Abstract
Electrophile-mediated post-translational modifications (PTMs) are known to cause tissue toxicities and disease progression. These effects are mediated via site-specific modifications and structural disruptions associated with such modifications. 1,4-Benzoquinone (BQ) and its quinone-thioether metabolites are electrophiles that elicit their toxicity via protein arylation and the generation of reactive oxygen species. Site-specific BQ-lysine adducts are found on residues in cytochrome c that are necessary for protein-protein interactions, and these adducts contribute to interferences in its ability to facilitate apoptosome formation. To further characterize the structural and functional impact of these BQ-mediated PTMs, the original mixture of BQ-adducted cytochrome c was fractionated by liquid isoelectric focusing to provide various fractions of BQ-adducted cytochrome c species devoid of the native protein. The fractionation process separates samples based on their isoelectric point (pI), and because BQ adducts form predominantly on lysine residues, increased numbers of BQ adducts on cytochrome c correlate with a lower protein pI. Each fraction was analyzed for structural changes, and each was also assayed for the ability to support apoptosome-mediated activation of caspase-3. Circular dichroism revealed that several of the BQ-adducted cytochrome c species maintained a slightly more rigid structure in comparison to native cytochrome c. BQ-adducted cytochrome c also failed to activate caspase-3, with increasing numbers of BQ-lysine adducts corresponding to a greater inability to activate the apoptosome. In summary, the specific site of the BQ-lysine adducts, and the nature of the adduct, are important determinants of the subsequent structural changes to cytochrome c. In particular, adducts at sites necessary for protein-protein interactions interfere with the proapoptotic function of cytochrome c.
Keywords: apoptosome formation; 1,4-benzoquinone; cytochrome c; fractionation; liquid IEF separation; post-translational modification
Proteins are known targets of chemical-induced post-translational modifications (PTMs). Protein covalent binding of chemicals is closely associated with increases in tissue toxicities and disease progression (Bolton and Thatcher, 2008; Cohen et al., 1997; Liebler, 2008; Price and Knight, 2007). Exposure to xenobiotic-derived electrophilic metabolites can lead to protein adducts or oxidations, oxidative damage to lipids, and altered metabolism of endogenous compounds and to changes in protein subcellular localization and cellular signaling events (Demozay et al., 2008; Go et al., 2007; Wang et al., 2007). Certain reactive electrophilic metabolites stimulate the production of reactive oxygen species (ROS), leading to the formation of secondary “endogenous” electrophiles, such as 4-hydroxynonenal and other reactive products of lipid peroxidation. Both xenobiotic-derived electrophiles and those formed as endogenous by-products can modify proteins with important functional consequences (Carbone et al., 2005; Sampey et al., 2007).
Quinones are a large class of electrophilic xenobiotics containing electron-deficient electrophilic carbon centers that react with specific nucleophilic sites within proteins. Quinones are also capable of redox cycling and consequently of producing ROS and subsequent oxidative stress (Bolton et al., 2000; Pagano, 2002; Peters et al., 1996, 1997; Ross, 2000; Verrax et al., 2005). Many natural products contain the quinone function, and the quinone moiety is incorporated into the structure of many drugs (Kamal et al., 2007; Sumi and Kumagai, 2007). 1,4-Benzoquinone (BQ) is a reactive quinone, formed via the oxidation of benzene, a ubiquitous aromatic hydrocarbon and environmental pollutant produced from the combustion of natural products. BQ binds to various nucleophilic sites on proteins, including cysteine thiols, lysine amines, histidine imidazoles, and protein N-terminal amines, all of which are targets of many other reactive endogenous electrophiles (Koen et al., 2006; Person et al., 2003, 2005; Poli et al., 2008).
We have utilized cytochrome c as a model protein to develop mass spectroscopy–based methods, and associated software, for the identification of specific chemical-induced PTMs (Fisher et al., 2007; Person et al., 2003, 2005). Cytochrome c is essential for formation of the multiprotein apoptosome complex during apoptosis. Following release from mitochondria, cytochrome c binds to cytosolic apoptosis protease-activating factor 1 (Apaf-1), creating the apoptosome, with subsequent recruitment and activation of initiator caspase-9 and the effector caspases-3 and -7 (Bratton and Salvesen, 2010; Bratton et al., 2001). BQ adduction of specific lysine residues in cytochrome c produces changes in the protein sufficient to inhibit its ability to promote Apaf-1 oligomerization into an apoptosome complex (Fisher et al., 2007). However, caspase-3 activity was only partially suppressed in this system either because certain adducted forms of cytochrome c remained capable of supporting apoptosome formation or the presence of unadducted, native cytochrome c contributed to the residual ability to process and activate caspase-3.
To address this question, we now report methodology that facilitates the separation of adducted from unadducted cytochrome c. Circular dichroism (CD) data reveal that BQ-adducted cytochrome c exhibits a more rigid secondary structure. Moreover, increasing numbers of BQ-lysine adducts in cytochrome c correlate with a decreased capacity to activate caspase-3. By identifying site-specific modifications that influence protein structure, chemical-induced PTMs can be associated with changes in biological function.
MATERIALS AND METHODS
Materials.
All chemicals and reagents were of the highest purity available and were obtained from Sigma-Aldrich (St Louis, MO) unless otherwise indicated.
BQ Modification on Cytochrome c.
Horse heart cytochrome c was dissolved in 10 mM Tris-HCl, pH 7.5, at a concentration of 1 mg/ml. BQ was dissolved in methanol at 5 mg/ml. Cytochrome c was reacted with BQ at a molar ratio of 1:10 at room temperature for 30 min. The mixture was extracted with three volumes of ethyl acetate to remove excess BQ. The control and treated cytochrome c samples were spotted onto the matrix-assisted laser desorption ionization (MALDI) target, and whole protein spectra were acquired.
Liquid Isoelectric Focusing for Separation of BQ-Adducted Cytochrome c.
