Of seven plastid genome-encoded ribosomal proteins analyzed by reverse genetics in tobacco (Nicotiana tabacum), two were found to be nonessential: S15 and L36. Elimination of ribosomal protein S15 produced normal plants, but elimination of L36 resulted in mutants with reduced apical dominance and strikingly altered leaf morphology, uncovering a role for plastid translational activity in plant development.
Abstract
Plastid genomes of higher plants contain a conserved set of ribosomal protein genes. Although plastid translational activity is essential for cell survival in tobacco (Nicotiana tabacum), individual plastid ribosomal proteins can be nonessential. Candidates for nonessential plastid ribosomal proteins are ribosomal proteins identified as nonessential in bacteria and those whose genes were lost from the highly reduced plastid genomes of nonphotosynthetic plastid-bearing lineages (parasitic plants, apicomplexan protozoa). Here we report the reverse genetic analysis of seven plastid-encoded ribosomal proteins that meet these criteria. We have introduced knockout alleles for the corresponding genes into the tobacco plastid genome. Five of the targeted genes (ribosomal protein of the large subunit22 [rpl22], rpl23, rpl32, ribosomal protein of the small subunit3 [rps3], and rps16) were shown to be essential even under heterotrophic conditions, despite their loss in at least some parasitic plastid-bearing lineages. This suggests that nonphotosynthetic plastids show elevated rates of gene transfer to the nuclear genome. Knockout of two ribosomal protein genes, rps15 and rpl36, yielded homoplasmic transplastomic mutants, thus indicating nonessentiality. Whereas Δrps15 plants showed only a mild phenotype, Δrpl36 plants were severely impaired in photosynthesis and growth and, moreover, displayed greatly altered leaf morphology. This finding provides strong genetic evidence that chloroplast translational activity influences leaf development, presumably via a retrograde signaling pathway.
INTRODUCTION
Translation in plastids, such as chloroplasts, occurs on bacterial-type 70S ribosomes that are similar in structure and composition to bacterial ribosomes (Yamaguchi and Subramanian, 2000; Yamaguchi et al., 2000; Manuell et al., 2007). All four RNA components (rRNAs) of chloroplast ribosomes are encoded by the plastid genome (plastome): the 23S, 5S, and 4.5S rRNAs of the large (50S) ribosomal subunit and the 16S rRNA of the small (30S) ribosomal subunit. By contrast, the protein components of the plastid ribosome, the ribosomal proteins, are partly encoded in the nuclear genome. In the model plant tobacco (Nicotiana tabacum), 12 out of the 21 proteins of the small ribosomal subunit that have homologs in Escherichia coli are encoded in the chloroplast genome, whereas the remaining 9 proteins are nuclear encoded. Similarly, 9 out of 31 proteins of the large ribosomal subunit are encoded by plastid genes, whereas the other 22 are encoded by nuclear genes. Plastid ribosomes also contain a small number of proteins that are not found in bacterial ribosomes, the plastid-specific ribosomal proteins. They are encoded by nuclear genes, and their functions in protein biosynthesis and/or ribosome assembly are still largely unknown (Yamaguchi and Subramanian, 2000; Yamaguchi et al., 2000; Manuell et al., 2007; Sharma et al., 2007).
The set of ribosomal protein genes retained in the plastid genome is highly conserved across seed plants. This suggests that the gene transfer of ribosomal protein genes to the nuclear genome was largely completed before the evolutionary diversification of seed plants and that a relatively stable situation has now been reached. Exceptions include rpl22 (transferred to the nuclear genome in legumes) (Gantt et al., 1991), rpl32 (transferred to the nuclear genome in poplar) (Ueda et al., 2007), and rpl23 in spinach (Spinacia oleracea) (replaced by a eukaryotic L23 protein version encoded in the nucleus and imported into plastids) (Bubunenko et al., 1994).
The most notable exceptions to the otherwise high degree of conservation of the set of ribosomal protein genes are the highly reduced plastomes of parasitic plants, which usually lack several plastid ribosomal protein genes (Wolfe et al., 1992; Funk et al., 2007; Delannoy et al., 2011). Whether these missing genes are dispensable under nonphotosynthetic conditions or, alternatively, have been transferred to the nuclear genome is currently unknown. Although most plastid genome-encoded proteins in nonparasitic plants function in photosynthesis, plastid gene expression is also important for many other cellular functions. These include, for example, tetrapyrrole biosynthesis (which requires the chloroplast tRNA-Glu) (Schön et al., 1986), fatty acid biosynthesis (which requires the plastid genome-encoded D subunit of the essential enzyme acetyl-CoA carboxylase) (Kode et al., 2005; Kahlau and Bock, 2008), and plastid protein homeostasis (which is critically dependent on the plastid-encoded ClpP1 protease subunit) (Shikanai et al., 2001; Kuroda and Maliga, 2003). The involvement of plastid gene expression in these essential functions is probably why the loss of plastid translational activity is fatal in most plants (Ahlert et al., 2003; Rogalski et al., 2006; Rogalski et al., 2008a). Known exceptions are the grasses (Han et al., 1992; Hess et al., 1994a) and some Brassicaceae (Zubko and Day, 1998).
Targeted inactivation of essential components of the plastid translational apparatus typically results in heteroplasmic plants that, in the presence of selective pressure, maintain a mix of mutant and wild-type plastomes (Ahlert et al., 2003; Rogalski et al., 2006). This is explained by a balancing selection, in which copies of the mutant plastome are needed to provide the antibiotic resistance (conferred by the selectable marker gene used for selection of transplastomic plants) and copies of the wild-type plastome are needed to provide the essential gene function disrupted in the mutant plastome (Drescher et al., 2000). Although random sorting of plastid genomes can lead to the appearance of mutant cells that are homoplasmic, these cells are unable to survive and divide. This then results in the loss of entire cell lineages, which becomes phenotypically apparent as severe defects in organ development. Typically, leaves and flowers of these plants are misshapen, because of the random loss of tissue sectors (Ahlert et al., 2003; Rogalski et al., 2006; Rogalski et al., 2008a). Growth on Suc does not rescue this phenotype, demonstrating that plastid gene functions unrelated to photosynthesis are what make plastid translation essential.
For this reason and because many components of the translational machinery are conserved in the otherwise highly reduced plastomes of nonphotosynthetic plastid-bearing organisms, it is generally assumed that at least some translational activity is present in the plastids of pathogenic protozoans (Apicomplexa) and holoparasitic plants. In view of the high synthesis rates of photosynthesis-related proteins in chloroplasts, it is conceivable that the lack of photosynthesis would result in a much lower demand for plastid translation. It, therefore, seems possible that the translational apparatus in plastids of nonphotosynthetic organisms is somewhat simpler and less efficient, because of the relaxed selection pressure for high translational capacity. Whether this makes some components of the translational machinery dispensable is currently not known. Alternatively, the relaxed pressure for high translational activity could make plastid ribosomal protein genes more prone to gene transfer into the nuclear genome, where at least initially their expression would be less efficient than in the plastid.
Here we have undertaken a systematic investigation of plastid ribosomal protein genes for their essentiality in the translation process. We focused on genes meeting at least one of two criteria that could indicate nonessentiality: (1) loss from at least one lineage of nonphotosynthetic plastid-bearing organisms, and/or (2) evidence of nonessentiality in bacteria.
RESULTS
Targeted Disruption of Seven Plastid Ribosomal Protein Genes
By applying the two criteria (nonessentiality in E. coli and gene loss from the plastomes of nonphotosynthetic plastid-bearing organisms, such as holoparasitic plants and apicoplast-containing protozoa) (Wilson, 2002; Barbrook et al., 2006; Krause, 2008), we identified seven plastid ribosomal proteins as candidates for being nonessential for translation (Table 1). As representatives of nonphotosynthetic plastid-containing species, we initially included the parasitic seed plants Cuscuta reflexa (Funk et al., 2007) and Epifagus virginiana (Wolfe et al., 1992), the colorless heterotrophic alga Euglena longa (Astasia longa) (Gockel et al., 1994), and the apicomplexan parasites Eimeria tenella (Cai et al., 2003), Theileria parva (Gardner et al., 2005), and Toxoplasma gondii (Wilson and Williamson, 1997; Wilson, 2002). Recently, the plastome of the parasitic orchid Rhizanthella gardneri was fully sequenced (Delannoy et al., 2011). It was found to be the most reduced plastid genome discovered to date in a seed plant, and its ribosomal protein gene content (kindly made available to us by the authors prior to publication) was also considered for the identification of potentially nonessential genes. Using the sequence information from these reduced plastomes and the information on essential and nonessential genes in the model bacterium E. coli (Baba et al., 2006), the following plastid ribosomal protein genes were identified as potentially nonessential: rpl22, rpl23, rpl32, rpl36, rps3, rps15, and rps16 (Table 1). For rpl32, rpl36, and rps15 (and the previously analyzed rpl33) (Rogalski et al., 2008b), evidence from mutant analyses in E. coli indicates that these could be nonessential ribosomal protein genes. rpl22, rpl23, rpl32, rps3, rps15, and rps16 are missing from at least one plastome of the parasitic or pathogenic reference species (Table 1). (Another potential candidate gene could be rpl14, which is present as a putative pseudogene in the plastome of E. virginiana, but was not investigated here.)
Table 1.
Essentiality and Evolutionary Conservation of Plastid Ribosomal Protein Genes in Parasitic and Pathogenic Plastid-Bearing Lineages
| Gene | Essential in E. colia | Essential in Tobacco Plastids | Present in Plastomes of Nongreen Lineages |
| rpl2 | Yes | NA | Yes |
| rpl14 | Yes | NA | Not in E. virginiana (Ψ) |
| rpl16 | Yes | NA | Yes |
| rpl20 | Yes | Yesb | Not in E. tenella, T. parva, and T. gondii |
| rpl22 | Yes | Yesc | Not in E. virginiana, R. gardneri, and T. gondii |
| rpl23 | Yes | Yesc | Not in C. reflexa |
| rpl32 | Nod | Yesc | Not in R. gardneri and C. reflexa (Ψ) |
| rpl33 | No | Nob | Not in T. gondii, E. longa, and R. gardneri (Ψ) |
| rpl36 | No | Noc | Yes |
| rps2 | Yes | Yesb | Not in T. gondii |
| rps3 | Yes | Yesc | Not in R. gardneri |
| rps4 | Yes | Yesb | Yes |
| rps7 | Yes | NA | Yes |
| rps8 | Yes | NA | Yes |
| rps11 | Yes | NA | Yes |
| rps12 | Yes | NA | Yes |
| rps14 | Yes | Yesb | Yes |
| rps15 | No | Noc | Not in E. virginiana and R. gardneri |
| rps16 | Yes | Yesc | Not in C. reflexa and R. gardneri |
| rps18 | Yes | Yesb | Not in E. longa and T. gondii |
| rps19 | Yes | NA | Yes |
Ψ, pseudogene; NA, not analyzed.
Data on E. coli ribosomal proteins are from Baba et al. (2006).
Data on plastid ribosomal proteins are from our previously published work (Ahlert et al., 2003; Rogalski et al., 2006; Rogalski et al., 2008b).
Data on plastid ribosomal proteins are from this study.
Could not be confirmed in this study (see text for details).
