Chromera velia is the only known eukaryotic phototroph that uses the C4 pathway to synthesize chlorophyll. The tetrapyrrole synthetic pathway of this alga is an evolutionary mosaic of the pathways of both heterotrophic and photosynthetic eukaryotes and is similar to the unusual heme biosynthetic pathway of the related apicomplexan parasites.
Abstract
Most photosynthetic eukaryotes synthesize both heme and chlorophyll via a common tetrapyrrole biosynthetic pathway starting from glutamate. This pathway was derived mainly from cyanobacterial predecessor of the plastid and differs from the heme synthesis of the plastid-lacking eukaryotes. Here, we show that the coral-associated alveolate Chromera velia, the closest known photosynthetic relative to Apicomplexa, possesses a tetrapyrrole pathway that is homologous to the unusual pathway of apicomplexan parasites. We also demonstrate that, unlike other eukaryotic phototrophs, Chromera synthesizes chlorophyll from glycine and succinyl-CoA rather than glutamate. Our data shed light on the evolution of the heme biosynthesis in parasitic Apicomplexa and photosynthesis-related biochemical processes in their ancestors.
INTRODUCTION
Apicomplexa is a diverse group of parasites, which infect various animals, including humans. Plasmodium, the causative agent of malaria kills millions of people every year (Alemu et al., 2011). Most of the Apicomplexa have been shown to contain a secondary four membrane–bound nonphotosynthetic plastid (apicoplast) that is essential for the parasite survival (reviewed in Oborník et al., 2009 or Kalanon and McFadden, 2010). The presence of this organelle suggested a photosynthetic ancestry for apicomplexans almost two decades ago; however, no close photosynthetic relative was identified until very recently (Moore et al., 2008). This novel alga, named Chromera velia, contains ultrastructural features typical for apicomplexans and related alveolates, such as cortical alveoli, underlying microtubular corset, and an apical microtubular cone (Moore et al., 2008; Oborník et al., 2011), but also possesses a single four membrane–bound plastid. Because Chromera and apicomplexans are sisters in both nuclear and plastid gene phylogenies and also share several unique plastid-related features (together with another alveolate known as CCMP3155), their extant plastids clearly share a common evolutionary origin (Janouškovec et al., 2010). Discovery of C. velia therefore gives an unprecedented opportunity to uncover the course of early apicomplexan evolution and improve our understanding of how a free-living alga transformed into an obligate parasite. Of particular interest is the transformation of the plastid organelle and metabolic pathways commonly found in photosynthetic eukaryotes that have been retained in the apicoplast. Three such pathways, mediating the synthesis of fatty acids, isoprenoids, and tetrapyrroles, have been shown to be indispensable for the apicomplexan parasites survival and are considered potential drug targets in treating malaria and other diseases (Ralph et al., 2004).
Tetrapyrrole biosynthesis ranks among the most fundamental pathways in living systems. It serves the synthesis of heme, a molecule central to the oxidative and energy metabolism, and chlorophyll formation in photoautotrophs. Synthesis of tetrapyrroles is conserved generally among all three domains of life, with the exception of the first step, the synthesis of δ-aminolevulinic acid (ALA) (Panek and O’Brian, 2002). Primarily heterotrophic eukaryotes, such as animals and fungi, synthesize ALA in the mitochondrion through the condensation of Gly and succinyl-CoA in a C4 pathway catalyzed by ALA-synthase (ALAS). This enzyme is otherwise found only in α-proteobacteria, which supports its mitochondrial origin (Figure 1A). ALA is exported to the cytosol, where the next four to five biosynthetic steps are catalyzed by enzymes originating from the ancient eukaryotic nucleus, with the possible exception of uroporphyrinogen synthase (UROS) (Figures 1B and 2; see Supplemental Figure 1 online). The final steps take place back in the mitochondrion, where the majority of heme is needed for respiratory cytochromes (Dailey et al., 2005) (for details, see summary in Figure 3).
Figure 1.
Phylogenetic Trees of Ferrochelatase and ALA-Synthase.
Bayesian phylogenetic trees as inferred from ALAS (A) and ferrochelatase (B) amino acid sequences, and the enzymes of the first and last step of the heme synthesis. Numbers above branches indicate Bayesian posterior probabilities. The trees demonstrate mitochondrial origin of ALAS (A) and uncovered enigmatic origins of the apicomplexan ferrochelatase (B).
[See online article for color version of this figure.]
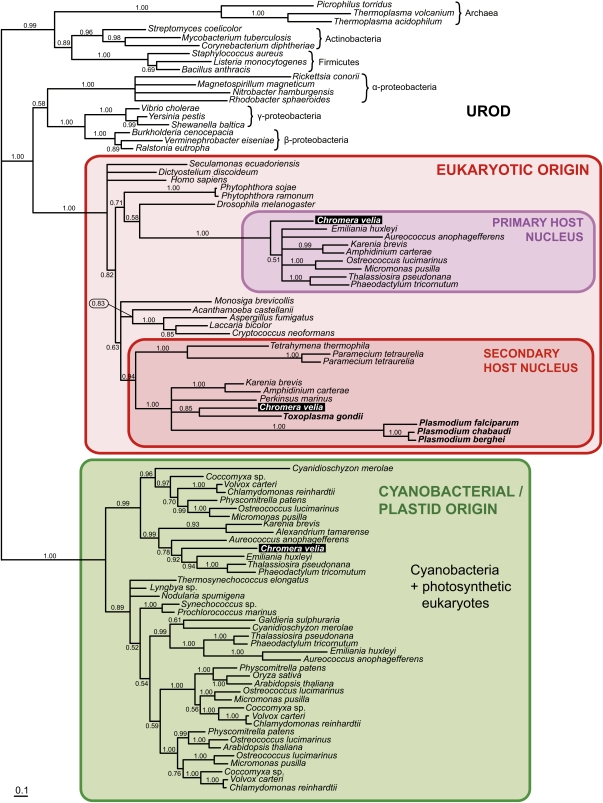
Figure 2.
