This study investigates the mechanisms underlying jasmonate-induced inhibition of primary root growth. Jasmonate inhibits the expression of two AP2-domain transcription factors, PLETHORA1 and 2, in a MYC2-dependent fashion. MYC2 is suggested to integrate the jasmonate and auxin pathways during the maintenance of the root stem cell niche.
Abstract
The root stem cell niche, which in the Arabidopsis thaliana root meristem is an area of four mitotically inactive quiescent cells (QCs) and the surrounding mitotically active stem cells, is critical for root development and growth. We report here that during jasmonate-induced inhibition of primary root growth, jasmonate reduces root meristem activity and leads to irregular QC division and columella stem cell differentiation. Consistently, jasmonate reduces the expression levels of the AP2-domain transcription factors PLETHORA1 (PLT1) and PLT2, which form a developmentally instructive protein gradient and mediate auxin-induced regulation of stem cell niche maintenance. Not surprisingly, the effects of jasmonate on root stem cell niche maintenance and PLT expression require the functioning of MYC2/JASMONATE INSENSITIVE1, a basic helix-loop-helix transcription factor that involves versatile aspects of jasmonate-regulated gene expression. Gel shift and chromatin immunoprecipitation experiments reveal that MYC2 directly binds the promoters of PLT1 and PLT2 and represses their expression. We propose that MYC2-mediated repression of PLT expression integrates jasmonate action into the auxin pathway in regulating root meristem activity and stem cell niche maintenance. This study illustrates a molecular framework for jasmonate-induced inhibition of root growth through interaction with the growth regulator auxin.
INTRODUCTION
Postembryonic root growth of higher plants is maintained by the root meristem, in which stem cells, including the mitotically inactive quiescent center (QC) and its surrounding stem cells, reside in a specialized microenvironment called the stem cell niche (Dolan et al., 1993; Watt and Hogan, 2000; Weigel and Jürgens, 2002; Laux, 2003; Aida et al., 2004). In Arabidopsis thaliana, the stem cell niche serves as the source of all differentiated cell types during root development (van den Berg et al., 1997; Dello Ioio et al., 2008). Although the structure of the primary root meristem is preestablished during embryo development, the meristematic activity can be altered by versatile developmental and environmental cues during postembryonic growth.
Several hormonal pathways are involved in the regulation of root growth, with auxin being the key player (Benjamins and Scheres, 2008; Benková and Hejátko, 2009; Santner et al., 2009; Vanneste and Friml, 2009). The whole process of root organogenesis, including initiation of the root pole (Friml et al., 2003), formation of the root stem cell niche (Sabatini et al., 1999; Blilou et al., 2005), maintenance of mitotic activity of the root meristem (Beemster and Baskin, 2000; Dello Ioio et al., 2007; Stepanova et al., 2008), and elongation and differentiation of cells leaving the root meristem (Rahman et al., 2007), has been demonstrated to be under the control of auxin, especially its featured gradient distribution (Tanaka et al., 2006; Benková and Hejátko, 2009; Petrásek and Friml, 2009). The PIN-FORMED (PIN) genes, which encode components of the auxin efflux machinery mediating polar auxin transport, are critical for the formation of a proper auxin distribution gradient and therefore direct root pattern formation and outgrowth (Blilou et al., 2005). It was proposed that the instructive auxin gradient should be translated into developmental information by molecular components. The auxin-inducible PLETHORA (PLT) genes, which encode the AP2 class of transcription factors that are essential for root stem cell niche patterning (Aida et al., 2004; Galinha et al., 2007), are good candidates for performing this translation (Benjamins and Scheres, 2008). Interestingly, the expression domain of PLT genes overlaps with the auxin maximum in the root. Recent work reveals an elegant interaction network of PINs and PLTs in regulating auxin-mediated root patterning and outgrowth; PIN proteins restrict PLT expression in the basal embryo region to initiate the root primordium. In turn, PLT genes maintain PIN transcription, which stabilizes the position of the root stem cell niche (Blilou et al., 2005; Grieneisen et al., 2007; Dinneny and Benfey, 2008).
Significant progress in our understanding of auxin signaling was achieved by the demonstration that the F-box protein TIR1 is an auxin receptor (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). Auxin binds to TIR1 and promotes the interaction of TIR1 with its auxin/indole-3-acetic acid (IAA) substrates. This interaction leads to the degradation of the auxin/IAA repressors by the 26S proteasome and therefore releases the transcriptional regulation activity of the AUXIN RESPONSE FACTOR proteins (Mockaitis and Estelle, 2008).
The jasmonate family of oxylipins, including jasmonic acid (JA) and its metabolites, play a well-established role in mediating defense responses and a wide range of developmental processes (Creelman and Mullet, 1997; Turner et al., 2002; Browse, 2005; Wasternack, 2007; Howe and Jander, 2008; Kazan and Manners, 2008). Among the first characterized physiological effects of jasmonate is growth inhibition (Dathe et al., 1981). It is proposed that JA-induced growth inhibition involves phase-specific disruption of cell cycle progression. Indeed, application of JA arrests synchronized tobacco (Nicotiana tabacum) BY-2 cells in both G1- and G2-phases (Swiatek et al., 2004). Whole-genome gene expression profiling of Arabidopsis cell cultures indicates that MeJA, the methyl ester of JA, represses the activation of M-phase genes and arrests the cell cycle in the G2-phase (Pauwels et al., 2008). Consistently, recent evidence suggests that wound-induced jasmonates stunt plant growth by inhibiting mitosis (Zhang and Turner, 2008). Even though the action mechanism governing JA-induced inhibition of plant growth remains elusive, characterization of JA-related mutants defective in JA-induced inhibition of root growth has significantly advanced our understanding of the molecular mechanism of JA-regulated gene expression (Browse, 2009). The coronatine insensitive1 (coi1) mutant is fully insensitive to JA both in terms of root growth inhibition and defense gene expression (Feys et al., 1994). COI1 encodes an F-box protein that interacts with SKP1 and CULLIN1 to assemble a functional SCFCOI1 ubiquitin ligase complex in vivo (Xie et al., 1998; Devoto et al., 2002; Xu et al., 2002). COI1, which shows high sequence homology with the auxin receptor TIR1, was shown recently to be the jasmonate receptor (Yan et al., 2009; Sheard et al., 2010). Compared with coi1, the jasmonate insensitive1 (jin1) (Berger et al., 1996; Lorenzo et al., 2004) mutant exhibits a relatively weak phenotype in JA-induced inhibition of root growth. JIN1 encodes a nuclear-localized basic helix-loop-helix–type transcription factor known as MYC2 (Boter et al., 2004; Lorenzo et al., 2004), which acts as both an activator and a repressor to regulate diverse aspects of JA-mediated gene expression (Dombrecht et al., 2007). A family of jasmonate ZIM domain (JAZ) proteins was identified as the target of the SCFCOI1 ubiquitin ligase complex in JA signaling (Chini et al., 2007; Thines et al., 2007). Recent structure-function studies indicated that the jasmonate receptor is a three-molecule complex consisting of COI1, JAZ transcriptional repressors, and inositol pentakisphosphate (Sheard et al., 2010). These studies together revealed that jasmonate and auxin show a similar signal perception and transduction paradigm in which F-box proteins (receptors) mediate the degradation of negative regulators (Mockaitis and Estelle, 2008). Jasmonoyl Ile, an active form of JA, promotes the degradation of JAZ proteins and, in turn, frees the transcriptional regulation activity of MYC2, the major transcription factor of jasmonate-mediated gene expression.
It has been shown that the jasmonate and auxin signaling pathways share important components in regulating root growth. For example, the first characterized auxin-resistant mutant, axr1 (Lincoln et al., 1990; Leyser et al., 1993), was later shown to be also resistant to JA in root growth (Tiryaki and Staswick, 2002). AXR1 encodes a subunit of the heterodimeric RUB-E1 enzyme, which is important for the modification of CUL1, a shared component of SCFTIR1 and SCFCOI1, demonstrating that the auxin and jasmonate proteasome pathways are directly connected through AXR1 (Tiryaki and Staswick, 2002). Recently, it was reported that JAZs interact with Novel Interactor of JAZ to recruit TOPLESS as a corepressor to repress JAZ-targeted transcription factors (Pauwels et al., 2010). Because TOPLESS also mediates auxin-dependent transcriptional repression (Szemenyei et al., 2008), these results provide another line of evidence for jasmonate/auxin interaction at the level of transcriptional repression.
In this study, we investigate the cellular and molecular mechanisms of jasmonate action on the inhibition of primary root growth. We show that JA modulates the cellular organization of the stem cell niche by promoting QC division and columella stem cell (CSC) differentiation. We also show that JA represses the expression of PLT1 and PLT2, and this effect of JA is executed by direct binding of MYC2 to the promoters of the PLT genes. Our results reveal that MYC2-mediated repression of PLT expression is involved in the long-standing observation that jasmonate inhibits primary root growth in Arabidopsis. PLT1 and PLT2 are key interaction nodes between jasmonate and auxin in the regulation of root stem cell niche maintenance and meristem activity.
RESULTS
JA-Induced Inhibition of Primary Root Growth Involves a Reduction of Root Meristem Activity
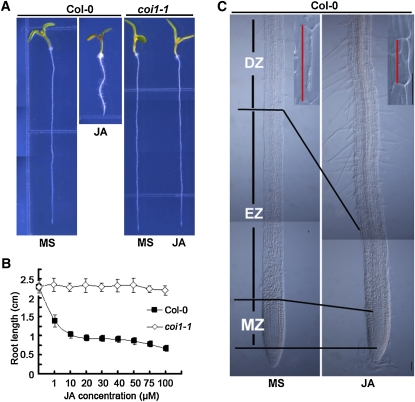
To understand how jasmonate inhibits root growth, we measured primary root length of Arabidopsis seedlings by germinating seeds in medium containing different concentrations of JA. As expected, root growth of wild-type plants was inhibited by JA in a dose-dependent manner, while root growth of the JA-insensitive coi1-1 mutants was largely unaffected by JA (Figures 1A and 1B). We then investigated the JA-induced cellular changes in the three morphologically distinguishable developmental zones along the longitudinal axis of the root: the differentiation zone (DZ), the elongation zone (EZ), and the meristem zone (MZ). As shown in Figure 1C, JA decreased the final cell length of the DZ and reduced the size of the EZ as well as the size of the MZ. Closer observation of the DZ and EZ of JA-treated roots indicated that JA reduced both cell number and cell length of these regions (see Supplemental Figure 1 online), indicating that JA reduces both cell proliferation and cell elongation, two basic cellular processes affecting primary root growth (Scheres et al., 2002).
