This work presents insight into how boron (B) toxicity develops at the molecular level in plants and suggests a role of condenisn II genes in tolerance to B toxicity. Experiments show that excess B induces DNA damage, and condensin II is involved in the amelioration of this damage.
Abstract
Although excess boron (B) is known to negatively affect plant growth, its molecular mechanism of toxicity is unknown. We previously isolated two Arabidopsis thaliana mutants, hypersensitive to excess B (heb1-1 and heb2-1). In this study, we found that HEB1 and HEB2 encode the CAP-G2 and CAP-H2 subunits, respectively, of the condensin II protein complex, which functions in the maintenance of chromosome structure. Growth of Arabidopsis seedlings in medium containing excess B induced expression of condensin II subunit genes. Simultaneous treatment with zeocin, which induces DNA double-strand breaks (DSBs), and aphidicolin, which blocks DNA replication, mimicked the effect of excess B on root growth in the heb mutants. Both excess B and the heb mutations upregulated DSBs and DSB-inducible gene transcription, suggesting that DSBs are a cause of B toxicity and that condensin II reduces the incidence of DSBs. The Arabidopsis T-DNA insertion mutant atr-2, which is sensitive to replication-blocking reagents, was also sensitive to excess B. Taken together, these data suggest that the B toxicity mechanism in plants involves DSBs and possibly replication blocks and that plant condensin II plays a role in DNA damage repair or in protecting the genome from certain genotoxic stressors, particularly excess B.
INTRODUCTION
The metalloid element boron (B) is an essential nutrient in plants and probably in animals (reviewed in Nielsen, 2008), but excess B is toxic to both plants and animals. In animals, exposure to high concentrations of B causes reproductive abnormalities, such as a decrease in the X:Y sperm ratio (Robbins et al., 2008). In plants, the biochemical and physiological effects of exposure to excess B have been well studied because B toxicity is an important agricultural problem (reviewed in Nable et al., 1997). Excess B has been shown to affect several developmental/biochemical processes in plants, altering metabolism (Lukaszewski et al., 1992), reducing root cell division (Liu et al., 2000), reducing shoot cell wall expansion (reviewed in Loomis and Durst, 1992), and generating reactive oxygen species (ROS) and subsequent oxidative damage (Cervilla et al., 2007). Furthermore, several genes involved in plant tolerance to excess B have been identified, including Arabidopsis thaliana BOR4 and TIP5;1 (Miwa et al., 2007; Pang et al., 2010) and barley (Hordeum vulgare) Bot1 (Sutton et al., 2007). These genes encode transport molecules that exclude excess B or regulate intracellular B homeostasis to prevent B toxicity. However, the identification of these transporters has not shed light on the molecular mechanisms underlying B toxicity, which remains poorly understood.
To obtain insight into the molecular mechanisms underlying B toxicity, we previously isolated seven Arabidopsis mutant lines that are hypersensitive to excess B (heb) and speculated that these mutations might involve genes essential for tolerance to B toxicity (Sakamoto et al., 2009). In this study, we characterized two of these mutants, heb1-1 and heb2-1, and found that HEB1 and HEB2 encode chromosomal-associated protein (CAP)-G2 and CAP-H2, respectively, two subunits of the chromosomal protein complex known as condensin II (Fujimoto et al., 2005).
Higher eukaryotes have two types of condensins, condensin I and II. Both types of condensin contain two structural maintenance of chromosome (SMC) subunits, CAP-C (SMC2) and CAP-E (SMC4), but each type has a unique set of non-SMC subunits. In condensin II, these non-SMC subunits are CAP-G2, -H2, and -D3 (reviewed in Hirano, 2005). Animal condensins are localized to chromosomes during mitosis and are required for proper chromosomal condensation and segregation (Ono et al., 2003; Hirota et al., 2004; Ono et al., 2004). Additionally, it has been suggested that condensins have roles in the repair of DNA double-strand breaks (DSBs) and single-stranded DNA (ssDNA) (Heale et al., 2006; Wood et al., 2008).
The subcellular localization of condensins and their function in chromosomal condensation and segregation have been demonstrated in plants (Siddiqui et al., 2003; Fujimoto et al., 2005), but their role in genotoxicity is unknown. In this study, we found that Arabidopsis condensin II may play a role in DNA damage repair or in protecting the Arabidopsis genome from abiotic stressors. We suggest that B toxicity induces genotoxicity that is ameliorated by condensin II.
RESULTS
Hypersensitivity of heb Mutants to Excess B
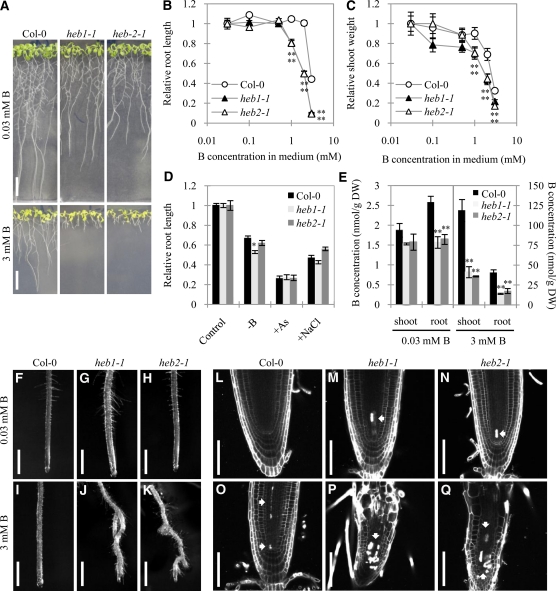
In medium containing excess B (i.e., 3 mM boric acid), root growth of wild-type (Columbia-0 [Col-0]) Arabidopsis plants is moderately inhibited (Figures 1A and 1B). In our previous study, we identified seven Arabidopsis mutants that exhibit defective root elongation when grown in 3 mM B-containing medium (hereafter, 3 mM B) using a genetic screen. Two of these mutants, heb1-1 and heb2-1, were selected for this study and backcrossed with Col-0 plants three times before use.
Figure 1.
Phenotypes of Arabidopsis heb1-1 and heb2-1 Mutants Under B Overload Stress.
(A) Col-0, heb1-1, and heb2-1 seedlings grown in medium containing 0.03 mM (top) or 3 mM B (bottom) for 14 d. Bars = 1 cm.
(B) and (C) Dose-dependent effects of B on growth of roots (B) and shoots (C) of Col-0, heb1-1, and heb2-1 seedlings. The primary root length and shoot flesh weight of 14-d-old seedlings grown in medium containing various concentrations of B (0.03, 0.1, 0.5, 1.0, 2.0, or 3.0 mM) are expressed as means ± se (n > 16) relative to control values (obtained in normal medium [0.03 mM B]), which are defined as 1.
(D) Sensitivity of Col-0, heb1-1, and heb2-1 seedlings to the mineral stress conditions of B deficiency (0.03 μM B), arsenite contamination (7.5 μM arsenite), and high salinity (75 mM NaCl). Primary root lengths of 14-d-old seedlings are expressed as means ± se (n > 12) relative to the values obtained in control conditions.
(E) B concentrations in tissues of Col-0, heb1-1, and heb2-1 seedlings grown as described in (A). Data are shown as means ± se (n = 3). Asterisks in (B) to (E) represent significant differences (*P < 0.05, **P < 0.01; Student’s t test) relative to Col-0. DW, dry weight.
(F) to (K) Stereomicroscope images showing effects of excess B on primary root morphology of Col-0 ([F] and [I]), heb1-1 ([G] and [J]), and heb2-1 ([H] and [K]) plants. Seedlings were grown in medium containing 0.03 mM B ([F] to [H]) or 3 mM B ([I] to [K]) for 14 d. Bars = 1 mm.
(L) to (Q) Confocal images showing the effects of excess B on root tips of Col-0 ([L] and [O]), heb1-1 ([M] and [P]), and heb2-1 ([N] and [Q]) seedlings grown in medium containing 0.03 mM ([L] to [N]) or 3 mM ([O] to [Q]) B for 14 d. PI staining was used for visualization of cell walls. Completely stained cells are dead (arrows). Bars = 50 μm.