Bayesian Phylogenetic Tree as Inferred from UROD Amino Acid Sequences.
The tree well demonstrates the complex symbiotic origin of C. velia. Particular UROD enzymes appear to originate from cyanobacteria, the primary host nucleus (nucleus of a eukaryotic alga involved in secondary endosymbiosis), and secondary host nucleus (nucleus of a heterotrophic eukaryote that engulfed the alga). Numbers above branches indicate Bayesian posterior probabilities.
[See online article for color version of this figure.]
Figure 3.
Tetrapyrrole Biosynthetic Pathways of Various Eukaryotes.
Distribution of different types of the tetrapyrrole biosynthetic pathway (B) among chromalveolates (A). (1) Canonical C4 pathway of heterotrophic eukaryotes; localization differs between animal cells (Dailey et al., 2005) and yeast (Camadro et al. 1986). (2) Canonical C5 pathway that is shared by most of the photosynthetic eukaryotes. (3) Tetrapyrrole pathway of C. velia with putative localizations based on in silico predictions using SignalP and TargetP, respectively (Emanuelsson et al., 2007). (4) Heme biosynthesis of Apicomplexa; localization differs between P. falciparum and Toxoplasma gondii (Sato et al., 2004; Nagaraj et al., 2009a, 2009b, 2010a, 2010b; van Dooren et al., 2006; Wu, 2006; Shanmugam et al., 2010). Numbers of the individual steps of the synthesis represent the enzymes: 1, ALAS; 1a, GTR; 1b, GSA-AT; 2, ALAD; 3, PBGD; 4, UROS; 5, UROD; 6, CPOX; 7, PPOX; 8, FeCH. The abbreviations of the enzyme names are explained in the text. The colors of the enzymatic steps stand for the origin of the genes, which are inferred from the phylogenetic trees (see Supplemental Figure 1 online). *Only incomplete information is available: Schizochytrium possesses ALAS, Perkinsus uses the C4 pathway as well as ALAD and UROD of the same origin as in primary heterotrophs. From Oxyrrhis marina, the sequence of the gene encoding PBGD shows a relation to the phototrophic PBGDs.
Photosynthetic eukaryotes synthesize ALA from Glu bound to tRNAGlu through the C5 pathway consisting of two steps catalyzed by glutamyl-tRNA reductase (GTR) and glutamate 1-semialdehyde aminotransferase (GSA-AT). The remaining steps are catalyzed by the same enzymes as in heterotrophs, but the entire pathway is localized in the plastid, and most of the involved enzymes originate from its cyanobacterial ancestor (Oborník and Green, 2005) (Figure 3B). This compartmentalization reflects the major need for the tetrapyrroles in the plastid, where chlorophyll and most of the synthesized heme are required for photosynthetic functions (Papenbrock et al., 1999; Castelfranco and Jones, 1975).
Heme synthesis in Apicomplexa follows a unique route distributed among three cellular compartments. ALA produced in the mitochondrion by the C4 pathway is exported to the apicoplast, where the next three to four steps take place. The sixth enzymatic reaction is situated in the cytosol, while the final two steps are catalyzed in the mitochondrion (Nagaraj et al., 2010a) (Figure 3B). The apicomplexan pathway is an evolutionary mosaic combining pathways from both heterotrophic and photosynthetic eukaryotes. ALAS, uroporphyrinogen decarboxylase (UROD), and coproporphyrinogen oxidase (CPOX) were present in the host cell prior to the endosymbiotic origin of the apicoplast. The remaining enzymes (apart from ferrochelatase) are related to homologs from photosynthetic eukaryotes, suggesting their origin in algal endosymbiont (Figure 3B). Ferrochelatase (FeCH) clusters with proteobacteria and probably comes from a nonendosymbiotic horizontal gene transfer (Sato and Wilson, 2003) (Figure 1B).
To shed more light on the evolution of the unusual tetrapyrrole synthesis in apicomplexan parasites, we investigated this pathway in C. velia, a closely related photosynthetic alga.
RESULTS AND DISCUSSION
Identification and Phylogeny of Tetrapyrrole Biosynthesis Genes in C. velia
Fragments of all tetrapyrrole biosynthesis enzymes, with the single exception of protoporphyrinogen oxidase (PPOX), were identified within the sequences of an ongoing 454 genome survey of C. velia (Janouškovec et al., 2010), and full-length transcripts were obtained by rapid amplification of cDNA ends (RACE). Interestingly, a gene encoding ALAS, the enzyme of the C4 pathway, was found (Figure 1A), but we failed to identify GTR or GSA-AT, the C5 pathway enzymes used by other photoautotrophs (Figure 3B).
