mass proteins can be generated during free-radical reactions via cross-linking of peptides or via hydrophobic interactions among oxidized proteins (Cervera and Levine, 1988; Stadtman and Berlett, 1997).
Figure 3.
Stability of soybean root GS polypeptides when the oxidized and nonoxidized forms of the enzyme were incubated in the root extract. A, Four-day-old root extract incubated in the DTT-mediated MCO system in the presence (DTT/Fe·EDTA) or absence of EDTA (DTT/FeCl3). Samples were taken at the indicated times, and the reaction was terminated by the addition of SDS and boiling. The samples were subjected to SDS-PAGE followed by electroblotting and immunodetection of GS polypeptides using anti-GS antibodies. B, After incubation in the MCO system for 2 h, the extract was passed over a Sephadex G-25 spinout column equilibrated with 50 mm imidazole, pH 7.4, 10 mm DTT, and 1 mm EDTA to remove the Fe3+ ion and then incubated for the indicated times. The samples were then subjected to SDS-PAGE followed by electroblotting and immunodetection of GS polypeptides using anti-GS antibodies. C, Bands in A and B were quantified using Intelligent Quantifier software and plotted. ○, DTT/Fe·EDTA treatment; •, DTT/FeCl3 treatment; ▵, DTT/FeCl3 system after removal of Fe3+. The figure represents typical results.
In another experiment the root extract was incubated in the DTT/FeCl3 system for 2 h, followed by gel filtration on Sephadex G-25 to remove all Fe3+, and then incubated for an additional 22 h. The samples were analyzed by SDS-PAGE and then by immunoblotting; the GS polypeptides were still susceptible to degradation (Fig. 3B), suggesting that the proteolysis step does not require the presence of Fe3+ and that oxidative modification and proteolysis do not have to occur concurrently. Most significant is that the high-molecular-mass immunoreactive bands seen in the nondialyzed samples (Fig. 3A) were not detected in the desalted samples.
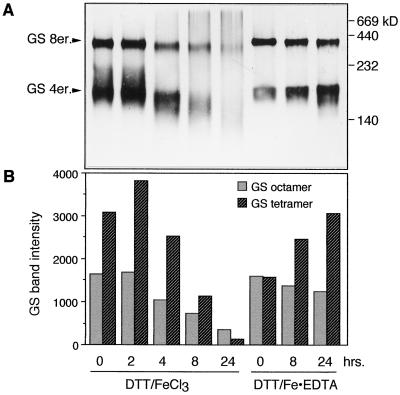
Effect of the MCO System on GS Holoenzyme Disassembly
To determine whether proteolysis of subunits was dependent or independent of holoenzyme disassembly, the extracts used in the previous experiment (Fig. 3A) were also subjected to native PAGE followed by activity staining or immunoblot analysis (Fig. 4A). Two immunoreactive bands were detected: a slower-migrating band and a faster-migrating, more diffused band. Based on standard molecular-mass markers included in the gel, we suggest that the faster-migrating band is the tetrameric form of GS, and the slower-migrating band is the octameric form. Activity staining of the native gels showed intense activity in the slower-migrating band in the samples incubated in the DTT/FeCl3 system in the presence of EDTA and a lower level of activity in the faster-migrating band. No activity was detected in the samples incubated in the DTT/FeCl3 system without EDTA (data not shown). The immunoreactive bands on native gels were quantified using Intelligent Quantifier software (Fig. 4B).
Figure 4.
Stability of soybean root GS holoenzyme in the native and the oxidized form incubated in root extract. The 4-d-old root extract was incubated in a DTT-mediated MCO system in the presence (DTT/Fe-EDTA) or absence of EDTA (DTT/FeCl3), and samples were removed at the indicated times. The reaction was terminated by the addition of glycerol to 20%, followed by immediate freezing. A, Results of native gel electrophoresis (at 4°C) followed by western blotting and immunodetection using anti-GS antibodies. 8er., Octamer; 4er., tetramer. B, Bands quantified using Intelligent Quantifier software and plotted. The figure represents typical results.
The 0-h time for samples incubated in the MCO system does not truly represent that time because the reaction was terminated only by the addition of glycerol (up to 20%) and placement of the sample in a −80°C freezer; therefore, there was a time lapse before the reaction was completely terminated, and the 0-h time more accurately represents an early time point. At the initial times of the experiment, the GS forms in the oxidized samples were more intensely stained than the nonoxidized sample (EDTA protected). This probably resulted from an increase in the hydrophobicity of the oxidized proteins. Moreover, the immunoreactive bands for both the octameric and tetrameric forms in the samples treated with the MCO system without EDTA were more intensely stained than the corresponding forms in the samples treated with EDTA. The intensity of immunostaining in the tetrameric and octameric forms was plotted for each time point to determine whether the tetramer was an intermediate in the turnover of the GS holoenzyme (Fig. 4B).
The tetramer:octamer ratio increased for the samples in the unprotected MCO system for the first 4 h, after which time the ratio decreased; in the EDTA-protected system the ratio showed an increase for the entire 24 h of incubation. A gradual loss of both immunoreactive bands was detected in the samples incubated in the DTT/FeCl3 system without EDTA after 2 h, and by about 24 h of incubation both bands were completely lost. In contrast, the sample incubated in the DTT/FeCl3 system in the presence of EDTA maintained both immunoreactive bands for the entire 24-h incubation period. However, the tetrameric form showed a gradual increase relative to the octameric form as the time of incubation in the MCO system was increased. This suggests that even in the protected system there is some loosening of interactions among the subunits and probably an increase in the hydrophobicity, resulting in an increase in the immunostaining of the tetramer as the time of incubation was increased.