We first examined the effect of various concentrations of B on the growth of the heb1-1 and heb2-1 mutants. In medium containing the control concentration of B (0.03 mM; hereafter, 0.03 B or normal B), the heb mutant roots were ~70% as long as the Col-0 roots, but in 3 mM B, the heb mutant roots were only 10% the length of the Col-0 roots (Figure 1A). Growth of heb1-1 and heb2-1 roots was reduced by B in a dose-dependent manner at B concentrations of 1 mM or more, whereas Col-0 root growth was not affected by B concentrations of up to 2 mM (Figure 1B). B at 1, 2, or 3 mM had a more severe effect on root length in the mutant plants than in the Col-0 plants (Figure 1B). High B concentrations also inhibited shoot growth more severely in the heb1-1 and heb2-1 plants than in Col-0 plants (Figure 1C). These results established that heb1-1 and heb2-1 are sensitive to excess B; relative to the wild-type line, these mutants exhibited growth inhibition at a lower B concentration, and their extent of B-induced growth inhibition was greater.
To examine whether the B-sensitive phenotype is specific to excess B, the growth of the heb mutants was tested under other conditions of mineral stress. Like excess B (Cervilla et al., 2007), B deficiency (Koshiba et al., 2009), arsenite treatment (reviewed in Verbruggen et al., 2009), and salinity stress (reviewed in Mahajan et al., 2008) induce the production of ROS. We reasoned that if the high sensitivity of the heb mutants to excess B was attributable to increased ROS sensitivity, treatments that generate ROS should also inhibit root growth in the heb mutants. However, the effects of arsenite treatment or high salinity on root elongation did not vary significantly between Col-0 and heb mutant plants, but B deficiency had a slightly greater effect on root elongation in heb1-1 plants than in Col-0 plants, suggesting that the heb1-1 phenotype is largely specific to B (Figure 1D) and that ROS are not involved in the hypersensitivity phenotype. The sensitivity of the mutants to other ROS-generating reagents (hydrogen peroxides and methyl viologen) was similar to that of Col-0 plants (see Supplemental Figure 1 online), providing further evidence that ROS are not involved in the response to excess B.
When we examined the B content of heb1-1, heb2-1, and Col-0 plants grown in medium containing 0.03 or 3 mM B, we found that the B concentration in the mutant shoots and roots was only 60 to 80% and 30%, respectively, of that in Col-0 shoots and roots (Figure 1E). This finding suggests that the growth defects observed in the heb mutants are not a result of overaccumulation of B.
Excess B Causes Severe Defects in the Meristematic Organization and Morphology of heb Mutant Roots
After growth under normal B conditions, the roots of Col-0, heb1-1, and heb2-1 plants exhibited a similar morphology (Figures 1F to 1H), but after growth in excess B, the heb1-1 and heb2-1 roots were twisted, with ectopic lateral root formation and dense root hairs, whereas the Col-0 roots retained their normal morphology (Figures 1I to 1K). Confocal microscopy of longitudinal root sections revealed that the meristematic zones of heb1-1 and heb2-1 root tips were shorter than those of Col-0 root tips, particularly after growth in excess B (see Supplemental Figure 2 online).
Propidium iodide (PI)–stained dead cells were observed in heb mutant root tips (15 of 19 roots examined for heb1-1 and 15 of 19 roots examined for heb2-1), but not in Col-0 root tips, after growth in normal B (Figures 1L to 1N). After growth in excess B, the number of PI-stained cells in heb1-1 and heb2-1 root tips increased (20 of 20 roots examined for heb1-1 and heb2-1), and the stained cells occupied a larger area; furthermore, some PI-stained cells were also found in Col-0 root tips (10 of 30 roots) (Figures 1O to 1Q). Enlarged cells of abnormal shape were observed in heb1-1 and heb2-1 root tips; additionally, in excess B, but not in normal B, the heb1-1 and heb2-1 root apical meristems were more disorganized than those of Col-0 plants (Figures 1L to 1Q). These observations suggest that HEB1 and HEB2 are involved in maintaining cell viability under normal conditions and that they are involved in cell division/elongation during B overload stress.
HEB1 and HEB2 Encode Subunits of the Chromosomal Protein Complex Condensin II
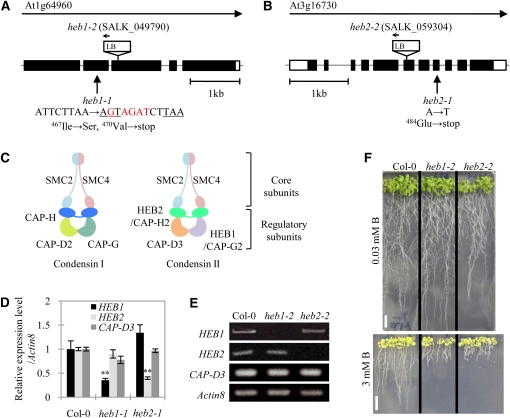
Genetic mapping limited the HEB1 and HEB2 loci to ~70- and 12-kb regions containing At1g64960 and At3g16730, respectively (see Supplemental Figure 3 online), and the mutations in the DNA regions containing the heb loci were identified (Figures 2A and 2B). The heb1-1 and heb2-1 mutant genes contain mutations in At1g64960 and At3g16730, respectively, which cause premature termination of the open reading frames. Database analysis identified HEB1 and HEB2 as CAP-G2 and CAP-H2, respectively, which together with CAP-D3, comprise the non-SMC subunits of condensin II (Ono et al., 2003) (Figure 2C).
Figure 2.
Molecular Characterization of HEB1 and HEB2.
(A) and (B) Genomic structures. Black and white boxes indicate coding regions and untranslated regions, respectively, of HEB1 (A) and HEB2 (B). Sites of the heb mutations and T-DNA insertion sites for SALK lines are shown.
(C) Subunit organization of the condensin complexes I and II.
(D) Real-time PCR analysis of condensin II gene expression in the roots of Col-0, heb1-1, and heb2-1 seedlings grown in medium containing 0.03 mM B. Total RNA was extracted from the whole roots of 14-d-old seedlings. At least 10 plants were used per replicate. Levels of condensin II mRNA were normalized to the actin8 mRNA levels in the same samples. The data are expressed as means ± se (n = 3) relative to the Col-0 value (defined as 1). Asterisks represent significant differences (**P < 0.01; Student’s t test) relative to Col-0.
(E) RT-PCR analysis of the expression of condensin II genes in the roots of Col-0, heb1-2, and heb2-2 seedlings. Total RNA was extracted from whole roots of 14-d-old seedlings. The actin8 mRNA levels were used as references.
(F) Excess B-dependent short root phenotype of heb1-2 and heb2-2 mutant seedlings. Col-0 and mutant seedlings were grown in medium containing 0.03 mM (top) or 3 mM (bottom) B for 10 d. Bars = 1 cm.
We isolated T-DNA insertion mutants of these alleles, heb1-2 (SALK_049790) and heb2-2 (SALK_059304) (Figures 2A and 2B). The transcripts of the corresponding genes accumulated in heb1-1 or heb2-1 roots to only 30 to 40% of the amount found in Col-0 roots (Figure 2D) and were not detected in heb1-2 or heb2-2 roots (Figure 2E). Both T-DNA insertion mutants showed short root phenotypes similar to those observed in heb1-1 and heb2-1 plants exposed to excess B (Figure 2F). Furthermore, we confirmed that At1g64960 and At3g16730 correspond to HEB1 and HEB2, respectively, by introducing HEB1pro:HEB1:GFP (for green fluorescent protein) and HEB2pro:HEB2:GFP into heb1-1 and heb2-1, respectively. After obtaining several independently transformed lines and confirming the expression of HEB1:GFP and HEB2:GFP, we demonstrated that all of the transgenic lines exhibited elongated roots similar to those of wild-type plants when grown in the presence of 3 mM B (see Supplemental Figure 4 online). These data established the identification of HEB1 and HEB2.
In our study, the heb1 and heb2 mutants, which have mutations in different subunits of the condensin II complex, had similar phenotypes, suggesting that the phenotype of the heb mutants is probably attributable to defects in condensin II.