Figure 1.
JA Represses Root Growth through Inhibition of both Cell Proliferation and Cell Elongation.
(A) Six-day-old seedlings of the wild type (Col-0) and coi1-1 were grown on medium without (MS) or with 20 μM JA.
(B) JA-mediated inhibition of root growth in Col-0 and coi1-1. Col-0 and coi1-1 seeds were germinated on medium containing different concentrations of JA, and seedling root length was measured at 6 d after germination. The effects of JA on Col-0 and coi1-1 were significantly different. Data shown are average and sd (n > 20) and are representative of at least three independent experiments.
(C) JA reduces the MZ, the EZ, and epidermal cell length of the DZ. A representative image of 6-d-old Col-0 seedlings grown in the absence or presence of JA. Insets show that JA reduces cell length (marked with red lines) of Col-0 epidermal cells in the DZ. Bars = 50 μm.
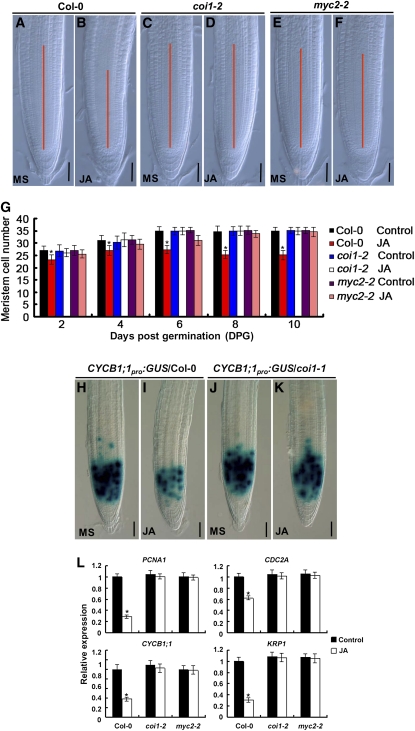
We then focused on the cellular and molecular mechanisms underlying the JA-induced reduction of root meristem size. The effect of JA on root meristem size was evaluated by determining the number of cortical cells in the region from the QC to the first elongated cell (Casamitjana-Martínez et al., 2003; Dello Ioio et al., 2007). As shown in Figures 2A to 2G, JA effectively reduced the root meristem cell number in the wild type, and this effect was substantially reduced in coi1-2 (Xu et al., 2002) and myc2-2 (Boter et al., 2004), indicating that JA reduces the number of the transit amplifying cells in a COI1- and MYC2-dependent manner. The reduction of root meristem cell number by JA could result from its negative effect on cell division. To test this idea, we monitored the expression of the transcriptional fusion reporter CYCB1;1pro:GUS (for β-glucuronidase), a widely used marker to indicate G2/M-phase of the cell cycle (Colón-Carmona et al., 1999). Without JA treatment, CYCB1;1pro:GUS was expressed in the actively dividing cells of the root meristem (Figures 2H and 2J). JA treatment markedly reduced the expression of CYCB1;1pro:GUS in the wild type (Figure 2I) but not in coi1-1 (Figure 2K), suggesting that the JA signaling pathway represses the cell division activity of the transit amplifying cells. It is noteworthy that the effect of JA on CYCB1;1pro:GUS expression was different from that of ethylene, which exerted little effect on CYCB1;1pro:GUS expression (Růzicka et al., 2007). Consistent with JA-induced reduction of CYCB1;1pro:GUS expression, our quantitative RT-PCR (qRT-PCR) assays revealed that exogenous JA markedly reduces the expression levels of several cell cycle–related genes, including CYCB1;1 (Fuerst et al., 1996), CDC2A (Martinez et al., 1992), KRP1 (Wang et al., 1997), and PCNA1 (Egelkrout et al., 2002) (Figure 2L). It is noteworthy that JA failed to repress the expression of cell cycle–related genes in coi1-2 and myc2-2 (Figure 2L), indicating that JA negatively regulates cell division activity of the root meristem through COI1 and MYC2. Consistently, using transgenic plants containing gene promoter and GUS fusions, we found that COI1pro:GUS and MYC2pro:GUS are richly expressed in the root meristem (see Supplemental Figures 2A to 2D online).
Figure 2.
JA Reduces Cell Division Activity of the Root Meristem in the Wild Type but Not in coi1-2 and myc2-2.
(A) to (F) Root meristems of 6-d-old Col-0, coi1-2, and myc2-2 seedlings grown on medium without (MS) or with 20 μM JA. The meristem zone was marked with red line. Bars = 50 μm.
(G) JA-induced reduction of root meristem cell number in Col-0, coi1-2, and myc2-2. Seedlings of the indicated genotypes were grown on medium without (MS) or with 20 μM JA, and cell number in the root meristem was examined at a 2-d interval. Data shown are average and sd (n = 20). Asterisks denote Student’s t test significance compared with untreated plants: *P < 0.05.
(H) to (K) JA-induced reduction of CYCB1;1pro:GUS expression in Col-0 and coi1-1. Five-day-old seedlings germinated on control medium were transferred to medium without (MS) or with 20 μM JA for 1 d, and the CYCB1;1pro:GUS expression was monitored. Bars = 50 μm.
(L) JA-regulated expression of cell cycle–related genes in Col-0, coi1-2, and myc2-2. Five-day-old Col-0 seedlings germinated on control medium were transferred to liquid medium without (control) or with 20 μM JA for 6 h, and 2-mm root tips were harvested for RNA extraction and qRT-PCR analysis. Transcript levels of cell cycle–related genes were normalized to ACTIN2 expression. The transcript levels of the indicated genes in Col-0 without JA treatment were arbitrarily set to 1. Error bars represent the sd of triplicate reactions of independent RNA samples prepared from three batches of Arabidopsis roots. Asterisks denote Student’s t test significance compared with untreated plants: *P < 0.05.
JA Promotes QC Division in a COI1- and MYC2-Dependent Manner
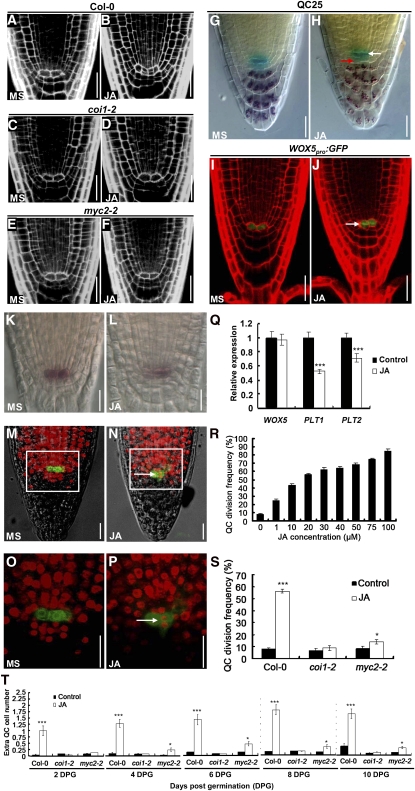
Our finding that JA alters root meristem activity prompted us to investigate its possible effect on the cellular organization of QC and its surrounding stem cells. In normal-grown wild-type roots, the well-defined QC cells rarely divide (Figures 3A, 3G, 3I, 3K, 3M, and 3O). However, obvious QC division was observed in a significant proportion of JA-treated roots (Figures 3B, 3H, 3J, 3L, 3N, and 3P). Using the QC-specific markers QC25 and WOX5pro:GFP (for green fluorescent protein; Blilou et al., 2005; Sarkar et al., 2007), we confirmed that the dividing cells were QC cells (Figures 3G to 3J). To test whether JA induces QC differentiation, we performed Lugol staining experiments using the QC25 marker line. As shown in Figure 3H, no starch accumulation was detected in the irregularly divided QC cells, suggesting that JA promotes QC division but does not induce QC differentiation. Considering that WOX5 plays an important role in regulating the maintenance of the root stem cell niche (Sarkar et al., 2007; Ding and Friml, 2010), we examined whether JA treatment affects WOX5 expression. RNA in situ assays indicated that JA treatment led to a slight expansion of the WOX5 expression domain (cf. Figures 3L and 3K), which could have resulted from the irregular QC division. However, as revealed by our qRT-PCR assays, JA treatment did not substantially alter the WOX5 transcript levels (Figure 3Q). To check whether cell cycle activity is altered in the QC of JA-treated roots, we cultured 2-d-old seedlings for 24 h in the presence of JA and EdU, a nucleoside analog of thymidine. EdU has been used to mark cell division in the root meristem because incorporation of this chemical in the nuclei is indicative of S-phase progression (Vanstraelen et al., 2009). In this assay, mitotically active cells show red fluorescent nuclei after coupling of the EdU with the Alexa Fluor 647 substrate. In untreated wild-type root meristems, most meristematic cells, including those QC-associated stem cells, incorporated EdU with red fluorescent nuclei, but only occasionally in the QC cells (marked with WOX5pro:GFP), indicating their low mitotic activity (Figures 3M and 3O). In JA-treated root meristems, however, the QC cells were able to incorporate EdU (Figures 3N and 3P; 13/15 in JA-treated roots versus 2/15 in control roots, n = 15), indicating that JA application induces high mitotic activity of the QC cells.
Figure 3.
JA-mediated Promotion of QC Division in the Wild Type, coi1-2, and myc2-2.
(A) to (F) Representative confocal images showing that JA promotes QC division in Col-0 but not in coi1-2 and myc2-2. Five-day-old seedlings were transferred to medium without (MS) or with 20 μM JA for another day before QC division was monitored. Bars = 20 μm.