Effect of Different Amino Acids and Substrates on MCO-Mediated Inactivation of GS
Oxidation of GS led to the rapid loss of enzymatic activity, suggesting that the modified site(s) was near the active site or in a region that affected the active site. In E. coli GS, the loss of activity was correlated with a loss of a single His and a single Arg residue per subunit (Farber and Levine, 1986; Climent and Levine, 1991; Liaw et al., 1993), situated at one of the two metal-binding sites on the enzyme (Yamashita et al., 1989). To determine whether the oxidation sites in the plant GS are similar to those of bacterial GS, we tested the effect of including some amino acids on the protection of the GS enzyme in a MCO system (Table II). Cys, Gln, and His showed the highest level of protection against inactivation, whereas Arg showed no protection. We also investigated the oxidative inactivation of GS in the presence of some of its substrates. As shown in Table II, ATP alone had a significant protective effect, and ATP plus Glu gave a still higher level of protection. Gln as a product had a higher protective activity, whereas Glu, which is the substrate, had no significant effect. Mg2+ and Mn2+ protect GS from inactivation, an indication of the role of metal ions in the generation of oxygen radicals. Both Mg2+ and Mn2+ protect the GS enzyme by competing with Fe2+ for the metal-binding sites on the enzyme. Mn2+ appeared to be a more potent inhibitor of inactivation than Mg2+, suggesting that the oxidative modification site on GS has a higher affinity for Mn2+ or that Mn2+ can act as an ·OH scavenger, reducing the amount of reactive free radicals (Stadtman and Oliver, 1991).
Effect of Hydrogen Peroxide and Nitrate Treatment on GS Activity and Level of GS Polypeptide and Transcript in Roots of Soybean
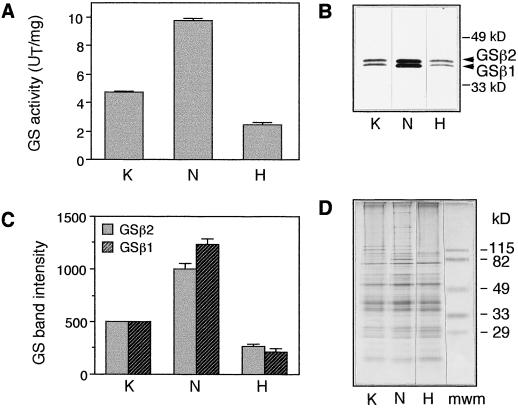
To determine whether oxidative modification of GS has any physiological relevance, the endogenous levels of oxygen radicals were increased by allowing soybean plants to take up hydrogen peroxide, and the effect of increased oxygen radicals on GS levels was measured. Plants (18–21 d old) were grown in hydrogen peroxide (0.1% or 29 mm) for 4 d, at which time visible symptoms in the form of necrotic lesions were seen in the leaves. The fact that Arabidopsis leaves treated with 10 mm hydrogen peroxide for 8 h were shown to have a 367% increase in the in vivo hydrogen peroxide levels compared with control (Rao et al., 1997), along with the visible symptoms of necrosis seen in the leaves of our hydrogen peroxide-treated soybean plants, suggests that growing soybean plants for 4 d in 29 mm hydrogen peroxide must have increased the endogenous hydrogen peroxide levels and therefore the level of oxygen radicals. GS activity was reduced by almost 50% in the hydrogen peroxide-treated roots compared with roots from untreated plants. Furthermore, in vitro inactivation studies have demonstrated that the GS enzyme from soybean roots is protected from MCO-mediated inactivation if the reaction is performed in the presence of either the substrate (ATP or Glu) or the product of GS activity (Gln). To determine whether this phenomenon holds true in vivo, roots of soybean plants that were treated with or without ammonium nitrate were analyzed for GS activity. Externally supplied nitrogen would eventually be converted into ammonia and then into Gln and Glu. Measurement of GS activity showed that there was an average 2-fold increase in the level of GS activity in the roots of plants that were fed ammonium nitrate compared with plants that were not fed nitrogen (Fig. 5A).
Figure 5.
Analysis of GS activity and polypeptide profile in roots of soybean plants treated with hydrogen peroxide and ammonium nitrate. Soybean plants (15–17 d after planting) were either fed 10 mm KCl (K) or 10 mm ammonium nitrate (N) or allowed to take up 0.1% hydrogen peroxide (H) for 4 d. The roots of the 19- to 21-d-old plants were harvested, and the soluble protein fraction was extracted. A, Average ± se (n ≥ 4) GS activity (transferase assay) from at least four independent experiments was calculated and the values were plotted. UT, Transferase units. B, Typical experiment in which 1 μg of the root-protein extract from the different treatments was subjected to SDS-PAGE followed by western blotting using anti-GS antibodies. The experiment was performed four times, and the results from a representative experiment are shown here. C, Immunoreactive bands (GSβ1 and GSβ2) were quantified using Intelligent Quantifier software, and the values were plotted. Values were standardized by using the total GS band intensity from the control KCl sample as the standard to compare between experiments. The average values ± se were plotted (n ≥ 4). D, Typical experiment in which 2.5 μg of the same root extracts were also subjected to SDS-PAGE in a 7% to 15% PAGE gradient gel, followed by silver staining. mwm, Standard molecular-mass markers included in the gel during electrophoresis.
To determine whether GS polypeptide levels are reflective of the GS activity levels in the hydrogen peroxide- and ammonium nitrate-treated plants, protein extracts from control roots, ammonium nitrate-treated roots, and hydrogen peroxide-treated roots were subjected to SDS-PAGE and then analyzed by western blotting using GS antibodies (Fig. 5B). The two immunoreactive bands (GSβ1 and GSβ2) were quantified using Intelligent Quantifier software, and the average values from the different experiments were plotted (Fig. 5C). In control roots the two GS1 polypeptide bands (GSβ1 and GSβ2) were found in equal amounts and both forms showed a dramatic increase after nitrate treatment; the GSβ1 form, however, showed a greater increase compared with that of the GSβ2 form. Both the GSβ1 and GSβ2 polypeptides showed decreased levels in the hydrogen peroxide-treated roots compared with control roots. The GS polypeptide levels in roots from the various treatments were truly reflective of the enzyme activity.