The heb Mutants Are Sensitive to DSB Induction
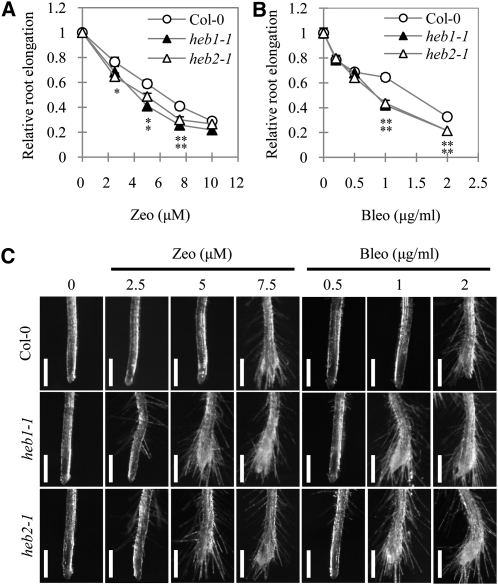
Human condensin II functions in homologous recombination repair after the generation of DSBs (Wood et al., 2008). To investigate whether Arabidopsis condensin II plays a role in the DNA damage response, we examined the effects of the radiomimetic reagents zeocin and bleomycin, which induce DSBs, on root elongation in the heb mutants. Plants were grown on normal medium for 5 d, transferred to medium containing various concentrations of the DSB-inducing reagents, and grown for another 4 d. Subsequent evaluation of root length showed that both zeocin (5 and 7.5 μM) and bleomycin (1 and 2 μg/mL) had more severe inhibitory effects on root elongation in the heb mutants than in the Col-0 plants (Figures 3A and 3B).
Figure 3.
Sensitivity of the heb Mutants Root Growth to DSBs Induced by Zeocin and Bleomycin.
(A) and (B) Effects of zeocin (Zeo; [A]) or bleomycin (Bleo; [B]) on root elongation in Col-0, heb1-1, and heb2-1 seedlings. Plants were treated with zeocin or bleomycin as described in Methods. Root elongation data are expressed as means ± se (n > 13) relative to values obtained under control conditions. Asterisks represent significant differences (*P < 0.05, **P < 0.01; Student’s t test) relative to Col-0.
(C) Effects of zeocin and bleomycin on root morphology of Col-0, heb1-1, and heb2-1 plants. Plants were treated with zeocin or bleomycin as described in Methods. Bars = 500 μm.
We also examined the effect of zeocin and bleomycin on root morphology (Figure 3C). Zeocin (2.5 μM) had no visible effect on root morphology in Col-0 plants, but slightly altered it in the heb mutants, causing some ectopically generated root hairs, and strongly enhanced cell death only in heb1-1 and heb2-1 root tips (see Supplemental Figure 6 online). Abnormal root morphology was evident in the heb mutants at >5 μM zeocin and in Col-0 plants at 7.5 μM zeocin. The effect of bleomycin on root morphology was similar to that of zeocin, and its effects were observed at a lower concentration in the heb mutants than in Col-0 plants. Thus, the heb mutants exhibited increased sensitivity to the effects of DSB-inducing reagents on both root elongation and root morphology, suggesting that HEB1 and HEB2 are involved in DSB repair and/or in tolerance to genotoxicity.
Excess B Induces DSBs in Root Tip Cells
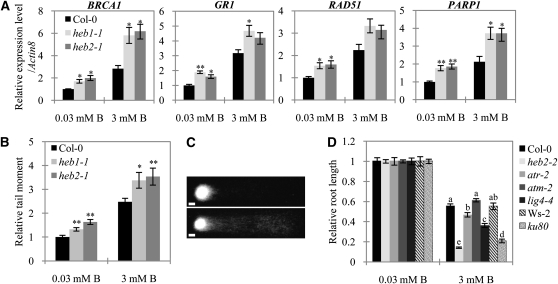
Based on our findings that the heb mutants are sensitive both to excess B and to the induction of DSBs, we hypothesized that excess B induces DSBs and that condensin II functions to ameliorate DSBs. To examine this hypothesis, we first conducted real-time RT-PCR analysis of the transcription of the DSB-inducible genes BRCA1 (Lafarge and Montané, 2003), GR1 (Deveaux et al., 2000), RAD51 (Doutriaux et al., 1998), and PARP1 (Doucet-Chabeaud et al., 2001). Growth of plants in excess B for 4 d upregulated transcript levels of these genes in root tips of both Col-0 and heb plants (Figure 4A), consistent with our hypothesis that excess B causes DNA damage and condensin II functions in the DNA damage response. When plants were grown in either normal or 3 mM B, transcript levels for all four genes were ~50 to 100% higher in the heb mutants than in Col-0 plants (Figure 4A), suggesting that levels of DSBs are increased in the heb mutants irrespective of the B conditions.
Figure 4.
DSB levels in the heb Mutants and Induction of DSBs by Excess B in Roots.
(A) Expression of DNA damage-inducible genes in root tips in response to B toxicity. Five-day-old Col-0, heb1-1, and heb2-1 seedlings were transferred to medium containing 0.03 or 3 mM B and incubated for 4 d. Total RNA was extracted from 1-cm distal root tip segments, and gene expression levels were determined by real-time PCR analysis. At least 50 plants were used per replicate. Data were normalized to actin8 mRNA levels in the same samples and are expressed as means ± se (n = 3) relative to the value for Col-0 grown in medium containing 0.03 mM B (defined as 1).
(B) DNA damage in root tips in response to B toxicity. Nuclei were extracted from root tips of plants grown as described in (A) and analyzed for DNA damage using the comet assay. The relative olive tail moment reflects the extent of DSBs in the nucleus relative to the extent measured for Col-0 plants grown in medium containing 0.03 mM B (defined as 1). Data are expressed as means ± se of at least 125 comets. The experiment was repeated three times. Asterisks in (A) and (B) represent significant differences (*P < 0.05 and **P < 0.01) relative to Col-0; Student’s t test.
(C) Examples of comets exhibiting nearly intact nuclei (top, short tail) and Col-0 nuclei severely damaged by B toxicity (bottom, long tail). Images were taken using a fluorescence microscope. Bars = 10 μm.
(D) Effect of excess B on DNA damage-sensitive plants. The primary root lengths of Col-0, heb2-2, atm-2, atr-2, lig4-4, Ws-2, and ku80 seedlings were measured after growth in medium containing 0.03 or 3 mM B for 10 d. Data shown are expressed as means ± se (n > 13) relative to the value obtained for Col-0 plants grown in medium containing 0.03 mM B (defined as 1). Means sharing the same letter within a column for each line are not significantly different at 5% probability by Tukey’s multiple range test.
We next investigated the levels of DSBs in the root tips of Col-0 and heb plants grown in normal and 3 mM B using comet assays (Menke et al., 2001). A 4-d incubation in 3 mM B significantly elevated the levels of DSBs in both Col-0 and heb plants (Figures 4B and 4C). A higher accumulation of DSBs was observed in root tips of heb mutants than in those of Col-0 plants, irrespective of the B concentration (Figure 4B). The difference in DSB levels between Col-0 and heb plants was larger at 3 mM B than at 0.03 mM B. The larger difference in DSB levels between Col-0 and heb plants at 3 mM implies that the effect of the condensin II mutations (heb1-1 and heb2-1) is not a simple addition of the DSB levels on top of the DSBs caused by high B treatment but that the mutants are more vulnerable to high B treatment. This synergy of B toxicity and lack of condensin II on DSB accumulation implies the involvement of condensin II in reverting the effects of B overload stress. Taken together, these results demonstrate that excess B promotes the formation of DSBs in root tip cells in Arabidopsis and that condensin II is involved in ameliorating this DNA damage.
atr, lig4, and ku80 Mutants Are Sensitive to Excess B
In eukaryotic cells, two related kinases, ataxia telangiectasia mutated (ATM) and ATM and Rad3-related (ATR), are well-known molecular players in different pathways in the DNA damage response: They are required for the detection of DSBs and ssDNA, respectively (reviewed in Cools and De Veylder, 2009). To investigate the possible involvement of these kinases and proteins in DSB repair through a nonhomologous end-joining (NHEJ) pathway such as LIG4 and Ku80 (West et al., 2002; Heacock et al., 2007) in B-induced DNA damage, we examined the growth response of the mutants atm-2, atr-2, lig4-4, and ku80, which have been established as hypersensitive to various types of genotoxicity (West et al., 2002; Garcia et al., 2003; Culligan et al., 2004, 2006; Heacock et al., 2007), to excess B. Unexpectedly, when grown on medium containing 3 mM B for 10 d, the root growth of atm-2 and Col-0 plants was similar, whereas the root growth of the atr-2, lig4-4, and ku80 mutants were reduced to a larger extent than that of Col-0 plants (Figure 4D), suggesting that ATR, LIG4, and Ku80, but not ATM, play a crucial role in the DNA damage response caused by excess B. However, we should note that the atm-2 mutant is not proven to be null (Garcia et al., 2003); thus, it remains possible that in this mutant, small amounts of ATM could be rendered and contribute to tolerance to B toxicity.
