This study reports the identification of a novel regulator of axillary meristem formation in rice, showing that LAX PANICLE2 (LAX2) likely acts in the maintenance of the axillary meristem. In addition, it reveals that LAX2 localizes to the nucleus and appears to form a dimer with LAX1, which is a basic helix-loop-helix transcriptional factor.
Abstract
Aerial architecture in higher plants is dependent on the activity of the shoot apical meristem (SAM) and axillary meristems (AMs). The SAM produces a main shoot and leaf primordia, while AMs are generated at the axils of leaf primordia and give rise to branches and flowers. Therefore, the formation of AMs is a critical step in the construction of plant architecture. Here, we characterized the rice (Oryza sativa) lax panicle2 (lax2) mutant, which has altered AM formation. LAX2 regulates the branching of the aboveground parts of a rice plant throughout plant development, except for the primary branch in the panicle. The lax2 mutant is similar to lax panicle1 (lax1) in that it lacks an AM in most of the lateral branching of the panicle and has a reduced number of AMs at the vegetative stage. The lax1 lax2 double mutant synergistically enhances the reduced-branching phenotype, indicating the presence of multiple pathways for branching. LAX2 encodes a nuclear protein that contains a plant-specific conserved domain and physically interacts with LAX1. We propose that LAX2 is a novel factor that acts together with LAX1 in rice to regulate the process of AM formation.
INTRODUCTION
The principal body plan of a plant is established during embryogenesis by the generation of an apical-basal axis. This bipolar organization is defined by a shoot apical meristem (SAM) and a root apical meristem (McSteen and Leyser, 2005). By contrast, the complexity of the adult plant architecture is generated by lateral growth, which is determined by the activity of postembryonically produced secondary meristems, called axillary meristems (AMs). In rice (Oryza sativa), an AM produced at the axil of the leaf generates a new shoot branch called the tiller, whereas an AM produced in the inflorescence generates a higher order of inflorescence branch, called the rachis branch, which bears a grass-specific structural unit of the inflorescence—the spikelet (Hoshikawa, 1989). More precisely, upon transition to the reproductive phase from the vegetative phase, the SAM of each tiller is transformed into an inflorescence meristem and forms an inflorescence called a panicle. During panicle development, the inflorescence meristem initiates primary branch meristems, each of which produces a primary panicle branch (PB). The PB also produces a certain number of secondary branch meristems, which produce secondary panicle branches (SBs) or differentiate into spikelets, depending on the time and position of their occurrence. Finally, the primary branch meristem and secondary branch meristem differentiate into a terminal spikelet. Thus, the pattern of AM formation is of special significance in crop production in various grass species because it regulates the number of shoot branches, inflorescence branches, and spikelets.
The pattern of lateral branching is affected by both the generation and growth of lateral shoots. Generation and outgrowth of lateral shoots are regulated by the combined action of hormones, such as auxin, cytokinin, and strigolactone, and environmental cues. Furthermore, a number of regulatory genes expressed in AM-specific and AM-nonspecific manners affect branching (McSteen, 2009).
Mutants in AM-nonspecific genes, including Arabidopsis thaliana revoluta and cup-shaped cotyledon3, show phenotypes not only in branching but also in various developmental aspects (SAM formation, leaf polarity, and vascular patterning), possibly derived from defects in general meristematic functions (Talbert et al.,1995; Otsuga et al., 2001; Hibara et al., 2006; Raman et al., 2008).
There is another type of mutant that exhibits AM-specific defects. In tomato (Solanum lycopersicum), lateral suppressor (ls) and blind (bl) mutants show defects in lateral branching (Schumacher et al., 1999; Schmitz et al., 2002). In Arabidopsis, lateral suppressor (las) and regulator of axillary meristem (rax) mutants are defective in AM formation (Greb et al., 2003; Müller et al., 2006). In maize (Zea mays), barren stalk1 (ba1) and barren inflorescence2 (bif2) mutants exhibit severe suppression of all types of AM (McSteen and Hake, 2001; Ritter et al., 2002; Gallavotti et al., 2004).
In rice, several mutants show AM-specific defects, including defects in panicle development. The mutants monoculm1/small panicle (moc1/spa), lax panicle1 (lax1), and frizzy panicle (fzp) have been shown to affect the patterning of AM (Komatsu et al., 2001, 2003; Li et al., 2003; Li et al., 2009). The genes underlying these mutations have been isolated, and most of these genes encode transcriptional regulators with expression patterns that evoke their involvement in the patterning of AMs. Thus, the transcriptional network operates during the initiation or maintenance of the AM, although the direct or indirect connections between these factors have not been precisely determined.