Protein extracts from control and treated roots were also subjected to SDS-PAGE and then silver staining to check for any other changes in the protein profile resulting from the treatment (Fig. 5D). The overall profile looked similar except that in the hydrogen peroxide-treated roots some of the higher-molecular-mass protein bands were not detectable. The roots fed nitrate also showed the presence of an 85-kD protein band, probably representing nitrate reductase, that was not seen in the control or hydrogen peroxide-treated roots.
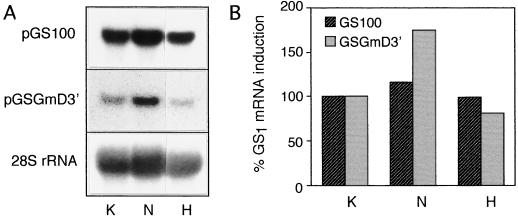
Total GS1 transcript levels were measured to determine whether changes in the GS polypeptide level associated with the uptake of hydrogen peroxide or treatment with ammonium nitrate were due to changes at the transcript level. Total RNA from the roots of control, hydrogen peroxide-treated, and ammonium nitrate-treated plants were subjected to RNA analysis using a conserved GS1 gene probe. The RNA loads were standardized by probing the blot with a probe for the 28S rRNA. Since the GSβ1 polypeptide showed a greater increase in roots treated with nitrate, the RNA blot was also probed with a probe specific for the gene encoding that polypeptide (pGSGmD3′; Roche, 1994). The hybridization signals were quantified using Intelligent Quantifier software, and the ratio of the signal with the two GS probes were standardized individually against the signal obtained with the rRNA probe. As shown in Figure 6, no appreciable difference in the level of total GS1 RNA was detected between control and hydrogen peroxide-treated roots. However, whereas the level of total GS1 transcript showed a 20% increase, the transcript for the specific gene corresponding to pGSGmD3′ showed a 75% increase in the ammonium nitrate-treated roots compared with the controls.
Figure 6.
Analysis of GS1 transcripts in the roots of soybean plants treated with hydrogen peroxide and ammonium nitrate. Soybean plants (18 d after planting) were either fed 10 mm KCl (K) or 10 mm ammonium nitrate (N) or allowed to take up 0.1% hydrogen peroxide (H) for 3 d. The roots of the 21-d-old plants were harvested, and the total RNA was isolated. The data from a representative experiment are shown here. A, RNA (20 μg) from each sample was then subjected to electrophoresis in a formaldehyde-agarose gel. The gel was blotted onto nitrocellulose and probed with the coding region of pGS100, the 3′-untranslated region of the ammonia-inducible gene (pGSGmD3′) or the 28S rRNA gene of soybean. B, Hybridization signals were quantified using Intelligent Quantifier software, and the values obtained with the two GS probes were standardized against the values obtained with the rRNA gene probe and plotted.
Taken together, our data suggest that the dramatic increase in GS1 polypeptides and activity in plants fed nitrate cannot be accounted for by just transcriptional regulation. There is specific induction of one gene member or subclass due to ammonium nitrate treatment at the transcriptional level, but, again, this was not adequate to account for the increase in the polypeptide level.
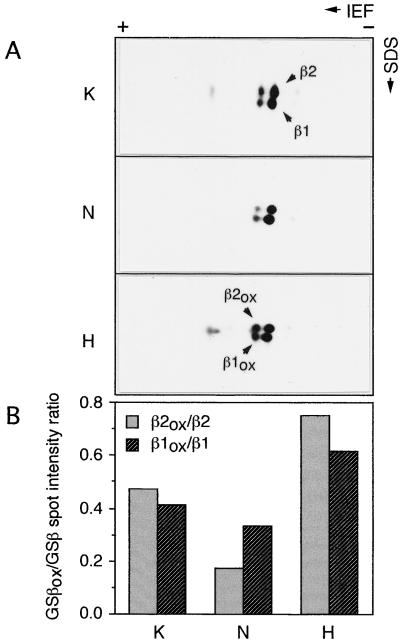
Effect of Feeding Hydrogen Peroxide and Ammonium Nitrate on the Oxidation State of the GS Polypeptides in the Roots of Soybean Plants
To implicate the oxidative modification step in the regulation of GS activity and enzyme levels associated with the hydrogen peroxide and ammonium nitrate treatments, root extracts from 21-d-old plants treated with water, hydrogen peroxide, or ammonium nitrate (see Methods) were subjected to two-dimensional SDS-PAGE followed by immunoblotting with GS antibodies (Fig. 7). The oxidized and nonoxidized forms of GS can be distinguished by their two-dimensional gel profile (Fig. 2). The GS1 subunits resolved into four major spots in all cases: β1, β2, β1ox, and β2ox (the latter two representing the oxidized forms of β1 and β2, respectively). The α1 spot seen in Figure 2 could not be detected in the control or the ammonium nitrate-treated samples because α1 is found only in young roots (K. Morey, J.L. Ortega, and C. Sengupta-Gopalan, unpublished data). However, α1 and α1ox were detectable in the hydrogen peroxide-treated samples, with a relatively higher level of the α1ox form. The spots were quantified using Intelligent Quantifier software, and the ratio of the signals in the β1 and β2 spots were compared with those of β1ox and β2ox, respectively, and the ratios were plotted (Fig. 7B). The level of the oxidized forms was higher in the hydrogen peroxide-treated roots than in the control sample. In the ammonium nitrate-treated samples, the β1 protein was more abundant than the β2 form, and, at the same time, the ratio of the oxidized form to the nonoxidized form for β2 was appreciably lower than in the KCl control sample, whereas there was a less appreciable difference (24%) in this ratio for the β1 form. These results suggest that hydrogen peroxide treatment promotes the oxidation of the two GS polypeptides, and ammonium nitrate treatment protects GS polypeptides from oxidative modification.