To functionally analyze these seven plastome-encoded ribosomal proteins and to clarify the possible relationship between gene loss in nongreen lineages of plastid evolution and nonessentiality, we used reverse genetics in tobacco, which is both a higher plant species amenable to plastid transformation (Svab and Maliga, 1993; Maliga, 2004; Bock, 2007) and a model plant for chloroplast functional genomics whose complete chloroplast genome sequence is available (Shinozaki et al., 1986; Ruf et al., 1997; Hager et al., 1999; Hager et al., 2002). We constructed knockout alleles for all seven genes by either replacing or disrupting the reading frame of the ribosomal protein gene of interest with aadA, the standard selectable marker gene for chloroplast transformation, in a cloned plastid DNA fragment (see Methods for details). The aadA gene product (the enzyme aminoglycoside 3′′-adenylyltransferase) confers resistance to the aminoglycoside antibiotics spectinomycin and streptomycin, which act as specific inhibitors of plastid translation. Knockout vectors for all genes of interest, which are part of operons, were produced by precisely replacing the coding region of the targeted ribosomal protein gene with the aadA coding region. This strategy ensures that selectable marker gene expression is driven by the endogenous expression signals of the ribosomal protein gene and thus avoids interference with the expression of neighboring genes in the operon.
All knockout alleles were then introduced into the tobacco plastid genome by particle gun-mediated (biolistic) transformation to replace the corresponding wild-type alleles by homologous recombination (Maliga, 2004; Maliga and Bock, 2011). For all constructs, selection on spectinomycin-containing plant regeneration medium produced multiple independent antibiotic-resistant lines. Successful transformation of the plastid genome was preliminarily confirmed by double resistance tests on tissue culture medium containing spectinomycin and streptomycin, a standard assay suitable for eliminating spontaneous spectinomycin-resistant mutants (Svab and Maliga, 1993; Bock, 2001). The primary transplastomic lines were subjected to two to four additional rounds of regeneration and selection for spectinomycin resistance to enrich the transgenic plastome and dilute out the residual wild-type plastomes. Unless a gene on the wild-type plastome is essential, this procedure typically results in homoplasmic transplastomic cell lines (i.e., lines that lack any residual wild-type plastome copies) after two to three rounds of selection and regeneration (Svab and Maliga, 1993; Bock, 2001; Maliga, 2004).
For each knockout construct, two to five independently generated transplastomic lines were selected for further analyses. The lines will be subsequently referred to as Δrpl22, Δrpl23, Δrpl32, Δrpl36, Δrps3, Δrps15, and Δrps16, respectively.
rpl22, rpl23, rpl32, rps3, and rps16 Are Essential Plastid Genes
Essentiality of plastid genes is typically revealed by two characteristic features: (1) the wild-type plastid genomes cannot be fully eliminated despite passage of the transplastomic lines through several rounds of stringent antibiotic selection, and (2) leaves of such transplastomic plants show characteristic deformations caused by the lack of large sectors of the leaf blade (due to the death of cell lines that segregated into homoplasmy) (Drescher et al., 2000; Rogalski et al., 2006; Rogalski et al., 2008a). To test for essentiality of the targeted ribosomal protein genes, we analyzed DNA samples from our transplastomic lines by restriction fragment length polymorphism (RFLP) analyses for homoplasmy of the knockout allele and also investigated the phenotypes of transplastomic lines upon growth in soil under standard greenhouse conditions.
When the Δrpl22, Δrpl23, Δrpl32, Δrps3, and Δrps16 transplastomic lines were tested by RFLP analysis, all of them showed hybridizing fragments that were larger than the hybridizing fragments in the wild-type sample and corresponded in size to the knockout allele carrying the aadA marker gene inserted into the respective ribosomal protein gene locus (Figures 1 to 5). However, all transplastomic lines had an additional hybridization signal for the wild-type restriction fragment even after three to four rounds of regeneration under spectinomycin selection (and supplementation with Suc), indicating that none of these five ribosomal protein genes can be eliminated from all plastome copies. To provide further evidence of these five genes being essential for cell survival, the phenotypes of plants grown in soil in the absence of antibiotic selection were analyzed. All transplastomic plants showed characteristic defects in leaf development caused by sectorial loss of leaf tissue (Figures 1D, 2D, 3E, 4D, and 5D) (Rogalski et al., 2006; Rogalski et al., 2008b), strongly suggesting that these five ribosomal protein genes are indispensable for cellular viability, despite their absence from the plastomes of some nonphotosynthetic plastid-containing lineages.
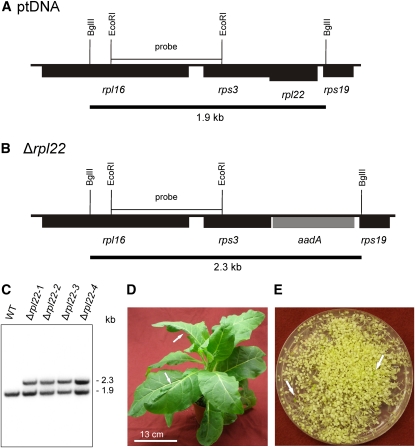
Figure 1.
Targeted Inactivation of the rpl22 Gene Encoding Plastid Ribosomal Protein L22.
(A) Physical map of the region in the tobacco plastid genome (ptDNA) (Shinozaki et al., 1986) containing the rpl22 gene. Genes below the line are transcribed from the right to the left.
(B) Map of the transformed plastid genome (transplastome) produced with plastid transformation vector pΔrpl22. The selectable marker gene aadA (Svab and Maliga, 1993) replaces rpl22 in the operon of ribosomal protein genes. Restriction sites used for cloning, RFLP analysis, and/or generation of hybridization probes are indicated. Expected sizes of restriction fragments detected by hybridization are indicated by black bars below the maps.
(C) RFLP analysis of four plastid transformants. All lines are heteroplasmic and show the 1.9-kb wild type–specific hybridization band in addition to the 2.3-kb band diagnostic of the transplastome. WT, wild type.
(D) Phenotype of a typical (heteroplasmic) Δrpl22 transplastomic plant. Arrows point to examples of misshapen leaves that lack part of the leaf blade.
(E) Segregation of the rpl22 knockout allele in the progeny from a parent plant grown without antibiotic selection as determined by seed germination assays on synthetic medium containing spectinomycin. The transplastome is lost from most seedlings as evidenced by their white (antibiotic-sensitive) phenotype. Occasional green spectinomycin-resistant seedlings are indicated by arrows.
Bar in (D) = 13 cm.
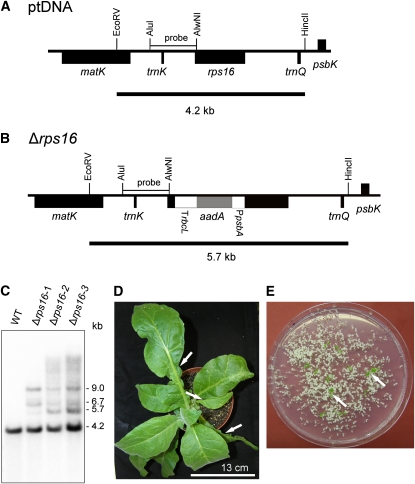
Figure 5.
Targeted Disruption of the rps16 Gene Encoding Plastid Ribosomal Protein S16.
(A) Physical map of the region in the tobacco plastid genome containing the rps16 gene. Transcriptional orientations and labeling of restriction sites, hybridization probes, and hybridizing fragments are as in Figure 2.
(B) Map of the transformed plastid genome produced with plastid transformation vector pΔrps16. The aadA cassette disrupts the rps16 gene.
(C) RFLP analysis of three plastid transformants. All lines are heteroplasmic and show the 4.2-kb wild type (WT)–specific hybridization band in addition to the 5.7-kb band diagnostic of the transplastome. Additional larger hybridizing bands are most likely the products of recombination events between homologous expression signals (Rogalski et al., 2006; Rogalski et al., 2008a). They were reproducibly observed in several independent experiments, do not match expected patterns for partial restriction enzyme digestion, and were not further characterized.
(D) Phenotype of a typical (heteroplasmic) Δrps16 transplastomic plant. Arrows point to examples of misshapen leaves that lack parts of their leaf blade.
(E) Segregation of the rps16 knockout allele in the progeny as determined by seed germination assays on synthetic medium containing spectinomycin. Two examples of green antibiotic-resistant seedlings (that still contain the transplastome) are indicated by arrows.
Bar in (D) = 13 cm.
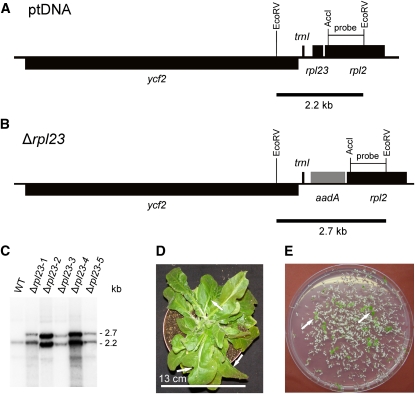
Figure 2.
Targeted Inactivation of the rpl23 Gene Encoding Plastid Ribosomal Protein L23.
(A) Physical map of the region in the tobacco plastid genome containing the rpl23 gene. Genes above the line are transcribed from the left to the right, genes below the line are transcribed in the opposite direction.
(B) Map of the transformed plastid genome produced with plastid transformation vector pΔrpl23. The selectable marker gene aadA replaces the rpl23 gene in the operon of ribosomal protein genes. Restriction sites used for cloning, RFLP analysis, and/or generation of hybridization probes are indicated. Expected sizes of restriction fragments detected by hybridization are indicated by black bars below the maps.
(C) RFLP analysis of five plastid transformants. All lines are heteroplasmic and show the 2.3-kb wild type–specific hybridization band in addition to the 2.7-kb band diagnostic of the transplastome. WT, wild type.
(D) Phenotype of a typical (heteroplasmic) Δrpl23 transplastomic plant. Arrows point to examples of misshapen leaves that lack part of the leaf blade.
(E) Segregation of the rpl23 knockout allele in the progeny as determined by seed germination assays on synthetic medium containing spectinomycin. Two examples of green antibiotic-resistant seedlings (that still contain the transplastome) are denoted by arrows.
Bar in (D) = 13 cm.
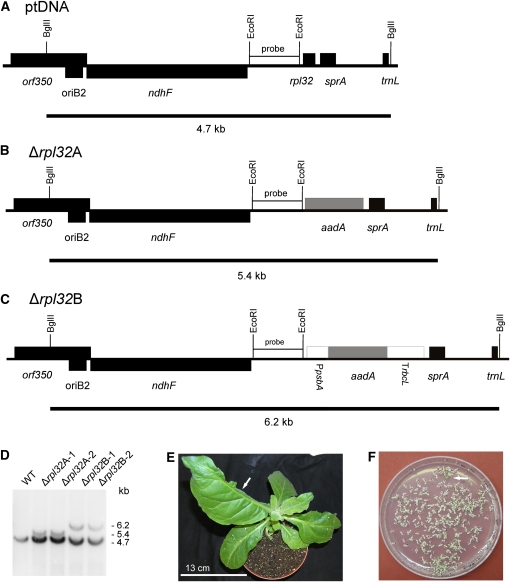
Figure 3.
Targeted Inactivation of the rpl32 Gene Encoding Plastid Ribosomal Protein L32.
(A) Physical map of the region in the tobacco plastid genome containing the rpl32 gene. Transcriptional orientations and labeling of restriction sites, hybridization probes, and hybridizing fragments are as in Figure 2.