BQ-cytochrome c (500 μg) in 100 μl of 10 mM Tris-HCl was diluted into 2.75 ml of focusing solution (7 M urea, 2 M thiourea, 10 mM DTT, 2% CHAPS, and 6% Bio-Lyte 8/10), which was then loaded into the focusing chamber. 2-(N-morpholino)ethanesulfonic acid (6 ml 0.25 M) was added to the anode electrolyte chamber and 6 ml 0.1 M NaOH was added to the cathode electrolyte chamber. The MicroRotofor Liquid-Phase Isoelectric Focusing (IEF) Cell (Bio-Rad) was then run at 1 W constant power until both the voltage and amps came to a plateau (∼1 h). Fractions were subsequently removed from the focusing chamber and the pH was determined for each fraction. A portion of each fraction was subsequently processed using ZipTips (Millipore) and analyzed using MALDI-time of flight (TOF). These fractions were then purified to isolate adducted and unadducted proteins from the focusing chamber solution by acetone precipitation.
Matrix-Assisted Laser Desorption Ionization–Time of Flight.
Prior to MALDI-MS analysis, samples were desalted using a C18 0.6 ml ZipTip (Millipore, Bedford, MA), with 0.1% formic acid in water as the equilibration buffer and 1% formic acid in 50% acetonitrile as the elution buffer, following the manufacturer's protocol. MALDI-TOF spectra were taken on an Applied Biosystems Voyager DE-STR instrument with a 2-m flight path in the positive ion mode. The instrument was equipped with a nitrogen laser operating at 337 nm. A laser intensity of 1200 was utilized with a delay time of 900 ns. The original BQ-cytochrome c mixture and the BQ-cytochrome c fractions following liquid IEF separation were diluted 1:1 in α-cyano-4-hydroxycinnamic acid and 1 μl was drop-dried on a target plate. Whole protein spectra were acquired in linear mode over the mass range 8000–40,000 Da.
CD Spectroscopy.
CD measurements of IEF-separated cytochrome c fractions were taken. Unadducted cytochrome c (fraction 9 from liquid IEF separation) and two separate BQ-adducted cytochrome c fractions (fractions 4 and 6 with different BQ adduction profiles following liquid IEF separation) were taken at 3.2 μm on a Jasco-810 spectropolarimeter (Jasco, Easton, MD) in 100 μM Tris-HCl at pH 7.5 using a quartz cell of 1.0 mm optical path length over a wavelength range of 180–260 nm. Each CD signal represents the average of three scans at room temperature. The CD spectra were baseline corrected and the signal contributions due to the buffer were subtracted.
Molecular Modeling.
The x-ray crystal structure coordinates for the horse cytochrome c structure from the Protein Data Bank data entry (1HRC) were used as a starting model to build various BQ-adducted cytochrome c models (Bushnell et al., 1990). Molecular modeling studies were performed with Biopolymer module of Insight II modeling software (Insight II 2005, Accelrys, Inc). BQ cyclized diquinone (196 Da) and the BQ (105 Da) molecules were built from the fragment library and the BQ adducts were formed on the appropriate residues of the protein. These residues were determined by conducting liquid chromatography with electrospray-tandem mass spectrometry analysis on the IEF-separated fractions of the BQ-adducted cytochrome c. The charges were assigned using extensible systematic force field parameters (Discover 3.0 ESFF-extensible systematic force field). The modified structure was then subjected to 1000 steps of minimization using Discover 3.0. The complex structure was then soaked with a 10 Å layer of TIP3P water molecules (Jorgensen et al., 1983). This assembly was then subjected to 40 ps equilibration and 100 ps of dynamic simulations. Trajectories were collected at every 0.1 ps. The lowest potential energy structure was selected and then minimized using 3000 steps of minimization. The final minimized structure was then used for analysis. The BQ-adducted cytochrome c structures were viewed and manipulated using the PyMOL freeware (DeLano Scientific, LLC). The lowest potential energy conformation of the BQ-adducted cytochrome c, constructed using Insight II, was then placed in the PyMOL software program. Spatial rearrangement of critical residues within cytochrome c as a consequence of BQ adduction was assessed by overlaying the adducted model with a native model of cytochrome c.
Caspase-3 Processing and Activation.
The ability of cytochrome c molecules carrying increasing numbers of BQ adducts, and unadducted cytochrome c, to initiate formation of the Apaf-1 apoptosome, and sequential activation of caspases-9 and -3 in a purely recombinant system was assessed by measuring caspase-3 DEVDase activity, as previously described (Fisher et al., 2007; Malladi et al., 2009).