Figure 7.
Two-dimensional PAGE analysis of the GS polypeptides from soybean roots treated with either hydrogen peroxide or ammonium nitrate. Soybean plants (17 d after planting) were either fed 10 mm KCl (K) or 10 mm ammonium nitrate (N) or allowed to take up 0.1% hydrogen peroxide (H) for 4 d. The roots of the 21-d-old plants were harvested, and the soluble protein fraction was extracted. A, Protein equivalent to 0.035 units of GS activity from each sample was then subjected to two-dimensional PAGE followed by immunoblotting using the anti-GS antibodies. The nonoxidized GS polypeptides are labeled β1 and β2 and their modified versions are labeled β1ox and β2ox, respectively. B, Blots from A were all quantified using Intelligent Quantifier software and the ratio of the intensity of the oxidized to the corresponding nonoxidized forms were calculated and plotted on a graph. Results shown are from a typical experiment.
DISCUSSION
The data presented in this paper clearly demonstrate that GS from plants is subject to MCO in a manner similar to GS from E. coli (Nakamura and Stadtman, 1984), Bacillus subtilis (Kimura and Sugano, 1992), N. crassa (Aguirre and Hansberg, 1986), Anabaena variabilis (Martin et al., 1997), and Monoraphidium braunii (Humanes et al., 1995). Furthermore, as is the case with bacterial GS, the oxidized form of plant GS is more susceptible to degradation than the nonoxidized form (Roseman and Levine, 1987). In E. coli GS, the loss of catalytic activity due to oxidative modification correlated well with the loss of a single His (His-269) and a single Arg residue (Arg-344) per subunit, and these residues are situated in one of the metal-binding sites of the enzyme that is conserved among all GS enzymes from different sources (Farber and Levine, 1986; Climent and Levine, 1991; Liaw et al., 1993).
Whereas we could demonstrate the protection of the soybean root GS from MCO-mediated oxidation by incubating the enzyme with excess His, Arg had no effect on the protection of plant GS. Protein-engineering studies have shown that His-269 but not Arg-344 is crucial for bacterial GS activity (Liaw et al., 1993). It is likely that, in spite of the strong conservation of the Arg-344 residue in all types of GS, there might be subtle differences in the active site of plant and bacterial GS. The soybean root GS is also protected from the MCO-mediated oxidation by Cys. It has been suggested that protection by Cys is due to a redox effect on the oxidase system (Levine, 1983b), but this may not be the case because GSH does not show any protective effect on GS inactivation (Table I). GS sequence alignment has shown strong conservation of Cys residues among GSs from different kingdoms (Shatters and Kahn, 1989; Sanangelantoni et al., 1990; Pesole et al., 1991).
Oxidative modification of Cys residues may prevent the formation of an active conformation of GS. This would explain the effect of DTT, GSH, and ascorbate on the activation of GS (Table II). Exogenous His and Cys might protect against oxidation by simple competition with the corresponding amino acid residue of the enzyme. It is intriguing that, although Gln protects the GS from soybean root and B. subtilis from MCO attack, GS from E. coli (Roseman and Levine, 1987) and N. crassa (Aguirre and Hansberg, 1986) are not protected by Gln, again suggesting some subtle differences in the active site among the different types of GS. Furthermore, our data also suggest that, as in bacterial systems (Fulks and Stadtman, 1985), under low-nitrogen conditions plant GS is subject to turnover and the process is mediated by oxidative modification of the protein.
Oxidative modification caused a rapid loss of the catalytic activity of GS within minutes, and by 2 h the inactivation was more than 90%. In fact, inactivation appeared to be instantaneous, since a 10% decline in activity could be detected soon after incubation of the enzyme extract in the MCO system. However, the loss of activity did not appear to be accompanied by disassembly of the holoprotein, as evidenced by the results of native gel electrophoresis (Fig. 4). The two immunoreactive bands on native gels corresponding to the octameric and the tetrameric forms during the early times of incubation in the MCO system showed a migration pattern essentially similar to that of the protected samples. However, the more intense staining of the bands and the diffused nature of the faster-migrating band in the nonprotected samples compared with samples that were protected by the presence of EDTA (Fig. 4) suggest that there were slight conformational changes that occurred in the octameric and tetrameric form of the holoprotein as a result of oxidation of the metal-binding sites. The changes in conformation and surface hydrophobicity probably resulted in increasing the immunoreactivity of the holoprotein and changing the migration pattern of the proteins. In GS from B. subtilis conformational changes in the enzyme were demonstrated by electrophoresis, crystal diffraction spectral studies, and electron micrograph studies (Kimura and Sugano, 1992). It is interesting that GS from E. coli showed no change in the migration pattern in gels after oxidative inactivation (Kimura and Sugano, 1992). The presence of oxidized forms of the GS subunits seen in the samples incubated in the MCO system in the presence of EDTA (Fig. 2) suggests that GS oxidation/degradation may take place in vivo or during the extraction procedure.
The oxidative inactivation of GS was not quantitatively related to the appearance of the oxidized forms of the GS subunits. Therefore, after 2 h of incubation in the MCO system the enzyme activity was decreased to 10%; at this time only about 50% of the β1 and β2 polypeptides were in the oxidized form (Fig. 2), suggesting that one or a few oxidized monomers in the oligomer bring about a conformational change that renders the enzyme inactive. Alternatively, it could indicate negative cooperation in the oligomer, e.g. one or a few oxidized monomers in the oligomer bring about a conformational change that renders the enzyme less active but inaccessible to further oxidation. It is interesting that the different GS polypeptides showed different degrees of oxidation when exposed to the MCO system (Fig. 2), suggesting differences in accessibility to oxidative modification among the different GS polypeptides. GS polypeptides have been shown to assemble into holoenzymes with different catalytic activities: the synthetase:transferase ratio can vary (Bennett et al., 1989), as can the affinity for Mg2+ (Cullimore et al., 1983), suggesting that differences in the active sites and the affinity for divalent cations may result in differences in oxidation of the GS subunits.