Rice MOC1, which encodes a transcriptional regulator of the GRAS family, is homologous to tomato LS and Arabidopsis LAS. Tomato BL and Arabidopsis RAX are orthologous genes encoding R2R3-type MYB family transcription factors. Rice LAX and maize BA1 are also orthologous; each encodes a transcription factor that contains a basic helix-loop-helix (bHLH) domain. The rice FZP gene, which encodes an ethylene-responsive element binding factor, is an ortholog of maize BD1. In many cases, transcriptional factors identified from AM-defective mutants are conserved in different plant species, suggesting that their functions in AM patterning are also conserved.
AM patterning can also be regulated at a level other than the transcriptional level. Rice SHORT PANICLE1 encodes a putative transporter that belongs to the peptide transporter family, while ERECT PANICLE2 encodes a novel plant-specific protein with unknown biochemical function that localizes to the endoplasmic reticulum (Li et al., 2009; Zhu et al., 2010). Maize BIF2 encodes a Ser/Thr protein kinase that is orthologous to PINOID. PINOID phosphorylates and regulates PIN-FORMED1 (PIN1) localization to the plasma membrane and mediates directional auxin transport (Michniewicz et al., 2007). Interestingly, BIF2 and BA1 are both required for AM patterning and have direct interaction with each other (Skirpan et al., 2008). These observations suggest the possibility that BIF2 regulates AM formation through posttranslational modifications of BA1. Another example of posttranslational regulation is the rice LAX1 protein. LAX1 is expressed in two to three layers of cells in the boundary between the SAM and the region of new meristem formation (Oikawa and Kyozuka, 2009). Interestingly, LAX1 movement between cells is required for the full functioning of LAX1.
Because the process of AM formation is regulated in multiple layers, elucidation of the genetic and biochemical relationship among the factors involved in this process is necessary to understand AM formation. Of particular interest is the analysis of mutants that show AM-specific defects and the clarification of genetic and biochemical interactions among the factors involved in AM formation. Here, we describe a novel gene, LAX2, that is involved in the maintenance of AM in rice. LAX2 regulates the branching of the aboveground parts of rice plants throughout plant development, except for the PB of the panicle. The lax2 mutant is similar to the lax1 mutant in that it lacks AMs for most of the lateral branching of the panicle (SBs and spikelets) and has reduced tillers at the vegetative stage. The synergistic enhancement of the branching phenotype of the lax1 lax2 double mutant suggests that there are multiple pathways for branching. We show that LAX2 encodes a nuclear protein with a plant-specific conserved domain and that LAX2 physically interacts with LAX1 through a region containing this conserved domain. Thus, we propose that LAX2 is a novel genetic component that is required for the process of AM formation and acts together with LAX1 in rice.
RESULTS
lax2 Affects Both Vegetative and Reproductive Branching
lax2 mutations were isolated while screening for mutants with abnormal panicle development. We found three independent mutant alleles, all of which exhibited a sparse-panicle phenotype. These alleles, recovered from ethyl methanesulfonate, tissue culture, and γ-ray irradiation mutagenesis populations, were designated as lax2-1, lax2-2, and lax2-3, respectively.
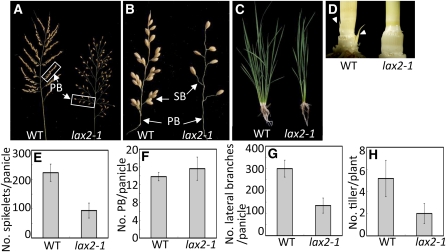
The panicle of wild-type rice is composed of PBs, SBs, and spikelets (the grass-specific structural unit of the inflorescence). There are ~12 PBs in a panicle; each PB bears several SBs and spikelets, and SBs also bear several spikelets. This multiple order of branching is the basis for the characteristic appearance of the rice panicle. To characterize the nature of the sparse appearance of the lax2-1 panicle, we counted the number of inflorescence branches in the wild type and mutant. The lax2-1 mutant panicle produced fewer spikelets compared with the wild-type panicle (Figures 1A and 1E). Quantitative analysis of the lax2-1 mutant panicle revealed that the number of PBs was not affected in the mutant (Figure 1F). However, the number of total lateral branches, which was the sum of the number of PBs, SBs, and spikelets in a panicle, was reduced in lax2-1 (Figure 1G). This indicates that the sparse panicle appearance and the reduction in spikelet number in the lax2-1 mutant are attributable both to reduced higher order branching, such as the number of SBs, and to a reduced number of spikelets borne directly on branches (Figure 1B). Because a spikelet other than a terminal spikelet is produced as a lateral branch from an inflorescence meristem, lax2-1 mutants seem to have a defect in producing all the lateral branches other than PBs in panicles.