Phylogenetic analyses show that the tetrapyrrole biosynthetic pathway of C. velia is a mosaic composed of genes of different origins. Although all the proteins are encoded in the nucleus of C. velia, only few of them display a phylogenetic pattern corresponding to the phylogeny of the host cell. The others seem to originate from distinct genomes (e.g., the plastid) and were horizontally transferred to the host nucleus. C. velia possesses genes that have the same origins as the genes of heme biosynthesis from Apicomplexa, indicating that these pathways share the same evolutionary history (Figures 1 to 3; see Supplemental Figure 1 online). However, in contrast with the pathway of apicomplexans where each step is catalyzed by a single enzyme, Chromera encodes four additional proteins (two URODs, CPOX, and FeCH), sharing an evolutionary history with other photosynthetic eukaryotes (Figure 3B). ALAS clusters with homologs from heterotrophic eukaryotes and α-proteobacteria, implying its origin in the mitochondrion (Figure 1A). ALAD, on the other hand, originates from the plastid as it groups with photosynthetic eukaryotes and cyanobacteria (see Supplemental Figure 1A online), which is also true for one UROD and one FeCH (Figures 1B and 2). PBGD of C. velia branches together with photosynthetic eukaryotes, suggesting its origin in the endosymbiont. However, this clade is not related to cyanobacteria but is nested within α-proteobacteria, which indicates its origin in the mitochondrion of the primary host (see Supplemental Figure 1B online). Similarly, UROS and one CPOX cluster with photosynthetic eukaryotes but not with cyanobacteria, and the primary origins of these particular enzymes are not clear (see Supplemental Figures 1C and 1D online). Apart from the cyanobacterial UROD, there are two other URODs encoded in the genome of C. velia, which are of eukaryotic origin (Figure 2). One comes from the nucleus of the primary alga as it groups with both primary and secondary algae, and the other one represents the original enzyme of the final host, which is also the case of one CPOX (see Supplemental Figure 1D online). The second FeCH is closely related to the same enzymes from Apicomplexa and was likely horizontally transferred from some proteobacteria to the ancestor of Chromera and Apicomplexa (Sato and Wilson, 2003; Figure 1B). Regardless of the primary origins of the enzymes, it is clear that ALAS, one UROD, one CPOX, and one FeCH represent the original heme pathway of the heterotrophic host, but ALAD, PBGD, UROS, two URODs, one CPOX, and one FeCH are derived from the endosymbiotic alga that was transformed into the plastid (see Figure 3B for the summary of gene origins).
Predicted Subcellular Location of C. velia Tetrapyrrole Biosynthesis Proteins
The analysis of 5′ ends of transcript sequences of all enzymes in question revealed that, unlike the others, ALAS possesses an N-terminal extension that resembles the mitochondrial transit peptide. This presequence is similar to those of other eukaryotic ALASs (see Supplemental Figure 2 online), which were experimentally shown to be present in mitochondria in various eukaryotes from different lineages including Apicomplexa (Volland and Felix, 1984; Dailey et al., 2005; Sato et al., 2004; Wu, 2006); thus, we believe the C. velia ALAS is most likely also located in the mitochondrion. Such localization corresponds well to the origin of ALAS in the α-proteobacterial predecessor of the mitochondrion (Schulze et al., 2006). This origin is shared by all eukaryotes, including C. velia (Figure 1A). Furthermore, two heme regulatory motifs in the N-terminal presequence of C. velia ALAS (see Supplemental Figure 3 online) suggest the same regulation of the tetrapyrrole synthesis as in heterotrophs, which is the heme-mediated inhibition of ALAS import into mitochondrion (Zhang and Guarente, 1995; Dailey et al., 2005). In addition to that, ALAS directly uses succinyl-CoA, the product of mitochondrially located citric acid cycle, as a substrate; thus, its localization to a different compartment would be rather hard to imagine. This corresponds to the fact that ALAS has never been localized elsewhere than to mitochondrion.
Except for ALAS, all the other enzymes of heme biosynthesis in C. velia possess typical N-terminal bipartite leader sequences that consist of a signal peptide (SP) and a transit peptide (TP) and are required for targeting to the complex plastids (Kroth, 2002). These leaders are highly similar to those found in heterokont algae and Apicomplexa; thus, all of these proteins seem to be targeted to the plastid of C. velia. SPs were easily recognized by in silico predictions with extremely high probabilities (from 0.966 to 1.000) in each of the proteins, using both approaches available to date, the neural networks (data not shown), and the hidden Markov models (Table 1). After removal of SPs, all the proteins still possess significantly long N-terminal presequences (compared with their bacterial counterparts), which display chemical properties typical for TPs (von Heijne et al., 1989), and most of them (eight out of 10) were recognized as TPs by the prediction tool (Table 1). Predictions of TPs are complicated by the fact that mitochondrial (mTP) and plastidial transit peptides (cTPs) tend to be similar in algae with complex plastids. However, it is well established that in secondary algae it is exclusively the combination of SPs and TPs that decides the plastid localization of a protein, whereas the mitochondrially located proteins are recognized by the presence of TP alone. This leads to a conclusion that there is no selection pressure to distinguish cTP from mTP in secondary algae (Jiroutová et al., 2007; Rao et al., 2008). This means that the SP can be followed by virtually any presequence that has the general chemical properties of a TP to target the proteins to the complex plastids.
Table 1.
In Silico Predictions for the Presence of SPs and TPs in the Protein Sequences of C. velia
| Protein/GenBank Accession No. | SP (SignalP) | TP (TargetP) |
| ALAS/HQ222925 | Not Predicted | Not Predicted (112 aa)a |
| ALAD/HQ222926 | 0.994 (15 aa) | 0.689 (cTP; 67 aa) |
| PBGD/HQ222927 | 0.996 (18 aa) | 0.638 (mTP; 22 aa) |
| UROS/HQ222928 | 0.985 (17 aa) | 0.771 (mTP; 16 aa) |
| URODa/HQ222929 | 1.000 (25 aa) | not predicted (43 aa)b |
| URODb/HQ222930 | 1.000 (22 aa) | 0.929 (cTP; 42 aa) |
| URODc/HQ222931 | 0.992 (21 aa) | 0.587 (mTP; 28 aa) |
| CPOXa/HQ222932 | 0.966 (25 aa) | 0.658 (mTP; 27 aa) |
| CPOXb/HQ222933 | 1.000 (23 aa) | 0.924 (cTP; 28 aa) |
| FeCHa/HQ222934 | 0.991 (32 aa) | 0.806 (mTP; 45 aa) |
| FeCHb/HQ222935 | 0.971 (31 aa) | not predicted (60 aa)b |
aa, amino acids.