Although inactivation was instantaneous following incubation in the MCO system, there was only about a 40% loss of the GS polypeptides after 2 h of incubation, suggesting that the quick loss in activity was due to conformational changes. Liaw et al. (1993) showed that the early modification in bacterial GS takes place at the n2 site, eliminating enzyme activity, and the later modification occurs at the n1 site, relaxing the GS structure and perhaps enabling proteolytic degradation. A gradual increase in the tetrameric form relative to the octameric form is seen during incubation in the MCO system in the presence or absence of EDTA, suggesting that disassembly of GS also takes place during the degradation process. The faster rate of loss of the GS polypeptides in the samples incubated in the MCO system over that incubated in the presence of EDTA suggests that the oxidized samples are more susceptible to proteolysis. In bacteria oxidation of GS holoenzyme increases its hydrophobicity, with a concomitant increase in its susceptibility to proteolysis (Cervera and Levine, 1988). Oxidized bacterial GS has been shown to be specifically degraded by an E. coli protease (Roseman and Levine, 1987) and by the proteasome complex from mammalian cells (Grune et al., 1997). Whether such a specific system of proteolysis that recognizes oxidized GS is in place in plant cells is not known. No immunoreactive peptides smaller than the authentic size GS peptides could be detected in any of the samples on SDS-PAGE, as has been detected for GS from E. coli and B. subtilis. This would suggest that either the proteases in soybean roots degrade the subunits completely to amino acids or the antibodies do not have affinity for the GS degradation products.
Our data strongly support the two-step mechanism involving oxidative modification for GS turnover in plants, and it appears that there are two modes of turnover: (a) conformational or hydrophobicity changes of the holoprotein followed by proteolysis and (b) disassembly followed by proteolysis. However, we have not ruled out the possibility that oxidative modification is the only route for GS turnover. It is interesting that, like the samples that had been inactivated by incubation in the MCO system, GS from samples incubated in the presence of EDTA also migrated as octamers, tetramers, and probably dimers and monomers (our gels were overrun). This observation has also been made with GS from other plant sources (Hopfner et al., 1988; Mack and Tischner, 1990). This would suggest that in vivo the octameric form of the GS holoenzyme is in equilibrium with lower-order oligomers in plants. We also cannot rule out the possibility that the lower-order oligomers are an artifact of extraction. GS from B. subtilis and E. coli migrates only as a dodecamer on native gels, suggesting that there may be some differences in the mechanism by which GS is turned over in plants compared with bacterial systems.
That oxidative modification is a marking step in the turnover of GS in plants is further supported by the fact that treatment with hydrogen peroxide, which would increase the production of ·OH radicals in the root cells, showed not only a decrease in enzyme activity but also a decrease in the GS polypeptides. Moreover, our data also showed that there was a dramatic increase in the level of the oxidized forms of GS subunits, suggesting that the oxidative modification may be an intermediate step in the turnover of GS in vivo. The gene for the α1 form was expressed only in the hydrogen peroxide-treated samples. The gene for the α1 form shows the highest homology to the gln-α gene of bean and therefore may be its ortholog (Morey, 1997). The gln-α gene of bean has been shown to be expressed in young roots and is induced by both biotic and abiotic stress (Watson and Cullimore, 1996). This may explain why the soybean α1 gene is expressed in young roots (Fig. 2) and induced in the stressed roots of hydrogen peroxide-treated plants (Fig. 7).
Furthermore, in accordance with the in vitro studies (Table II) demonstrating that the GS enzyme could be protected from MCO-mediated turnover by incubating the enzyme with either the substrates or the product of the reaction, GS activity in vivo could be greatly increased by feeding the plants ammonium nitrate. This enhancement was not just at the level of transcription or transcript stability but also at the level of polypeptide stability. Although there was induction of the ammonium nitrate-inducible GS1 subclass (Miao et al., 1991; Roche et al., 1993) at the level of transcription, the increase in the level of the transcript (approximately 1.5-fold) could not account for the 2.5-fold increase in the GS polypeptide corresponding to this gene and the more than 2-fold increase in the overall GS activity level. We attribute this to the protection of the GS holoenzyme from oxidative modification by the substrates of the enzyme. Two-dimensional gel analysis of the GS1 polypeptides showed that the oxidized form of the polypeptide β2 was 2- to 3-fold lower in the ammonium nitrate-fed plants than in the sample from plants that were not fed nitrogen. This suggests that in the presence of the substrate the active site of the enzyme is less accessible to oxidative modification or the production of oxygen radicals is enhanced under low-nitrogen conditions. The β1 form, however, showed only a moderate change in the ratio between the oxidized and nonoxidized form. It is possible that the increased synthesis of β1 polypeptides resulting from ammonium nitrate induction of the corresponding genes was accompanied by an increased turnover of the holoenzyme containing the β1 subunits.
The observations made in this paper may explain the results obtained by Hoelzle et al. (1992) that feeding nitrogen to nonnodulated soybean plants resulted in a significant increase in GS activity without affecting the overall GS1 polypeptide concentration. The increase in activity in the plants fed nitrogen was probably due to a lowering in the level of oxidatively modified GS subunits. On a similar note, we demonstrated that the nodule-specific GS isozyme is unstable in ineffective soybean nodules, probably because of the absence of fixed nitrogen (in the form of ammonia) in the infected cells of these ineffective nodules (Temple et al., 1996). Our findings from this study may also explain the discrepancy between GS activity and polypeptide level in dark-stressed bean nodules (Gogorcena et al., 1997).