RESULTS
Cytochrome c-BQ IEF Separation Followed by MALDI-TOF Analysis
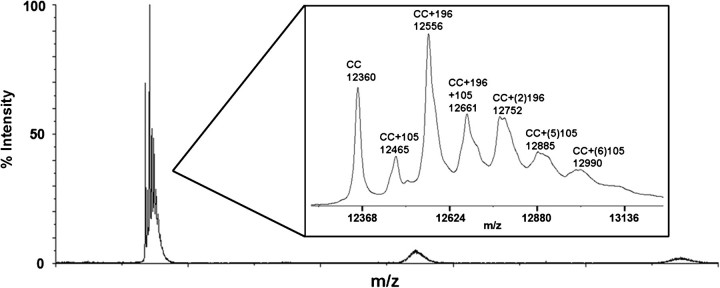
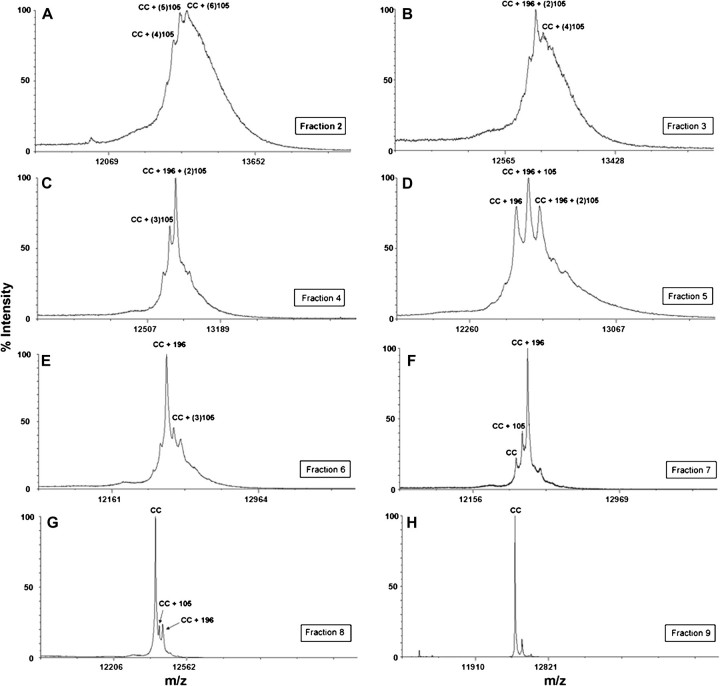
Cytochrome c was reacted with BQ as described in the Materials and Methods. MALDI-TOF analysis was conducted on this sample to confirm BQ adduction and the consequent adduction profile as a result of reaction with BQ (Fig. 1). This sample was then separated into various BQ-adducted cytochrome c species based on isoelectric point (pI). Because BQ adducts predominantly form on lysine residues on cytochrome c, the pI of the protein will decrease concomitant with increasing numbers of BQ adducts. This permits the separation of BQ-adducted cytochrome c from the native cytochrome c in the original reacted sample and for the isolation of cytochrome c species with increasing amounts of BQ adducts. The more BQ adducts present on cytochrome c, the lower the pI of the protein. Following liquid IEF separation, 10 fractions were collected, with increasing pH, from fraction 1 to fraction 10. Initial MALDI-TOF analysis was conducted on the different fractions to determine the BQ adduct profile of each (Fig. 2). The results reveal decreasing amounts of BQ adducts per molecule of cytochrome c from fraction 2 to fraction 9, with fraction 2 containing the most BQ adducts on cytochrome c and fraction 9 containing unadducted native cytochrome c (Table 1). Fractions 1 and 10 contained undetectable amounts of protein.
FIG. 1.
MALDI-TOF whole protein spectra for cytochrome c reacted with BQ. Cytochrome c was incubated in 10 mM Tris-HCl at pH 7.5 and then reacted with a 1:10 molar ratio of BQ. The resulting MALDI spectrum shows several BQ additions of 105 Da to cytochrome c as well as several BQ additions of 196 Da to cytochrome c. These BQ additions correspond to one addition at 12,465 m/z, two additions at 12,556 m/z, three additions at 12,661 m/z, four additions at 12,752 m/z, five additions at 12,885 m/z, and six additions at 12,990 m/z. The 196-Da additions correspond to a cyclized diquinone adduct as a result of postadduction chemistry following addition of two BQ to adjacent lysine residues of cytochrome c. Additionally, the MALDI analysis shows the native cytochrome c at 12,360 m/z. The inset is a magnified region of the spectrum. This original sample was then subjected to liquid IEF separation to allow for fractionation of the BQ-adducted species away from the native cytochrome c.
FIG. 2.
MALDI-TOF analysis of BQ-cytochrome c fractions following liquid IEF separation. Following reaction of cytochrome c with BQ and MicroRotofor separation, 10 fractions were collected. These fractions were purified using C18 packed tips (ZipTips) and then spotted on the MALDI target plate. The eight panels shown here represent the MALDI spectrum from each fraction containing protein were collected and identified. (A) Fraction 2 containing four to six BQ adducts, (B) fraction 3 containing four to five BQ adducts, (C) fraction 4 containing three to four BQ adducts, (D) fraction 5 containing two to four BQ adducts, (E) fraction 6 containing two to three BQ adducts, (F) fraction 7 containing one to two BQ adducts with small amounts of native cytochrome c present, (G) fraction 8 containing one to two BQ adducts, with large amounts of native cytochrome c present, and (H) corresponds to fraction 9 showing native cytochrome c with no BQ adducts.
TABLE 1.
IEF Separation and MALDI-TOF Analysis on Cytochrome c-BQ Adducts
| Fraction | Adduct description following MALDI-TOF analysis |
| 2 | 4, 5, and 6 BQ adducts, no native cytochrome c |
| 3 | 4 and 5 BQ adducts, no native cytochrome c |
| 4 | 3 and 4 BQ adducts, no native cytochrome c |
| 5 | 2, 3, and 4 BQ adducts, no native cytochrome c |
| 6 | 2 and 3 BQ adducts, no native cytochrome c |
| 7 | 1 and 2 BQ adduct(s), small amounts of native cytochrome c |
| 8 | 1 and 2 BQ adduct(s), mostly native cytochrome c |
| 9 | Native cytochrome c, no BQ adducts |
Note. Numbers in bold indicate most predominant BQ adduct formation in each fraction.
CD Spectroscopy of BQ-adducted Cytochrome c
Following the reaction of cytochrome c with BQ, and liquid IEF separation (Materials and Methods), far UV spectroscopy was used to establish whether the secondary structure of various BQ-adducted cytochrome c samples was significantly different than that of native cytochrome c. Similar structural analysis of BQ-adducted cytochrome c in a mixed population of unadducted protein species and BQ-adducted protein species has been performed in our laboratory. Those results indicated that the structural features of the mixed population of BQ-adducted cytochrome c and native cytochrome c species were quite similar to that of the native protein (Fisher et al., 2007). IEF-mediated separation of native cytochrome c from various BQ-adducted cytochrome c species permitted the structural analysis of the various BQ-adducted protein species without interference from native protein, providing a more accurate representation of the impact of BQ PTM on cytochrome c.