Simultaneous DSB Induction and Replication Blocks Mimic the Effect of Excess B on Root Growth in the heb Mutants
The atr mutants of Arabidopsis are hypersensitive to replication blocks induced by aphidicolin, an inhibitor of DNA polymerase α, or hydroxyurea, which depletes deoxynucleotide triphosphate pools (Culligan et al., 2004). Therefore, the high sensitivity of the atr-2 mutant to excess B led us to speculate that excess B interferes with DNA replication and that condensin II is involved in maintaining replication processes. To investigate these possibilities, we evaluated the sensitivity of the heb mutants to aphidicolin and hydroxyurea. Exposure to 8 or 12 μg/mL aphidicolin inhibited root elongation more severely in the heb mutants than in Col-0 plants (Figure 5A). On the other hand, hydroxyurea appeared to have no effect on the heb mutants (Figure 5B). However, the elongation of atr-2 mutant roots was inhibited at lower concentrations of aphidicolin and hydroxyurea than were the heb mutants (Figures 5A and 5B). In terms of alteration of root morphology, the heb mutants had distinct responses to the two types of replication blocking reagents (Figure 5C). Exposure to 8 μg/mL aphidicolin caused ectopic root hairs to appear in the heb mutants, but not in the Col-0 plants, and this difference in root morphology was eliminated by an increase in the aphidicolin concentration to 12 μg/mL. Aphidicolin treatment (12 μg/mL) inhibited root elongation by 50 to 70% (Figure 5A). By contrast, exposure to 3 mM hydroxyurea (Figure 5C), which inhibited root elongation by 70% (Figure 5B), had no effect on the root morphology of either Col-0 or heb plants. These results indicate that both the elongation and morphology of heb roots are sensitive to replication blocks induced by aphidicolin, but not hydroxyurea.
Figure 5.
Comparative Sensitivity of Root Growth of Col-0 and the heb Mutants to the Stressors of DNA Damage and B Overload.
(A) and (B) Effects of the replication-blocking reagents aphidicolin (Aph) (A) and hydroxyurea (HU) (B) on root elongation of Col-0, heb1-1, heb2-1, and atr-2 seedlings (see details in Methods). Data shown are expressed as means ± se (n > 11) relative to the control condition.
(C) Root morphology of Col-0, heb1-1, and heb2-1 mutant plants treated with aphidicolin or hydroxyurea as in (A) and (B), respectively. Images were taken using a stereomicroscope. Bars = 500 μm.
(D) Comparison of the root elongation–inhibiting effects of zeocin (Zeo), aphidicolin (Aph), and excess B on Col-0, heb1-1, and heb2-1 seedlings. After 5 d in normal MGRL medium (control condition), seedlings were transferred to medium containing zeocin (1 μM) and/or aphidicolin (4 μg/mL) or 3 mM B as indicated and grown for a further 4 d or 6 d. Root elongation data are expressed as means ± se (n > 13) relative to values obtained under control conditions. Asterisks in (A), (B), and (D) represent significant differences (*P < 0.05 and **P < 0.01) relative to Col-0; Student’s t test.
(E) Root morphology of Col-0, heb1-1, and heb2-1 plants grown in zeocin (Zeo) and/or aphidicolin (Aph) or 3 mM B (4 and 6 d) as described in (D). Bars = 500 μm.
If HEB1 and HEB2 are involved in the cellular response to DSBs and replication blocks, then both stressors might be responsible for the B hypersensitivity of heb mutants. In root elongation assays, growth in 3 mM B reduced root elongation in Col-0 plants by ~20%, but this effect was 3 times greater in the heb mutants (Figure 5D). By contrast, the DSB-inducing (Figures 3A and 3B) and replication-blocking (Figure 5A) reagents were associated with smaller differences in the heb mutants relative to the Col-0 plants. In contrast with the effect of 3 mM B, the inhibitory effect of zeocin on root elongation was only 1.5 times greater in the heb mutants than in Col-0 plants (Figure 3A), and the effect of aphidicolin on the heb mutants was similar (Figure 5A). These observations suggest that the hypersensitivity of the heb mutants to excess B cannot be due to DSBs or replication blocks alone.
We next examined the combined effect of DSB induction and replication block on root elongation using zeocin and aphidicolin at 1 μM and 4 μg/mL, respectively. At these concentrations, root elongation in Col-0 plants was inhibited by ~20%, which is the same effect as that of 3 mM B. When these reagents were used separately, their differential effect on root elongation in the heb mutants versus Col-0 plants was less pronounced than the differential effect of 3 mM B (Figure 5D), and neither reagent severely altered root morphology in the heb mutants or Col-0 plants (Figure 5E). On the other hand, when both of these reagents were used together, a severe reduction in root growth (Figure 5D) and severe defects in root morphology (Figure 5E) were observed in the heb mutants but not in Col-0 plants. In the heb mutants, the effect of zeocin/aphidicolin combination treatment on root growth was comparable to that observed for 3 mM B, but its effect on root morphology differed from that of 3 mM B, as 3 mM B did not severely alter root morphology in Col-0 or heb plants (Figure 5E). At 6 d after treatment with 3 mM B, the heb mutants exhibited severe defects in root morphology, and the relative root length was reduced by ~80% (Figures 5D and 5E). Overall, these results suggest that both DSB induction and impairment of replication processes are involved in B toxicity and that HEB1 and HEB2 are required for the response to both types of DNA damage.
B Toxicity Affects Cell Cycle Progression in Roots
In plants, checkpoint control systems respond to a variety of genotoxicities by delaying or arresting the progression of the cell cycle (reviewed in Cools and De Veylder, 2009). Here, we observed that excess B caused DNA damage in Col-0 and heb mutants and affected root growth of the atr-2 mutant (Figure 4D), which has defective G2 checkpoint regulation, suggesting that excess B might impair cell cycle progression in roots. It is also possible that excess B increases premature endoreduplication as enlarged cells were seen in the heb mutants in excess B (Figures 1P and 1Q). Exposure to excess B for 4 d reduced the number of cells in the meristematic zones in Col-0 and heb plants by ~20 and 50%, respectively (Figure 6A), suggesting that excess B impairs cell division, particularly in the heb mutants. We examined the cell ploidy in the root tips of plants grown in 3 mM B for 4 d (Figure 6B), as 2C and 4C counts in mitotic cells generally correspond to the number of cells in G1 and G2 phase, respectively (López-Juez et al., 2008). We should note that endoreduplicative cells also contain 4C counts (Yoshizumi et al., 2006). Exposure to excess B for 4 d increased the 4C/2C ratio in Col-0 plants from 0.35 to 0.42, in heb2-1 plants from 0.50 to 0.73, and in heb2-2 plants from 0.58 to 0.73, suggesting the existence of a prolonged G2 phase and/or premature endoreduplication in the heb mutants. Thus, B overload appears to alter cell cycle progression, which could reduce the number of cells in the meristematic zone.
Figure 6.
Effects of Excess B on Mitotic Activity in Roots of the heb Mutants.
(A) Effect of excess B on the number of cortex cells in the root meristematic zone. Five-day-old Col-0, heb1-1, and heb2-1 seedlings were transferred to medium containing 0.03 or 3 mM B and grown for 4 d. The roots were stained with PI, and the number of cortex cells in the meristematic zone was counted under a confocal microscope. Data shown are means ± se (n = 5).
(B) Effect of excess B on cell ploidy in root tips of Col-0, heb2-1, and heb2-2 seedlings grown as described in (A). Data shown are means ± se for ~5000 nuclei per group (n = 3). In heb2-1 and heb2-2 plants, the 0.03 and 3 mM B conditions differ significantly in their 4C:2C ratios (P < 0.05; Student’s t test).
(C) Effect of B toxicity on expression of cell cycle–related genes in root tips. Plants were grown as described in (A). Total RNA was extracted from 1-cm distal root tip segments, and mRNA levels for the indicated gene levels were determined by real-time PCR analysis. At least 50 plants were used per replicate. Data shown are normalized to level of actin8 mRNA in the same samples and are expressed as means ± se (n = 3) relative to the value for Col-0 grown in medium containing 0.03 mM B (defined as 1). Asterisks in (A) and (C) represent significant differences (*P < 0.05 and **P < 0.01) relative to Col-0; Student’s t test.