The possibility that MCO modification of enzymes can be used for selective regulation of enzyme degradation is suggested by the demonstration that substrates of enzymes can protect them from oxidative modifications. Such metabolite effects could account for the differential responses of various enzyme levels to nutritional deficiencies. Thus, nitrogen starvation in bacterial cultures results in a decrease in the intracellular level of Glu and ATP, substrates that protect GS against oxidative modification and subsequent degradation. In the absence of the substrate an enzyme is biologically inactive and, therefore, its selective degradation can have little effect on its biological functions. However, by degradation it can yield amino acids needed for the synthesis of other proteins. In the same context, it can be argued that in plants the GS enzyme is always available to metabolize any ammonia that may be produced, thus avoiding toxic buildup of ammonia. It would follow that in plants the process of GS turnover and synthesis is a continuous process: When the substrate is limiting, the enzyme is turned over, and when the substrate is available, the enzyme is stabilized.
ACKNOWLEDGMENTS
We thank Stephen Temple and Tom Knight for critical reading of the manuscript.
Abbreviations:
- GS
Gln synthetase
- MCO
metal-catalyzed oxidation
Footnotes
This work was supported by U.S. Department of Agriculture grant no. 9237305-7941 and by the Agricultural Experiment Station at New Mexico State University, Las Cruces.
LITERATURE CITED
- Aguirre J, Hansberg W. Oxidation of Neurospora crassa glutamine synthetase. J Bacteriol. 1986;166:1040–1045. doi: 10.1128/jb.166.3.1040-1045.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RD. Dissection of oxidative stress tolerance using transgenic plants. Plant Physiol. 1995;107:1049–1050. doi: 10.1104/pp.107.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MJ, Lightfoot DA, Cullimore JV. cDNA sequence and differential expression of the gene encoding the glutamine synthetase γ polypeptide of Phaseolus vulgaris L. Plant Mol Biol. 1989;12:553–565. doi: 10.1007/BF00036969. [DOI] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cervera J, Levine RL. Modulation of the hydrophobicity of glutamine synthetase by mixed-function oxidation. FASEB J. 1988;2:2591–2595. doi: 10.1096/fasebj.2.10.2898411. [DOI] [PubMed] [Google Scholar]
- Climent I, Levine RL. Oxidation of the active site of glutamine synthetase: conversion of arginine-344 to γ-glutamyl semialdehyde. Arch Biochem Biophys. 1991;289:371–375. doi: 10.1016/0003-9861(91)90425-i. [DOI] [PubMed] [Google Scholar]
- Cock JM, Brock IW, Watson AT, Swarup R, Morby AP, Cullimore JV. Regulation of glutamine synthetase genes in leaves of Phaseolus vulgaris. Plant Mol Biol. 1991;17:761–771. doi: 10.1007/BF00037059. [DOI] [PubMed] [Google Scholar]
- Cock JM, Hermon P, Cullimore JV. Characterization of a gene encoding the plastid-located glutamine synthetase of Phaseolus vulgaris: regulation of β-glucuronidase gene fusions in transgenic tobacco. Plant Mol Biol. 1992;18:1141–1149. doi: 10.1007/BF00047717. [DOI] [PubMed] [Google Scholar]
- Cullimore JV, Lara M, Lea PJ, Miflin BJ. Purification and properties of two forms of glutamine synthetase from the plant fraction of Phaseolus root nodules. Planta. 1983;157:245–253. doi: 10.1007/BF00405189. [DOI] [PubMed] [Google Scholar]
- Dalton DA. Antioxidant defenses of plants and fungi. In: Ahmad S, editor. Oxidative Stress and Antioxidant Defenses in Biology. New York: Chapman & Hall; 1995. pp. 298–355. [Google Scholar]
- De Vries SC, Springer J, Wessels JGH. Diversity of abundant mRNA sequences and patterns of protein synthesis in etiolated and greened pea seedlings. Planta. 1982;156:120–135. doi: 10.1007/BF00395427. [DOI] [PubMed] [Google Scholar]
- del Rio LA, Pastori GM, Palma JM, Sandalio LM, Sevilla F, Corpas FJ, Jiménez A, López-Huertas E, Hernández JA. The activated oxygen: role of peroxisomes in senescence. Plant Physiol. 1998;116:1195–1200. doi: 10.1104/pp.116.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JW, Walker EL, Coruzzi GM. Cell-specific expression in transgenic plants reveals nonoverlapping roles for chloroplast and cytosolic glutamine synthetase. Proc Natl Acad Sci USA. 1990;87:3459–3463. doi: 10.1073/pnas.87.9.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber JM, Levine RL. Sequence of a peptide susceptible to mixed-function oxidation. Probable cation binding site in glutamine synthetase. J Biol Chem. 1986;261:4574–4578. [PubMed] [Google Scholar]
- Ferguson AR, Sims AP. Inactivation in vivo of glutamine synthetase and NAD-specific glutamate dehydrogenase. Its role in the regulation of glutamine synthesis in yeast. J Gen Microbiol. 1971;69:423–427. doi: 10.1099/00221287-69-3-423. [DOI] [PubMed] [Google Scholar]
- Forde BG, Day HM, Turton JF, Shen W-J, Cullimore JV, Oliver JE. Two glutamine synthetase genes from Phaseolus vulgaris L. display contrasting developmental and spatial patterns of expression in transgenic Lotus corniculatus plants. Plant Cell. 1989;1:391–401. doi: 10.1105/tpc.1.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Descourvieres P, Kunert KJ. Protection against oxygen radicals: an important defense mechanism studied in transgenic plants. Plant Cell Environ. 1994;17:507–523. [Google Scholar]
- Fulks RM, Stadtman ER. Regulation of glutamine synthetase, aspartokinase and total protein turnover in Klebsiella arogenes. Biochim Biophys Acta. 1985;843:214–229. doi: 10.1016/0304-4165(85)90142-4. [DOI] [PubMed] [Google Scholar]