Figure 1.
Plant Morphology of the lax2-1 Mutant.
(A) Mature panicles. The lax2-1 mutant has a sparse appearance due to the production of fewer branches and spikelets. WT, wild type.
(B) Enlarged view of boxes in (A).
(C) Whole plants at the vegetative growth stage. lax2-1 mutant has fewer tillers than the wild type.
(D) Culms of the wild type and lax2-1 after removal of the surrounding leaves. White arrowheads indicate the prophyll, which encloses the AM.
(E) Quantification of the number of spikelets in a panicle.
(F) Quantification of the number of PBs per panicle.
(G) Quantification of the number of lateral branches, which is the sum of the number of PBs, SBs, and spikelets in a panicle.
(H) Quantification of the number of tillers per plant.
Error bars in (E) to (H) represent sd. The sample size for (E) to (H) is n = 9.
lax2-1 mutants also had defects in vegetative development. In lax2-1 mutants, the number of tillers produced during vegetative growth was reduced (Figures 1C and 1H). Because there was no trace of tiller buds at the base of the leaves in lax2-1 mutants (Figure 1D), the reduction in the number of tillers is caused by a defect in AM formation and not by enhanced apical dominance or growth arrest of tiller buds. As a result of the reduction in the number of tillers in lax2-1, mature lax2-1 plants had fewer panicles than wild-type plants. Thus, the lax2-1 mutant failed to develop lateral branches during both vegetative and reproductive development, although the number of PBs in the panicle was not affected.
The other lax2 mutant alleles, lax2-2 and lax2-3, had similar branching defects, as observed in lax2-1 mutants in panicles, except that the number of PBs in lax2-2 and lax2-3 was slightly increased (Table 1). Since lax2 mutations reduce branching at most of the developmental stages, the increase in the number of PBs in these mutants could be due to an indirect effect to compensate for the reduction of the higher order branching and spikelets. The number of tillers in lax2-2 and lax2-3 was reduced but the degree of the reduction in lax2-3 was not as evident as lax2-1 or lax2-2 (Table 1). Considering that all three lax2 alleles carry mutations that are presumably null (discussed below) and all three lax2 mutants originated from different genetic backgrounds (see Methods), the vegetative branching phenotype might be affected by genetic background.
Table 1.
Phenotypic Characterization of lax2 Mutants and Their Respective Parental Wild Types in Rice
| Genotype | n | No. of Tillers per Plant | No. of Primary Branches per Panicle | No. of Secondary Branches per Panicle | No. of Spikelets per Panicle |
| Wild type (Zhonghua 11) | 9 | 5.2 ± 1.6 | 13.8 ± 0.9 | 43.8 ± 5.9 | 223.2 ± 29.4 |
| lax2-1 | 9 | 2.1 ± 0.9*** | 15.6 ± 2.6 | 18.3 ± 4.6*** | 93.8 ± 25.6*** |
| Wild type (Nipponbare) | 60 | 19.6 ± 3.6 | 8.9 ± 0.6 | 10.0 ± 2.0 | 76.8 ± 8.8 |
| lax2-2 | 60 | 17.8 ± 2.8*** | 9.9 ± 0.9*** | 0.0 ± 0.0*** | 36.3 ± 4.0*** |
| Wild type (Norin 8) | 60 | 18.7 ± 4.3 | 9.4 ± 0.7 | 13.6 ± 2.2 | 92.5 ± 8.8 |
| lax2-3 | 60 | 17.8 ± 3.2* | 10.7 ± 0.8*** | 0.1 ± 0.2*** | 54.0 ± 4.3*** |
Average values ± sd are shown. Asterisks (*** and *) indicate that the differences between the mutants and their corresponding parental wild types are statistically significant at P < 0.001 and < 0.05, respectively, according to the Student's t test.