(G) and (H) JA induces QC division and CSC differentiation as revealed by Lugol staining of the QC25 marker line. Five-day-old seedlings of the QC25 marker line were transferred to medium without (MS) or with 100 μM JA for 1 d before GUS (blue) and Lugol (dark brown) double stainings were performed. White arrow indicates no starch accumulation in the irregular QC cells. Red arrow shows starch accumulation in the irregular CSCs. Bars = 20 μm.
(I) and (J) JA-induced QC division in the QC marker line WOX5pro:GFP. Five-day-old seedlings of WOX5pro:GFP were transferred to medium without (MS) or with 20 μM JA for another day before QC division was monitored. White arrows mark the cell divisions in the QC. Bars = 20 μm.
(K) and (L) Whole-mount in situ hybridization with a WOX5-specific probe showing that JA causes supernumerary QC cells. Roots of seedlings 5 d after germination grown on medium without (MS) or with 20 μM JA were used for whole-mount in situ hybridization. Bars = 20 μm.
(M) to (P) EdU incorporation assays showing that JA promotes QC division. Two-day-old WOX5pro:GFP seedlings grown on medium without (MS) ([M] and [O]) or with 100 μM JA treatment ([N] and [P]) were cultured with EdU for 1 d before EdU incorporation in the QC was examined. (O) represents the outlined area in (M), and (P) represents the outlined area in (N). Red fluorescence of EdU-positive nuclei in QC cells of JA-treated roots indicates that QC is in a state of active division (white arrow). Bars = 20 μm.
(Q) JA-induced WOX5 expression revealed by qRT-PCR. Five-day-old Col-0 seedlings germinated on control medium were transferred to liquid medium without (control) or with 20 μM JA for 6 h, and 2-mm root tips were harvested for RNA extraction and qRT-PCR analysis. WOX5, PLT1, and PLT2 transcription levels without JA treatment were arbitrarily set to 1. Error bars represent the sd of triplicate reactions of independent RNA samples prepared from three batches of Arabidopsis roots. Asterisks denote Student’s t test significance compared with untreated plants: ***P < 0.001.
(R) Dose-dependent effect of JA on QC division in Col-0 roots. Col-0 seedlings were grown on medium containing indicated concentrations of JA for 6 d before QC division frequency (percentage of seedlings with obvious QC division) was determined. At least 50 seedlings were examined for each concentration for each biological repeat. Data shown are average and sd and are representative of at least three independent experiments.
(S) JA-induced QC division frequency in Col-0, coi1-2, and myc2-2. Seedlings were grown on medium without (Control) or with 20 μM JA for 6 d before QC division frequency was determined. At least 50 seedlings were examined for each biological repeat. Data shown are average and sd and are representative of at least three independent experiments. Asterisks denote Student’s t test significance compared with untreated plants: *P < 0.05 and ***P < 0.001.
(T) Time course of JA-induced extra QC cells in Col-0, coi1-2, and myc2-2. Seeds of the indicated genotypes were germinated on medium without (Control) or with 20 μM JA for 10 d, and extra QC cells were quantified at a 2-d interval. The effects of JA on Col-0 compared with coi1-2 and myc2-2 were significantly different. At least 50 seedlings were examined for each biological repeat. Data shown are average and sd and are representative of at least three independent experiments. Asterisks denote Student’s t test significance compared with untreated plants: *P < 0.05 and ***P < 0.001.
Similar to JA-induced root growth inhibition, we found that JA-induced QC division was dose dependent (Figure 3R). We followed the time course of the onset of the JA-induced QC phenotype by quantifying extra QC cells for 10 d after germination on medium containing 20 μM JA. Quantification of extra QC cells has been successfully used to characterize ethylene-induced QC division (Ortega-Martínez et al., 2007). In the wild type, obvious QC division was observed as early as 2 d after germination, and the number of extra QC cells increased gradually with time (Figure 3T). Importantly, our parallel experiments revealed that the effect of JA on QC division was largely abolished in the JA signaling mutants, including coi1-2 and myc2-2 (Figures 3C to 3F, 3S, and 3T), indicating that the promotional effect of JA on QC division requires the function of COI1 and MYC2.
To test whether the above-described effect of JA on the promotion of QC division is independent of that of ethylene, which has also been shown to modulate stem cell division in Arabidopsis roots (Ortega-Martínez et al., 2007), we examined the effect of JA on QC division in the previously described ethylene signaling mutants ein2-5 and ein3-1 eil1-1 (Guo and Ecker, 2003). As shown in Supplemental Figure 3 online, the ein2-5 and ein3-1 eil1-1 mutants did not affect the promotional effect of JA on QC division. Similarly, the addition of AgNO3, which effectively blocks the ethylene signaling pathway, did not alter the effect of JA on the promotion of QC division (see Supplemental Figure 3 online). These results confirmed that the effect of JA on the promotion of QC division is independent of that of ethylene.
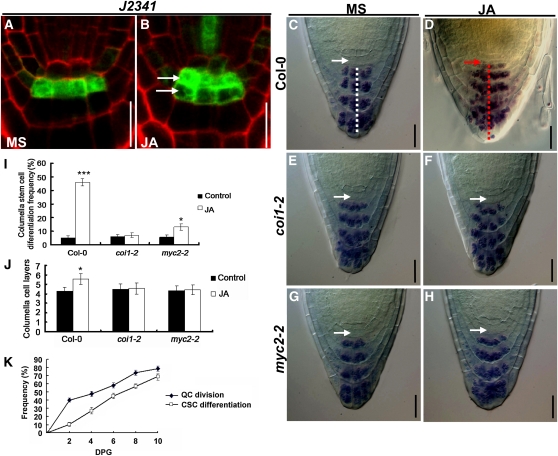
JA Induces Deregulated Differentiation of CSCs
In the root stem cell niche, the mitotically inactive QC cells act to maintain the identity and function of the surrounding stem cells. To assess the possible effect of JA-induced QC division on stem cell maintenance, we examined the cell layer immediately below the QC, at the position of the CSCs. The J2341 marker, which is specifically expressed in the single layer of CSCs (Figure 4A), expanded to more than one cell tier upon JA treatment (cf. Figures 4A and 4B). We then asked whether the distal stem cells might have undergone premature differentiation in JA-treated roots, which in the columella can be visualized by the accumulation of starch granule-containing organelles. In the absence of JA, 6-d-old wild-type roots do not show starch granule accumulation in the CSCs (Figure 4C, white arrow), which are distinct from the underlying well-organized four tiers of columella cells that accumulate starch granules (Figures 4C, white dashed line). In the presence of JA, however, irregular cells containing starch granules were observed immediately below the QC cells (Figure 4D, red arrow), indicating that the CSCs were in a state of differentiation. Our time-course study revealed that, in JA-treated roots, the appearance of QC division is generally earlier than that of CSC differentiation (Figure 4K), suggesting that the JA-induced irregular QC divisions may lead to a failure to maintain the stem cell fate of CSCs. In addition, JA treatment led to the formation of extra columella cell layers, and the general cellular organization of columella tiers was disturbed (Figures 4D, red dashed line, 4I, and 4J). The above-described effect of JA on CSC differentiation and extra columella layer formation was substantially reduced in coi1-2 (Figures 4E, 4F, 4I, and 4J) and myc2-2 (Figures 4G to 4J), suggesting that the effect of JA on CSC differentiation and columella organization requires the function of COI1 and MYC2.
Figure 4.
JA-Mediated Promotion of CSC Differentiation in the Wild Type, coi1-2, and myc2-2.
(A) and (B) JA induces deregulated division of CSCs, as revealed by the CSC-specific marker J2341. Five-day-old seedlings of the J2341 marker line were transferred to medium without (MS) or with 20 μM JA for 1 d before GFP expression in CSCs was examined. White arrows indicate the presence of two CSC layers in JA-treated roots. Bars = 20 μm.
(C) to (H) Lugol staining showing that JA induces CSC differentiation in Col-0 ([C] and [D]) but not in coi1-2 ([E] and [F]) and myc2-2 ([G] and [H]). Five-day-old seedlings were transferred to medium without (MS) or with 20 μM JA for 1 d before Lugol staining was performed. Nondifferentiated CSCs (white arrows) below the QC are characterized by the absence of starch granules, whereas starch granules are visible in differentiated CSCs (red arrow). The white dashed line in (C) indicates the well-organized columella cell layers of untreated wild-type roots. The red dashed line in (D) indicates increased and disorganized columella cell layers of JA-treated wild-type roots. Bars = 20 μm.
(I) JA-induced CSC differentiation frequency in Col-0, coi1-2, and myc2-2. Seedlings were grown on medium without (Control) or with 20 μM JA for 6 d before CSC differentiation frequency was determined. At least 50 seedlings were examined for each genotype for each experiment. Data shown are average and sd and are representative of at least three independent experiments. Asterisks denote Student’s t test significance compared with untreated plants: *P < 0.05 and ***P < 0.001.
(J) JA induces extra columella cell layers in Col-0 but not in coi1-2 and myc2-2. Seedlings were grown on medium without (Control) or with 20 μM JA for 6 d before columella cell layers were determined. At least 50 seedlings were examined for each genotype for each experiment. Data shown are average and sd and are representative of at least three independent experiments. Asterisks denote Student’s t test significance compared with untreated plants: *P < 0.05.
(K) Kinetics of JA-induced QC division and CSC differentiation. Col-0 seeds were germinated on medium containing 20 μM JA (JA), and QC division frequency and CSC differentiation frequency were determined at a 2-d interval. At least 50 seedlings were examined for each biological repeat. Data shown are average and sd and are representative of at least three independent experiments.
To test whether endogenous JA has a similar effect as exogenous JA (i.e., to reduce root meristem activity and to disturb the maintenance of the root stem cell niche), we sought to obtain mutants with elevated endogenous JA levels or with increased JA sensitivity. The constitutive expression of VSP1 (cev1) mutant has increased production of both JA and ethylene and exhibits a short root phenotype (Ellis and Turner, 2001; Ellis et al., 2002). When grown on control medium, cev1 seedlings show reduced meristem cell number (see Supplemental Figure 4A online), increased QC division (see Supplemental Figures 4B and 4C online), and CSC differentiation (see Supplemental Figures 4D to 4I online) than wild-type seedlings. When grown on medium containing 2-aminoethoxyvinyl glycine or AgNO3, which can effectively block ethylene biosynthesis and perception, respectively, cev1 roots still exhibit reduced meristem size, exaggerated QC division, and CSC differentiation compared with wild-type roots (see Supplemental Figure 4 online). These results support that endogenous JA exerts a similar effect as exogenous JA on stem cell niche maintenance.