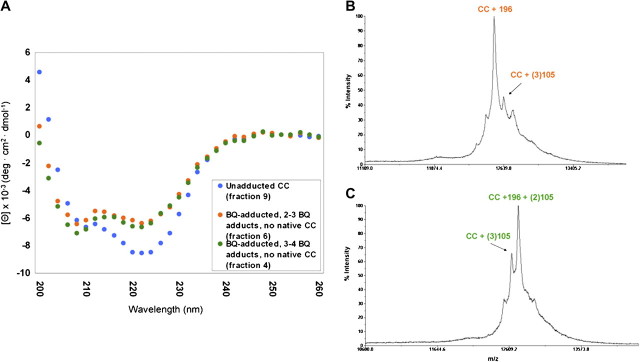
The CD spectra of BQ-adducted cytochrome c appear similar to that of native cytochrome c with respect to an intact secondary structure, although there appears to be more α-helical structure associated with BQ-adducted cytochrome c, as indicated by increased CD signals at 208 nm, a characteristic of α-helices (Fig. 3A). The samples used for CD analysis were analyzed via MALDI-TOF (as discussed above) to determine more precise modifications of cytochrome c by BQ. The two samples chosen for CD analysis were those that corresponded to a BQ adduct profile containing two or three BQ adducts per molecule of cytochrome c (fraction 6) (Fig. 3B) and a BQ adduct profile containing three or four BQ adducts (fraction 4) (Fig. 3C), with the most prominent adduct corresponding to that of the 196-Da diquinone BQ adduct in each case. Therefore, the CD results reveal that BQ-induced PTMs likely induce a more rigid cytochrome c structure, with a higher population of α-helical structures, which inhibits protein flexibility necessary for critical protein-protein interactions.
FIG. 3.
Far UV CD of BQ-adducted cytochrome c. (A) The CD spectra of the BQ-adducted cytochrome c with three to four BQ adducts (fraction 4) (green circles) and the BQ-adducted cytochrome c with two to three BQ adducts (fraction 6) (orange circles) are shown with the native cytochrome c structure (fraction 9) (blue circles). The BQ-adducted cytochrome c species maintain a more alpha-helical structure in comparison to the native cytochrome c. (B) MALDI-TOF spectrum of BQ-adducted cytochrome c with two and three BQ adducts (fraction 6), with the most prominent adduct as the 196-Da BQ adduct. (C) MALDI-TOF spectrum of BQ-adducted cytochrome c with three and four BQ adducts (fraction 4), again with the most predominant adduct being the 196-Da BQ adduct.
Predicted Protein Conformational Changes Associated with BQ Modification
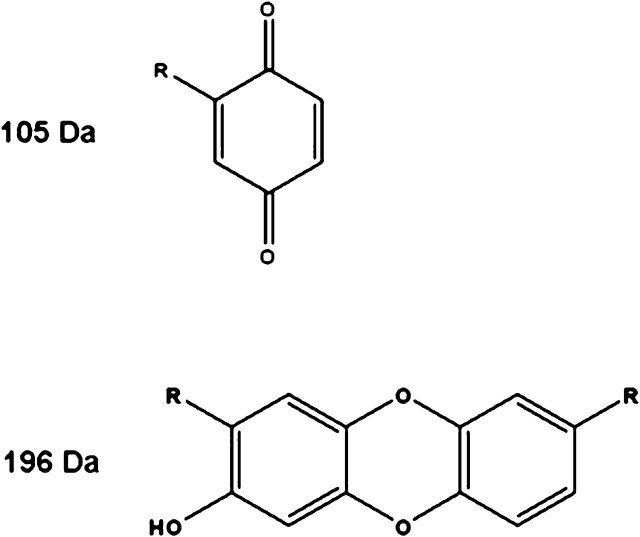
BQ may induce conformational changes in cytochrome c that alter residues involved in Apaf-1 binding, thereby inhibiting apoptosome formation. Site-directed mutagenesis studies have shown that cytochrome c forms complexes with Apaf-1 via 62ETLM65 and an annulus of lysine residues on K7, K25, K39, and K72 (Yu et al., 2001), and many of the BQ-mediated PTMs form on residues necessary for cytochrome c protein-protein interactions (Fisher et al., 2007) (Fig. 4). In addition to 196-Da mass modifications, BQ also gives rise to 105-Da mass modifications (Fisher et al., 2007; Person et al., 2003, 2005). As noted above, the 196-Da adduct represents a cyclized diquinone adduct formed via adduction of one BQ molecule on a lysine residue with the concomitant formation of a second BQ molecule on an adjacent lysine. The two BQ molecules subsequently form a cyclic product and oxidize to form a stable ring structure with a mass of 196 Da (Person et al., 2003). Collectively, our studies have provided solid evidence that the most targeted residues on cytochrome c are K25–K27 and K86–K87, where the cyclized diquinone adduct is formed with a 196-Da mass modification, and K7, K25, K39, K72, and K87 with a 105-Da mass modification (data not shown). These sites are the most consistent and most prevalent sites of adduction by BQ.
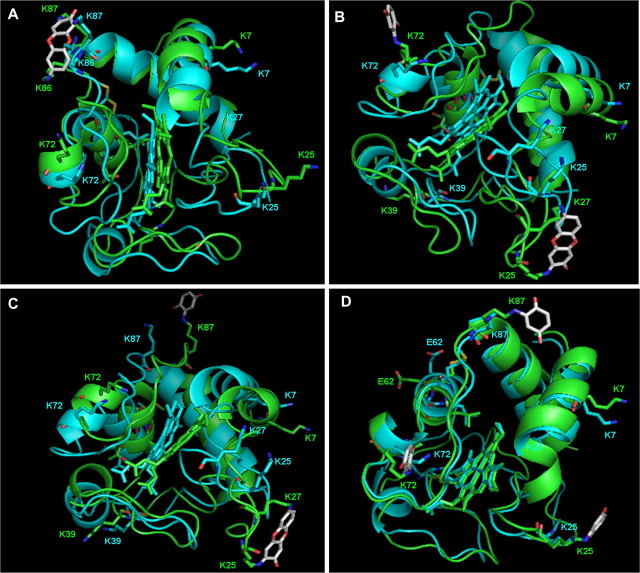
FIG. 4.