- Gogorcena Y, Gordon AJ, Escuredo PR, Minchin FR, Witty JF, Moran JF, Becana M. N2 fixation, carbon metabolism, and oxidative damage in nodules of dark-stressed common bean plants. Plant Physiol. 1997;113:1193–1201. doi: 10.1104/pp.113.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grune T, Reinheckel T, Davies KJ. Degradation of oxidized proteins in mammalian cells. FASEB J. 1997;11:526–534. [PubMed] [Google Scholar]
- Harding SA, Oh S-H, Roberts DM. Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO J. 1997;16:1137–1144. doi: 10.1093/emboj/16.6.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel B, Bouet C, King B, Layzell D, Jacobs F, Verma DPS. Glutamine synthetase genes are regulated by ammonia provided externally or by symbiotic nitrogen fixation. EMBO J. 1987;6:1167–1171. doi: 10.1002/j.1460-2075.1987.tb02350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzle I, Finer JJ, McMullen MD, Streeter JG. Induction of glutamine synthetase activity in nonnodulated roots of Glycine max, Phaseolus vulgaris, and Pisum sativum. Plant Physiol. 1992;100:525–528. doi: 10.1104/pp.100.1.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner M, Reifferscheid G, Wild A. Molecular composition of glutamine synthetase of Sinapis alba L. Z Naturforsch. 1988;43C:194–198. [Google Scholar]
- Humanes L, Garcia-Fernandez JM, Lopez-Ruiz A, Diez J. Glutamine synthetase from the green alga Monoraphidium braunii is regulated by oxidative modification. Plant Sci. 1995;110:269–277. [Google Scholar]
- Hunt JB, Ginsburg A. Mn2+ and substrate interactions with glutamine synthetase from Escherichia coli. J Biol Chem. 1980;255:590–593. [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I, Escuredo PR, Arrese-Igor C, Becana M. Oxidative damage in pea plants exposed to water deficit or paraquat. Plant Physiol. 1998;116:173–181. [Google Scholar]
- Kim KH, Rhee SG. Sequence of peptides from Saccharomyces cerevisiae glutamine synthetase. N-terminal peptide and ATP-binding domain. J Biol Chem. 1988;263:833–838. [PubMed] [Google Scholar]
- Kimura K, Sugano S. Inactivation of Bacillus subtilis glutamine synthetase by metal-catalyzed oxidation. J Biochem. 1992;112:828–833. doi: 10.1093/oxfordjournals.jbchem.a123984. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;27:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Ohmann E, Tschiersch H. Light induced oxidative stress in Euglena gracilis. Photosynthetica. 1997;33:433–442. [Google Scholar]
- Lara M, Porta H, Padilla J, Folch J, Sánchez F. Heterogeneity of glutamine synthetase polypeptides in Phaseolus vulgaris L. Plant Physiol. 1984;76:1019–1023. doi: 10.1104/pp.76.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RL. Oxidative modification of glutamine synthetase. I. Inactivation is due to loss of one histidine residue. J Biol Chem. 1983a;258:11823–11827. [PubMed] [Google Scholar]
- Levine RL. Oxidative modification of glutamine synthetase. II. Characterization of the ascorbate model system. J Biol Chem. 1983b;258:11828–11833. [PubMed] [Google Scholar]
- Levine RL, Oliver CV, Fulks RM, Stadtman ER. Turnover of bacterial glutamine synthetase: oxidative inactivation precedes proteolysis. Proc Natl Acad Sci USA. 1981;78:2120–2124. doi: 10.1073/pnas.78.4.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw S-H, Villafranca JJ, Eisenberg D. A model for oxidative modification of glutamine synthetase, based on crystal structures of mutant H269N and the oxidized enzyme. Biochemistry. 1993;32:7999–8003. doi: 10.1021/bi00082a022. [DOI] [PubMed] [Google Scholar]
- Low PS, Merida JR. The oxidative burst in plant defense: function and signal transduction. Physiol Plant. 1996;96:533–542. [Google Scholar]
- Mack G, Tischner R. Glutamine synthetase oligomers and isoforms in sugarbeet (Beta vulgaris) Planta. 1990;181:10–17. doi: 10.1007/BF00202319. [DOI] [PubMed] [Google Scholar]
- Martin G, Haehnel W, Böger P. Oxidative inactivation of glutamine synthetase from the cyanobacterium Anabaena variabilis. J Bacteriol. 1997;179:730–734. doi: 10.1128/jb.179.3.730-734.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao GH, Hirel B, Marsolier MC, Redge RW, Verma DPS. Ammonia-regulated expression of a soybean gene encoding cytosolic glutamine synthetase in transgenic Lotus corniculatus. Plant Cell. 1991;3:11–22. doi: 10.1105/tpc.3.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey KJ (1997) Transcriptional regulation of the glutamine synthetase gene family in soybean: characterization of cis-acting elements involved in the differential expression of individual GS gene family members. PhD dissertation. New Mexico State University, Las Cruces
- Morrissey JH. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Stadtman ER. Oxidative inactivation of glutamine synthetase subunits. Proc Natl Acad Sci USA. 1984;81:2011–2015. doi: 10.1073/pnas.81.7.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netto LES, Stadtman ER. The iron-catalyzed oxidation of dithiothreitol is a biphasic process: hydrogen peroxide is involved in the initiation of a free radical chain of reactions. Arch Biochem Biophys. 1996;333:233–242. doi: 10.1006/abbi.1996.0386. [DOI] [PubMed] [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Pell EJ, Schlagnhaufer CD, Arteca RN. Ozone-induced oxidative stress: mechanism of action and reaction. Physiol Plant. 1997;100:264–273. [Google Scholar]