(B) Map of the transformed plastid genome produced with plastid transformation vector pΔrpl32A. In this vector, the selectable marker gene aadA replaces the rpl32 gene and is driven by the native promoter upstream of rpl32.
(C) Map of the transformed plastid genome produced with plastid transformation vector pΔrpl32B. In this vector, a chimeric selectable marker gene cassette was used to replace the rpl32 gene. It consists of the promoter of the plastid psbA gene, the coding region of the aadA marker and the 3′ UTR derived from the rbcL gene.
(D) RFLP analysis of four plastid transformants. All lines are heteroplasmic and show the 4.7-kb wild type–specific hybridization band. The transplastomes give the expected 5.4-kb band (Δrpl32A) or 6.2-kb band (Δrpl32B). WT, wild type.
(E) Phenotype of a typical (heteroplasmic) Δrpl32 transplastomic plant. The arrow points to a misshapen leaf that lacks almost half of the leaf blade.
(F) Segregation of the rpl32 knockout allele in the progeny as determined by seed germination assays on synthetic medium containing spectinomycin. A single green antibiotic-resistant seedling (that still contains the transplastome) is marked by the arrow.
Bar in (E) = 13 cm.
Figure 4.
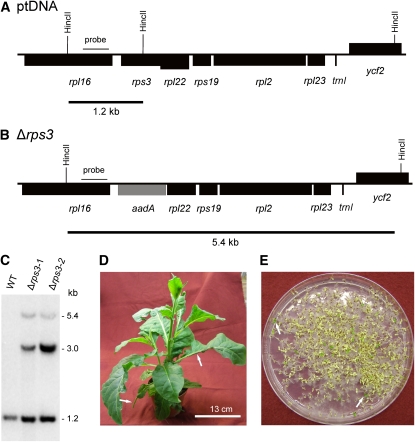
Targeted Inactivation of the rps3 Gene Encoding Plastid Ribosomal Protein S3.
(A) Physical map of the region in the tobacco plastid genome containing the rps3 gene. Transcriptional orientations and labeling of restriction sites, hybridization probes, and hybridizing fragments are as in Figure 2.
(B) Map of the transformed plastid genome produced with plastid transformation vector pΔrps3. The selectable marker gene aadA replaces the rps3 gene (residing in an operon with many other ribosomal protein genes).
(C) RFLP analysis of two plastid transformants. Both lines are heteroplasmic and show the 1.2-kb wild type–specific hybridization band in addition to the 5.4-kb band diagnostic of the transplastome. The 3.0-kb band corresponds in size to the fragment expected for the co-integrate (resulting from single crossover integration) and was not further characterized. WT, wild type.
(D) Phenotype of a typical (heteroplasmic) Δrps3 transplastomic plant. Arrows point to examples of misshapen leaves that lack parts of the leaf blade.
(E) Segregation of the rps3 knockout allele in the progeny as determined by seed germination assays on synthetic medium containing spectinomycin. Two examples of green antibiotic-resistant seedlings (that still contain the transplastome) are indicated by arrows.
Bar in (D) = 13 cm.
In the absence of antibiotic selection, the knockout alleles in the transplastomic lines confer a selective disadvantage and, therefore, are usually rapidly lost due to random segregation of plastid genomes during cell division (Drescher et al., 2000). Consistent with this expectation, transmission of the transplastome into the next generation was very low when parent plants were grown without selection for spectinomycin resistance, as revealed by seed germination assays on spectinomycin-containing culture medium (Figures 1E, 2E, 3F, 4E, and 5E).
Essentiality of rpl32 in plastids was somewhat surprising, because the gene has been identified as nonessential in E. coli (Baba et al., 2006). This prompted us to order the E. coli knockout strain from the Keio collection (Baba et al., 2006) and reinvestigate it. Sequencing of the rpl32 locus in the mutant strain that we received revealed that the gene was undisrupted. Whether this means that the E. coli data on rpl32 are incorrect or, alternatively, a mix-up in strains has occurred upon storage or shipping remains to be clarified by the curators of the Keio collection.
Rps15 Is a Nonessential Plastid Ribosomal Protein
In contrast with the stable heteroplasmy observed in the Δrpl22, Δrpl23, Δrpl32, Δrps3, and Δrps16 transplastomic lines, no hybridization signal for the wild-type plastome was detectable in digested DNA samples from Δrps15 plants after the lines had passed the third regeneration round (Figures 6A to 6C). This tentatively suggested that the lines are homoplasmic and that the S15 protein may not be essential for maintenance of plastid translation. Homoplasmy of the Δrps15 knockout lines was ultimately confirmed by inheritance assays. Germination of seeds from Δrps15 plants on spectinomycin-containing medium yielded a uniform population of homogeneously green seedlings (Figure 6D). Lack of segregation in the T1 generation provides strong genetic proof of homoplasmy of the transplastomic lines and thus of the nonessentiality of the plastid rps15 gene.
Figure 6.
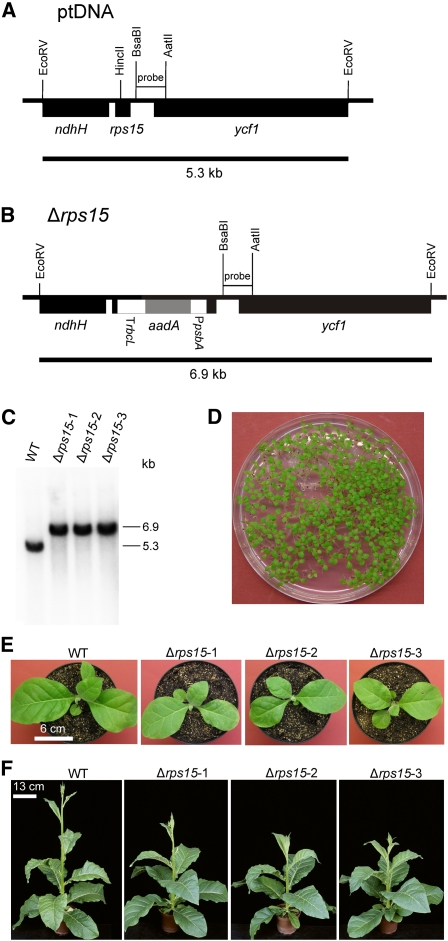
Knockout of the rps15 Gene Encoding Plastid Ribosomal Protein S15.
(A) Physical map of the region in the tobacco plastid genome containing the rps15 gene. Transcriptional orientations and labeling of restriction sites, hybridization probes, and hybridizing fragments are as in Figure 2.
(B) Map of the transformed plastid genome produced with plastid transformation vector pΔrps15. The aadA cassette disrupts the rps15 gene.
(C) RFLP analysis of three plastid transformants. All transplastomic lines are homoplasmic and show exclusively the 6.9-kb band diagnostic of the transplastome. WT, wild type.
(D) Confirmation of homoplasmy by segregation assays. Germination of seeds on synthetic medium containing spectinomycin results in a homogeneous population of green antibiotic-resistant seedlings.
(E) Phenotype of Δrps15 transplastomic plants after growth for 3 weeks at 100 μE m−2 s−1.
(F) Phenotype of Δrps15 transplastomic plants after growth for 8 weeks at 100 μE m−2 s−1 followed by growth for 2 weeks at 350 μE m−2 s−1.
Bar in (E) = 6 cm; bar in (F) = 13 cm.
When Δrps15 knockout plants were raised under a variety of different light intensities (ranging from 100 to 1000 μE m−2 s−1), they displayed no obvious mutant phenotype (Figures 6E and 6F). In fact, their growth rates were almost identical to the wild-type plants, although young plants grew slightly more slowly (Figure 6E), and flowering was a bit delayed (Figure 6F).
To confirm that chloroplast ribosomes in the Δrps15 mutants indeed function without an S15 subunit, we purified plastid ribosomes and subjected them to mass spectrometric protein identification (Rogalski et al., 2008b). Although S15 was readily detectable in the wild-type ribosomes, no S15 protein was found in ribosomes from Δrps15 knockout plants (see Supplemental Table 1 online). These data confirm that the knockout leads to a complete loss of Rps15 and, moreover, demonstrate that the loss of the plastid-encoded S15 is not compensated by import of a nuclear-encoded S15 protein. This is consistent with our failure to identify putative nuclear genes for a chloroplast S15 protein when we searched the available genome and EST databases for tobacco and other Solanaceous plants.
Photosynthesis in Δrps15 Plants
The efficiency of photosynthetic electron transport and the accumulation levels of the protein complexes in the thylakoid membrane are highly sensitive indicators of plastid translational capacity (Rogalski et al., 2008b). Because of the very mild growth phenotype of the Δrps15 knockout lines, we decided to measure photosynthetic performance and protein complex accumulation in the wild-type and transplastomic plants. Because the demand for de novo synthesis of the proteins of the photosynthetic apparatus decreases with leaf development, we analyzed a developmental series of leaves ranging from young expanding leaves (leaves number 3 and 4, the youngest leaves of sufficient size to be measured fluorometrically) to fully expanded mature leaves (Figure 7).
Figure 7.
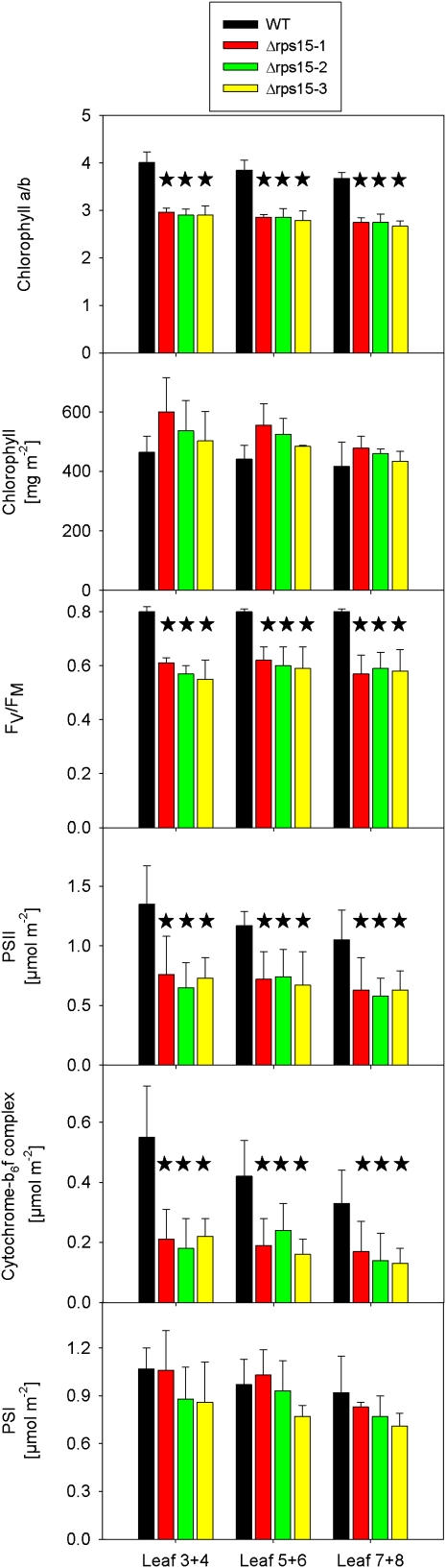
Analysis of Chlorophyll Contents and Various Photosynthetic Parameters in Δrps15 Transplastomic Knockout Mutants and Wild-Type Control Plants.