The mitochondrial TP of ALAS is not predictable by TargetP in C. velia as well as in other eukaryotes, including apicomplexans, in which the mitochondrial location has been experimentally proven; however, the protein possesses a long N-terminal extension, when compared to bacterial sequences, which resembles the mitochondrial TPs of other eukaryotic ALAS enzymes.
Although the TPs were not predicted for some of the proteins that have SPs, they possess significant N-terminal extensions (after removal of SP) compared to bacterial sequences. Their approximate length is noted in parentheses.
It has been suggested that it is the number of membranes surrounding the plastids that decides the mechanism of protein targeting (Nassoury et al., 2003). In contrast with double membrane plastids of primary algae and plants, the complex plastids of secondary algae are surrounded by additional membranes, and the proteins are therefore imported into this organelle by a distinct route: SP targets the proteins to endoplasmic reticulum and consequently across the outermost plastid membrane that is topologically identical with the endoplasmic reticulum. When the SP is cleaved off, the TP is recognized by the plastid import machinery and transfers the proteins through the two innermost membranes of the complex plastid, which correspond to the primary plastid envelope (Lang et al., 1998; Waller et al., 1998; McFadden, 1999; Foth et al., 2003). Although the particulars of the import mechanism slightly differ among taxa, the use of bipartite leader sequence seems to be a universal mark that guides proteins into the complex plastids of all secondary algae. Such protein import has been already experimentally proven in apicomplexans (Waller et al., 1998), diatoms (Lang et al., 1998), euglenozoans (Shashidhara et al., 1992), dinoflagellates (Nassoury et al., 2003), cryptophytes (Gould et al., 2006), and chlorarachniophytes (Hirakawa et al., 2009). Since phylogenies based on both nuclear and plastid genes congruently place C. velia within chromalveolates close to apicomplexan parasites (Janouškovec et al., 2010), it is bordering on certainty that the same targeting mechanism operates in C. velia as well.
Our predicted localizations of these enzymes into the plastid of C. velia in most cases reflect their origin in the algal symbiont. Most of them are most closely related to the enzymes of other photosynthetic eukaryotes, where the entire pathway is located to the plastid (Oborník and Green, 2005). The proteobacteria-like FeCH of C. velia that is not present in the genomes of other phototrophs but is shared with apicomplexan parasites seems to be located in the plastid of C. velia as well. This is rather surprising since this enzyme was shown to localize to the mitochondrion of Apicomplexa. However, in C. velia, it possesses clear bipartite N-terminal presequence (~90 amino acids long), whereas its homolog in Plasmodium falciparum bears much shorter presequence (~20 amino acids), which represents the mTP alone. Similarly, the cytosolic CPOX of P. falciparum lacks any N-terminal presequence when compared with bacterial homologs, while the N terminus of the CPOX of the same origin (secondary host nucleus) in C. velia is 65 amino acids longer, and both the SP and TP were predicted in this presequence (Table 1). These marked differences in the N-terminal sequences of the closely related enzymes between C. velia and P. falciparum are very likely the result of a selection pressure for different localizations. The presence of two distinct ferrochelatases in the plastid is not unprecedented; it was experimentally demonstrated in plants (Woodson et al., 2011).
Although the presence of a bipartite targeting presequence is generally considered as an evidence of localization to complex plastids, dual targeting to other compartment cannot be ruled out in some cases. However, testing this possibility would require direct experimental localization of these enzymes, which is not available at the moment due to the lack of transfection system for C. velia.
The multiple lines of evidence involving presence of putative targeting sequences, molecular phylogeny of the studied enzymes, as well as arrangement of the pathway in other phototrophic eukaryotes examined so far lead us to the prediction of intracellular location of enzymes involved in tetrapyrrole biosynthesis in C. velia, as shown in Figure 3B.
C. velia Synthesizes Tetrapyrroles from Gly and Succinyl-CoA
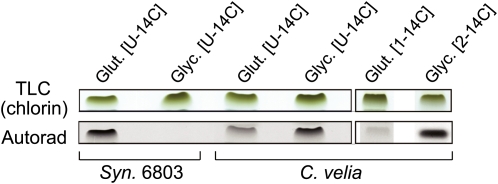
Our analysis of genomic information leads to the prediction that C. velia is unique in being able to synthesize chlorophyll from Gly and succinyl-CoA via the C4 pathway. Other photoautotrophs synthesize chlorophylls using the C5 pathway. To test this prediction, we supplied C. velia with either 14C-Gly or 14C-Glu and monitored the incorporation of labeled amino acids into the chlorophyll. The cyanobacterium Synechocystis 6803, with a well-characterized C5 pathway, was used as a control. As expected, C. velia uses Gly as a substrate for chlorophyll synthesis in contrast with Synechocystis, which only incorporates Glu into chlorophyll (Figure 4). We also found that C. velia metabolizes U-14C-Glu into chlorophyll, which could be due to the parallel use of the C5 pathway but could also be due to the easy conversion of this amino acid into succinyl-CoA in the mitochondrion. To distinguish between these two possibilities, C. velia was grown in the presence of 1-14C-Glu labeled specifically on the carboxyl group, which is metabolized to eight carbons of chlorophyll through C5 pathway but is released as CO2 when Glu enters the citric acid cycle. In this case, we detected only traces of labeled chlorophyll when compared with 2-14C-Gly used as a control (Figure 4).