Given that JAZ proteins serve as negative regulators of JA signaling, it is reasonable to expect that loss-of-function mutants of JAZ genes exhibit increased sensitivity to JA in root growth inhibition. Indeed, it has been shown that RNA interference (RNAi) lines of JAZ10 show a hypersensitive response to JA (Yan et al., 2007). We found that, like the JAZ10 RNAi lines, a T-DNA insertion line (CS872819, SAIL_92_D08) that disrupts the expression of JAZ10 also exhibits increased sensitivity to JA in root growth inhibition (see Supplemental Figure 5A online). Consistently, JA-induced meristem cell number reduction (see Supplemental Figure 5B online), QC division (see Supplemental Figure 5C online), CSC differentiation (see Supplemental Figures 5E to 5H online), and columella layer disorganization (see Supplemental Figure 5D online) are all increased in jaz10 relative to the wild type. These results confirm that, like exogenous JA, endogenous JA also modulates the cellular organization of the root stem cell niche and reduces root meristem activity.
JA Reduces the Expression of PLT1 and PLT2 in a MYC2-Dependent Manner
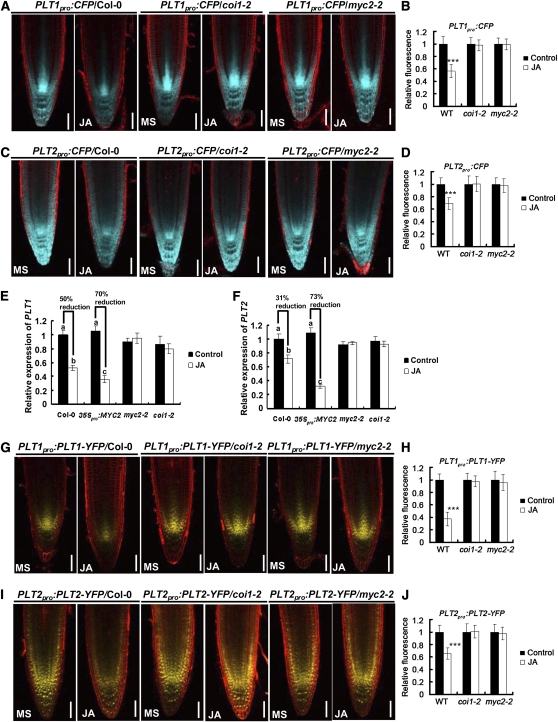
The PLT family of transcription factors provides important patterning information for the root stem cell niche and determines the number of the transit-amplifying daughter cells that make up the meristem (Aida et al., 2004; Galinha et al., 2007). We show that Arabidopsis roots grown on JA-containing medium exhibit ectopic cell division in the QC, irregular differentiation of CSCs, and loss of transit-amplifying cells, defects that are also observed in the plt1-4 plt2-2 double mutant roots (Aida et al., 2004). The root phenotype similarity between JA-treated roots and mutants of the PLT genes prompted us to test the possible link between JA and the PLT network in regulating root development. We first examined JA-induced expression of PLT1 and PLT2 at the transcription level. Using transgenic plants containing the promoters of PLT1 and PLT2 fused to the cyan fluorescent protein (CFP) gene (Galinha et al., 2007), we found that JA significantly reduced the expression levels of PLT1pro:CFP (Figures 5A and 5B) and PLT2pro:CFP (Figures 5C and 5D) in the wild type but not in coi1-2 and myc2-2, suggesting that JA represses PLT1 and PLT2 expression in a COI1- and MYC2-dependent manner. Next, using qRT-PCR assays, we compared the repression effect of JA on PLT expression in the wild type, coi1-2, myc2-2, and the MYC2 overexpression line 35Spro:MYC2 (see Supplemental Figures 2E and 2F online). In line with previous reports (Boter et al., 2004; Lorenzo et al., 2004), our 35Spro:MYC2 plants did not show constitutive activation of JA responses, although enhanced inhibition of root growth was observed in these 35Spro:MYC2 seedlings after treatment with JA (see Supplemental Figure 2F online). Consistently, we showed that, in the absence of JA, PLT1 and PLT2 expression was largely similar among the genotypes compared (Figures 5E and 5F). JA treatment markedly downregulated the transcript levels of PLT1 and PLT2 in the wild type but not in coi1-2 and myc2-2 (Figures 5E and 5F). Significantly, JA-induced downregulation of PLT1 and PLT2 expression was substantially exaggerated in 35Spro:MYC2 compared with the wild type (Figures 5E and 5F), indicating an important role of MYC2 in mediating JA-induced repression of PLT1 and PLT2 expression. Importantly, the observation that MYC2 represses PLT gene expression only in the presence of JA suggests that this action of MYC2 may require JA perception or other upstream signaling events.
Figure 5.
JA Reduces the Expression of PLT1 and PLT2 through COI1 and MYC2.
(A) JA reduces PLT1pro:CFP expression levels in Col-0, but not in coi1-2 and myc2-2.
(B) Quantification of CFP fluorescence shown in (A).
(C) JA reduces PLT2pro:CFP expression levels in Col-0, but not in coi1-2 and myc2-2.
(D) Quantification of CFP fluorescence shown in (C).
For (A) to (D), 5-d-old seedlings were transferred to medium without (MS) or with 20 μM JA for 1 d before CFP fluorescence was monitored. Data shown are average and sd (n = 15 to 20). Asterisks denote Student’s t test significance compared with untreated plants: ***P < 0.001. Bars = 50 μm.
(E) and (F) qRT-PCR assay showing that JA downregulates the transcription of PLT1 (E) and PLT2 (F) in a COI1- and MYC2-dependent manner. Five-day-old seedlings germinated on MS medium were transferred to medium without (Control) or with 20 μM JA for 1 d, and 2-mm root tips were harvested for RNA extraction and qRT-PCR analysis. The transcript levels of PLT1 and PLT2 were normalized to the ACTIN2 expression. PLT1 and PLT2 transcription levels of Col-0 without JA treatment were arbitrarily set to 1. Data presented are mean values of four biological repeats with sd. Samples with different letters are significantly different: P < 0.01.
(G) JA reduces PLT1pro:PLT1-YFP expression levels in Col-0, but not in coi1-2 and myc2-2.
(H) Quantification of YFP fluorescence shown in (G). Data shown are average and sd (n = 15 to 20). Asterisks denote Student’s t test significance compared with untreated plants: ***P < 0.001.
(I) JA reduces PLT2pro:PLT2-YFP expression levels in Col-0, but not in coi1-2 and myc2-2.
(J) Quantification of YFP fluorescence shown in (I). Data shown are average and sd (n = 15 to 20). Asterisks denote Student’s t test significance compared with untreated plants: ***P < 0.001.
For (G) to (J), 5-d-old seedlings were transferred to medium without (MS) or with 20 μM JA for 1 d before YFP fluorescence was monitored. Bars = 50 μm.
Next, using transgenic plants containing the promoters of PLT1 and PLT2 combined with PLT-YFP (for yellow fluorescent protein) protein fusions (Galinha et al., 2007), we found that JA also reduced the expression levels of PLT1pro:PLT1-YFP (Figures 5G and 5H) and PLT2pro:PLT2-YFP (Figures 5I and 5J) in the wild type but not in coi1-2 and myc2-2. Together, our results support the hypothesis that JA represses the transcriptional expression of PLT1 and PLT2 through the COI1- and MYC2-mediated jasmonate signaling pathway.
Given that JA promotes auxin biosynthesis (Dombrecht et al., 2007; Sun et al., 2009) and auxin itself regulates PLT expression (Aida et al., 2004), we designed experiments to test whether the above-described effect of JA on PLT expression is achieved through auxin. To this end, we compared JA-induced expression of PLT1 and PLT2 in the wild type and the auxin biosynthesis mutants asa1-1 (Sun et al., 2009) and yuc1D (previously known as yucca; Zhao et al., 2001). Our recent work demonstrates that the asa1-1 mutant contains a mutation in the Anthranilate Synthase α1 (ASA1/WEI2) gene and therefore is defective in JA-induced auxin biosynthesis (Sun et al., 2009). The dominant yuc1D mutant contains elevated free IAA levels, resulting from the overexpression of a rate-limiting auxin biosynthesis gene YUCCA1 (YUC1) (Zhao et al., 2001; Zhao, 2010). As revealed by our qRT-PCR assays, in wild-type roots, a slight reduction of PLT1 and PLT2 transcripts was observed 2 h after JA treatment and, a marked reduction of these transcripts was observed at 6 and 12 h after JA treatment (see Supplemental Figures 6A and 6B online). Importantly, JA-induced reduction levels and kinetics of PLT1 and PLT2 expression were largely similar among asa1-1, yuc1D, and the wild type (see Supplemental Figures 6A and 6B online), indicating that the effect of JA on PLT expression is not achieved through the ASA1- and YUC1-dependent auxin biosynthesis. Consistently, the asa1-1 and yuc1D mutations did not affect JA-induced inhibition of root growth (see Supplemental Figure 6C online) and JA-induced reduction of root meristem cell number (see Supplemental Figure 6D online). Together, these data support the notion that the effect of JA on PLT expression is independent of the auxin pathway.