Data sets are shown for plants grown under 100 μE m−2 s−1 for 8 weeks followed by 2 weeks of growth under 350 μE m−2 s−1. To capture possible changes during development, pairs of leaves (numbered from the top to the bottom of the plant) were analyzed. For each plant line, three different plants were measured, and data were subjected to one-way analysis of variance using a pair-wise multiple comparison procedure (Holm-Sidak method) in SigmaPlot. Highly significant differences are indicated by stars (P < 0.001). Error bars represent the sd. FV/FM represents the maximum quantum efficiency of PSII in the dark-adapted state. Photosynthetic complexes were quantified from difference absorbance measurements of cytochrome b559 (PSII), cytochromes b6 and f, and P700 (PSI) in isolated thylakoids. WT, wild type.
Although the chlorophyll content in the mutant lines was not significantly different from the wild-type control, the mutants had a clearly lower chlorophyll a:b ratio than the wild type (Figure 7). This points to a reduced accumulation of the (largely plastid-encoded) reaction center proteins in the Δrps15 plants, whereas the nuclear-encoded antenna proteins are unaffected or may even overaccumulate, thus explaining the unchanged total chlorophyll content. Interestingly, the decreased chlorophyll a:b ratio did not ameliorate over the developmental time span examined (Figure 7), suggesting that deficits in photosynthetic complex biogenesis cannot be compensated over time.
The maximum quantum efficiency of photosystem II (PSII), FV/FM, was also significantly reduced in the Δrps15 plants and, similar to the chlorophyll a:b ratio, this reduction was largely independent of leaf age and developmental stage (Figure 7). When we measured the contents of the photosynthetic protein complexes by difference absorption spectroscopy (Schöttler et al., 2007a), the amounts of PSII and the cytochrome b6f complex were found to be significantly reduced in the Δrps15 lines, whereas the amount of photosystem I (PSI) was much less affected (Figure 7). Because most of the reaction center subunits of the protein complexes involved in photosynthetic electron transport are encoded in the plastid genome, these data tentatively suggested that plastid gene expression is somewhat less efficient in the absence of the ribosomal protein S15. PSII is known to require a particularly high translation capacity due to the constant requirement for repair synthesis of the D1 protein (Takahashi and Badger, 2011). By contrast, PSI is very stable, which may explain why it is less affected in the mutant plants.
Efficient chloroplast protein biosynthesis is particularly important in very young developing leaves (Albrecht et al., 2006; Rogalski et al., 2008a; Rogalski et al., 2008b), most likely because the de novo assembly of the photosynthetic machinery absorbs very high translational capacity. In addition, cold stress has recently been identified as a condition aggravating the phenotype of plants with subtle defects in chloroplast translation (Rogalski et al., 2008b). We therefore tested whether the mild growth phenotype observed in the Δrps15 plants under standard growth conditions is more apparent in very young leaves or becomes more severe under chilling stress. Indeed, very young developing leaves of the mutant plants (of sizes smaller than what can be measured fluorometrically) (Figure 7) were light green compared with leaves of the wild-type plants, suggesting that the synthesis of the photosynthetic apparatus is delayed in Δrps15 plants (Figure 8A). This phenotype became much more pronounced under chilling stress (Figure 8B), suggesting that, in the Δrps15 mutant, chloroplast translational capacity limits the biogenesis of the photosynthetic apparatus in young leaves and under stress conditions.
Figure 8.
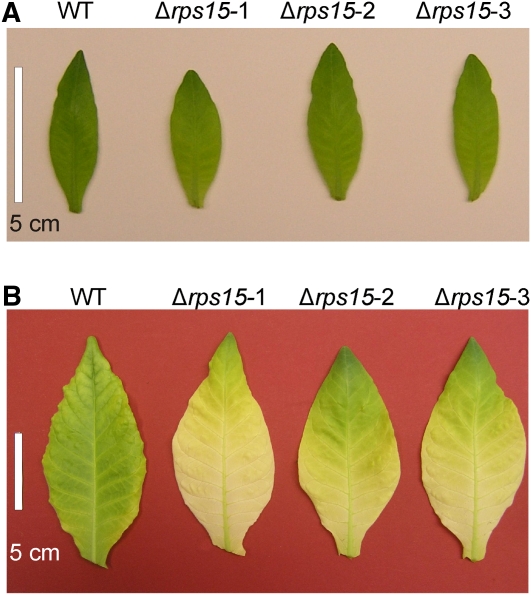
Leaf Pigmentation Phenotypes in Δrps15 Transplastomic Plants.
(A) Young leaves of Δrps15 transplastomic plants are slightly paler than the wild-type (WT) leaves when grown under 100 μE m−2 s−1 at 26°C.
(B) Leaves of Δrps15 plants grown under cold stress conditions for 65 d show a much more severe pigment loss than the wild-type leaves.
Bars = 5 cm.
Plastid Ribosomal Accumulation in Δrps15 Plants
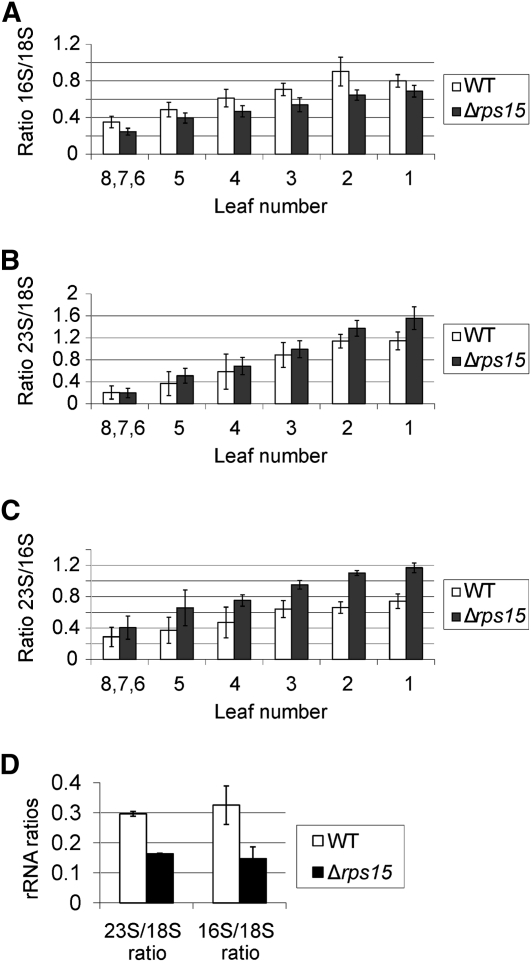
We next wanted to assess the consequences of the lack of the S15 protein at the level of translation and plastid ribosome biogenesis. To this end, we used a microfluidics-based platform for sizing and quantifying the rRNA species. If not assembled in ribosomal subunits, rRNAs are not stable and, therefore, rRNA accumulation can serve as a proxy for ribosomal subunit accumulation (Walter et al., 2010). We first determined the relative abundance of chloroplast to cytosolic ribosomal subunits and calculated the ratios 16S rRNA:18S rRNA and 23S rRNA:18S rRNA. (16S and 23S represent plastid rRNA species, whereas 18S is the rRNA of the small subunit of the cytosolic ribosome.) The data revealed that the Δrps15 plants have reduced amounts of small (30S) subunits of the plastid ribosome, as evidenced by a decreased 16S rRNA:18S rRNA ratio (Figure 9A). Interestingly, the accumulation of large (50S) subunits of the chloroplast ribosome was even slightly higher than in the wild type (evidenced by an increased 23S rRNA:18S rRNA ratio) (Figure 9B), possibly suggesting that the cell senses the chloroplast ribosome deficiency and attempts to compensate for it by an up-regulation of de novo synthesis. Analysis of the 23S rRNA:16S rRNA ratio confirmed that the knockout of the plastid rps15 gene leads to a specific reduction in small ribosomal subunits (Figure 9C), in line with S15 being a component of the 30S subunit of the plastid ribosome.
Figure 9.
Accumulation of rRNAs as a Proxy for the Corresponding Ribosomal Subunits in the Wild-Type and Δrps15 Plants.
(A) Ratio of the plastid 16S rRNA to the cytosolic 18S rRNA. To capture possible changes during development, a developmental series of leaves (numbered from the bottom to the top of the plant) was analyzed for each plant. Leaves numbered 6, 7, and 8 correspond to the youngest leaves, which had to be pooled due to their small size. For each line, three different plants were measured with two technical replicates each. The error bars indicate the sd.
(B) Ratio of the plastid 23S rRNA to the cytosolic 18S rRNA.
(C) Ratio of the plastid 23S rRNA to the plastid 16S rRNA.
(D) 23S rRNA:18S rRNA ratio and 16S:18S rRNA ratio in leaves of Δrps15 transplastomic plants and the wild-type plants grown under cold stress conditions (Figure 8).
WT, wild type.
To examine how chilling stress affects plastid ribosomes in Δrps15 transplastomic plants, we compared the 23S rRNA:18S rRNA and 16S:18S rRNA ratios in young leaves of mutant and wild-type plants grown at 4°C. Consistent with their severe phenotype in the cold (Figure 8), Δrps15 plants showed a drastic reduction in plastid ribosomes. In contrast with growth under unstressed conditions (Figure 9B), now the 50S ribosomal subunit of the plastid ribosome was also strongly affected (evidenced by a severely decreased 23S rRNA:18S rRNA ratio in the mutant compared with the wild type) (Figure 9D), indicating that a secondary loss of plastid ribosomes occurs in Δrps15 plants during chilling stress.
Plastid Translation in Δrps15 Plants
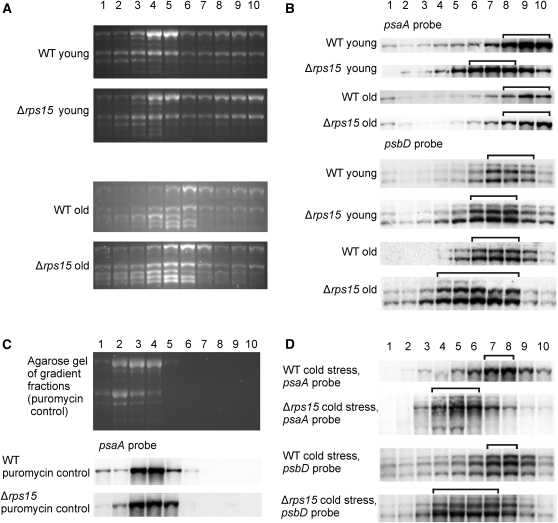
In view of the mutant phenotype (Figures 6 and 8) and the deficiency in plastid ribosomes, it seemed reasonable to assume that Δrps15 transplastomic plants suffer from reduced levels of chloroplast translation. To directly investigate plastid translational activity in the absence of the S15 protein, we conducted polysome loading analyses (Figure 10). These assays determine the coverage of mRNAs with translating ribosomes and thus represent a measure of translational activity (Barkan, 1988; Barkan, 1998; Kahlau and Bock, 2008). We comparatively analyzed polysome association of two plastid transcripts: the dicistronic psaA/B mRNA encoding the two reaction center proteins of PSI and the psbD mRNA encoding the reaction center protein D2 of PSII (which is co-transcribed with psbC, the gene encoding the CP43 inner antenna protein of PSII). To explore a possible correlation with the leaf age-dependent phenotype, the analyses were performed in parallel for young leaves and fully expanded leaves (“old” leaves in Figure 10; comparable to leaves 7 and 8 in Figure 7).