Structural modification of cytochrome c as a result of BQ adduction. The models were derived from the coordinates of the protein structure of Protein Data Bank entry 1HRC, and the software program Insight II was used to build the adducted proteins. Following molecular dynamics and energy minimizations, the lowest potential energy structure was placed in PyMOL. The models were built to reflect BQ-adducted cytochrome c following separation into various fractions with slightly different BQ adduction profiles. Mass spectral analysis has provided information corresponding to mass and site of BQ modification in each case. (A) BQ 196-Da adduct located on K86–K87 following reaction, where this is the addition of two BQ molecules to cytochrome c, followed by postadduction chemistry. (B) BQ 196-Da adduct located on K25–K27 and BQ 105-Da adduct located on K72 corresponding to three BQ additions to cytochrome c. (C) BQ 196-Da adduct located on K25–K27 and BQ 105-Da adduct located on K87 corresponding to three BQ additions to cytochrome c. These sites of modification are the most targeted sites of BQ modification on cytochrome c according to liquid chromatography with electrospray-tandem mass spectrometry analysis. (D) BQ 105-Da adducts located on K25, K87, and K72, respectively, corresponding to three BQ additions to cytochrome c. The modified protein (green) is overlaid with the native protein (blue) to observe spatial rearrangement of critical residues involved in cytochrome c function. The residues shown in the modified and native proteins are those crucial for cytochrome c binding to Apaf-1. These residues are colored by atom for clarification, where red corresponds to oxygen atoms and dark blue corresponds to nitrogen atoms within each residue. Following adduction with the BQ molecule, many of these residues adjusted for the addition(s). These models were built to complement CD structural analysis corresponding to two and three BQ modifications on cytochrome c that result in a slightly more rigid protein structure.
Because the CD data provided additional structural information following IEF separation of the BQ-adducted species, and the exact sites of BQ adduction are known from mass spectral analysis, we subsequently modeled several of these modifications to further analyze their structural impact on cytochrome c. The CD data were acquired from IEF-derived fractions 4 and 6, containing cytochrome c with two to four BQ adducts. Several models were constructed to mimic these BQ modifications on cytochrome c. With the cyclized BQ addition to K86–K87 on cytochrome c, the protein conformation changed to accommodate this modification, such that addition of cyclized BQ substantially modified the bond distance of the lysine residues (Fig. 4A). The residues in cytochrome c that interact with Apaf-1 are highlighted in this model, and following cyclic BQ adduction to K86–87, the specific residues involved in the interaction with Apaf-1 undergo spatial rearrangement. The model represents the addition of two BQ molecules to cytochrome c. Next, cytochrome c modeled at K25–K27 with the 196-Da BQ adduct, with an additional 105-Da BQ adduct on either K72 or K87 was investigated. Both these models represent the addition of three BQ molecules to cytochrome c (Figs. 4B–C). Finally, a model was built representing three single 105-Da BQ modifications on K25, K72, and K87, respectively (Fig. 4D). Both the sites and the chemical nature of the modifications have all been confirmed by mass spectral analysis and correspond to those fractions used for CD analysis. Following molecular dynamic simulations, there are obvious structural rearrangements of critical cytochrome c residues in each of these models, with the 196-Da BQ adducts inducing the most notable structural alterations. In each cytochrome c model, the BQ-adducted cytochrome c is overlayed with native cytochrome c to emphasize the structural differences.
Caspase Activation
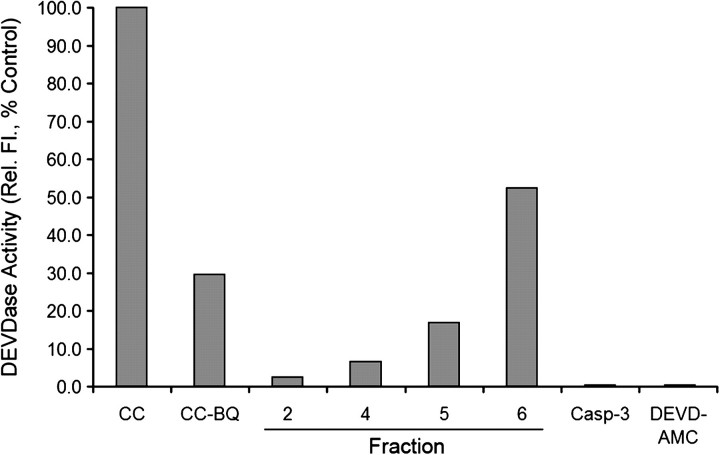
We finally examined the ability of IEF-separated fractions, containing varying degrees of BQ-adducted cytochrome c (fractions 2–6), as well as native cytochrome c, to initiate formation of the Apaf-1 apoptosome, as determined by the sequential activation of caspases-9 and -3. We have previously shown that cytochrome c samples containing a mixture of native and BQ-adducted cytochrome c exhibited a reduced capacity to activate the apoptosome, but some activity was observed. We surmised that removal of native cytochrome c from various BQ-adducted species should highlight the inability of modified cytochrome c to induce formation of the Apaf-1•caspase-9 apoptosome and activate caspase-3. The relationship between the quantity of BQ-modified amino acid residues within cytochrome c and the protein's ability to support caspase-3 activation was also determined. To assess the effects of BQ-adducted cytochrome c fractions, we reconstituted the apoptosome complex with recombinant Apaf-1, procaspase-9, procaspase-3, and either native cytochrome c or various BQ-adducted cytochrome c (fractions 2, 4, 5, and 6). As anticipated, native cytochrome c induced oligomerization of Apaf-1 (data not shown) and consequently activated caspases-9 and -3, resulting in caspase-3 DEVDase activity (Fig. 5). However, the various BQ-cytochrome c adducts failed in varying degrees to activate caspase-3. Fraction 6, which contained cytochrome c with a minimum (two to three) number of BQ adducts, was still capable of activating caspases, but only to about 50% of control levels (Fig. 5). Cytochrome c in fraction 5 contained between two and four BQ adducts and was significantly restricted in its ability to support caspase-3 activation (17% of controls). Finally, fractions 4 and 2, which contained cytochrome c with three to four or four to six BQ adducts, respectively, were essentially devoid of activity (7% and 2.5% of control values) (Fig. 5). Analysis of the original mixture of BQ-adducted cytochrome c, prior to IEF fractionation, supported caspase activation to a level 30% of control values (data not shown), which approximately represents the average of all the individual fractions assayed for caspase activity. In summary, the higher the number of BQ adducts that exist on cytochrome c, the lower the caspase-3 activity.