The Effect of Excess B and Condensin II on Expression of Cell Cycle–Related Genes
We next investigated the effect of excess B on the expression of cell cycle–related genes in root tips. Using real-time RT-PCR, we analyzed the levels of mRNA of cyclin genes, including CYCA1;1, CYCA2;1, and CYCB1;1, and the cyclin-dependent kinase (CDK) genes CDKA1 and CDKB2;1 (Figure 6C) in root tips (1-cm proximal segments) of heb and Col-0 plants under normal conditions and after a 4-d exposure to 3 mM B.
First, we examined the expression of CYCA2;1, CYCA1;1, and CDKA1, which are expressed at G2/M phase (Menges et al., 2005). Under normal conditions, the level of CYCA2;1 mRNA accumulation in the heb mutants was almost 6 times that in the Col-0 plants. Growth in 3 mM B increased accumulation of this transcript dramatically (by 8.4- and 17.9- to 34.3-fold in Col-0 plants and heb plants, respectively). By contrast, levels of transcript accumulation for three other A2-type cyclin genes (CYCA2;2, CYCA2;3, and CYCA2;4) did not differ by more than twofold between the Col-0 and heb plants, and excess B did not result in a drastic induction of these genes. For CYCA1;1, excess B increased the mRNA level in the heb mutants but decreased it in Col-0 plants. For CDKA1, levels of transcript accumulation were higher in the heb mutants than in Col-0 plants under both normal and excess B conditions, and excess B led to a significant increase in transcript accumulation in all plants.
We also examined the expression of other G2/M- markers, CYCB1;1 and CDKB2;1 (Menges et al., 2005; Culligan et al., 2006). Under normal conditions, the level of CYCB1;1 mRNA accumulation was slightly lower in the Col-0 plants than in the heb2-1 mutant but similar to that in the heb1-1 mutant. Excess B treatment increased levels of CYCB1;1 mRNA accumulation in both heb mutants but not in Col-0 plants. By contrast, CDKB2;1 mRNA expression levels were similar in Col-0 and heb plants and were unaffected by excess B.
Taken together, these results show that excess B affects expression of cell cycle–related genes in a unique manner. In other words, not all of the genes involved in G2/M progression are coordinately regulated. Furthermore, the expression of the cyclin gene CYCA2;1 appears to be affected by the defects in condensin II.
DISCUSSION
In plants, an inverse correlation between the level of insensitivity to B toxicity and the level of B accumulation in tissue has been well established; e.g., tissue levels of B are lower in B-tolerant barley cultivars than in B-sensitive barley cultivars (Nable 1991). However, in our two Arabidopsis heb mutants that exhibit hypersensitivity to B toxicity, heb1-1 and heb2-1, a reduction in the cellular B concentration was not critical for tolerance of excess B. While the heb mutants contained less B than wild-type plants, their sensitivity to excess B was much greater (Figure 1E), indicating that Arabidopsis has a cellular mechanism that is particularly sensitive to excess B.
In this study of the heb mutants, we established that the genes encoding the CAP-G2 and CAP-H2 subunits of condensin II are essential for tolerance to excess B. Through a forward genetics analysis, we demonstrated that Arabidopsis condensin II acts in DSB amelioration and may act to maintain the replication process, two functions that are considered to be required for plant tolerance of B toxicity. As condensin II is conserved in animals, the mechanism of B toxicity examined herein may be common among plants and animals.
DNA Damage Is a Major Cause of B Toxicity in Arabidopsis Roots
Using a comet assay, we established that excess B causes DSBs in Arabidopsis root tip cells (Figure 4B). We also demonstrated that several genes known to be associated with increased DNA damage (BRCA1, GR1, RAD51, and PARP1) (Doutriaux et al., 1998; Deveaux et al., 2000; Doucet-Chabeaud et al., 2001; Lafarge and Montané, 2003) are induced in roots of Arabidopsis plants grown in medium containing excess B (Figure 4A). Moreover, roots of wild-type plants grown in excess B conditions have dead cells in the stem cell niche around the quiescent root center (Figure 1O); this region is the primary site of cell death caused by treatment of Arabidopsis roots with DNA-damaging reagents (Fulcher and Sablowski, 2009), suggesting that DNA damage induced by excess B is the cause of B overload-induced cell death.
We observed a negative correlation between the extent of DNA damage in roots of Arabidopsis plants and root elongation (see Supplemental Figure 5 online), suggesting that the inhibition of root elongation results from DNA damage in these plants. Furthermore, in the heb1-1 and heb2-1 mutants, we observed that the combined effects of zeocin and aphidicolin, reagents that directly and indirectly damage DNA, respectively, mimicked the effects of excess B (Figure 5D), which is consistent with our conclusion that DNA damage is the underlying basis of B toxicity. Moreover, we observed that the roots of wild-type plants treated with high concentrations of DSB-inducing agents or aphidicolin were short and had severe morphological defects (Figures 3C and 5C), similar to those observed in the roots of heb1-1 and heb2-1 plants grown in excess B conditions (Figures 1J and 1K). In addition, atr-2, lig4-4, and ku80 mutants, which have increased sensitivity to DNA damage (West et al., 2002; Culligan et al., 2004, 2006; Heacock et al., 2007), were sensitive to excess B. These findings confirm DNA damage as the major cause of B toxicity in roots.
How does excess B induce DNA damage? Although ROS cause DNA damage (Roldán-Arjona and Ariza, 2009) and are found in the aerial parts of plants exposed to excess B (Cervilla et al., 2007), the heb1-1 and heb2-1 mutants were not sensitive to direct or indirect ROS-generating agents (Figure 1D; see Supplemental Figure 1 online), suggesting that ROS are unlikely to be a major factor in B toxicity in roots and are therefore unlikely to be the cause of B-dependent DNA damage. The mechanism by which B induces DNA damage will be a key focus of our future investigations of B toxicity.
B-Induced DNA Damage Has Special Characteristics
We found that the DNA damage-sensitive mutant atm-2, unlike heb2-2 and atr-2, was not sensitive to excess B (Figure 4D). The transcriptional regulation of BRCA1, GR1, RAD51, and PARP1 is ATM dependent (Garcia et al., 2003; Culligan et al., 2006). Moreover, cell death after DNA damage in the stem cell niche requires both ATM and ATR (Fulcher and Sablowski, 2009), suggesting that these genes are required for excess B tolerance. However, we found that only ATR is crucial for B tolerance. As a possible explanation for this apparent inconsistency, we point out the possibility that the growth defect caused by excess B arises downstream of the perception of DSBs mediated by ATM. In this case, atm-2 mutant could not be sensitive to excess B, and the atm-2 heb double mutant could show better root elongation than in the heb mutants. On the other hand, atm-2 is unlikely to be null, so that atm-2 has residual ATM activity sufficient for B tolerance. Involvement of ATM in B tolerance is to be confirmed.
Surprisingly, combination of zeocin and aphidicolin treatment caused a severe reduction in root elongation comparable to that observed in excess B in the heb mutants (Figure 5D). In addition, we found the atr-2, lig4-4, and ku80 mutants were highly sensitive to excess B (Figure 4D). ATR is required for sensing of ssDNA caused by replication blocks (reviewed in Cools and De Veylder, 2009), whereas LIG4 and Ku80 specifically act in DSBs repair (West et al., 2002; Heacock et al., 2007). Thus, these findings suggest that B toxicity might be caused by combination of DSBs and replication stress.
Aluminum (Al) is also known to be a genotoxic factor in plant cells (Rounds and Larsen, 2008). However, in contrast with the case of excess B, Arabidopsis does not require ATR function in Al toxicity tolerance, since atr mutants are tolerant to Al toxicity compared with the wild-type plants (Rounds and Larsen, 2008). This implies that although genotoxicities are common feature of both B and Al toxicity, the process of how genotoxicity develops is different. In other words, plants may have particular systems to attenuate genotoxicities induced by each abiotic stress.