Figure 4.
Incorporation of Radiolabeled Amino Acids into Chlorophyll in C. velia and Cyanobacterium Synechocystis 6803.
Cells were incubated with 350 μM 14C-Glu or 14C-Gly (specific activity ~50 mCi/mmol) for 2 h at 30°C and at 80 μmol of photons s−1 m−2. Chlorophyll was than extracted and converted into chlorin lacking both the phytol and methyl group at isocyclic E ring; thus, contains carbons from ALA precursors only. Prepared chlorin was separated by thin layer chromatography and exposed to an x-ray film.
[See online article for color version of this figure.]
Implications of the Evolution of Tetrapyrrole Synthesis
Tetrapyrrole biosynthesis in C. velia represents an evolutionary intermediate between the homologous pathways in other photosynthetic eukaryotes and parasitic apicomplexans. Targeting presequences of C. velia enzymes suggest that the bulk of the tetrapyrrole biosynthesis takes place in the plastid, as is the case in other photosynthetic eukaryotes. This localization is in agreement with the primary need for tetrapyrrole products in plastidic chlorophyll and heme biosynthesis, which most likely represents the state of the photosynthetic ancestor of apicomplexans. Moreover, C. velia contains multiple enzyme isoforms in at least three steps of the pathway, and evolutionary histories of these proteins suggest they were also present in the ancestor of apicomplexans. When photosynthesis was lost in the ancestor of apicomplexans, all enzyme isoforms were reduced to a single copy, and the last steps of the pathway catalyzed in the plastid were relocated to the cytosol (CPOX) and the mitochondrion (PPOX and FeCH).
This recompartmentalization is especially interesting because it readily illustrates how different cellular demands resulting from a life mode change can dramatically reshape organelle metabolism. In photosynthetic eukaryotes, chlorophyll is synthesized at a much higher rate than heme, and most of the heme is used for photosynthetic functions in the plastid. It is used in the cytochromes of photosystems and also serves as a precursor for the synthesis of billin pigments (Papenbrock et al., 1999; Castelfranco and Jones, 1975), while the mitochondrial heme requirements comprises only a fraction of the bulk tetrapyrrole product needed in the chloroplast. In heterotrophic eukaryotes and parasites, however, the heme biosynthesis in mitochondria is the principal tetrapyrrole sink. Tetrapyrrole biosynthesis in apicomplexans therefore provides a nice example how a metabolic pathway and trophic mode directly link through compartmentalization of the last biosynthetic steps, which generate products where most needed.
Interestingly, this pathway also gives us a unique opportunity to understand the recompartmentalization process, which is intimately linked to acquisition of a second ferrochelatase isoform from proteobacteria by the ancestor of apicomplexans and C. velia. The shared presence of this enzyme in C. velia and apicomplexans shows that its uptake was not an adaptive response to switch to parasitism in apicomplexans but a more ancestral condition that may have facilitated such a lifestyle change. More specifically, it is likely that presence of two functionally overlapping ferrochelatases lead to mitochondrial retargeting of the proteobacterial form in the apicomplexan ancestor without harmfully affecting tetrapyrrole biosynthesis in the plastid. After the acquisition of parasitism, the need for plastid tetrapyrroles became redundant and the canonical plastidic ferrochelatase was lost, while the proteobacterial ferrochelatase took over the tetrapyrrole supply for mitochondrial heme. In conclusion, this process illustrates how functional enzyme duplication mediated by horizontal gene transfer can facilitate relocation of a biosynthetic step between organelles.
To gain broader understanding of the evolution of the unusual pathways in Apicomplexa and C. velia, we examined the tetrapyrrole pathways of other chromalveolates. The whole-genome searches and our phylogenetic data revealed that heterotrophic chromalveolates in which plastids have not been found, such as ciliates and oomycetes, use the C4 pathway to synthesize ALA, and all the genes involved in the tetrapyrrole synthesis have the same origins as in primary heterotrophs. By contrast, the photosynthetic stramenopiles, cryptophytes, and haptophytes synthesize tetrapyrroles using the C5 pathway, as do primary algae (Figure 3). The same is probably true for dinoflagellates, as only the C5 pathway genes were found in the available expressed sequence tags of eight different species of this taxon. Since plastids of alveolates and heterokont algae are believed to share a common ancestry (Janouškovec et al., 2010), the presence of a heterotrophic-like pathway in some chromalveolates (Figure 3A) can be explained by the early loss of the plastid and the coexistence of separate tetrapyrrole pathways in these lineages before this event. In the lineage represented by C. velia and apicomplexan parasites, both pathways were combined to yield their unique pathways (Figure 3). Both tetrapyrrole pathways that are suggested to operate in the ancestral lineages of Chromalveolates are still found in photosynthetic euglenids, which arose through an independent secondary symbiosis involving a green alga. Euglena gracilis still possesses the original C4 pathway of the host cell that produces heme in the mitochondrion, while the plastid-localized C5 pathway that was derived from the algal endosymbiont serves only for the synthesis of chlorophyll and plastidial heme (Weinstein and Beale, 1983; Kořený and Oborník, 2011). Consequently, Euglena seems to be the only known eukaryotic phototroph that can lose its plastid and live as a heterotroph without external source of heme (Osafune and Schiff, 1983). As mentioned elsewhere (Oborník et al., 2009), the plastid could be lost only during the early stages of secondary endosymbiosis before the host cell delegated essential functions to this organelle. However, when the plastid took over the tetrapyrrole synthesis and the original pathway disappeared (as happened in most of photosynthetic eukaryotes), the plastid could no longer be lost without first acquiring some other source of tetrapyrroles. This corresponds well to the findings that the only apicomplexans without detectable plastids are those that do not synthesize their own heme (Mather and Vaidya, 2008; Mogi and Kita, 2010; Templeton et al., 2010).