Figure 10.
Analysis of Translation in Δrps15 Plants.
(A) to (D) Analysis of rRNA distribution in polysome preparations from young leaves (numbers 8, 7, and 6; Figure 9) and old leaves (number 1) of the wild-type (WT) and mutant plants. Polysomes were separated in Suc gradients, and each gradient was fractionated into 10 fractions (numbered from the top to the bottom) as indicated above the panels.
(A) Ethidium bromide-stained agarose gels prior to blotting.
(B) Comparison of polysome association of psaA and psbD transcripts in the wild-type and Δrps15 transplastomic plants. Young and old leaves are compared for both genes.
(C) As a control, polysomes were isolated in the presence of the polysome-dissociating antibiotic puromycin, blotted, and hybridized to the psaA probe (Bottom). For analysis of rRNA distribution, an ethidium bromide-stained agarose gel prior to blotting is also shown (Top).
(D) Analysis of polysome association of psaA and psbD transcripts in leaves of plants grown under cold stress conditions. The gradient fractions containing the bulk of the transcripts are indicated by brackets.
Comparison of polysome association in young leaves of the wild-type and mutant plants revealed that plastid translation is indeed less efficient in the Δrps15 mutant. For both transcripts investigated, the maximum in the mRNA distribution is shifted toward lighter fractions in the Suc density gradient in the mutant. For example, the maximum in the psaA/B mRNA distribution is in gradient fractions 8–10 in the wild type, but in fractions 6–8 in the mutant (Figure 10B). Consistent with the observed leaf age-dependent phenotype, older leaves were somewhat less affected than younger leaves.
Having observed a more severe phenotype of Δrps15 plants under chilling stress, we also examined plastid translational efficiency in the cold. As expected, chilling stress affected plastid translation in both the wild type and the mutant, as evidenced by a shift in the maximum in mRNA distribution across the Suc density gradient. Consistent with the temperature-dependence of the mutant phenotype, the difference in ribosome association between the wild-type and the Δrps15 transplastomic plants was even more pronounced in the cold (Figure 10D).
The Plastid rpl36 Gene Is Nonessential, but Its Knockout Leads to a Severe Mutant Phenotype
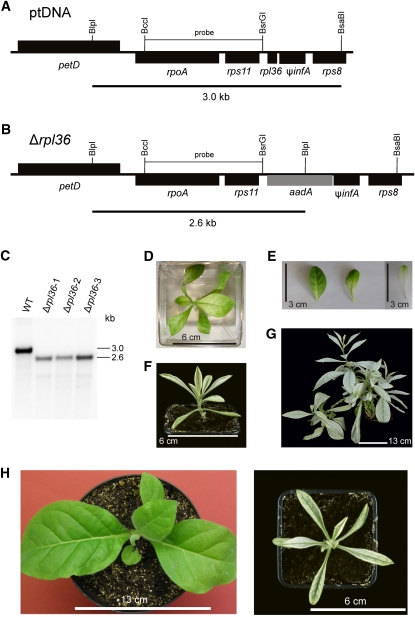
Although experimental evidence suggests that the ribosomal protein gene rpl36 may not be essential in E. coli (Baba et al., 2006), the gene is conserved in the plastid genomes of all nongreen lineages (Table 1). To test whether rpl36 encodes an essential component of the plastid ribosome, we constructed a knockout allele by precisely replacing the rpl36 reading frame with the aadA coding region within the rpoA operon (Figures 11A and 11B). Consequently, in the knockout construct, the aadA marker is driven by the endogenous expression signals of rpl36. Analysis of transplastomic Δrpl36 plants by DNA gel blotting suggested homoplasmy of all three lines investigated (Figure 11C). During growth in sterile culture on Suc-containing medium, the Δrpl36 lines displayed a strong phenotype. Mutant leaves were pale and more elongated than the wild-type leaves. Growth under extreme low-light conditions resulted in partial greening of Δrpl36 leaves (Figures 11D and 11E), suggesting that the mutant plants are highly susceptible to photo-oxidative damage. When plants were transferred to soil, they grew extremely slowly, showed severe pigment deficiency (Figure 11F) even under low-light conditions (50–70 μE m−2 s−1), and did not produce seeds. In addition, lateral branching, which is not normally seen in the wild-type tobacco plants, occurred extensively in the mutant, giving the plants a bushy appearance (Figure 11G) and suggesting that impaired plastid translation reduces apical dominance.
Figure 11.
Knockout of the rpl36 Gene Encoding Plastid Ribosomal Protein L36.
(A) Physical map of the region in the tobacco plastid genome containing the rpl36 gene. Transcriptional orientations and labeling of restriction sites, hybridization probes, and hybridizing fragments are as in Figure 2.
(B) Map of the transformed plastid genome produced with plastid transformation vector pΔrpl36. The aadA gene replaces rpl36 in the large operon of ribosomal protein genes.
(C) RFLP analysis of three plastid transformants. All transplastomic lines are homoplasmic and show exclusively the 2.6-kb band diagnostic of the transplastome. WT, wild type.
(D) Phenotype of a Δrpl36 transplastomic plant grown on Suc-containing synthetic medium for 3 months under low-light conditions (5 μE m−2 s−1).
(E) Greening of Δrpl36 leaves after plant transfer from medium-light to low-light conditions. The left two leaves are from a plant 4 weeks after transfer from 55 to 5 μE m−2 s−1, and the right leaf is from a plant continuously grown under 55 μE m−2 s−1.
(F) Phenotype of a Δrpl36 transplastomic plant growing in soil 2 months after transfer to the greenhouse.
(G) The same plant after 1.3 years. Note the atypical extensive branching.
(H) Severely altered leaf shape in Δrpl36 transplastomic knockout mutants. A wild-type plant (left) is compared with a transplastomic plant at approximately the same developmental stage.
Bars in (D) and (F) = 6 cm; bar in (E) = 3 cm; bar in (G) 13 cm; bar in (H) at left = 13 cm and at right = 6 cm.
The most dramatic aspect of the mutant phenotype was a strong alteration in leaf morphology. Mutant leaves were much more slender than the wild-type leaves (Figure 11H), because of an extreme reduction in the width of the leaf blade. Because similar changes in leaf shape are not seen in mutants affected in photosynthesis (Bock et al., 1994; Hager et al., 2002), this finding suggests that leaf morphology is specifically influenced by the translational activity in the plastid.
DISCUSSION
Although knockout of plastid protein biosynthesis is fatal in tobacco (Ahlert et al., 2003), this does not mean that each individual component of the chloroplast ribosome is essential. Previous work has established that the L33 protein of the plastid ribosome is not required for translation under normal growth conditions but is important under cold stress conditions (Rogalski et al., 2008b). In this study, we have systematically investigated seven plastid-encoded ribosomal protein genes that were lost from plastid genomes of nongreen plastid-bearing lineages and/or have been suggested to be nonessential in bacteria (Table 1).
Our work has identified two additional nonessential protein components of the plastid ribosome: S15 and L36. In contrast with the previously analyzed rpl33 gene (Rogalski et al., 2008b), knockout of rps15 and rpl36 is not phenotypically neutral under standard growth conditions. rps15 knockout plants have only a mild growth phenotype, despite a significant reduction in photosynthetic complex accumulation, ribosome content, and plastid translational activity (Figures 7, 9, and 10). The phenotypic effects are most strongly pronounced in young developing leaves and under chilling stress. This finding is consistent with the demand for plastid translational capacity being highest in developing leaves, when the photosynthetic machinery needs to be synthesized and assembled. By contrast, in mature leaves, lower levels of plastid translation seem to be sufficient to ensure maintenance and repair of the photosynthetic apparatus. This is in agreement with polysome loading in older leaves of the Δrps15 knockout mutant being less affected than in young leaves (Figure 10B). In the assembly of the E. coli 30S ribosomal subunit, ribosomal protein S15 binds early to the 16S rRNA and is possibly involved in triggering conformational changes that may facilitate downstream assembly steps (Talkington et al., 2005; Kaczanowska and Rydén-Aulin, 2007; Woodson, 2008; Connolly and Culver, 2009; Sykes et al., 2010). It is, therefore, conceivable that the loss of S15 partially impairs biogenesis of 30S ribosomal particles in plastids, thus explaining the preferential loss of small ribosomal subunits in our transplastomic Δrps15 knockout mutants (Figure 9).
Knockout of rpl36, a gene not lost from any of the sequenced plastid genomes in nonphotosynthetic lineages, results in nearly white plants that show severe morphological aberrations and hardly grow photo-autotrophically. This indicates that the low translational efficiency in the absence of the L36 protein is insufficient to sustain the (presumably relatively low) protein biosynthesis levels required in the nongreen plastids of the nonphotosynthetic plants listed in Table 1. L36 is also highly conserved in bacteria, but is not present in the ribosomes of archaea and eukaryotes. In E. coli, conflicting data have been published on the importance of L36 in translation and ribosome assembly. Whereas one study reported that deletion of the rpl36 gene did not cause a mutant phenotpye (Ikegami et al., 2005), another study reported that strains lacking L36 are severely impaired in growth and concluded that L36 may play a significant role in organizing the 23S rRNA structure (Maeder and Draper, 2005). Our data obtained in plastids are more consistent with the latter report, but cannot ultimately explain the reasons for the conflicting data obtained in E. coli.
Taken together with previous data, we now have a comprehensive overview of the essentiality of plastome-encoded ribosomal protein genes that were lost in nonphotosynthetic lineages of parasitic plants and pathogenic protozoa (Table 1). A ribosomal protein gene can be absent from the plastome of a nonphotosynthetic organism for two possible reasons: (1) the gene was lost permanently, because it is functionally dispensable due to the low demand for plastid translational capacity in nongreen plastids, or (2) the gene was transferred from the plastid genome to the nuclear genome. Our data reveal no obvious correlation between the loss of plastid ribosomal protein genes and the dispensability of these genes under heterotrophic conditions. Instead, given that most of these genes are essential in both E. coli and plastids (Table 1), they have most likely been transferred to the nuclear genome in the nonphotosynthetic lineages (Timmis et al., 2004; Bock and Timmis, 2008). Because many of these ribosomal protein genes are highly conserved in the plastid genomes of the green lineages (with only occasional examples of gene transfer or gene replacement—see Introduction), this in turn suggests that the loss of photosynthesis and the transition to a heterotrophic lifestyle results in an increased transfer frequency of ribosomal protein genes to the nucleus.
What could be the evolutionary driving force for such an increased gene transfer rate to the nucleus? A reasonable hypothesis could be that the loss of photosynthesis and the concomitantly reduced demand for plastid translational capacity increase the success rate of functional gene transfer to the nucleus. Recent work has shown that the transfer of plastid DNA into the nuclear genome occurs at high frequency (Huang et al., 2003; Stegemann et al., 2003). However, these transferred DNA pieces are usually not expressed in the nucleus, due to the prokaryotic nature of their expression signals (promoter, 5′ untranslated region [UTR]). Their functional activation in the nuclear genome requires rearrangements that change the prokaryotic expression signals into eukaryotic ones (e.g., by promoter capture) (Stegemann and Bock, 2006; Bock and Timmis, 2008). Very likely, whether these transferred DNA pieces regain the full expression level from the nuclear gene copy depends on additional mutations that gradually cause the transferred plastid gene to resemble a typical eukaryotic gene. Thus, it seems conceivable that the success rate of functional gene transfer is correlated to the required expression level of the gene in the nucleus. Genes for which relatively low expression levels suffice have a higher probability of establishing a functional nuclear copy before mutational degeneration of the transferred DNA sets in (Stegemann and Bock, 2006; Sheppard and Timmis, 2009). We therefore, propose that the low demand for plastid translational activity in nonphotosynthetic plastids increases the chances of transferred ribosomal protein genes becoming functional nuclear genes, facilitating their subsequent loss from the plastid genome.