MYC2 Directly Regulates the Expression of PLT1 and PLT2
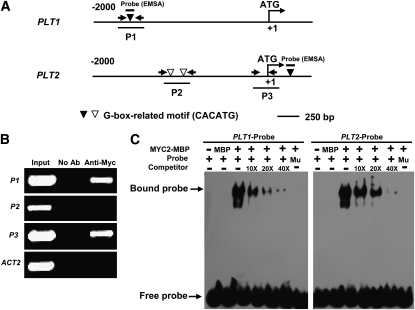
The basic helix-loop-helix–type transcription factor MYC2 acts as a master regulator of JA-mediated gene transcription and regulates versatile aspects of JA responses, including defense gene expression and root growth inhibition (Dombrecht et al., 2007; Memelink, 2009). Our finding that JA represses the transcriptional expression of PLT1 and PLT2 in a MYC2-dependent manner suggests that MYC2 may associate with promoters of PLT1 and PLT2. It is well known that MYC2 preferably binds to the G-box–related hexamer 5′-CACATG-3′ of its target genes (Dombrecht et al., 2007). Our sequence analysis revealed one 5′-CACATG-3′ motif in the P1 region (located −1609 to −1614 bp upstream of the translational start codon; Figure 6A) of the PLT1 promoter. Sequence analysis also identified two 5′-CACATG-3′ motifs in the P2 region (located −940 to −945 and −1098 to −1103 bp upstream of the translational start codon; Figure 6A) and one 5′-CACATG-3′ motif close to the P3 region (located +288 to +293 bp downstream of the translational start codon; Figure 6A) of PLT2. To test the in vivo interaction between MYC2 and the chromatin regions of PLT1 and PLT2, we performed chromatin immunoprecipitation (ChIP) assays using the 35Spro:MYC2 transgenic plants (containing the 35Spro:MYC2-4Myc construct) and anti-Myc antibodies (Roche). As shown in Figure 6B, chromatin immunoprecipitated with anti-Myc antibodies was profoundly enriched in the P1 region of the PLT1 promoter and in the P3 region of the PLT2 promoter. These results indicate a specific association of MYC2 with the promoters of PLT1 and PLT2.
Figure 6.
MYC2 Associates with PLT1 and PLT2 Promoters.
(A) Schematic diagram of potential MYC2 binding sites (white and black triangles), DNA fragments (P1, P2, and P3) used for ChIP, and probes used for EMSA. The sequence 2 kb upstream of the start site and part of the coding sequence for PLT1 and PLT2 are shown. The translational start site (ATG) is shown at position +1.
(B) Enrichment of the indicated DNA fragments (P1 and P3) following ChIP using anti-Myc antibodies. Chromatin of transgenic plants expressing 35Spro:MYC2-4Myc was immunoprecipitated with anti-Myc antibodies, and the presence of the indicated DNA in the immune complex was determined by RT-PCR. The Actin2 promoter fragment was used as a negative control. The experiment was repeated three times with similar results.
(C) EMSA showing that the MYC2-MBP fusion protein binds to the DNA probes of PLT1 and PLT2 in vitro. Biotin-labeled probes were incubated with MYC2-MBP protein, and the free and bound DNAs (arrows) were separated in an acrylamide gel. As indicated, unlabeled probes were used as competitors. Mu, mutated probe in which the 5′-CACATG-3′ motif was deleted.
A DNA electrophoretic mobility shift assay (EMSA) was conducted to confirm that MYC2 directly binds these G-box–related motifs presented in the upstream regions of PLT1 and PLT2. Full-length MYC2 protein was expressed as a maltose binding protein (MBP) fusion protein in Escherichia coli and affinity purified. As shown in Figure 6C, the MYC2-MBP fusion proteins were able to bind DNA probes containing the 5′-CACATG-3′ motif located in the P1 region of the PLT1 promoter and in the P3 region of the PLT2 gene. Furthermore, additional unlabeled DNA probes competed for binding in a dose-dependent manner (Figure 6C). Parallel experiments indicated that MYC2-MBP failed to bind DNA probes containing the mutant form of the 5′-CACATG-3′ motif (Figure 6C). Together, these results reveal that MYC2 regulates PLT1 and PLT2 expression through direct association with their promoters.
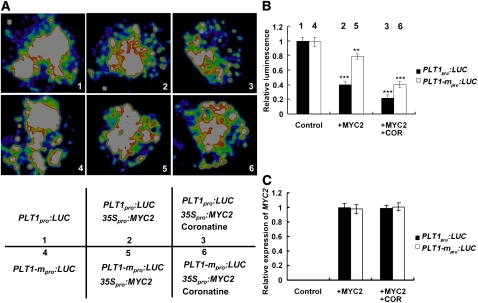
Next, using the well-established transient expression assay of Nicotiana benthamiana leaves, we verified the repression effect of MYC2 on the expression of a reporter containing the PLT1 promoter fused with the firefly luciferase gene (LUC). When the PLT1pro:LUC reporter was infiltrated into N. benthamiana, a substantial amount of LUC activity could be detected (Figures 7A and 7B), indicating that endogenous factors of N. benthamiana may activate the expression of the PLT1pro:LUC reporter. Coexpression of PLT1pro:LUC with the 35Spro:MYC2 construct led to an obvious reduction in luminescence intensity (Figure 7), suggesting that the manipulation process of the assay could have simulated the JA signaling of the infiltrated N. benthamiana leaf area, which, in turn, activated the repression effect 35Spro:MYC2 on PLT1pro:LUC expression. Significantly, exogenous application of coronatine (COR), a jasmonoyl Ile mimic molecule that can potently activate JA signaling (Katsir et al., 2008; Fonseca et al., 2009; Sheard et al., 2010), onto the N. benthamiana leaf area coexpressing PLT1pro:LUC and 35Spro:MYC2, led to a more dramatic reduction effect of 35Spro:MYC2 on PLT1pro:LUC expression (Figure 7). In a parallel experiment, PLT1-mpro:LUC, in which the G-box of the PLT1 promoter was deleted and fused with LUC, was coexpressed with 35Spro:MYC2 in N. benthamiana leaves. As shown in Figure 7, in the absence or presence of COR, the repression effect of 35Spro:MYC2 on PLT1-mpro:LUC expression was largely attenuated. The observation that the PLT1-mpro:LUC reporter was still responsive to the repression of 35Spro:MYC2 could be explained by the presence of the 5′-GAGTA-3′ motif in the PLT1 promoter (see Supplemental Figure 7 online), which has been shown to be responsive to JA (Boter et al., 2004). Together, our transient assays of N. benthamiana leaves confirmed that MYC2 represses PLT1 expression in vivo.
Figure 7.
MYC2 Represses PLT1 Expression, as Revealed by Transient Assays of N. benthamiana leaves.
(A) Transient expression assays showing that MYC2 represses the expression of PLT1. Representative images of N. benthamiana leaves 72 h after infiltration are shown. The bottom panel indicates the infiltrated constructs and treatments.
(B) Quantitative analysis of luminescence intensity in (A). Five independent determinations were assessed. Error bars represent sd. Asterisks denote Student’s t test significance compared with control plants: **P < 0.01 and ***P < 0.001.
(C) qRT-PCR analysis of MYC2 expression in the infiltrated leaf areas shown in (A). Total RNAs were extracted from leaves of N. benthamiana coinfiltrated with the constructs. Five independent determinations were assessed. Error bars represent sd.
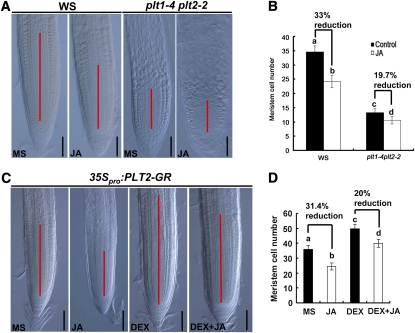
The above-described results suggest that MYC2-mediated repression of PLT1 and PLT2 expression is involved in JA-induced modulation of root meristem activity and stem cell niche maintenance. To determine the genetic relationship between JA and PLT proteins, we assessed the effect of JA on root meristems of the wild type and the plt1-4 plt2-2 double mutant. As shown in Figures 8A and 8B, JA reduced the root meristem cell number by 33% in the wild type but only by 19.7% in plt1-4 plt2-2, indicating that the double mutations significantly attenuated the reduction effect of JA on the root meristem cell number. To test whether overexpression of PLT genes could somehow rescue the reduction effect of JA on root meristem cell number, we used transgenic plants containing the inducible construct 35Spro:PLT2-GR (Galinha et al., 2007). Figures 8C and 8D indicated that, without dexamethasone (DEX) induction (i.e., without PLT2 overexpression), JA reduced the root meristem cell number of 35Spro:PLT2-GR plants by 31.4%. In the presence of DEX induction; however, JA reduced the root meristem cell number of the PLT2 overexpressor by 20% (Figures 8C and 8D). The finding that the JA-induced reduction of root meristem cell number is reduced in plt1-4 plt2-2 and PLT2 overexpression plants supports the scenario that PLT proteins act genetically downstream of JA in regulating root meristem maintenance. The fact that plt1-4 plt2-2 double mutants and PLT2 overexpression plants are still responsive to JA could be explained by at least two possibilities: first, the existence of PLT family members other than PLT1 and PLT2 (i.e., PLT3 and BABY BOOM) (Galinha et al., 2007); second, in addition to the PLT family of transcription factors, JA could employ other mechanisms to regulate root meristem maintenance.
Figure 8.
JA-induced Root Meristem Cell Number in plt1-4 plt2-2 and PLT2-overexpression Plants.
(A) JA-induced reduction of meristem size in wild-type (Wassilewskija [WS]) and plt1-4 plt2-2 plants. Seedlings were grown on medium without (MS) or with 20 μM JA for 4 d before meristem cell number was determined. The MZ is marked with a red line. Bars = 50 μm.
(B) Quantification of meristem cell number in (A). Data shown are average and sd (n = 20). Samples with different letters are significantly different: P < 0.05. These experiments were repeated at least three times with similar results.
(C) JA-induced reduction of root meristem size in transgenic plants expressing 35Spro:PLT2-GR. Bars = 50 μm.
(D) Statistics of the meristem cell number shown in (C). Data shown are average and sd (n = 15 to 20). Samples with different letters are significantly different: P < 0.01. These experiments were repeated at least three times with similar results.
For (C) and (D), 4-d-old seedlings germinated on MS medium were transferred to control medium (MS) or medium containing 2 μM DEX for another 2 d before root meristem cell number was determined. Four-day-old seedlings germinated on medium with 20 μM JA were transferred to medium with 20 μM JA + 2 μM DEX (DEX + JA) or 20 μM JA only (JA) for another 2 d before root meristem cell number was determined.