Excess B Affects Cell Cycle Progression
We found that the growth in short-term excess B decreased the number of mitotic cells in root tips and increased the fraction of 4C cells of wild-type plants and that these effects of excess B were even more pronounced in the heb mutants (Figures 6A and 6B). There are two possible causes of the B-induced reduction in the mitotic cell count: the severe delay in G2 progression, which would reduce the rate of cell division, and the premature differentiation. Given that 4C cells mostly correspond to G2 phase cells, it is likely that excess B delays cell cycle progression during G2 phase. The involvement of ATR in B tolerance supports this idea and indicates that the G2 checkpoint and subsequent G2 delay to allow repair of B-induced DNA damage are required to prevent the damaged DNA from being carried through into M phase. Alternatively, excess B might trigger premature endoreduplication, resulting in the increase in 4C cells. Recently, it was established that DSBs induce endoreduplicative cells that initiate differentiation in roots of Arabidopsis (Adachi et al., 2011). This process is likely to be applicable to the heb mutants in excess B because excess B induces DSBs even upon short-term exposure (Figure 4B) and the roots of the heb mutants upon long-term exposure exhibited a higher proportion of differentiated cells in the tips than those in normal conditions (Figures 1M, 1N, 1P, and 1Q). Taken together, it is conceivable that excess B initially retards the progression of the mitotic cycle and subsequently converts the mitotic cycle to endocycles with the accumulation of DNA damage.
To confirm that excess B alters cell cycle progression, we examined cyclin expression. In wild-type Arabidopsis, genotoxic exposure, such as to aphidicolin or ionizing radiation, strongly induces expression of CYCB1;1, which is thus considered to be a marker for DNA damage, as well as for defective G2/M progression (Culligan et al., 2004, 2006). However, among the G2/M-specific cell cycle–related genes examined, only CYCA2;1 was strongly induced by excess B; CYCB1;1 was not induced (Figure 6C). Thus, some cyclins exhibit dissimilar expression patterns in response to DNA damage and/or G2 delay, suggesting that cyclin expression patterns may depend on the type of stress that is affecting cell cycle progression. Molecular characterization of CYCA2;1 in plants grown under B overload stress may aid our understanding of the effect of B toxicity on cell cycle progression.
Condensin II Plays a Role in Reducing DNA Damage
In this study, we demonstrated that HEB1 and HEB2 encode CAP-G2 and CAP-H2, respectively, two non-SMC subunits of condensin II (Figures 2A and 2B). Through characterization of heb mutants, we found that condensin II is required for tolerance to DSB induction (Figures 3A to 3C) and replication blocks in Arabidopsis roots (Figures 5A and 5C), indicating that condensin II maintains genomic stability by reducing DNA damage. This interpretation is in agreement with the high levels of DSB accumulation and DNA repair gene transcription in the heb mutant root tips under normal conditions (Figures 4A and 4B). We also found that condensin II is required for tolerance to UV-C–induced DNA damage in plant aerial parts (see Supplemental Figure 7 online). Analysis of HEB2/CAP-H2pro:GUS (for β-glucuronidase) indicated that condensin II is expressed in both roots and shoots, including root apical meristems and shoot apical domes (see Supplemental Figures 8A to 8E online) and is induced by excess B in root tips (see Supplemental Figures 8F and 8G online), where DNA damage appeared to accumulate. Therefore, we propose that condensin II contributes to genomic stability by reducing DNA damage, particularly in mitotically active regions.
The mechanism by which condensin II ameliorates DNA damage is unknown; however, animal and yeast studies provide two hypotheses: Condensin II might physically protect the genome from the attack of genotoxic substrates, as suggested by the observation that yeast condensin (a type I condensin) responds to nutrient starvation by compacting the genome in the nucleolus during interphase and thereby stabilizing it (Tsang et al., 2007a, 2007b). Alternatively, condensin II might be involved in the repair of DSBs and damaged replication forks. As recombination-mediated repair plays important roles in correcting both types of DNA damage in eukaryotic cells, condensin II might mediate homologous recombination. Indeed, human condensin II has been reported to function in homologous recombination repair after DSB generation (Wood et al., 2008). Furthermore, Ide et al. (2010) proposed that yeast condensin is crucial for the initiation of recombinational repair of DSBs and damaged replication forks by facilitating the cohesion of sister chromatids in the ribosomal DNA array. Additionally, as lig4-4 and ku80 mutants, which are defective in the NHEJ pathway, another system for DSB repair, were susceptible to B toxicity as is the case of heb mutants (Figure 4D), it is also possible that condensin II acts in the NHEJ pathway in cooperation with LIG4 and Ku80. Taken together, the above findings suggest that Arabidopsis condensin II plays a role in preventing and/or repairing DNA damage. The crossing of the heb mutants with other mutants defective in the DNA damage response (e.g., lig4 mutant) might help further our understanding of condensin II function.
During plant mitosis, condensin I and condensin II are localized on chromosomes (Fujimoto et al., 2005); like animal condensins (Ono et al., 2003), they might function together in chromosomal condensation and segregation during the mitotic phase. By contrast, the condensins might play distinct roles during interphase, during which condensin I is localized in the cytosol and condensin II is localized in the nucleus (Fujimoto et al., 2005). However, we cannot rule out the possibility that Arabidopsis condensin I contributes to the maintenance of genomic stability during interphase. During interphase in animal cells, some condensin I remains in the nucleus (Schmiesing et al., 2000) and is involved in ssDNA repair, but not DSB repair (Heale et al., 2006). Yeast condensin has been established as playing a role in tolerance to replication stress (Aono et al., 2002; Chen et al., 2004). In our experiment, exposure to excess B upregulated the non-SMC subunit genes of condensins I and II (see Supplemental Figure 8 online), suggesting that, like condensin II, Arabidopsis condensin I plays a role in reducing B-induced DNA damage. Unfortunately, we were unable to further investigate this hypothesis using condensin I non-SMC mutants because we were unable to obtain such mutants, which are probably embryonic lethal.
METHODS
Plant Materials and Growth Conditions
The heb1-1 and heb2-1 mutants of Arabidopsis thaliana (ecotype Col-0) were isolated as described previously (Sakamoto et al., 2009). The T-DNA–inserted alleles (ecotype Col-0) of HEB1 (heb1-2; SALK_049790) and HEB2 (heb2-2; SALK_059304), and atr-2 (SALK_032841) were obtained from the ABRC. Lines carrying T-DNA in the homozygote were established, and the presence of T-DNA was determined by PCR using the primer sets listed in Supplemental Table 1 online. The atm-2 (ecotype Col-0) mutant was a kind gift from Anne Britt (University of California, Davis). The lig4-4 (ecotype Col-0) and ku80 (ecotype Wassilewskija) mutants were kind gifts from Robert Sablowski (John Innes Centre).
In all experiments, plant seeds were sown on plates containing MGRL solution (Fujiwara et al., 1992), 1% (w/v) sucrose, 1.5% (w/v) gellan gum, and the indicated concentrations of boric acid. After a 3-d incubation at 4°C, plates were placed vertically in the growth chamber until analyzed (16-h-light/8-h-dark cycle; 22°C).
Positional Mapping of HEB Genes
The heb1-1 and heb2-1 mutants (M3 generation; Col-0 background) were crossed with Landsberg erecta wild-type plants. DNA was extracted from 1152 (for heb1-1) and 1134 (for heb2-1) F2 plants and analyzed using simple sequence length polymorphism and cleaved-amplified polymorphic sequence markers (see Supplemental Table 1 online) that were generated based on the Cereon Arabidopsis Polymorphism Collection (http://www.Arabidopsis.org/Cereon). F3 progeny of F2 recombinants were grown in high-B medium (3 mM B), and the short-root phenotype was used to map the heb mutations.
Determination of B Content
Whole roots and shoots of 14-d-old seedlings were harvested from individual plants grown in medium containing 0.03 mM B and from three to five plants grown in medium containing 3 mM B. The samples were dried at 60°C for 2 d, digested with concentrated (13 M) nitric acid (Wako Pure Chemical Industries), and analyzed for B content using inductively coupled plasma–mass spectrometry (SPQ-9700; Seiko Instrument) as previously described (Nozawa et al., 2006).
Analysis of Root Morphology
After the exposure of plants to excess B or DNA-damaging reagents (see below), their root morphology was observed and recorded using a stereomicroscope (SZH10; Olympus) equipped with a digital camera (Olympus). To observe the root tip structures, roots of 14-d-old seedlings were stained with PI (10 μg/mL; Molecular Probes) for 5 to 10 min, and their images were captured using a confocal fluorescence microscope (FV-1000; Olympus) with excitation and emission wavelengths of 619 and 559 nm, respectively. At least 10 plants per line were observed.
Gene Expression Analysis
Total RNA was extracted using an RNeasy plant mini kit (Qiagen) and treated with RNase-free DNase (Qiagen). A 500-ng aliquot of the total RNA was then used for cDNA synthesis using a PrimeScript RT reagent kit (Takara) and subjected to RT-PCR and quantitative real-time PCR with actin8 mRNA as an internal control.