An intriguing aspect of the phenotype of our Δrpl36 mutants was the drastic change in two traits that are not typically associated with plastid function and plastid gene expression: apical dominance and leaf shape (Figure 11). Apical dominance is mainly determined by concentration gradients of the phytohormone auxin. Auxin can be synthesized by two biochemical pathways: a Trp-dependent and a Trp-independent pathway (Normanly et al., 1993; Cohen et al., 2003; Woodward and Bartel, 2005). Although the Trp-dependent pathway of auxin biosynthesis is reasonably well understood, the biochemistry of the Trp-independent pathway is still a mystery. However, evidence has been obtained that the plastid compartment is crucially involved in the Trp-independent pathway of auxin biosynthesis (Rapparini et al., 1999; Rapparini et al., 2002). Hence, it seems possible that the drastically reduced levels of plastid translation in the Δrpl36 mutants directly or indirectly affect the chloroplast-localized enzymes involved in Trp-independent auxin biosynthesis. This could either occur by a plastome-encoded gene product influencing the activity or turnover of the (nuclear-encoded) auxin biosynthetic enzymes or, alternatively, by a retrograde signaling pathway emanating from plastid gene expression (Larkin and Ruckle, 2008; Pogson et al., 2008; Kleine et al., 2009) and regulating the expression level(s) of these enzyme(s). Consistent with a possible defect in auxin metabolism or transport in the Δrpl36 mutants, rooting of stem cuttings and regenerated shoots was very inefficient and severely delayed compared with the wild type.
Retrograde signaling is also the most likely cause of the leaf shape phenotype in the Δrpl36 mutants. Early genetic work in evening primroses (involving reciprocal crosses between two sexually compatible species, Oenothera odorata and Oenothera berteriana), provided evidence for an influence of the plastid genotype on leaf shape (especially on the width of the leaf blade and the intensity of serration of the leaf margin) (Schwemmle, 1941; Schwemmle, 1943). Because of biparental plastid transmission in evening primroses, hybrid plants with identical nuclear genomes could be produced that had either O. odorata plastids or O. berteriana plastids. Interestingly, the two hybrids differed markedly in leaf morphology. Plants with O. odorata plastids had narrower leaf blades than leaves with O. berteriana plastids, and, strikingly, the leaf phenotypes of the hybrids very closely resembled the leaf phenotypes of the parent who donated the plastids (Schwemmle, 1941; Schwemmle, 1943). Because none of the known factors determining leaf development is encoded in the plastid genome, it seems reasonable to assume that this is the result of a retrograde signaling pathway from the plastid to the nucleus. The strongly altered leaf shape in the Δrpl36 mutants uncovers a role of plastid translation in this process, in that the generation of the retrograde signal that determines leaf shape is dependent on, or at least modulated by, plastid translational activity. Previous work has established that the fidelity of plastid gene expression influences the expression of a subset of genes in the nucleus (Börner et al., 1986; Hess et al., 1994b; Koussevitzky et al., 2007), but until now, no morphological phenotypes have been firmly associated with plastid gene expression-dependent retrograde signaling. Our work identifies leaf shape as a morphological output of retrograde signaling and plastid translational activity as a factor involved in signal generation. More work will be needed to determine the nuclear target genes of this signaling pathway and to pinpoint its components that act downstream of plastid translation.
In summary, our work presented here has (1) identified essential and nonessential plastid ribosomal proteins, (2) suggested that the transfer of plastid ribosomal protein genes to the nucleus is greatly accelerated in nonphotosynthetic lineages, and (3) revealed a previously unrecognized role of plastid translational fidelity in two developmental processes: shoot branching and leaf morphogenesis.
METHODS
Plant Material, Growth Conditions, and Phenotypical Assays
To generate leaf material for chloroplast transformation experiments, tobacco plants (Nicotiana tabacum cv Petit Havana) were grown under aseptic conditions on agar-solidified Murashige and Skoog medium containing 30 g/L Suc (Murashige and Skoog, 1962). Transplastomic lines were rooted and propagated on the same medium in the presence of spectinomycin (500 mg/L). For seed production and analysis of plant phenotypes, transplastomic plants were grown in soil under standard greenhouse conditions (relative humidity 55%, day temperature 25°C, night temperature 20°C, diurnal cycle 16 h light and 8 h darkness, light intensity 300–600 μE m−2 s−1). Inheritance patterns and seedling phenotypes were analyzed by germination of surface-sterilized seeds on Murashige and Skoog medium without or with spectinomycin (500 mg/L).
Growth tests under different light conditions were performed by raising the wild-type and Δrps15 mutant plants in soil at 22°C under the following light intensities: 100, 350, and 1000 μE m−2 s−1. For cold stress treatments, plants were transferred after 3 weeks of growth under standard conditions to 4°C for ~2 months (at 80 μE m−2 s−1).
Construction of Plastid Transformation Vectors
Vectors for the targeted knockout of rps15 and rps16, both transcribed as monocistronic mRNAs, were constructed by inserting an aadA cassette conferring spectinomycin resistance (including promoter, 5′ UTR and 3′ UTR) into the coding region of the ribosomal protein genes (insertion mutagenesis). Knockout vectors for all other genes of interest, which are part of operons, were produced by replacing the coding region of the targeted ribosomal protein gene with the aadA coding region, thus employing the endogenous expression signals of the ribosomal protein gene to drive selectable marker gene expression. In the case of rpl32, which is co-transcribed with the sprA gene, both strategies were used in parallel.
An aadA cassette was constructed by digesting plasmid pSK.Kmr (Bateman and Purton, 2000) with XhoI and PstI and inserting the excised aphA-6 cassette into the similarly cut vector pKCZ (Zou et al., 2003), generating plasmid pKCZaphA-6. The transgene expression cassette in pKCZ consists of the psbA promoter, the psbA 5′ UTR and the rbcL 3′ UTR from Chlamydomonas reinhardtii. Subsequently, pKCZaphA-6 was cut with PstI, and the recessed ends were converted to blunt ends by a fill-in reaction with the Klenow fragment of DNA polymerase I from Escherichia coli. Following digestion with NcoI, the aadA coding region (Svab and Maliga, 1993) was inserted as an NcoI/XbaI fragment (with the XbaI overhang filled in), generating plasmid pLS1.
For construction of vector pΔrps15, a fragment from the N. tabacum plastid genome (corresponding to nucleotide positions 124,662 to 126,561; GenBank accession number Z00044) was amplified with the primers P5′ndhH and P3′ycf1 (all primer sequences are listed in Supplemental Table 2 online). The PCR product was cut with BamHI and HindIII (sites underlined in primer sequences) and inserted into the cloning vector pUC18 digested with the same enzymes. This resulting plasmid clone was partially digested with HincII to linearize it within the rps15 gene and was ligated to the aadA cassette excised from pLS1 with SmaI. A clone harboring the aadA cassette in the desired orientation was selected, and the correctness of all manipulations was confirmed by complete sequencing.
Vector pΔrps16 was constructed by amplifying the rps16–containing region (corresponding to plastome positions 3999 to 6244) with primers P5′rps16 and P3′rps16. The obtained PCR fragment was cloned into pUC18 digested with SmaI. The resulting plasmid was linearized with BglII and treated with Klenow DNA polymerase to blunt the overhanging ends. Subsequently, the aadA cassette (obtained by digestion of pLS1 with SmaI) was ligated into the blunted BglII site, resulting in disruption of the rps16 reading frame. A plasmid clone containing the aadA cassette in the desired orientation was identified and sequenced.
For construction of vector pΔrpl23, the genomic region surrounding the rpl23 gene (corresponding to nucleotide positions 88,162 to 89,596) was amplified with primers P5′rpl23 and P3′rpl23. The PCR product was cloned into pUC18 digested with SmaI, generating plasmid pSA10. To replace the rpl23 coding region with the aadA coding region, a PCR strategy was used. An initial PCR amplification was done with P5′rpl2 and P3′rpl2 with the wild-type DNA as template. Another PCR was performed with the primers P5′trnI and P3′trnI. A third PCR amplified the aadA coding region (adding short overhangs that are complementary to the flanking plastome sequences) using the primers P5′rpl2aadA and P3′trnIaadA and pLS1 as template. In the final PCR reaction, the three PCR products were used as templates for amplification with primers P5′rpl2 and P3′trnI. The resulting amplification product was treated with AccI and XcmI and was cloned into pSA10 digested with the same enzymes.
Vector pΔrpl36 was constructed by amplifying the rpl36 genomic region (corresponding to nucleotide positions 80,921 to 83,276) with the primers P5′rpl36 and P3′rpl36. The PCR fragment was cloned into the SmaI site of pUC18, resulting in vector pSA8. To replace rpl36 with aadA, the aadA coding region was amplified from pLS1 plasmid DNA with primers carrying overhangs complementary to the plastome sequences surrounding rpl36, P5′rpl36aadA and P3′rpl36aadA. The adjacent part of the plastome was amplified with primers P5′rpl36infA and P3′rpl36infA. The two PCR products were combined in amplification reactions with primers P5′rpl36aadA and P3′rpl36infA. The resulting product was then digested with BsrGI and BglII and ligated into the similarly cut pSA8, generating transformation vector pΔrpl36.
To generate vectors for targeted inactivation of the rpl32 gene (pΔrpl32A and pΔrpl32B), the rpl32 genomic region (corresponding to nucleotide positions 114,021 to 116,232) was amplified with primers P5′rpl32 and P3′rpl32. The introduced BamHI and PstI restriction sites are underlined in the primer sequences. The obtained PCR fragment was digested with BamHI and PstI and cloned into the similarly cut vector pUC18, producing plasmid pSA11. Vector pΔrpl32A, in which the coding region of rpl32 is replaced by the aadA coding region, was produced by a PCR strategy. First, the coding region of aadA was amplified from pLS1 using the primers P5′aadA and P3′aadA. In a second PCR, the region upstream of rpl32 was amplified with primers P5′ndhF and P3′ndhF usingpSA11 as template. The two PCR products were then combined by performing an amplification reaction with primers P3′ndhF and P5′aadA. The resulting fragment was digested with BamHI and BstBI and was inserted into the similarly cut plasmid pSA11, generating plastid transformation vector pΔrpl32A. Vector pΔrpl32B, in which an aadA cassette disrupts the rpl32 gene, was constructed by partial digestion of pSA11 with SspI (to linearize the vector in the rpl32 gene) followed by insertion of the aadA cassette (obtained as described above for vector pΔrps15).