DISCUSSION
Postembryonic root growth of higher plants exhibits remarkable plasticity to adapt to the ever-changing environmental conditions. The adaptation ability of plant root growth is determined by the postembryonic activity of cells within the root meristem that is coordinated by several phytohormones, including jasmonate (Benková and Hejátko, 2009; Petrásek and Friml, 2009; Wolters and Jürgens, 2009). Here, we address the cellular and molecular mechanisms underlying JA-induced inhibition of primary root growth of Arabidopsis plants. We show that, at the cellular level, JA reduces cell number and cell elongation of the DZ and EZ, inhibits cell division activity of the root meristem, and alters the cellular organization of the stem cell niche. We also show that, at the molecular level, MYC2 directly represses the expression of PLT1 and PLT2 upon JA treatment, suggesting that MYC2-mediated repression of PLT expression is important for JA-induced modulation of root meristem activity and stem cell niche maintenance. This study demonstrates a mode of jasmonate-auxin crosstalk, in which a major transcription factor of the JA signaling pathway directly represses master transcription factors known to act in the auxin pathway. Our results extend the view of how plants can translate stress cues into growth response and thus retain growth plasticity during postembryonic development.
JA Modulates Maintenance of the Root Stem Cell Niche
Although jasmonate-induced root growth inhibition has been employed as the most prominent genetic assay to identify jasmonate-related mutants in Arabidopsis (Wasternack, 2007; Browse, 2009), the cellular and molecular basis of jasmonate action on root growth is hitherto not clear. We show here that JA reduces both cell number and cell length of the root, indicating that JA-induced inhibition of primary root growth is a complex process that involves multiple cellular processes in distinct root tissues.
JA-induced reduction of root meristem activity is evidenced by reduced meristem size and decreased expression levels of the CYCB1;1pro:GUS reporter gene and a group of cell cycle–related genes. The reduced cell proliferation rate in the JA-treated root meristem could contribute to the observed reduction of cell number in the EZ. Our observations reveal that the mechanism by which JA inhibits primary root growth is distinct from that of ethylene, an important stress hormone showing extensive crosstalk with JA (Lorenzo et al., 2003). Several groups showed that ethylene does not affect cell cycle activity of the root meristem but strongly reduces cell elongation in the EZ (Růzicka et al., 2007; Stepanova et al., 2007; Swarup et al., 2007).
Consistent with a reduced root meristem activity by JA, we found that JA disturbs the cellular organization of the root stem cell niche. For example, JA triggers the division of QC cells and differentiation of CSCs. Our time-course study revealed that, upon JA treatment, the occurrence of QC division is generally earlier than that of CSC differentiation. In the context that proper expression of WOX5 in the QC is required to maintain CSCs as undifferentiated (Sarkar et al., 2007), we examined the possible effect of JA on WOX5 expression and found that JA treatment has little, if any, effect on WOX5 expression. Given that ethylene has been shown to be able to induce QC division (Ortega-Martínez et al., 2007), it is important to determine whether this hormone and JA might affect the mitotic activity of QC through similar mechanisms. Two lines of evidence favor that JA and ethylene modulate the balance between proliferation and quiescence of QC cells through distinct mechanisms. First, whereas the supernumerary QC cells produced by ethylene-induced QC division still display their QC function to maintain the stem cell identity of CSCs (Ortega-Martínez et al., 2007), JA-induced supernumerary QC cells partially lose their ability to maintain the stem cell identity of CSCs. Second, blocking of the ethylene pathway by genetic or chemical manipulation does not affect JA-induced QC division.
Interestingly, our results reveal that, depending on the tissue context, JA exerts opposite effects on cell division. For example, JA represses cell division in the proximal root meristem, as evidenced by reduced meristem cell number, reduced expression of the mitotic cell cycle marker CYCB1;1pro:GUS, and reduced expression levels of several cell cycle–related genes in JA-treated roots. These results are consistent with previous observations showing that, as a growth regulator, JA generally inhibits cell division and therefore represses growth (Dathe et al., 1981; Swiatek et al., 2004; Pauwels et al., 2008; Zhang and Turner, 2008). On the other hand, JA promotes cell division in the QC, as evidenced by the presence of supernumerary QC cells in JA-treated roots, QC-specific marker gene expression, and EdU staining assays. It will be interesting in future studies to unravel the molecular mechanisms underlying these distinct aspects of JA action. In the context that JA plays an important role in transmitting environmental cues into developmental responses, our finding that JA impacts QC division presents an attractive scenario on how plants adapt their growth to environments by modulating stem cell division and activity.
MYC2-Mediated Repression of PLT Expression Integrates Jasmonate and Auxin Pathways in Regulating Maintenance of the Root Stem Cell Niche
It is noteworthy that the above-described effects of JA on the root meristem and stem cell niche require the function of MYC2, a major transcription factor that positively or negatively regulates different aspects of jasmonate responses, including defense gene expression and root growth inhibition. Therefore, our investigation hints at the involvement of MYC2-mediated transcriptional regulation in JA-induced root growth inhibition.
PLT genes encode proteins that act as dose-dependent regulators for root development (Aida et al., 2004; Galinha et al., 2007). For example, high levels of PLT2 maintain the stem cells, and intermediate levels facilitate transit amplifying cell divisions that define the meristem region (Galinha et al., 2007). In line with the JA-induced reduction of root meristem activity and disturbance of stem cell niche maintenance, we found that JA downregulates the expression levels of PLT1 and PLT2 in a MYC2-dependent manner. In the context that JA affects auxin biosynthesis (Dombrecht et al., 2007; Sun et al., 2009) and that auxin itself regulates the transcriptional expression of PLT genes (Aida et al., 2004), we provided several lines of evidence that the effect of JA on PLT expression and, therefore, on root stem cell niche maintenance is independent of the effect of JA on auxin biosynthesis. First, our previous work revealed that JA promotes auxin biosynthesis through transcriptional activation of the auxin biosynthetic gene ASA1/WEI2 (Sun et al., 2009; Stepanova et al., 2005). We show here that the asa1-1 mutant, which harbors a loss-of-function mutation of ASA1/WEI2 and is therefore defective in JA-induced auxin biosynthesis, shows normal response to JA in terms of root growth inhibition and meristem cell number reduction. Second, the asa1-1 (Stepanova et al., 2005; Sun et al., 2009) and yuc1D (Zhao et al., 2001) mutations, which are defective in auxin biosynthesis, do not affect JA-induced repression levels and kinetics of PLT1 and PLT2 expression. Third, the action mode of JA on PLT expression is distinct from that of auxin. Whereas auxin positively regulates PLT1 and PLT2 expression (Aida et al., 2004), we show here that JA negatively regulates their expression. These, together with other data described in this work, support that JA-mediated regulation of PLT1 and PLT2 expression is independent of the auxin pathway.
Our finding that JA represses PLT1 and PLT2 expression in a MYC2-dependent manner raises the interesting possibility that MYC2 may directly regulate PLT expression. Indeed, our EMSA and ChIP assays indicate that MYC2 associates with the promoters of both PLT1 and PLT2. Using the well-established transient assay of N. benthamiana leaves, we show that MYC2 indeed represses PLT1pro:LUC expression and, importantly, that the G-box-related motif CACATG in the promoter of PLT1 is important for this MYC2 action. Consistently, the reduction effect of JA on root meristem cell number was substantially reduced in the plt1-4 plt2-2 double mutant and transgenic plants overexpressing PLT2. Collectively, these results support the notion that the PLT1 and PLT2 transcription factors act downstream of MYC2 in the JA-mediated regulation of stem cell niche maintenance and root meristem activity. Considering the established role of the PLT transcription factors in mediating developmental response to auxin in the root meristem (Aida et al., 2004; Dinneny and Benfey, 2008), we propose that MYC2-induced repression of PLT1 and PLT2 expression integrates JA action into the auxin signaling pathway in regulating postembryonic root growth. It has been shown that auxin upregulates PLT1 and PLT2 transcripts and positively regulates stem cell niche maintenance and meristem activity (Aida et al., 2004). In contrast with auxin, this study reveals that JA downregulates PLT1 and PLT2 expression and negatively regulates stem cell niche maintenance and meristem activity. PLT1 and PLT2 therefore represent a key junction for crosstalk between the JA and auxin signaling pathways in the regulation of root stem cell niche maintenance.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotypes Columbia-0 (Col-0), C24, and Wassilewskija were used. Some of the plant materials used in this study were previously described: J2341 and QC25 (Sabatini et al., 1999), PLT1pro:PLT1-YFP, PLT2pro:PLT2-YFP, PLT1pro:CFP, and PLT2pro:CFP (Galinha et al., 2007), coi1-1 (Xie et al., 1998), coi1-2 (Xu et al., 2002), cev1 (Ellis and Turner, 2001), ein3-1 eil1-1 (Alonso et al., 2003a), ein2-5 (Alonso et al., 1999), CYCB1;1pro:GUS (Colón-Carmona et al., 1999), WOX5pro:GFP (Blilou et al., 2005), myc2-2 (Boter et al., 2004), plt1-4 plt2-2 (Aida et al., 2004), and 35Spro:PLT2-GR (Galinha et al., 2007). jaz10 (CS872819, SAIL_92_D08) was identified from the SIGnAL T-DNA collection (Alonso et al., 2003b).
MYC2pro:GUS and COI1pro:GUS plants were obtained from transformation of Col-0 plants with the MYC2pro:GUS or COI1pro:GUS constructs. 35Spro:MYC2 plants were obtained from transformation of Col-0 with the 35Spro:MYC2-4Myc constructs.
Arabidopsis seeds were surface sterilized for 15 min in 10% bleach, washed four times with sterile water, and plated on half-strength Murashige and Skoog (MS) medium. Plants were stratified at 4°C for 2 d in darkness and then transferred to a phytotrone set at 22°C with a 16-h-light/8-h-dark photoperiod (light intensity 120 μmol m−2 s−1). Nicotiana benthamiana was grown under a 16-h-light (28°C)/8-h-dark (22°C) photoperiod. All experiments were repeated at least three times.