For semiquantitative RT-PCR, HEB1, HEB3, CAP-D3, and actin8 were PCR amplified using the optimum number of cycles determined for each gene. Quantitative real-time RT-PCR for condensin, DSB-inducible, and cell cycle–related genes was performed using SYBR Premix ExTaq II (Takara) on a Thermal Cycler Dice real-time system (Takara). Threshold cycle (Ct) values were calculated using the second derivative maximum method, and relative transcript levels were calculated based on a standard curve generated with serial dilutions of cDNA. The primer sets are listed in Supplemental Table 1 online.
Determination of Sensitivity to Genotoxic Stressors
To test the sensitivity of plants to DNA-damaging reagents, 5-d-old seedlings preincubated on vertical MGRL plates were transferred to plates containing various concentrations of zeocin (Invitrogen), bleomycin, aphidicolin, or hydroxyurea (the latter three from Wako Pure Chemical Industries). They were incubated for an additional 4 d, and the lengths of the newly elongated primary roots were determined using ImageJ (http://rsb.info.nih.gov/ij/).
Comet Assay
DSBs were detected using comet assays with a neutral electrophoresis without alkaline denaturation protocol (N/N protocol) as previously described (Menke et al., 2001). Images of ethidium bromide–stained comets were captured using a fluorescence microscope (BX50WI; Olympus) equipped with a digital CCD camera (Olympus). The comets were analyzed using CASP software (http://www.casp.of.pl/). “Olive tail moment” was used as an index of DNA damage. In assessing DSBs induced by excess B and UV-C exposure, 50 root tips and 10 aerial parts, respectively, were used per sample.
Cell Ploidy Analysis
Protoplasts were isolated from the root tips (2- to 3-mm segments) of 50 plants according to a method optimized for roots (Birnbaum et al., 2005). After cell wall digestion, protoplasts were collected by centrifugation at 500g for 2 min at room temperature. After the supernatant was carefully removed, the prepared protoplasts were analyzed by flow cytometry as previously described (Yoshizumi et al., 2006).
Cloning for Complementation and GUS Activity Analyses
For complementation analysis, the genomic regions harboring CAP-G2/HEB1/At1g64960 and CAP-H2/HEB2/At3g16730 were amplified from Arabidopsis (ecotype Col-0) genomic DNA. These fragments contained the 2.0- and 2.5-kb regions upstream of the HEB1/At1g64960 and HEB2/At3g16730 start codons, respectively. For GUS activity analysis, a 2.45-kb region upstream of the CAP-H2/HEB2/At3g16730 start codon was amplified from Col-0 genomic DNA. Primers sets are described below. The amplified DNA fragments were purified with a gel extraction kit (Qiagen) and then subcloned into the pENTR-D/TOPO vector (Invitrogen) according to the manufacturer’s protocol. The accuracy of the DNA sequences was confirmed by sequencing. The cloned genomic fragments were subsequently subcloned into the Gateway plant transformation destination vector pMDC107, which harbors the gene for GFP, and into pMDC162, which harbors the GUS gene, for complementation and GUS activity analyses, respectively, using an attL-attR (LR) recombination reaction. The constructs were mobilized into Agrobacterium tumefaciens (GV3101) and used to transform Col-0 plants by the floral dip method (Clough and Bent, 1998). Transformants were selected on half-strength Murashige and Skoog plates containing 2% Suc and 20 μg/mL hygromycin. T3 transformants harboring homozygous T-DNA inserts were used for the complementation and GUS activity analyses.
To detect GUS activity, seedlings were stained with a solution containing 100 mM Na2HPO4, pH 7.0, 0.1% Triton X-100, 2 mM K3Fe[CN]6, 2 mM K4Fe[CN]6, and 0.5 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid for 1 h at 37°C. The GUS-stained seedlings were treated with 70 and 99.5% ethanol ,and, immediately before microscopy, they were embedded in clearing solution (80% chloral hydrate and 10% glycerol).
Determination of UV-C Sensitivity
UV-C treatment was conducted on a clean bench (PCV-800TPG; Hitachi) equipped with a UV-C lamp (GL15; Hitachi). Five-day-old seedlings were exposed to UV-C on opened plates as follows: First, the plates were placed vertically at a 40-cm distance from the UV-C lamp and incubated for various lengths of time under UV-C light. As a control, other plates were placed at the same position and incubated for 2 h without UV-C light. Plates were then incubated under normal light for 4 d to allow the plants to recover. The shoot fresh weights were then determined as an inverse measure of plant UV-C sensitivity.
Accession Numbers
Sequence data from this study can be found in the Arabidopsis Genome Initiative data libraries under accession numbers At1g64760 (HEB1), At3g16730 (HEB2), At4g15890 (CAP-D3), At5g37630 (CAP-G), At2g32590 (CAP-H), and At3g57060 (CAP-D2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sensitivity of the heb Mutants to Oxidative Stress.
Supplemental Figure 2. Effects of Excess B on the Length of the Meristematic Zone.
Supplemental Figure 3. Mapping of heb1 and heb2 Mutations.
Supplemental Figure 4. Complementation Analysis of heb1-1 and heb2-1 Mutants.
Supplemental Figure 5. Effect of Zeocin Treatment on Cell Viability in the Root Apical Meristem.
Supplemental Figure 6. Relationship between DSB Level and Root Property in Arabidopsis.
Supplemental Figure 7. DNA Damage by UV-C in the Shoots of the heb Mutants.
Supplemental Figure 8. Expression Pattern of a Condensin II Gene and Responsiveness of Condensins Genes to Excess B.
Supplemental Table 1. List of Primers Used in This Study.
Acknowledgments
We thank Y. Kawara for great technical assistance, T. Kamiya and K. Kasai for critical reading of the manuscript, A. Britt for donating the atr-2 mutant, R. Sablowski for donating lig4-4 and ku80 mutants, and the ABRC for providing the T-DNA inserted SALK lines. This work was supported in part by grants from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation Grant IPG-0005 to T.F.), a Grant-in-Aid for Scientific Research (to T.F.), and a Grant-in-Aid for Scientific Research Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T.F.).
AUTHOR CONTRIBUTIONS
T.S. and T.F. designed the research. T.S., Y.T.I., and T.Y. performed the research. T.S., Y.T.I., T.Y., M.M., S.M., M.U., K.F., and T.F. analyzed data. T.S., S.U., and T.F. wrote the article.