METHODS
Sequence Retrieval and Phylogenetic Analyses
Partial sequences of the genes of the heme biosynthesis pathway were searched in the sequence data of the 454 genome sequencing surveys of Chromera velia and CCMP3155 (Janouškovec et al., 2010) by BLAST 2.2.18 (http://blast.ncbi.nlm.nih.gov). Complete coding sequences were amplified from the cDNA of C. velia by 3′ RACE and 5′ RACE using the FirstChoice RLM-RACE kit (Ambion), cloned, and sequenced. For each enzyme, appropriate homologs were identified by BLAST and downloaded from Web sources (see Supplemental Table 1 online), aligned using MAFFT (Katoh and Toh, 2008), manually edited using BioEdit (Hall, 1999), and used for further phylogenetic analyses (see Supplemental Data Sets 1 to 3 online). Bayesian trees were computed using PhyloBayes 3.2d under CAT-GTR evolutionary model (Lartillot and Philippe, 2004). For each analysis, two independent chains were run for a sufficient number of steps after the chains converged. The convergence of two independent chains was assessed based on bpcomp analysis (PhyloBayes 3.2d). The burnin value was chosen according to the same analysis (usually 20% of the trees were discarded). It should be noted that the presence of Plasmodium sequences in our analyses did not support any other method of tree construction because of their high divergence. Only the use of the advanced CAT model supported the expected placement of Plasmodium into the same clade with other apicomplexans in many of the analyses. Thus, the trees show only Bayesian posterior probabilities and not any other support, such as maximum likelihood bootstraps. We preferred to use all apicomplexan sequences, including those that are highly divergent, before the use of other additional phylogenetic methods on the reduced data set.
Putative Localizations of Enzymes of the Tetrapyrrole Synthesis in C. velia
Putative localizations of nuclear encoded enzymes of the tetrapyrrole pathway were tested by in silico predictions using CBS prediction servers (http://www.cbs.dtu.dk/services/). In particular, programs SignalP and TargetP (Emanuelsson et al., 2007) were used to predict SPs and TPs, respectively.
In Vivo Radiolabeling of Chlorophyll
To perform in vivo radiolabeling of chlorophyll, 50 mL of C. velia or Synechocystis 6803 cells at OD750 ~0.4 were concentrated into 350 μL of growth media supplemented with 20 mM TES/NaOH buffer, pH 8.0, transferred into a glass tube, and incubated on a shaker for 1 h at 30°C and at 80 μmol of photons s−1 m−2. Then, 350 μM 14C-Glu or 14C-Gly was added (specific activity ~50 mCi/mmol; American Radiolabeled Chemicals), and cells were further incubated under the same conditions. Incorporation of labeled amino acid into cells was monitored by measuring decrease of the radioactivity from the supernatant every 30 min on liquid scintillation analyzer (Packard Tri-Carb 1500). After 2 h, cells were washed three times with water and then pigments were extracted from pelleted cells using 1.3 mL of methanol/0.2% NH4. This solution was mixed with 140 μL of 1 M NaCl and 400 μL of hexane and vortexed. Hexane phase containing chlorophyll was removed and evaporated on a vacuum concentrator, resuspended in 100 μL of methanol, and mixed with 100 μL of methanol containing 10% KOH. After incubation for 15 min at room temperature, the solution was extracted twice with 200 μL hexane and once by 200 μL of petroleum ether (boiling range 40 to 65°C) to remove phytol and unprocessed chlorophyll, and finally acidified by 20 μL of concentrated HCl. The resulting solution containing the chlorophyll derivate chlorin was cleared by centrifugation, the supernatant transferred into a new tube, dried on a vacuum concentrator, and resuspended in a drop of methanol. Reverse-phase thin layer chromatography was performed on silica-gel plate (Macherey-Nagel); chloroform:methanol:water (65:25:3) was used as the mobile phase. After separation, the plate was dried and the signal captured on an x-ray film (exposure time of 5 d).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: HQ222925 to HQ222935 for the complete genes of the tetrapyrrole biosynthesis of C. velia and HQ245653 to HQ245656 for the partial gene sequences of the strain CCMP3155. Accession numbers for sequences used in phylogenetic analysis are presented in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Trees of δ-Aminolevulinic Acid Dehydratase, Porphobilinogen Deaminase, Uroporphyrinogen Synthase, Coproporphyrinogen Oxidase, and Protoporphyrinogen Oxidase.
Supplemental Figure 2. Comparison of ALAS N-Terminal Presequences of Various Eukaryotes, Including C. velia.
Supplemental Figure 3. Presence of Conserved Heme Regulatory Motifs (CP Motifs) in the N-Terminal Presequences of δ-Aminolevulinic Acid Synthases of Various Eukaryotes, Including C. velia.
Supplemental Table 1. Sequences Used in the Analyses.
Supplemental Data Set 1. Text File of Alignment Corresponding to Figure 1.
Supplemental Data Set 2. Text File of Alignment Corresponding to Figure 2.
Supplemental Data Set 3. Text File of Alignment Corresponding to Supplemental Figure 1.