JA (Sigma-Aldrich) was dissolved with ethanol and prepared as a 200 mM stock solution. The 1-amino-1-cyclopropane carboxylic acid (Sigma-Aldrich) was dissolved with water and prepared as a 10 mM stock solution.
DNA Constructs and Plant Transformation
The COI1 promoter region was PCR amplified with the primers COI1pro-F, 5′-CCGCTGCAGGAGTCAACACCAAGTACAATGAC-3′, and COI1pro-R, 5′-CACCTCTTGATATCAGGATCCTCCATCGGATCG-3′. The PCR product was cloned into the PstI-BamHI sites of the binary vector pCAMBIA1391-Z (CAMBIA) to generate the COI1pro:GUS construct. The MYC2 promoter region was PCR amplified with the primers MYC2pro-F, 5′-CACCGACCCTTCCTAAAAAGTAAG-3′, and MYC2pro-R, 5′-CATTCCATAAACCGGTGACCGGT-3′. The PCR product was cloned using a pENTR Directional TOPO cloning kit (Invitrogen) and then recombined with the plant binary vector pGWB3 (Nakagawa et al., 2007) to generate the MYC2pro:GUS construct. MYC2 cDNA was PCR amplified from the reverse transcription product with primers 5′-CACCATGACTGATTACCGGCTACA-3′ and 5′-ACCGATTTTTGAAATCAAACTTGC-3′. The PCR product was cloned using the pENTR Directional TOPO cloning kit and then recombined with the binary vector pGWB17 (35S promoter, C-4Myc) to generate the 35Spro:MYC2-4Myc construct.
The above constructs were then transformed into Agrobacterium tumefaciens strain GV3101 (pMP90), which was used for transformation of Arabidopsis plants by vacuum infiltration (Bechtold and Pelletier, 1998).
Phenotypic Analysis, Statistics, and Microscopy
Seedlings were photographed and root length was measured (Image J; National Institutes of Health; http://rsb.info.nih.gov/ij). Root meristem size and QC division were analyzed on seedlings mounted in HCG solution (chloroacetaldehyde:water:glycerol = 8:3:1). Microscopy was performed on a Leica Microsystems DM5000B microscope and DFC490 charge-coupled device (CCD) camera. Root meristem size was assessed as the cell number between the QC and the first elongating cell in the cortex cell file (Casamitjana-Martínez et al., 2003; Dello Ioio et al., 2007). The statistical significance was evaluated by Student’s t test analysis. For multiple comparisons, an analysis of variance followed by Fisher’s LSD mean separation test (SPSS) was performed on the data. Samples with different letters are significantly different at P < 0.01 or P < 0.05. Data presented are mean values of at least three biological repeats with sd. Images were processed with Adobe Photoshop CS and Spot Flex software.
GFP, YFP, CFP, and propidium iodide fluorescence was imaged under a Leica confocal laser scanning microscope (Leica Microsystems). For imaging GFP, YFP, CFP, and propidium iodide, the 488-, 514-, 458-, and 543-nm lines of the laser were used for excitation, and emission was detected at 510, 530, 480, and 620 nm, respectively. Fluorescence was quantified with the LAS AF Lite program on confocal sections acquired with the same microscope settings (Růzicka et al., 2007; Zhou et al., 2010). Approximately 10 seedlings/images were examined, and at least three independent experiments were performed, giving the same statistically significant results.
Whole seedlings or different tissues were cleared in HCG solution for several minutes before microscopy analysis. For Lugol staining of roots, tissues were incubated in Lugol solution (Sigma-Aldrich) for 3 to 5 min, washed in water once, and mounted in HCG solution for microscopy analysis (Zhou et al., 2010). Histochemical staining for GUS activity in transgenic plants was performed as described previously (Jefferson et al., 1987).
An EdU incorporation assay was performed as previously described (Vanstraelen et al., 2009; Zhou et al., 2010). Seedlings were grown in liquid MS containing 10 μM EdU (Click-iT EdU Alexa Fluor 647 imaging kit; Invitrogen). After growth, seedlings were fixed in 3.7% formaldehyde in PBS, pH 7.4, for 15 min. Samples were then washed in 3% BSA in PBS, treated with 0.5% Triton in PBS for 20 min, and washed again. Coupling of EdU to the Alexa fluor substrate occurred in the dark in the Click-iT reaction mixture, prepared according to the manufacturer’s instructions, and observations were made using confocal microscopy.
Whole-Mount in Situ Hybridization
Whole-mount in situ hybridization was performed according to the reported protocols (Weigel and Glazebrook, 2002; Hejátko et al., 2006). Antisense and sense probes were synthesized with digoxigenin-11-UTP (Roche Diagnostics) using T7 and SP6 RNA polymerases, respectively. The following primers were used to amplify the DNA template for the probe synthesis: WOX5 primers 5′-AAACTCGAGAGGCAGAAACGTCGTAAAATCT-3′ and 5′-TCAGGATCCTTAAAGAAAGCTTAATCGAAGATCTA-3′.
Gene Expression Analysis
For qRT-PCR analysis, seedling roots were harvested and frozen in liquid nitrogen for RNA extraction. RNA was extracted with the RNeasy kit (Qiagen). Poly(dT) cDNA was prepared from 10 μg of total RNA with Superscript III reverse transcriptase (Invitrogen) and quantified with a cycler apparatus (Bio-Rad) with the Real Master Mix kit (SYBR Green; Tiangen) according to the manufacturer’s instructions. PCR was performed in 96-well optical reaction plates heated for 5 min at 95°C to activate hot start Taq DNA polymerase, followed by 40 cycles of denaturation for 30 s at 95°C, annealing for 30 s at 59°C, and extension for 20 s at 68°C. Expression levels of target genes were normalized to those of ACTIN2 or ACTIN7. The statistical significance was evaluated by Student’s t test. For multiple comparisons, an analysis of variance followed by Fisher’s LSD mean separation test (SPSS) was performed on the data. Samples with different letters are significantly different at P < 0.01 or P < 0.05. Primers used to quantify gene expression levels are listed in Supplemental Table 1 online.
ChIP-PCR Assay
One gram of 8-d-old 35Spro:MYC2 (35Spro:MYC2-4Myc) seedlings was used for ChIP experiments. ChIP was performed as previously described (Gendrel et al., 2005). The enrichment of DNA fragments was determined by semiquantitative PCR using the following primer pairs: ACT2pro F, 5′-CGTTTCGCTTTCCTTAGTGTTAGCT-3′; ACT2proR, 5′-CACAACGCATGCTAAACAGATCTAG-3′; PLT1pro(P1)F, 5′-GCCCCCTTATTGAATTGGCTCTT-3′; PLT1pro(P1)R, 5′-GCATACTTGATCCAGTATATGCA-3′; PLT2pro(P2)F, 5′-CACATGCAATGAACTCGGGGATC-3′; PLT2pro(P2)R, 5′-CCCTCTGATCTCTACATACTAAC-3′; PLT2pro(P3)F, 5′-CAGTCCCTCCTTGATGAGATCAT-3′; and PLT2pro(P3)R, 5′-CATTTCCCTTTTCTTGGAATCAA-3′.
EMSA Assay
To construct a plasmid for the expression of recombinant MYC2 protein in Escheichia coli, the full-length cDNA fragment was amplified by PCR using primers 5′-GCTAGCATGACTGATTACCGGCTACAAC-3′ and 5′-CTGCAGGACCGATTTTTGAAATCAAACTTGC-3′ and cloned into the pMAL-c2 vector via XbaI and PstI restriction sites. Oligonucleotide probes were synthesized and labeled with biotin at the 3′ end (Invitrogen). EMSA was performed using a LightShift Chemiluminescent EMSA kit (Thermo Scientific). Briefly, biotin-labeled probes were incubated in 1× binding buffer, 2.5% glycerol, 50 mM KCl, 5 mM MgCl2, and 10 mM EDTA with or without proteins at room temperature for 20 min. For nonlabeled probe competition, nonlabeled probes were added to the reactions. The probe sequences were as follows: PLT1 probe sequence, 5′-GAATTGGCTCTTATAATATTGCATGAACGTACACATGCAGAAGTGAGAGAATTCGGGGATCTACAATAA-3′; PLT2 probe sequence, 5′-AAGACATGTAAAATCGAAAGCATCATTGCACATGTTTTAAGATTGTTATGTTCATATATTTTCTTG-3′; PLT1 probe without the CACATG motif (Mu), 5′-GAATTGGCTCTTATAATATTGCATGAACGTACAGAAGTGAGAGAATTCGGGGATCTACAATAA-3′; PLT2 probe without the CACATG motif (Mu), 5′-AAGACATGTAAAATCGAAAGCATCATTGTTTTAAGATTGTTATGTTCATATATTTTCTTG-3′.
Transinhibition of PLT1 Promoter Activity by MYC2 in N. benthamiana Leaves
The transient expression assays were performed in N. benthamiana leaves as previously described (Matsui et al., 2008; Shang et al., 2010; Qi et al., 2011). The PLT1 promoter was amplified with the primer pairs 5′-CACCCCTAGCACAATACAATGCAAAGGAC-3′ and 5′-CTACCACTTTGGTATGATCAATATACC-3′ and cloned into pENTR using the pENTR Directional TOPO cloning kit (Invitrogen). To generate PLT1 promoter with mutations, site-directed mutagenesis was used to delete the CACATG motif in the P1 region of the PLT1 promoter (Figure 6A) using the TaKaRa MutanBEST kit. Then, both PLT1 promoter versions were fused with the luciferase reporter gene LUC through the Gateway reactions into the plant binary vector pGWB35 (Nakagawa et al., 2007) to generate the reporter constructs PLT1pro:LUC and PLT1-mpro:LUC. The MYC2 effector construct was the above-described 35Spro:MYC2-4Myc (35Spro:MYC2).
Agrobacterium-mediated infiltration of N. benthamiana leaves was performed as described (Shang et al., 2010; Qi et al., 2011). Infiltrated plants were incubated at 22°C for 72 h before CCD imaging. For COR treatment, the infiltration buffer containing 5 μM COR (+COR) (or solvent buffer as control) was infiltrated into the N. benthamiana leaf area coexpressing the indicated constructs 10 h before CCD imaging.