- Pesole G, Bozzetti MP, Lanave C, Preparata G, Saccone C. Glutamine synthetase gene evolution: a good molecular clock. Proc Natl Acad Sci USA. 1991;88:522–526. doi: 10.1073/pnas.88.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MV, Paliyath G, Ormod DP, Murr DP, Watkins CB. Influence of salicylic acid on hydrogen peroxide production, oxidative stress and hydrogen peroxide-metabolizing enzymes. Plant Physiol. 1997;115:137–149. doi: 10.1104/pp.115.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SG, Kim K, Kim IH, Stadtman ER. Protein that prevents mercaptan-mediated protein oxidation. Methods Enzymol. 1990;186:478–485. doi: 10.1016/0076-6879(90)86142-i. [DOI] [PubMed] [Google Scholar]
- Rivett AJ, Levine RL. Metal-catalyzed oxidation of Escherichia coli glutamine synthetase: correlation of structural and functional changes. Arch Biochem Biophys. 1990;278:26–34. doi: 10.1016/0003-9861(90)90226-o. [DOI] [PubMed] [Google Scholar]
- Roche D (1994) Regulation of glutamine synthetase activity in roots and nodules of soybeans—synthesis, assembly and turnover. PhD dissertation. New Mexico State University, Las Cruces
- Roche D, Temple SJ, Sengupta-Gopalan C. Two classes of differentially regulated glutamine synthetase genes are expressed in the soybean nodule: a nodule-specific class and a constitutively expressed class. Plant Mol Biol. 1993;22:971–983. doi: 10.1007/BF00028970. [DOI] [PubMed] [Google Scholar]
- Roseman JE, Levine RL. Purification of a protease from Escherichia coli with specificity for oxidized glutamine synthetase. J Biol Chem. 1987;262:2101–2110. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanangelantoni AM, Barbarini D, Di Pasquale G, Cammarano P, Tiboni O. Cloning and nucleotide sequence of an archaebacteria glutamine synthetase gene: phylogenetic implications. Mol Gen Genet. 1990;221:187–194. doi: 10.1007/BF00261719. [DOI] [PubMed] [Google Scholar]
- Shatters RG, Kahn ML. Glutamine synthetase II in Rhizobium: reexamination of the proposed horizontal transfer of DNA from eukaryotes to prokaryotes. J Mol Evol. 1989;29:422–428. doi: 10.1007/BF02602912. [DOI] [PubMed] [Google Scholar]
- Stadtman ER. Discovery of glutamine synthetase cascade. Methods Enzymol. 1990;182:793–809. doi: 10.1016/0076-6879(90)82062-7. [DOI] [PubMed] [Google Scholar]
- Stadtman ER, Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Chem Res Toxicol. 1997;10:485–494. doi: 10.1021/tx960133r. [DOI] [PubMed] [Google Scholar]
- Stadtman ER, Oliver CN. Metal-catalyzed oxidation of proteins. Physiological consequences. J Biol Chem. 1991;266:2005–2008. [PubMed] [Google Scholar]
- Stewart GR, Mann AF, Fentem PA (1980) Enzymes of glutamate formation: glutamate dehydrogenase, glutamine synthetase and glutamate synthase. In BJ Miflin, ed, The Biochemistry of Plants, Vol 5. Academic Press, New York, pp 271–327
- Sukanya R, Li M-G, Snustad DP. Root- and shoot-specific responses of individual glutamine synthetase genes of maize to nitrate and ammonium. Plant Mol Biol. 1994;26:1935–1946. doi: 10.1007/BF00019504. [DOI] [PubMed] [Google Scholar]
- Tate SS, Meister A. Regulation of rat liver glutamine synthetase: activation by alpha-ketoglutarate and inhibition by glycine, alanine, and carbamyl phosphate. Proc Natl Acad Sci USA. 1971;68:781–785. doi: 10.1073/pnas.68.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple SJ, Heard J, Ganter G, Dunn K, Sengupta-Gopalan C. Characterization of a nodule-enhanced glutamine synthetase from alfalfa: nucleotide sequence, in situ localization and transcript analysis. Mol Plant-Microbe Interact. 1995;8:218–227. doi: 10.1094/mpmi-8-0218. [DOI] [PubMed] [Google Scholar]
- Temple SJ, Knight TJ, Unkefer PJ, Sengupta-Gopalan C. Modulation of glutamine synthetase gene expression in tobacco by the introduction of an alfalfa glutamine synthetase gene in sense and antisense orientation: molecular and biochemical anaylsis. Mol Gen Genet. 1993;236:315–325. doi: 10.1007/BF00277128. [DOI] [PubMed] [Google Scholar]
- Temple SJ, Kunjibettu S, Roche D, Sengupta-Gopalan C. Total glutamine synthetase activity during soybean nodule development is controlled at the level of transcription and holoprotein turnover. Plant Physiol. 1996;112:1723–1733. doi: 10.1104/pp.112.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple SJ, Sengupta-Gopalan C (1997) Manipulating amino acid biosynthesis. In CH Foyer, WP Quick, eds, A Molecular Approach to Primary Metabolism in Higher Plants. Taylor & Francis, London, pp 155–177
- Tischer E, Das Sarma S, Goodman HM. Nucleotide sequence of an alfalfa glutamine synthetase gene. Mol Gen Genet. 1986;203:221–229. [Google Scholar]
- Villafranca JJ, Ranson SC, Gibbs EJ. Biophysical studies of Escherichia coli glutamine synthetase. Curr Top Cell Regul. 1985;26:207–219. doi: 10.1016/b978-0-12-152826-3.50023-1. [DOI] [PubMed] [Google Scholar]
- Walker EL, Coruzzi GM. Developmentally regulated expression of the gene family for cytosolic glutamine synthetase in Pisum sativum. Plant Physiol. 1989;91:702–708. doi: 10.1104/pp.91.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AT, Cullimore JV. Characterization of the expression of glutamine synthetase gln-α gene of Phaseolus vulgaris using promoter-reporter gene fusions in transgenic plants. Plant Sci. 1996;120:139–151. [Google Scholar]
- Yamashita MM, Almassey RJ, Janson CA, Cascio D, Eisenberg D. Refined atomic model of glutamine synthetase at 3.5 Å resolution. J Biol Chem. 1989;264:17681–17690. doi: 10.2210/pdb2gls/pdb. [DOI] [PubMed] [Google Scholar]