Acknowledgments
This work was supported by the Czech Science Foundation (GA206/08/1423), the Grant Agency of the Academy of Sciences of the Czech Republic (IAA601410907), by Award IC/2010/09 made by King Abdullah University of Science and Technology, by the project Algatech (CZ.1.05/2.1.00/03.0110), and by the Grant Agency of University of South Bohemia (GAJU 146/2010/P) to M.O.; by Institutional Research Concept AV0Z50200510 to R.S.; and by the Canadian Institutes for Health Research (MOP42517) to P.J.K. P.J.K. is a Fellow of the Canadian Institute for Advanced Research and a Senior Scholar of the Michael Smith Foundation for Health Research.
AUTHOR CONTRIBUTIONS
L.K., R.S., P.J.K., and M.O. designed research. L.K. and R.S. performed research. L.K. and J.J. analyzed data. L.K. and M.O. wrote the article.
References
- Alemu A., Tsegaye W., Golassa L., Abebe G. (2011). Urban malaria and associated risk factors in Jimma town, south-west Ethiopia. Malar. J. 10: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camadro J.M., Chambon H., Jolles J., Labbe P. (1986). Purification and properties of coproporphyrinogen oxidase from the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 156: 579–587 [DOI] [PubMed] [Google Scholar]
- Castelfranco P.A., Jones O.T.G. (1975). Protoheme turnover and chlorophyll synthesis in greening barley tissue. Plant Physiol. 55: 485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey T.A., Woodruff J.H., Dailey H.A. (2005). Examination of mitochondrial protein targeting haem synthetic enzymes: in vivo identification of three functional haem-responsive motifs in 5-aminolaevulinate synthase. Biochem. J. 386: 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Foth B.J., Ralph S.A., Tonkin C.J., Struck N.S., Fraunholz M., Roos D.S., Cowman A.F., McFadden G.I. (2003). Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science 299: 705–708 [DOI] [PubMed] [Google Scholar]
- Gould S.B., Sommer M.S., Hadfi K., Zauner S., Kroth P.G., Maier U.G. (2006). Protein targeting into the complex plastid of cryptophytes. J. Mol. Evol. 62: 674–681 [DOI] [PubMed] [Google Scholar]
- Hall T.A. (1999). BioEdit: User-friendly biological sequence alignmet editor and analysis program for Windows 95/98/NT. Nucleic Acids Res. 41: 95–98 [Google Scholar]
- Hirakawa Y., Nagamune K., Ishida K. (2009). Protein targeting into secondary plastids of chlorarachniophytes. Proc. Natl. Acad. Sci. USA 106: 12820–12825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouškovec J., Horák A., Oborník M., Lukeš J., Keeling P.J. (2010). A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc. Natl. Acad. Sci. USA 107: 10949–10954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiroutová K., Horák A., Bowler C., Oborník M. (2007). Tryptophan biosynthesis in stramenopiles: eukaryotic winners in the diatom complex chloroplast. J. Mol. Evol. 65: 496–511 [DOI] [PubMed] [Google Scholar]
- Kalanon M., McFadden G.I. (2010). Malaria, Plasmodium falciparum and its apicoplast. Biochem. Soc. Trans. 38: 775–782 [DOI] [PubMed] [Google Scholar]
- Katoh K., Toh H. (2008). Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9: 286–298 [DOI] [PubMed] [Google Scholar]
- Kořený L., Oborník M. (2011). Sequence evidence for the presence of two tetrapyrrole pathways in Euglena gracilis. Genome Biol. Evol. 3: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroth P.G. (2002). Protein transport into secondary plastids and the evolution of primary and secondary plastids. Int. Rev. Cytol. 221: 191–255 [DOI] [PubMed] [Google Scholar]
- Lang M., Apt K.E., Kroth P.G. (1998). Protein transport into “complex” diatom plastids utilizes two different targeting signals. J. Biol. Chem. 273: 30973–30978 [DOI] [PubMed] [Google Scholar]
- Lartillot N., Philippe H. (2004). A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21: 1095–1109 [DOI] [PubMed] [Google Scholar]
- Mather M.W., Vaidya A.B. (2008). Mitochondria in malaria and related parasites: Ancient, diverse and streamlined. J. Bioenerg. Biomembr. 40: 425–433 [DOI] [PubMed] [Google Scholar]
- McFadden G.I. (1999). Plastids and protein targeting. J. Eukaryot. Microbiol. 46: 339–346 [DOI] [PubMed] [Google Scholar]
- Mogi T., Kita K. (2010). Diversity in mitochondrial metabolic pathways in parasitic protists Plasmodium and Cryptosporidium. Parasitol. Int. 59: 305–312 [DOI] [PubMed] [Google Scholar]
- Moore R.B., et al. (2008). A photosynthetic alveolate closely related to apicomplexan parasites. Nature 451: 959–963 [DOI] [PubMed] [Google Scholar]
- Nagaraj V.A., Arumugam R., Chandra N.R., Prasad D., Rangarajan P.N., Padmanaban G. (2009a). Localisation of Plasmodium falciparum uroporphyrinogen III decarboxylase of the heme-biosynthetic pathway in the apicoplast and characterisation of its catalytic properties. Int. J. Parasitol. 39: 559–568 [DOI] [PubMed] [Google Scholar]
- Nagaraj V.A., Arumugam R., Prasad D., Rangarajan P.N., Padmanaban G. (2010a). Protoporphyrinogen IX oxidase from Plasmodium falciparum is anaerobic and is localized to the mitochondrion. Mol. Biochem. Parasitol. 174: 44–52 [DOI] [PubMed] [Google Scholar]