We used a low-light cooled CCD imaging apparatus (NightOWL II LB983 with indigo software) to capture the LUC image and to count luminescence intensity. The leaves were sprayed with 100 mM luciferin and were placed in darkness for 3 min before luminescence detection. Five independent determinations were assessed. Error bars represent sd. The experiments were repeated at least five times with similar results.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: At3g18780 (ACTIN2), At5g09810 (ACTIN7), At4g37490 (CYCB1;1), At3g48750 (CDC2A), At2g23430 (KRP1), At1g07370 (PCNA1), At3g20840 (PLT1), At1g51190 (PLT2), At3g11260 (WOX5), At1g32640 (MYC2), and At2g39940 (COI1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. JA Reduces Both Cell Number and Cell Length of the DZ and EZ.
Supplemental Figure 2. COI1 and MYC2 Are Involved in JA-Induced Inhibition of Root Growth.
Supplemental Figure 3. JA-Mediated Promotion of QC Division in the Ethylene Signaling Mutants ein2-5 and ein3-1 eil1-1.
Supplemental Figure 4. The JA Overproduction Mutant cev1 Is Defective in Maintenance of the Root Stem Cell Niche.
Supplemental Figure 5. JA-Induced Modulation of Root Meristem Size and Stem Cell Niche Maintenance in the JA-Hypersensitive Mutant jaz10.
Supplemental Figure 6. JA-Induced Repression of PLT Expression in the Auxin Biosynthesis Mutants asa1-1 and yuc1D.
Supplemental Figure 7. Presence of the JA-Responsive 5′-GAGTA-3′ Motif in Promoter Regions of PLT1 and PLT2.
Supplemental Table 1. DNA Primers Used for qRT-PCR Assays in This Study.
Acknowledgments
We thank Ben Scheres and Daoxin Xie for kindly providing seeds used in this study. This work was supported by grants from The Ministry of Science and Technology of China (2011CB915400 and 2007CB948200) and The National Natural Science Foundation of China (31030006, 90717007, and 31070251).
AUTHOR CONTRIBUTIONS
C.L. designed the research, analyzed the data, and wrote the article. Q.C. and J.S. designed and performed the research and analyzed the data. Q.Z., W.Z., L.Q., L.X., B.W., R.C., H.J., J.Q., and X.L. performed the research and analyzed the data. K.P. analyzed the data.
References
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.S., Amasino R., Scheres B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003b). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova A.N., Solano R., Wisman E., Ferrari S., Ausubel F.M., Ecker J.R. (2003a). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N., Pelletier G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Beemster G.T., Baskin T.I. (2000). Stunted plant 1 mediates effects of cytokinin, but not of auxin, on cell division and expansion in the root of Arabidopsis. Plant Physiol. 124: 1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: The looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465 [DOI] [PubMed] [Google Scholar]
- Benková E., Hejátko J. (2009). Hormone interactions at the root apical meristem. Plant Mol. Biol. 69: 383–396 [DOI] [PubMed] [Google Scholar]
- Berger S., Bell E., Mullet J.E. (1996). Two methyl jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 111: 525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2005). Jasmonate: An oxylipin signal with many roles in plants. Vitam. Horm. 72: 431–456 [DOI] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Casamitjana-Martínez E., Hofhuis H.F., Xu J., Liu C.M., Heidstra R., Scheres B. (2003). Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Creelman R.A., Mullet J.E. (1997). Oligosaccharins, brassinolides, and jasmonates: Nontraditional regulators of plant growth, development, and gene expression. Plant Cell 9: 1211–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe W., Rönsch H., Preiss A., Schade W., Sembdner G., Schreiber K. (1981). Endogenous plant hormones of the broad bean, Vicia faba L. (-)-jasmonic acid, a plant growth inhibitor in pericarp. Planta 153: 530–535 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F.S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., Sabatini S. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17: 678–682 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M.T., Aoyama T., Costantino P., Sabatini S. (2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Devoto A., Nieto-Rostro M., Xie D., Ellis C., Harmston R., Patrick E., Davis J., Sherratt L., Coleman M., Turner J.G. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32: 457–466 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Ding Z., Friml J. (2010). Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 107: 12046–12051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny J.R., Benfey P.N. (2008). Plant stem cell niches: Standing the test of time. Cell 132: 553–557 [DOI] [PubMed] [Google Scholar]
- Dolan L., Janmaat K., Willemsen V., Linstead P., Poethig S., Roberts K., Scheres B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelkrout E.M., Mariconti L., Settlage S.B., Cella R., Robertson D., Hanley-Bowdoin L. (2002). Two E2F elements regulate the proliferating cell nuclear antigen promoter differently during leaf development. Plant Cell 14: 3225–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C., Karafyllidis I., Wasternack C., Turner J.G. (2002). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C., Turner J.G. (2001). The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Fuerst R.A., Soni R., Murray J.A., Lindsey K. (1996). Modulation of cyclin transcript levels in cultured cells of Arabidopsis thaliana. Plant Physiol. 112: 1023–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R., Scheres B. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218 [DOI] [PubMed] [Google Scholar]
- Grieneisen V.A., Xu J., Marée A.F.M., Hogeweg P., Scheres B. (2007). Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Guo H., Ecker J.R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Hejátko J., Blilou I., Brewer P.B., Friml J., Scheres B., Benková E. (2006). In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat. Protoc. 1: 1939–1946 [DOI] [PubMed] [Google Scholar]
- Howe G.A., Jander G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2008). Jasmonate signaling: Toward an integrated view. Plant Physiol. 146: 1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Laux T. (2003). The stem cell concept in plants: A matter of debate. Cell 113: 281–283 [DOI] [PubMed] [Google Scholar]
- Leyser H.M., Lincoln C.A., Timpte C., Lammer D., Turner J., Estelle M. (1993). Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364: 161–164 [DOI] [PubMed] [Google Scholar]
- Lincoln C., Britton J.H., Estelle M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sánchez-Serrano J.J., Solano R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M.C., Jørgensen J.E., Lawton M.A., Lamb C.J., Doerner P.W. (1992). Spatial pattern of cdc2 expression in relation to meristem activity and cell proliferation during plant development. Proc. Natl. Acad. Sci. USA 89: 7360–7364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K., Umemura Y., Ohme-Takagi M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55: 954–967 [DOI] [PubMed] [Google Scholar]
- Memelink J. (2009). Regulation of gene expression by jasmonate hormones. Phytochemistry 70: 1560–1570 [DOI] [PubMed] [Google Scholar]
- Mockaitis K., Estelle M. (2008). Auxin receptors and plant development: A new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24: 55–80 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Ortega-Martínez O., Pernas M., Carol R.J., Dolan L. (2007). Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317: 507–510 [DOI] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Morreel K., De Witte E., Lammertyn F., Van Montagu M., Boerjan W., Inzé D., Goossens A. (2008). Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 105: 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J., Friml J. (2009). Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Qi T.C., Song S.S., Ren Q.C., Wu D.W., Huang H., Chen Y., Fan M., Peng W., Ren C.M., Xie D.X. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A., Bannigan A., Sulaman W., Pechter P., Blancaflor E.B., Baskin T.I. (2007). Auxin, actin and growth of the Arabidopsis thaliana primary root. Plant J. 50: 514–528 [DOI] [PubMed] [Google Scholar]
- Růzicka K., Ljung K., Vanneste S., Podhorská R., Beeckman T., Friml J., Benková E. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Santner A., Calderon-Villalobos L.I., Estelle M. (2009). Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5: 301–307 [DOI] [PubMed] [Google Scholar]
- Sarkar A.K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Scheres B., Benfey P., Dolan L. (2002). Root development. In The Arabidopsis Book. 1: e0101, doi/10.1199/tab.0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Hoyt J.M., Hamilton A.A., Alonso J.M. (2005). A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17: 2230–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Dolezal K., Schlereth A., Jürgens G., Alonso J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Yun J., Likhacheva A.V., Alonso J.M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19: 2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., et al. (2009). Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21: 1495–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Perry P., Hagenbeek D., Van Der Straeten D., Beemster G.T., Sandberg G., Bhalerao R., Ljung K., Bennett M.J. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19: 2186–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatek A., Azmi A., Stals H., Inzé D., Van Onckelen H. (2004). Jasmonic acid prevents the accumulation of cyclin B1;1 and CDK-B in synchronized tobacco BY-2 cells. FEBS Lett. 572: 118–122 [DOI] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Dhonukshe P., Brewer P.B., Friml J. (2006). Spatiotemporal asymmetric auxin distribution: A means to coordinate plant development. Cell. Mol. Life Sci. 63: 2738–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Tiryaki I., Staswick P.E. (2002). An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol. 130: 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J.G., Ellis C., Devoto A. (2002). The jasmonate signal pathway. Plant Cell 14(suppl.): S153–S164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C., Willemsen V., Hendriks G., Weisbeek P., Scheres B. (1997). Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390: 287–289 [DOI] [PubMed] [Google Scholar]
- Vanneste S., Friml J. (2009). Auxin: A trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Vanstraelen M., Baloban M., Da Ines O., Cultrone A., Lammens T., Boudolf V., Brown S.C., De Veylder L., Mergaert P., Kondorosi E. (2009). APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 106: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Fowke L.C., Crosby W.L. (1997). A plant cyclin-dependent kinase inhibitor gene. Nature 386: 451–452 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007). Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F.M., Hogan B.L. (2000). Out of Eden: Stem cells and their niches. Science 287: 1427–1430 [DOI] [PubMed] [Google Scholar]
- Weigel D., Glazebrook J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Weigel D., Jürgens G. (2002). Stem cells that make stems. Nature 415: 751–754 [DOI] [PubMed] [Google Scholar]
- Wolters H., Jürgens G. (2009). Survival of the flexible: Hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 10: 305–317 [DOI] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Turner J.G. (2008). Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Christensen S.K., Fankhauser C., Cashman J.R., Cohen J.D., Weigel D., Chory J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
- Zhou W., Wei L., Xu J., Zhai Q., Jiang H., Chen R., Chen Q., Sun J., Chu J., Zhu L., Liu C.M., Li C. (2010). Arabidopsis Tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 22: 3692–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]