FIG. 5.
BQ-cytochrome c adduction prevents apoptosome formation. Apoptosome-mediated activation of procaspase-3 was measured using the caspase-3 substrate, N-acetyl-aspartate-glutamate-valine-aspartate-7-amino-4-methylcoumarin. Fractions 2, 4, 5, and 6, corresponding to four to six BQ adducts, three to four BQ adducts, two to four BQ adducts, and two to three BQ adducts, respectively, and also native cytochrome c were examined for their abilities to bind Apaf-1XL, induce formation of the apoptosome, and sequentially activate caspases-9 and -3. The data represent the average of two determinations and were plotted as % control compared with caspase-3 activation using native cytochrome c (also known as fraction 9).
DISCUSSION
Functional cytochrome c is necessary for initiation of stress-induced apoptosis, where it induces formation of a multiprotein apoptosome complex with Apaf-1, facilitated via many residues on the surface of cytochrome c, including K7, K25, K39, 62ETLM65, and K72 (Yu et al., 2001). BQ-induced PTMs on K7, K25, K39, E62, and K72 have been identified on these residues (Fisher et al., 2007), which are crucial contributors to protein-protein interactions. Consequently, their modification by BQ or 2-(N-acetylcystein-Syl)-1,4-benzoquinone caused a decrease in cytochrome c binding, Apaf-1 oligomerization, caspase-9 activation, and caspase-3 processing (Fisher et al., 2007). These initial studies utilized a mixture of cytochrome c molecules containing variations in the number of BQ-lysine adducts in addition to residual unadducted (native) cytochrome c. The presence of native cytochrome c, and perhaps adducted forms of cytochrome c that do not impact protein function, likely contributed to the residual ability of the reconstituted system to process and activate caspase-3. To address this question, we separated the various BQ-adducted cytochrome c species to determine the precise effects of BQ-lysine adducts on the ability of cytochrome c to support caspase-3 processing and activation.
A liquid IEF system was used to separate protein samples in their native state based on their pI (D'Amici et al., 2008; Sugiyama et al., 2006). Because BQ adducts form predominantly on lysine residues within cytochrome c, the pI of the protein progressively decreases concomitant with increases in BQ adduction. This facilitates separation of the various BQ-cytochrome c adducts into species that contain either many BQ adducts, fewer BQ adducts, or completely unadducted cytochrome c. Such differentially adducted cytochrome c fractions were then assayed for structural changes via CD analysis and for their ability to promote caspase activation. Studies using a mixture of BQ-adducted and native cytochrome c suggested that the secondary structure was identical to that of the native cytochrome c (Fisher et al., 2007). IEF fractions 4, 6, and 9, corresponding to three to four BQ adducts, two to three BQ adducts, and unadducted cytochrome c, respectively, were selected for CD analysis, which suggested that BQ-lysine adducts on cytochrome c create a more rigid protein structure than that of the native protein (Fig. 3). In particular, fraction 4, which contains cytochrome c molecules containing either three or four BQ-lysine adducts, exhibited a more disrupted CD spectra than fraction 6, which contains cytochrome c molecules containing only two or three BQ-lysine adducts. The most predominant BQ adduct is the 196-Da diquinone adduct, which represents an intramolecular cross-link between adjacent or neighboring lysines and likely contributes significantly to the resulting structural rigidity. Thus, the protein flexibility necessary to sustain cytochrome c protein-protein interactions may be compromised following BQ adduction, with consequent loss of protein function. Structural rigidity following adduction by quinones suggests one possible mechanism by which the cytochrome c protein-protein interactions are inhibited.
Protein modeling studies were conducted to gain additional structural insight regarding various BQ-adducted cytochrome c species. Mass spectral results indicated that the most predominant BQ adducts are those on K86–K87 and K25–K27 corresponding to the cyclized diquinone 196-Da adduct and those of the 105-Da adduct on K7, K25, K39, K72, and K87. Several models were built to correspond to the fractionated BQ-cytochrome c species, with models having two and/or three BQ adducts on the targeted residues described above. The most significant structural alterations are observed following 196-Da BQ adduction, and even more dramatic structural rearrangements occur following the addition of a 105-Da BQ adduct to the same cytochrome c molecule, resulting in a total of three BQ-lysine adducts on cytochrome c. For comparison, a separate model was built to predict the structural impact of three 105-Da BQ-lysine adducts, all on the same cytochrome c model. The structural alterations following these 105-BQ modifications do indeed rearrange critical residues within cytochrome c. However, the overall structural alterations appear to be less dramatic than those arising as a consequence of 196-Da adduct formation on cytochrome c. The 196-Da BQ adduct forms as a result of two BQ molecules adducting neighboring lysine residues, followed by postadduction chemistry linking the two molecules together (Fig. 6). The modeling studies therefore appear consistent with the CD data and confirm the likelihood that the 196-Da BQ adducts play a major role in altering the structural integrity of cytochrome c (Fig. 4).