- Nagaraj V.A., Prasad D., Arumugam R., Rangarajan P.N., Padmanaban G. (2010b). Characterization of coproporphyrinogen III oxidase in Plasmodium falciparum cytosol. Parasitol. Int. 59: 121–127 [DOI] [PubMed] [Google Scholar]
- Nagaraj V.A., Prasad D., Rangarajan P.N., Padmanaban G. (2009b). Mitochondrial localization of functional ferrochelatase from Plasmodium falciparum. Mol. Biochem. Parasitol. 168: 109–112 [DOI] [PubMed] [Google Scholar]
- Nassoury N., Cappadocia M., Morse D. (2003). Plastid ultrastructure defines the protein import pathway in dinoflagellates. J. Cell Sci. 116: 2867–2874 [DOI] [PubMed] [Google Scholar]
- Oborník M., Green B.R. (2005). Mosaic origin of the heme biosynthesis pathway in photosynthetic eukaryotes. Mol. Biol. Evol. 22: 2343–2353 [DOI] [PubMed] [Google Scholar]
- Oborník M., Janouškovec J., Chrudimský T., Lukeš J. (2009). Evolution of the apicoplast and its hosts: From heterotrophy to autotrophy and back again. Int. J. Parasitol. 39: 1–12 [DOI] [PubMed] [Google Scholar]
- Oborník M., Vancová M., Lai D.H., Janouškovec J., Keeling P.J., Lukeš J. (2011). Morphology and ultrastructure of multiple life cycle stages of the photosynthetic relative of apicomplexa, Chromera velia. Protist 162: 115–130 [DOI] [PubMed] [Google Scholar]
- Osafune T., Schiff J.A. (1983). W10BSmL, a mutant of Euglena gracilis var. bacillaris lacking plastids. Exp. Cell Res. 148: 530–535 [DOI] [PubMed] [Google Scholar]
- Panek H., O’Brian M.R. (2002). A whole genome view of prokaryotic haem biosynthesis. Microbiology 148: 2273–2282 [DOI] [PubMed] [Google Scholar]
- Papenbrock J., Mock H.P., Kruse E., Grimm B. (1999). Expression studies in tetrapyrrole biosynthesis: inverse maxima of magnesium chelatase and ferrochelatase activity during cyclic photoperiods. Planta 208: 264–273 [Google Scholar]
- Ralph S.A., van Dooren G.G., Waller R.F., Crawford M.J., Fraunholz M.J., Foth B.J., Tonkin C.J., Roos D.S., McFadden G.I. (2004). Tropical infectious diseases: Metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat. Rev. Microbiol. 2: 203–216 [DOI] [PubMed] [Google Scholar]
- Rao A., Yeleswarapu S.J., Srinivasan R., Bulusu G. (2008). Localization of heme biosynthesis pathway enzymes in Plasmodium falciparum. Indian J. Biochem. Biophys. 45: 365–373 [PubMed] [Google Scholar]
- Sato S., Clough B., Coates L., Wilson R.J.M. (2004). Enzymes for heme biosynthesis are found in both the mitochondrion and plastid of the malaria parasite Plasmodium falciparum. Protist 155: 117–125 [DOI] [PubMed] [Google Scholar]
- Sato S., Wilson R.J.M. (2003). Proteobacteria-like ferrochelatase in the malaria parasite. Curr. Genet. 42: 292–300 [DOI] [PubMed] [Google Scholar]
- Shanmugam D., Wu B., Ramirez U., Jaffe E.K., Roos D.S. (2010). Plastid-associated porphobilinogen synthase from Toxoplasma gondii: Kinetic and structural properties validate therapeutic potential. J. Biol. Chem. 285: 22122–22131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashidhara L.S., Lim S.H., Shackleton J.B., Robinson C., Smith A.G. (1992). Protein targeting across the three membranes of the Euglena chloroplast envelope. J. Biol. Chem. 267: 12885–12891 [PubMed] [Google Scholar]
- Schulze J.O., Schubert W.D., Moser J., Jahn D., Heinz D.W. (2006). Evolutionary relationship between initial enzymes of tetrapyrrole biosynthesis. J. Mol. Biol. 358: 1212–1220 [DOI] [PubMed] [Google Scholar]
- Templeton T.J., Enomoto S., Chen W.J., Huang C.G., Lancto C.A., Abrahamsen M.S., Zhu G. (2010). A genome-sequence survey for Ascogregarina taiwanensis supports evolutionary affiliation but metabolic diversity between a Gregarine and Cryptosporidium. Mol. Biol. Evol. 27: 235–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dooren G.G., Stimmler L.M., McFadden G.I. (2006). Metabolic maps and functions of the Plasmodium mitochondrion. FEMS Microbiol. Rev. 30: 596–630 [DOI] [PubMed] [Google Scholar]
- Volland C., Felix F. (1984). biosynthesisIsolation and properties of 5-aminolevulinate synthase from the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 142: 551–557 [DOI] [PubMed] [Google Scholar]
- von Heijne G., Steppuhn J., Herrmann R.G. (1989). Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem. 180: 535–545 [DOI] [PubMed] [Google Scholar]
- Waller R.F., Keeling P.J., Donald R.G.K., Striepen B., Handman E., Lang-Unnasch N., Cowman A.F., Besra G.S., Roos D.S., McFadden G.I. (1998). Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 95: 12352–12357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J.D., Beale S.I. (1983). Separate physiological roles and subcellular compartments for two tetrapyrrole biosynthetic pathways in Euglena gracilis. J. Biol. Chem. 258: 6799–6807 [PubMed] [Google Scholar]
- Woodson J.D., Perez-Ruiz J.M., Chory J. (2011). Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr. Biol. 21: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B. (2006). Heme Biosynthetic Pathway in Apicomplexan Parasites. PhD dissertation (Philadelphia, PA: University of Pennsylvania) [Google Scholar]
- Zhang L., Guarente L. (1995). Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 14: 313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]