The knockout vectors for rpl22 and rps3 are both based on a cloned PCR fragment generated with primers P5′Rpl22 and P3′Rpl22. The amplified region of the tobacco plastid genome (corresponding to positions 84,183 to 87,454) contains both genes and was cloned into a SmaI-digested pUC18 vector. The resulting plasmid clone was named pTF11. The rpl22 gene overlaps with rps3 by 16 nucleotides. To sustain translation of rps3 upon knockout of rpl22, the psbE/psbF spacer region (5′ GAGGCCCTA 3′) was inserted downstream of the aadA marker gene to create (in combination with the TAG stop codon of the aadA) a perfect Shine-Dalgarno sequence upstream of rps3. As a control, the rps3 knockout was constructed including the same spacer sequence. In addition, the stop codon of rpl22 was mutated from TAA to TAG to provide a consensus Shine-Dalgarno sequence for the downstream aadA gene. To construct a knockout allele for rpl22, its coding region was replaced with the aadA coding region. To this end, the sequence downstream of rpl22 was amplified by PCR with tobacco DNA as template and the primers P5′ClaI and P3′ClaI. The sequence upstream of rpl22 was amplified with the primers P5′NcoIfor and P3′NcoIrev. A third PCR to amplify the aadA coding region using pLS1 as template was performed with the primers P5′Rpl22aadA and P3′Rpl22aadA. The fourth PCR used the products of the first three PCRs as templates and the primers P5′ClaI and P3′NcoI. The product was treated with NcoI and ClaI and cloned into pTF11 digested with the same enzymes, generating plastid transformation vector pΔrpl22.
To exchange the rps3 coding region with aadA, an analogous strategy was pursued. The two flanking regions were amplified from tobacco DNA with the primer pairs P5′EagI/P3′EagI and P5′BamHI/P3′BamHI. A third PCR was performed to amplify the aadA coding region with the primers P5′Rps3aadA and P3′Rps3aadA using plasmid pLS1 as template. The final PCR combined the three PCR products by amplification with the primers P5′BamHI and P3′EagI. The obtained PCR product was cut with EagI and AflII and ligated into the similarly digested plasmid pTF11, generating plastid transformation vector pΔrps3.
Plastid Transformation and Selection of Transplastomic Lines
Young leaves from aseptically grown tobacco plants were bombarded with plasmid-coated 0.6-μm gold particles using a helium-driven biolistic gun (PDS1000He; BioRad). Primary spectinomycin-resistant lines were selected on plant regeneration medium containing 500 mg/L spectinomycin (Svab and Maliga, 1993). Spontaneous spectinomycin-resistant plants were eliminated by double selection tests on medium containing both spectinomycin and streptomycin (500 mg/L each) (Svab and Maliga, 1993; Bock, 2001). Several independent transplastomic lines were generated for each construct and were subjected to three to four additional rounds of regeneration on spectinomycin-containing plant regeneration medium to enrich the transplastome and select for homoplasmy.
Isolation of Nucleic Acids and Hybridization Procedures
Total plant DNA was extracted from plants grown under spectinomycin selection in vitro by a cetyltrimethylammoniumbromide-based method (Doyle and Doyle, 1990). For RFLP analysis, DNA samples were treated with restriction enzymes and separated on 0.8 to 1.2% agarose gels and blotted onto Hybond N nylon membranes (GE Healthcare). For hybridization, [α32P]dCTP-labeled probes were produced by random priming (Multiprime DNA labeling kit; GE Healthcare). Restriction fragments (generated as indicated in the corresponding figures) were used as hybridization probes. The probe for analysis of the rps3 knockout was produced by PCR amplification with the primers P5′rpl16 and P3′rpl16. Hybridizations were performed at 65°C using standard protocols. Total plant RNA was isolated with the NucleoSpin RNA Plant Kit (Macherey-Nagel) or, alternatively, by a guanidine isothiocyanate/phenol-based method (peqGOLD TriFast; Peqlab). RNA samples were denatured, separated in denaturing formaldehyde-containing agarose gels (1 to 1.2%) and blotted onto Hybond N nylon membranes (GE Healthcare).
rRNA and Polysome Analysis
Polysomes were purified as described previously (Rogalski et al., 2008a), except that gradients were fractionated into 10 fractions to obtain higher resolution. RNA pellets were dissolved in 30 μL of water, 5 μL of which was heat denatured and loaded onto a denaturing formaldehyde-containing 1.2% agarose gel. Specific probes for detection of psaA and psbD transcripts were generated by PCR using the following primers: P5′psaA, P3′psaA, P5′psbD, and P3′psbD. For rRNA quantitation, RNA samples were analyzed in an Agilent 2100 Bioanalyzer with the Agilent RNA 6000 nano Kit according to the instructions of the manufacturer (Agilent Technologies). rRNA ratios were determined as described previously (Walter et al., 2010). To follow ribosome accumulation during leaf development, three biological replicates and two technical replicates were measured for each plant line. In the cold stress assays, two technical replicates were measured.
Ribosome Isolation and Mass Spectrometry
Ribosomes were isolated according to the polysome purification protocol (Rogalski et al., 2008a), except that the preparations were not loaded onto Suc gradients but layered onto a 1 M Suc cushion containing 10 mM Tris-HCl, pH 7.6, 50 mM KCl, 10 mM magnesium acetate, and 7 mM β-mercaptoethanol. The ribosomes were pelleted by centrifugation at 86.000 g for 17 h, and the pellet was resuspended in 40 mM Tris-HCl, 20 mM KCl, 10 mM MgCl2 (pH adjusted to 8.5), followed by precipitation with three volumes of 90% acetone, 10% methanol, 10 mM DDT overnight at −20°C. Subsequently, the ribosomes were collected by centrifugation at 14.000 g for 15 min, washed twice with acetone, and air-dried for 10 min. The pellet was resuspended in T25K100M5D5T buffer (Rogalski et al., 2008b) and frozen at −80°C until further use. Mass spectrometric protein identification and estimation of protein abundance were performed exactly as described previously (Rogalski et al., 2008b).
Physiological Measurements
Chlorophyll contents were determined in 80% (v/v) acetone (Porra et al., 1989). Chlorophyll fluorescence was recorded with a pulse-amplitude modulated fluorimeter (Dual-PAM-100; Heinz Walz) on intact plants (grown under 350 μE m−2 s−1) at room temperature after dark adaptation for 20 min. The contents of PSII, the cytochrome b6f complex, and PSI were determined by difference absorption spectroscopy as described previously (Schöttler et al., 2007a; Schöttler et al., 2007b). Significance analyses were performed using a one-way analysis of variance with a pair-wise multiple comparison procedure (Holm-Sidak method) in SigmaPlot.
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank database under the following accession numbers:
Cuscuta reflexa chloroplast genome, NC_009766; Epifagus virginiana chloroplast genome, NC_001568; Euglena longa plastid genome, NC_002652; Eimeria tenella apicoplast genome, NC_004823; Theileria parva apicoplast genome, NC_007758; Toxoplasma gondii apicoplast genome, NC_001799; Rhizanthella gardneri plastid genome, NC_014874), N. tabacum chloroplast genome, NC_001879; N. tabacum chloroplast ribosomal gene rpl22, GeneID:800419; N. tabacum chloroplast ribosomal gene rpl23, GeneID:800421; N. tabacum chloroplast ribosomal gene rpl32, GeneID:800466; N. tabacum chloroplast ribosomal gene rpl33, GeneID:800444; N. tabacum chloroplast ribosomal gene rpl36, GeneID:7564684; N. tabacum chloroplast ribosomal gene rps3, GeneID:800454; N. tabacum chloroplast ribosomal gene rps15, GeneID:800489; N. tabacum chloroplast ribosomal gene rps16, GeneID:800493; N. tabacum chloroplast photosystem gene psbD, GeneID:800525; N. tabacum chloroplast photosystem gene psaA, GeneID:800453; C. reinhardtii chloroplast photosystem gene psbA, GeneID:2716987; E. coli gene aadA, GeneID:1446561; E. coli knockout strain for rpl32 from the Keio collection CGSC#: 9028, JW 1075-1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Comparative Mass Spectrometric Analysis of Plastid Ribosomal Proteins in the Wild Type and the Δrps15 Knockout Mutant.
Supplemental Table 2. Sequences of Primers Used in This Study.
Acknowledgments
We thank Ian Small (University of Western Australia, Perth) for making available information on the gene content of the Rhizanthella plastome prior to publication. We are grateful to Stephanie Ruf, Stefanie Seeger, Claudia Hasse, and Yvonne Weber for help with chloroplast transformation, Annemarie Matthes for help with mass spectrometry, Wolfram Thiele for technical assistance with the physiological measurements (all with the Max-Planck-Institut für Molekulare Pflanzenphysiologie), and the Max-Planck-Institut für Molekulare Pflanzenphysiologie Green Team for plant care and cultivation. This work was supported by the Max Planck Society and by a grant from the Deutsche Forschungsgemeinschaft to R.B. (FOR 804; BO 1482/15-2).
AUTHOR CONTRIBUTIONS
T.T.F., L.B.S., S.A., M.A.S., and R.B. designed the research; T.T.F., S.A., S.H., and M.A.S. performed research; T.T.F., L.B.S., S.H., M.A.S., and R.B. analyzed data; R.B. wrote the article with significant input from T.T.F., L.B.S., and M.A.S.