FIG. 6.
Chemical structure of postulated BQ adducts on cytochrome c lysine residues. The 105-Da BQ results following Michael addition of the lysine residue to the BQ ring. The 196-Da BQ adduct is a result of two BQ molecules adducting neighboring lysine residues and subsequent postadduction chemistry linking the two BQ molecules together. In both structures, the R groups correspond to the lysine residues on cytochrome c. These electrophiles were detected as M-2 additions to the lysine residues, with unknown mechanisms.
The number of BQ-lysine adducts on cytochrome c also plays a critical role in influencing the structure and function of cytochrome c. These increases in the number of BQ-lysine adducts result in decreased protein flexibility (Fig. 3) and thus less potential for protein-protein interactions. Increases in the number of BQ-lysine adducts on cytochrome c also result in concomitant decreases in the ability of adducted cytochrome c to support caspase-3 activation (Fig. 5) via the inability to facilitate apoptosome formation (Fisher et al., 2007). The data confirm that site-specific chemical-induced PTMs impact the structure and function of susceptible proteins. Attempts to determine whether cytochrome c is a cellular target of hydroquinone/BQ are ongoing.
In summary, the electrophile BQ forms adducts with selective nucleophilic residues within cytochrome c, resulting in changes to protein structure and function. The structural and functional impact of chemical-induced PTMs may be masked in the presence of native protein. IEF-mediated separation of variously BQ-lysine adducted cytochrome c molecules and the removal of native protein permitted the direct examination of the effects of BQ-induced PTMs on cytochrome c. Model proteins such as cytochrome c will continue to provide guidance in developing strategies for identifying and predicting the effects of chemical-induced PTMs. Finally, molecular modeling provides further insight into possible structural and functional consequences resulting from site-specific PTMs on target proteins.
FUNDING
Grants GM070890 (S.S.L.), ES006694 (S.S.L.), ES007091 (A.A.F.), P30 ES007784 (S.B.B.), and CA129521 (S.B.B.).
Acknowledgments
The authors acknowledge the support of the P30 ES006694 Southwest Environmental Health Sciences Center in particular the Arizona Proteomics Consortium and the Molecular Modeling and Synthetic Chemistry Facility Core. We are also grateful to Dr Lawrence Hurley and Dr Danzhou Yang, Division of Medicinal Chemistry, for use and interpretation, respectively, of CD spectroscopy instrumentation and data. We would also like to thank Dr Vijay Gokhale, Division of Medicinal Chemistry, for assistance in molecular modeling.
References
- Bolton JL, Thatcher GR. Potential mechanisms of estrogen quinone carcinogenesis. Chem. Res. Toxicol. 2008;21:93–101. doi: 10.1021/tx700191p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton JL, Trush MA, Penning TM, Dryhurst G, Monks TJ. Role of quinones in toxicology. Chem. Res. Toxicol. 2000;13:135–160. doi: 10.1021/tx9902082. [DOI] [PubMed] [Google Scholar]
- Bratton SB, Salvesen GS. Regulation of the Apaf-1-caspase-9 apoptosome. J. Cell Sci. 2010;123:3209–3214. doi: 10.1242/jcs.073643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton SB, Walker G, Srinivasula SM, Sun X M, Butterworth M, Alnemri E S, Cohen G M. Recruitment, activation and retention of caspases-9 and -3 by Apaf-1 apoptosome and associated XIAP complexes. EMBO J. 2001;20:998–1009. doi: 10.1093/emboj/20.5.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell GW, Louie GV, Brayer GD. High-resolution three-dimensional structure of horse heart cytochrome c. J. Mol. Biol. 1990;214:585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- Carbone DL, Doorn JA, Kiebler Z, Petersen DR. Cysteine modification by lipid peroxidation products inhibits protein disulfide isomerase. Chem. Res. Toxicol. 2005;18:1324–1331. doi: 10.1021/tx050078z. [DOI] [PubMed] [Google Scholar]
- Cohen SD, Pumford NR, Khairallah EA, Boekelheide K, Pohl L R, Amouzadeh H R, Hinson J A. Selective protein covalent binding and target organ toxicity. Toxicol. Appl. Pharmacol. 1997;143:1–12. doi: 10.1006/taap.1996.8074. [DOI] [PubMed] [Google Scholar]
- D'Amici GM, Timperio AM, Zolla L. Coupling of native liquid phase isoelectrofocusing and blue native polyacrylamide gel electrophoresis: a potent tool for native membrane multiprotein complex separation. J. Proteome. Res. 2008;7:1326–1340. doi: 10.1021/pr700613r. [DOI] [PubMed] [Google Scholar]
- Demozay D, Mas JC, Rocchi S, Van Obberghen E. Faldh reverses the deleterious action of oxidative stress induced by the lipid peroxidation product, Hne, on insulin signaling in 3t3–L1 adipocytes. Diabetes. 2008;57:1216–1226. doi: 10.2337/db07-0389. [DOI] [PubMed] [Google Scholar]
- Discover 3.0 ESFF (extensible systematic force field). Molecular Mechanics Force Fields, Insight II 2005, Molecular Modeling Software. Accelrys, Inc., San Diego, CA. [Google Scholar]