References
- Adachi S., et al. (2011). Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 10004–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono N., Sutani T., Tomonaga T., Mochida S., Yanagida M. (2002). Cnd2 has dual roles in mitotic condensation and interphase. Nature 417: 197–202 [DOI] [PubMed] [Google Scholar]
- Birnbaum K., Jung J.W., Wang J.Y., Lambert G.M., Hirst J.A., Galbraith D.W., Benfey P.N. (2005). Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods 2: 615–619 [DOI] [PubMed] [Google Scholar]
- Cervilla L.M., Blasco B.A., Ríos J.J., Romero L., Ruiz J.M. (2007). Oxidative stress and antioxidants in tomato (Solanum lycopersicum) plants subjected to boron toxicity. Ann. Bot. (Lond.) 100: 747–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.S., Sutani T., Yanagida M. (2004). Cti1/C1D interacts with condensin SMC hinge and supports the DNA repair function of condensin. Proc. Natl. Acad. Sci. USA 101: 8078–8083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cools T., De Veylder L. (2009). DNA stress checkpoint control and plant development. Curr. Opin. Plant Biol. 12: 23–28 [DOI] [PubMed] [Google Scholar]
- Culligan K., Tissier A., Britt A. (2004). ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan K.M., Robertson C.E., Foreman J., Doerner P., Britt A.B. (2006). ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48: 947–961 [DOI] [PubMed] [Google Scholar]
- Deveaux Y., Alonso B., Pierrugues O., Godon C., Kazmaier M. (2000). Molecular cloning and developmental expression of AtGR1, a new growth-related Arabidopsis gene strongly induced by ionizing radiation. Radiat. Res. 154: 355–364 [DOI] [PubMed] [Google Scholar]
- Doucet-Chabeaud G., Godon C., Brutesco C., de Murcia G., Kazmaier M. (2001). Ionising radiation induces the expression of PARP-1 and PARP-2 genes in Arabidopsis. Mol. Genet. Genomics 265: 954–963 [DOI] [PubMed] [Google Scholar]
- Doutriaux M.P., Couteau F., Bergounioux C., White C. (1998). Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol. Gen. Genet. 257: 283–291 [DOI] [PubMed] [Google Scholar]
- Fujimoto S., Yonemura M., Matsunaga S., Nakagawa T., Uchiyama S., Fukui K. (2005). Characterization and dynamic analysis of Arabidopsis condensin subunits, AtCAP-H and AtCAP-H2. Planta 222: 293–300 [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Hirai M.Y., Chino M., Komeda Y., Naito S. (1992). Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol. 99: 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher N., Sablowski R. (2009). Hypersensitivity to DNA damage in plant stem cell niches. Proc. Natl. Acad. Sci. USA 106: 20984–20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V., Bruchet H., Camescasse D., Granier F., Bouchez D., Tissier A. (2003). AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock M.L., Idol R.A., Friesner J.D., Britt A.B., Shippen D.E. (2007). Telomere dynamics and function of critically shortened telomeres in plants lacking DNA ligase IV. Nucleic Acids Res. 35: 6490–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heale J.T., Ball A.R., Jr, Schmiesing J.A., Kim J.S., Kong X., Zhou S., Hudson D.F., Earnshaw W.C., Yokomori K. (2006). Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol. Cell 21: 837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. (2005). Condensins: Organizing and segregating the genome. Curr. Biol. 15: R265–R275 [DOI] [PubMed] [Google Scholar]
- Hirota T., Gerlich D., Koch B., Ellenberg J., Peters J.M. (2004). Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 117: 6435–6445 [DOI] [PubMed] [Google Scholar]
- Ide S., Miyazaki T., Maki H., Kobayashi T. (2010). Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327: 693–696 [DOI] [PubMed] [Google Scholar]
- Koshiba T., Kobayashi M., Matoh T. (2009). Boron nutrition of tobacco BY-2 cells. V. oxidative damage is the major cause of cell death induced by boron deprivation. Plant Cell Physiol. 50: 26–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarge S., Montané M.H. (2003). Characterization of Arabidopsis thaliana ortholog of the human breast cancer susceptibility gene 1: AtBRCA1, strongly induced by gamma rays. Nucleic Acids Res. 31: 1148–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Jiang W., Zhang L., Li L. (2000). Effects of boron Ions on root growth and cell division of broadbean (Vicia faba L.). Isr. J. Plant Sci. 48: 47–51 [Google Scholar]
- Loomis W.D., Durst R.W. (1992). Chemistry and biology of boron. Biofactors 3: 229–239 [PubMed] [Google Scholar]
- López-Juez E., Dillon E., Magyar Z., Khan S., Hazeldine S., de Jager S.M., Murray J.A., Beemster G.T., Bögre L., Shanahan H. (2008). Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. Plant Cell 20: 947–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewski K.M., Blevins D.G., Randall D.D. (1992). Asparagine and boric acid cause allantoate accumulation in soybean leaves by inhibiting manganese-dependent allantoate amidohydrolase. Plant Physiol. 99: 1670–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S., Pandey G.K., Tuteja N. (2008). Calcium- and salt-stress signaling in plants: Shedding light on SOS pathway. Arch. Biochem. Biophys. 471: 146–158 [DOI] [PubMed] [Google Scholar]
- Menges M., de Jager S.M., Gruissem W., Murray J.A. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Menke M., Chen I., Angelis K.J., Schubert I. (2001). DNA damage and repair in Arabidopsis thaliana as measured by the comet assay after treatment with different classes of genotoxins. Mutat. Res. 493: 87–93 [DOI] [PubMed] [Google Scholar]
- Miwa K., Takano J., Omori H., Seki M., Shinozaki K., Fujiwara T. (2007). Plants tolerant of high boron levels. Science 318: 1417. [DOI] [PubMed] [Google Scholar]
- Nable R.O. (1991). Distribution of boron within barley genotypes with differing susceptibilities to boron toxicity. J. Plant Nutr. 14: 453–461 [Google Scholar]
- Nable R.O., Baňuelos G.S., Paull J.G. (1997). Boron toxicity. Plant Soil 193: 181–198 [Google Scholar]
- Nielsen F.H. (2008). Is boron nutritionally relevant? Nutr. Rev. 66: 183–191 [DOI] [PubMed] [Google Scholar]
- Nozawa A., Miwa K., Kobayashi M., Fujiwara T. (2006). Isolation of Arabidopsis thaliana cDNAs that confer yeast boric acid tolerance. Biosci. Biotechnol. Biochem. 70: 1724–1730 [DOI] [PubMed] [Google Scholar]
- Ono T., Fang Y., Spector D.L., Hirano T. (2004). Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell 15: 3296–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Losada A., Hirano M., Myers M.P., Neuwald A.F., Hirano T. (2003). Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115: 109–121 [DOI] [PubMed] [Google Scholar]
- Pang Y., Li L., Ren F., Lu P., Wei P., Cai J., Xin L., Zhang J., Chen J., Wang X. (2010). Overexpression of the tonoplast aquaporin AtTIP5;1 conferred tolerance to boron toxicity in Arabidopsis. J. Genet. Genomics 37: 389–397, 1–2 [DOI] [PubMed] [Google Scholar]
- Robbins W.A., Wei F., Elashoff D.A., Wu G., Xun L., Jia J. (2008). X:Y sperm ratio in boron-exposed men. J. Androl. 29: 115–121 [DOI] [PubMed] [Google Scholar]
- Roldán-Arjona T., Ariza R.R. (2009). Repair and tolerance of oxidative DNA damage in plants. Mutat. Res. 681: 169–179 [DOI] [PubMed] [Google Scholar]
- Rounds M.A., Larsen P.B. (2008). Aluminum-dependent root-growth inhibition in Arabidopsis results from AtATR-regulated cell-cycle arrest. Curr. Biol. 18: 1495–1500 [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Inui-Tsujimoto Y., Fujiwara T. (2009). Isolation of mutants sensitive to excess boron. In The Proceedings of the International Plant Nutrition Colloquium XVI. (Davis, CA: University of California; ), pp. 1126–1129 [Google Scholar]
- Schmiesing J.A., Gregson H.C., Zhou S., Yokomori K. (2000). A human condensin complex containing hCAP-C-hCAP-E and CNAP1, a homolog of Xenopus XCAP-D2, colocalizes with phosphorylated histone H3 during the early stage of mitotic chromosome condensation. Mol. Cell. Biol. 20: 6996–7006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui N.U., Stronghill P.E., Dengler R.E., Hasenkampf C.A., Riggs C.D. (2003). Mutations in Arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development 130: 3283–3295 [DOI] [PubMed] [Google Scholar]
- Sutton T., Baumann U., Hayes J., Collins N.C., Shi B.J., Schnurbusch T., Hay A., Mayo G., Pallotta M., Tester M., Langridge P. (2007). Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science 318: 1446–1449 [DOI] [PubMed] [Google Scholar]
- Tsang C.K., Li H., Zheng X.S. (2007a). Nutrient starvation promotes condensin loading to maintain rDNA stability. EMBO J. 26: 448–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang C.K., Wei Y., Zheng X.F. (2007b). Compacting DNA during the interphase: Condensin maintains rDNA integrity. Cell Cycle 6: 2213–2218 [DOI] [PubMed] [Google Scholar]
- Verbruggen N., Hermans C., Schat H. (2009). Mechanisms to cope with arsenic or cadmium excess in plants. Curr. Opin. Plant Biol. 12: 364–372 [DOI] [PubMed] [Google Scholar]
- West C.E., Waterworth W.M., Story G.W., Sunderland P.A., Jiang Q., Bray C.M. (2002). Disruption of the Arabidopsis AtKu80 gene demonstrates an essential role for AtKu80 protein in efficient repair of DNA double-strand breaks in vivo. Plant J. 31: 517–528 [DOI] [PubMed] [Google Scholar]
- Wood J.L., Liang Y., Li K., Chen J. (2008). Microcephalin/MCPH1 associates with the Condensin II complex to function in homologous recombination repair. J. Biol. Chem. 283: 29586–29592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizumi T., Tsumoto Y., Takiguchi T., Nagata N., Yamamoto Y.Y., Kawashima M., Ichikawa T., Nakazawa M., Yamamoto N., Matsui M. (2006). Increased level of polyploidy1, a conserved repressor of CYCLINA2 transcription, controls endoreduplication in Arabidopsis. Plant Cell 18: 2452–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]