References
- Ahlert D., Ruf S., Bock R. (2003). Plastid protein synthesis is required for plant development in tobacco. Proc. Natl. Acad. Sci. USA 100: 15730–15735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht V., Ingenfeld A., Apel K. (2006). Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: The impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol. Biol. 60: 507–518 [DOI] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2: 2006, 0008 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbrook A.C., Howe C.J., Purton S. (2006). Why are plastid genomes retained in nonphotosynthetic organisms? Trends Plant Sci. 11: 101–108 [DOI] [PubMed] [Google Scholar]
- Barkan A. (1988). Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 7: 2637–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1998). Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 297: 38–57 [Google Scholar]
- Bateman J.M., Purton S. (2000). Tools for chloroplast transformation in Chlamydomonas: Expression vectors and a new dominant selectable marker. Mol. Gen. Genet. 263: 404–410 [DOI] [PubMed] [Google Scholar]
- Bock R. (2001). Transgenic plastids in basic research and plant biotechnology. J. Mol. Biol. 312: 425–438 [DOI] [PubMed] [Google Scholar]
- Bock R. (2007). Plastid biotechnology: Prospects for herbicide and insect resistance, metabolic engineering and molecular farming. Curr. Opin. Biotechnol. 18: 100–106 [DOI] [PubMed] [Google Scholar]
- Bock R., Timmis J.N. (2008). Reconstructing evolution: Gene transfer from plastids to the nucleus. Bioessays 30: 556–566 [DOI] [PubMed] [Google Scholar]
- Bock R., Kössel H., Maliga P. (1994). Introduction of a heterologous editing site into the tobacco plastid genome: The lack of RNA editing leads to a mutant phenotype. EMBO J. 13: 4623–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner T., Mendel R.R., Schiemann J. (1986). Nitrate reductase is not accumulated in chloroplast-ribosome-deficient mutants of higher plants. Planta 169: 202–207 [DOI] [PubMed] [Google Scholar]
- Bubunenko M.G., Schmidt J., Subramanian A.R. (1994). Protein substitution in chloroplast ribosome evolution. A eukaryotic cytosolic protein has replaced its organelle homologue (L23) in spinach. J. Mol. Biol. 240: 28–41 [DOI] [PubMed] [Google Scholar]
- Cai X., Fuller A.L., McDougald L.R., Zhu G. (2003). Apicoplast genome of the coccidian Eimeria tenella. Gene 321: 39–46 [DOI] [PubMed] [Google Scholar]
- Cohen J.D., Slovin J.P., Hendrickson A.M. (2003). Two genetically discrete pathways convert tryptophan to auxin: More redundancy in auxin biosynthesis. Trends Plant Sci. 8: 197–199 [DOI] [PubMed] [Google Scholar]
- Connolly K., Culver G. (2009). Deconstructing ribosome construction. Trends Biochem. Sci. 34: 256–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy E., Fujii S., Colas des Francs-Small C., Brundrett M., Small I. (2011). Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol. Biol. Evol. 28: 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J., Doyle J.L. (1990). A rapid total DNA preparation procedure for fresh plant tissue. Focus 12: 13–15 [Google Scholar]
- Drescher A., Ruf S., Calsa T., Jr, Carrer H., Bock R. (2000). The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 22: 97–104 [DOI] [PubMed] [Google Scholar]
- Funk H.T., Berg S., Krupinska K., Maier U.G., Krause K. (2007). Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 7: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt J.S., Baldauf S.L., Calie P.J., Weeden N.F., Palmer J.D. (1991). Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 10: 3073–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.J., Bishop R., Shah T., de Villiers E.P., Carlton J.M., Hall N., Ren Q., Paulsen I.T., Pain A., Berriman M., Wilson R.J., Sato S., et al. (2005). Genome sequence of Theileria parva, a bovine pathogen that transforms lymphocytes. Science 309: 134–137 [DOI] [PubMed] [Google Scholar]
- Gockel G., Hachtel W., Baier S., Fliss C., Henke M. (1994). Genes for components of the chloroplast translational apparatus are conserved in the reduced 73-kb plastid DNA of the nonphotosynthetic euglenoid flagellate Astasia longa. Curr. Genet. 26: 256–262 [DOI] [PubMed] [Google Scholar]
- Hager M., Biehler K., Illerhaus J., Ruf S., Bock R. (1999). Targeted inactivation of the smallest plastid genome-encoded open reading frame reveals a novel and essential subunit of the cytochrome b(6)f complex. EMBO J. 18: 5834–5842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager M., Hermann M., Biehler K., Krieger-Liszkay A., Bock R. (2002). Lack of the small plastid-encoded PsbJ polypeptide results in a defective water-splitting apparatus of photosystem II, reduced photosystem I levels, and hypersensitivity to light. J. Biol. Chem. 277: 14031–14039 [DOI] [PubMed] [Google Scholar]
- Han C.D., Coe E.H., Jr, Martienssen R.A. (1992). Molecular cloning and characterization of iojap (ij), a pattern striping gene of maize. EMBO J. 11: 4037–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess W.R., Hoch B., Zeltz P., Hübschmann T., Kössel H., Börner T. (1994a). Inefficient rpl2 splicing in barley mutants with ribosome-deficient plastids. Plant Cell 6: 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess W.R., Müller A., Nagy F., Börner T. (1994b). Ribosome-deficient plastids affect transcription of light-induced nuclear genes: Genetic evidence for a plastid-derived signal. Mol. Gen. Genet. 242: 305–312 [DOI] [PubMed] [Google Scholar]
- Huang C.Y., Ayliffe M.A., Timmis J.N. (2003). Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature 422: 72–76 [DOI] [PubMed] [Google Scholar]
- Ikegami A., Nishiyama K.-i., Matsuyama S.-i., Tokuda H. (2005). Disruption of rpmJ encoding ribosomal protein L36 decreases the expression of secY upstream of the spc operon and inhibits protein translocation in Escherichia coli. Biosci. Biotechnol. Biochem. 69: 1595–1602 [DOI] [PubMed] [Google Scholar]
- Kaczanowska M., Rydén-Aulin M. (2007). Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 71: 477–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlau S., Bock R. (2008). Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: Chromoplast gene expression largely serves the production of a single protein. Plant Cell 20: 856–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T., Voigt C., Leister D. (2009). Plastid signalling to the nucleus: Messengers still lost in the mists? Trends Genet. 25: 185–192 [DOI] [PubMed] [Google Scholar]
- Kode V., Mudd E.A., Iamtham S., Day A. (2005). The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 44: 237–244 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S., Nott A., Mockler T.C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Krause K. (2008). From chloroplasts to “cryptic” plastids: Evolution of plastid genomes in parasitic plants. Curr. Genet. 54: 111–121 [DOI] [PubMed] [Google Scholar]
- Kuroda H., Maliga P. (2003). The plastid clpP1 protease gene is essential for plant development. Nature 425: 86–89 [DOI] [PubMed] [Google Scholar]
- Larkin R.M., Ruckle M.E. (2008). Integration of light and plastid signals. Curr. Opin. Plant Biol. 11: 593–599 [DOI] [PubMed] [Google Scholar]
- Maeder C., Draper D.E. (2005). A small protein unique to bacteria organizes rRNA tertiary structure over an extensive region of the 50 S ribosomal subunit. J. Mol. Biol. 354: 436–446 [DOI] [PubMed] [Google Scholar]
- Maliga P. (2004). Plastid transformation in higher plants. Annu. Rev. Plant Biol. 55: 289–313 [DOI] [PubMed] [Google Scholar]
- Maliga P., Bock R. (2011). Plastid biotechnology: Food, fuel, and medicine for the 21st century. Plant Physiol. 155: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuell A.L., Quispe J., Mayfield S.P. (2007). Structure of the chloroplast ribosome: Novel domains for translation regulation. PLoS Biol. 5: e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue culture. Physiol. Plant. 15: 473–497 [Google Scholar]
- Normanly J., Cohen J.D., Fink G.R. (1993). Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc. Natl. Acad. Sci. USA 90: 10355–10359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B.J., Woo N.S., Förster B., Small I.D. (2008). Plastid signalling to the nucleus and beyond. Trends Plant Sci. 13: 602–609 [DOI] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394 [Google Scholar]
- Rapparini F., Cohen J.D., Slovin J.P. (1999). Indole-3-acetic acid biosynthesis in Lemna gibba studied using stable isotope labeled anthranilate and tryptophan. Plant Growth Regul. 27: 139–144 [Google Scholar]
- Rapparini F., Tam Y.Y., Cohen J.D., Slovin J.P. (2002). Indole-3-acetic acid metabolism in Lemna gibba undergoes dynamic changes in response to growth temperature. Plant Physiol. 128: 1410–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski M., Ruf S., Bock R. (2006). Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res. 34: 4537–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski M., Karcher D., Bock R. (2008a). Superwobbling facilitates translation with reduced tRNA sets. Nat. Struct. Mol. Biol. 15: 192–198 [DOI] [PubMed] [Google Scholar]
- Rogalski M., Schöttler M.A., Thiele W., Schulze W.X., Bock R. (2008b). Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell 20: 2221–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S., Kössel H., Bock R. (1997). Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J. Cell Biol. 139: 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön A., Krupp G., Gough S., Berry-Lowe S., Kannangara C.G., Söll D. (1986). The RNA required in the first step of chlorophyll biosynthesis is a chloroplast glutamate tRNA. Nature 322: 281–284 [DOI] [PubMed] [Google Scholar]
- Schöttler M.A., Flügel C., Thiele W., Bock R. (2007a). Knock-out of the plastid-encoded PetL subunit results in reduced stability and accelerated leaf age-dependent loss of the cytochrome b6f complex. J. Biol. Chem. 282: 976–985 [DOI] [PubMed] [Google Scholar]
- Schöttler M.A., Flügel C., Thiele W., Stegemann S., Bock R. (2007b). The plastome-encoded PsaJ subunit is required for efficient Photosystem I excitation, but not for plastocyanin oxidation in tobacco. Biochem. J. 403: 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwemmle J. (1941). Weitere Untersuchungen an Eu-Oenotheren über die genetische Bedeutung des Plasmas und der Plastiden. Z. indukt. Abstammungs- u. Vererbungslehre 79: 321–333 [Google Scholar]
- Schwemmle J. (1943). Plastiden und Genmanifestation. Flora 137: 61–72 [Google Scholar]
- Sharma M.R., Wilson D.N., Datta P.P., Barat C., Schluenzen F., Fucini P., Agrawal R.K. (2007). Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc. Natl. Acad. Sci. USA 104: 19315–19320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard A.E., Timmis J.N. (2009). Instability of plastid DNA in the nuclear genome. PLoS Genet. 5: e1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T., Shimizu K., Ueda K., Nishimura Y., Kuroiwa T., Hashimoto T. (2001). The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 42: 264–273 [DOI] [PubMed] [Google Scholar]
- Shinozaki K., et al. (1986). The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 5: 2043–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann S., Bock R. (2006). Experimental reconstruction of functional gene transfer from the tobacco plastid genome to the nucleus. Plant Cell 18: 2869–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann S., Hartmann S., Ruf S., Bock R. (2003). High-frequency gene transfer from the chloroplast genome to the nucleus. Proc. Natl. Acad. Sci. USA 100: 8828–8833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z., Maliga P. (1993). High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA 90: 913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes M.T., Shajani Z., Sperling E., Beck A.H., Williamson J.R. (2010). Quantitative proteomic analysis of ribosome assembly and turnover in vivo. J. Mol. Biol. 403: 331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Badger M.R. (2011). Photoprotection in plants: A new light on photosystem II damage. Trends Plant Sci. 16: 53–60 [DOI] [PubMed] [Google Scholar]
- Talkington M.W.T., Siuzdak G., Williamson J.R. (2005). An assembly landscape for the 30S ribosomal subunit. Nature 438: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis J.N., Ayliffe M.A., Huang C.Y., Martin W. (2004). Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5: 123–135 [DOI] [PubMed] [Google Scholar]
- Ueda M., Fujimoto M., Arimura S.-i., Murata J., Tsutsumi N., Kadowaki K.-i. (2007). Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene 402: 51–56 [DOI] [PubMed] [Google Scholar]
- Walter M., Piepenburg K., Schöttler M.A., Petersen K., Kahlau S., Tiller N., Drechsel O., Weingartner M., Kudla J., Bock R. (2010). Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. Plant J. 64: 851–863 [DOI] [PubMed] [Google Scholar]
- Wilson R.J.M. (2002). Progress with parasite plastids. J. Mol. Biol. 319: 257–274 [DOI] [PubMed] [Google Scholar]
- Wilson R.J.M., Williamson D.H. (1997). Extrachromosomal DNA in the Apicomplexa. Microbiol. Mol. Biol. Rev. 61: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K.H., Morden C.W., Palmer J.D. (1992). Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA 89: 10648–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson S.A. (2008). RNA folding and ribosome assembly. Curr. Opin. Chem. Biol. 12: 667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A.W., Bartel B. (2005). Auxin: Regulation, action, and interaction. Ann. Bot. (Lond.) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K., Subramanian A.R. (2000). The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 275: 28466–28482 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., von Knoblauch K., Subramanian A.R. (2000). The plastid ribosomal proteins. Identification of all the proteins in the 30 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 275: 28455–28465 [DOI] [PubMed] [Google Scholar]
- Zou Z., Eibl C., Koop H.-U. (2003). The stem-loop region of the tobacco psbA 5’UTR is an important determinant of mRNA stability and translation efficiency. Mol. Genet. Genomics 269: 340–349 [DOI] [PubMed] [Google Scholar]
- Zubko M.K., Day A. (1998). Stable albinism induced without mutagenesis: A model for ribosome-free plastid inheritance. Plant J. 15: 265–271 [DOI] [PubMed] [Google Scholar]