- Fisher AA, Labenski MT, Malladi S, Gokhale V, Bowen M E, Milleron R S, Bratton S B, Monks T J, Lau S S. Quinone electrophiles selectively adduct “electrophile binding motifs” within cytochrome c. Biochemistry. 2007;46:11090–11100. doi: 10.1021/bi700613w. [DOI] [PubMed] [Google Scholar]
- Go YM, Halvey PJ, Hansen JM, Reed M, Pohl J, Jones DP. Reactive aldehyde modification of thioredoxin-1 activates early steps of inflammation and cell adhesion. Am. J. Pathol. 2007;171:1670–1681. doi: 10.2353/ajpath.2007.070218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insight II. Molecular Modeling Software. San Diego, CA: Accelrys Inc; 2005. [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- Kamal A, Ramu R, Tekumalla V, Khanna G B, Barkume M S, Juvekar A S, Zingde S M. Synthesis, DNA binding, and cytotoxicity studies of pyrrolo[2,1-c][1,4]benzodiazepine-anthraquinone conjugates. Bioorg. Med. Chem. 2007;15:6868–6875. doi: 10.1016/j.bmc.2007.08.026. [DOI] [PubMed] [Google Scholar]
- Koen YM, Yue W, Galeva NA, Williams TD, Hanzlik RP. Site-specific arylation of rat glutathione S-transferase A1 and A2 by bromobenzene metabolites in vivo. Chem. Res. Toxicol. 2006;19:1426–1434. doi: 10.1021/tx060142s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebler DC. Protein damage by reactive electrophiles: targets and consequences. Chem. Res. Toxicol. 2008;21:117–128. doi: 10.1021/tx700235t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malladi S, Challa-Malladi M, Fearnhead HO, Bratton SB. The Apaf-1*procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J. 2009;28:1916–1925. doi: 10.1038/emboj.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano G. Redox-modulated xenobiotic action and ROS formation: a mirror or a window? Hum. Exp. Toxicol. 2002;21:77–81. doi: 10.1191/0960327102ht214oa. [DOI] [PubMed] [Google Scholar]
- Person MD, Mason DE, Liebler DC, Monks TJ, Lau SS. Alkylation of cytochrome c by (glutathion-S-yl)-1,4-benzoquinone and iodoacetamide demonstrates compound-dependent site specificity. Chem. Res. Toxicol. 2005;18:41–50. doi: 10.1021/tx049873n. [DOI] [PubMed] [Google Scholar]
- Person MD, Monks TJ, Lau SS. An integrated approach to identifying chemically induced posttranslational modifications using comparative MALDI-MS and targeted HPLC-ESI-MS/MS. Chem. Res. Toxicol. 2003;16:598–608. doi: 10.1021/tx020109f. [DOI] [PubMed] [Google Scholar]
- Peters MM, Jones TW, Monks TJ, Lau SS. Cytotoxicity and cell-proliferation induced by the nephrocarcinogen hydroquinone and its nephrotoxic metabolite 2,3,5-(tris-glutathion-S-yl)hydroquinone. Carcinogenesis. 1997;18:2393–2401. doi: 10.1093/carcin/18.12.2393. [DOI] [PubMed] [Google Scholar]
- Peters MM, Rivera MI, Jones TW, Monks TJ, Lau SS. Glutathione conjugates of tert-butyl-hydroquinone, a metabolite of the urinary tract tumor promoter 3-tert-butyl-hydroxyanisole, are toxic to kidney and bladder. Cancer Res. 1996;56:1006–1011. [PubMed] [Google Scholar]
- Poli G, Biasi F, Leonarduzzi G. 4-Hydroxynonenal-protein adducts: a reliable biomarker of lipid oxidation in liver diseases. Mol. Aspects Med. 2008;29:67–71. doi: 10.1016/j.mam.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Price CL, Knight SC. Advanced glycation: a novel outlook on atherosclerosis. Curr. Pharm. Des. 2007;13:3681–3687. doi: 10.2174/138161207783018608. [DOI] [PubMed] [Google Scholar]
- Ross D. The role of metabolism and specific metabolites in benzene-induced toxicity: evidence and issues. J. Toxicol. Environ. Health A. 2000;61:357–372. doi: 10.1080/00984100050166361. [DOI] [PubMed] [Google Scholar]
- Sampey BP, Carbone DL, Doorn JA, Drechsel DA, Petersen DR. 4-Hydroxy-2-nonenal adduction of extracellular signal-regulated kinase (Erk) and the inhibition of hepatocyte Erk-Est-like protein-1-activating protein-1 signal transduction. Mol. Pharmacol. 2007;71:871–883. doi: 10.1124/mol.106.029686. [DOI] [PubMed] [Google Scholar]
- Sugiyama Y, Sueyoshi N, Kameshita I. Two-dimensional expression pattern analysis of protein kinases after separation by MicroRotofor/SDS-PAGE. Anal. Biochem. 2006;359:271–273. doi: 10.1016/j.ab.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Sumi D, Kumagai Y. Chemical biology of 1,2-naphthoquinone, a novel air pollutant that affects signal transduction pathways. Yakugaku Zasshi. 2007;127:1949–1956. doi: 10.1248/yakushi.127.1949. [DOI] [PubMed] [Google Scholar]
- Verrax J, Delvaux M, Beghein N, Taper H, Gallez B, Buc Calderon P. Enhancement of quinone redox cycling by ascorbate induces a caspase-3 independent cell death in human leukaemia cells. An in vitro comparative study. Free Radic. Res. 2005;39:649–657. doi: 10.1080/10715760500097906. [DOI] [PubMed] [Google Scholar]
- Wang G, Ansari GA, Khan MF. Involvement of lipid peroxidation-derived aldehyde-protein adducts in autoimmunity mediated by trichloroethene. J. Toxicol. Environ. Health A. 2007;70:1977–1985. doi: 10.1080/15287390701550888. [DOI] [PubMed] [Google Scholar]
- Yu T, Wang X, Purring-Koch C, Wei Y, McLendon GL. A mutational epitope for cytochrome C binding to the apoptosis protease activation factor-1. J. Biol. Chem. 2001;276:13034–13038. doi: 10.1074/jbc.M009773200. [DOI] [PubMed] [Google Scholar]