Genetic Interactions of lax2 with lax1 or moc1
To clarify the genetic interactions of lax2 with other rice mutants that exhibit a defect in AM formation, we made double mutants containing lax2 and either lax1 or moc1. lax1 had a sparse panicle with reduced SBs and spikelets, which was similar to the lax2 panicle, and lax1 had tillers with slightly reduced numbers at the vegetative stage (Figures 2A to 2C and 2G to 2I) (Oikawa and Kyozuka, 2009). We made lax1 lax2 double mutants using null alleles of lax1, lax1-1, and lax1-6. lax1-1 is a deletion allele of lax1 (Komatsu et al., 2003), and lax1-6 has a single nucleotide deletion at the 5′ end of the region encoding the conserved bHLH domain. The phenotype of the lax1 lax2 double mutant was more extreme than that of the single mutants. In the double mutant panicle, all SBs and spikelets except terminal spikelets were absent, whereas the number of PBs was not affected compared with the single mutants (Figure 2K, Table 2). At the vegetative stage, both single mutants shared the similar phenotype of a slightly reduced number of tillers (Figures 2B and 2C); however, there was a strong reduction in tiller branches in the double mutant (Figure 2E, Table 2). Thus, the dramatic effect on branching of the spikelets and the lack of tiller branches in the lax1 lax2 double mutant implies that there are multiple pathways for branching and that LAX1 and LAX2 consistently function in these pathways both at the vegetative stage and reproductive stage, except during PB formation.
Figure 2.
Morphology of Double Mutants of lax2 with lax1 or moc1.
(A) to (F) Mature plants. WT, wild type.
(G) to (L) Panicle.
(A) and (G) The wild type.
(B) and (H) lax1-1.
(C) and (I) lax2-1.
(D) and (J) moc1-4.
(E) and (K) lax1-1 lax2-1 double mutant.
(F) and (L) lax2-2 moc1-4 double mutant. The inset in (L) is a higher magnification of the double mutant panicle. Bar = 2 cm.
White and red arrowheads indicate the position of nodes in the panicle and terminal spikelets, respectively.
Table 2.
Quantification of the Double Mutants of lax1 and lax2
| Linea | Genotype | n | No. of Tillers per Plant | No. of Primary Branches per Panicle | No. of Secondary Branches per Panicle | No. of Spikelets per Panicle |
| F3 No. 1 | LAX2/± lax1-6/lax1-6 | 21 | 15.2 ± 4.1 | 10.5 ± 1.0 | 0.1 ± 0.3 | 38.0 ± 6.4 |
| lax2-3/lax2-3 lax1-6/lax1-6 | 5 | 1.6 ± 0.9* | 11.3 ± 1.6 | 0.0 ± 0.0 | 12.6 ± 2.4* | |
| F3 No. 2 | lax2-3/lax2-3 LAX1/± | 16 | 20.6 ± 5.6 | 9.3 ± 0.9 | 0.0 ± 0.1 | 39.5 ± 6.6 |
| lax2-3/lax2-3 lax1-6/lax1-6 | 10 | 2.6 ± 1.2* | 9.6 ± 1.8 | 0.0 ± 0.0 | 9.9 ± 2.3* |
Average values ± sd are shown. Asterisks indicate that the differences between the double and single mutants are statistically significant at P < 0.001, according to the Student's t test.
F3 plants were derived from self-pollination of an F2 plant that was either heterozygous for lax2-3 and homozygous for lax1-6 or homozygous for lax2-3 and heterozygous for lax1-6. The F2 plants originated from crossing lax2-3 and lax1-6 mutants.
The allele of moc1 used in our analysis, moc1-4, also has a sparse panicle; however, the branching pattern of the moc1-4 panicle is different from that of the wild type, lax1, or lax2. moc1-4 has only a few PBs, the reduction of which is mostly due to the loss of the PBs in the lower part of the panicle. The numbers of SBs and spikelets on the PB are also reduced in moc1-4 (Figure 2J). At the vegetative stage, moc1-4 did not affect tillering (Figure 2D), although moc1-4 contains a nonsense mutation at a conserved Trp residue within the GRAS domain and appears to be a null allele. Because the vegetative branching phenotype in moc1 is strongly affected by the genetic background, we tried to make a lax2 moc1 double mutant in the same genetic background as the corresponding single mutants. We used lax2-2 and moc1-4, both of which were derived from Nipponbare. The phenotype of the lax2-2 moc1-4 double mutant was more extreme than that of either single mutant. In the double mutant panicle, all PBs, SBs, and spikelets were absent (Figure 2L), and there were no tiller branches at the vegetative stage (Figure 2F). Thus, the dramatic effect on branching of the panicle and tiller branches in the lax2 moc1 double mutant implies that there is another pathway for branching in which MOC1 is involved and that LAX2 and MOC1 function in vegetative and reproductive branching, including during PB formation.
LAX2 Is Involved in the Maintenance of the AM
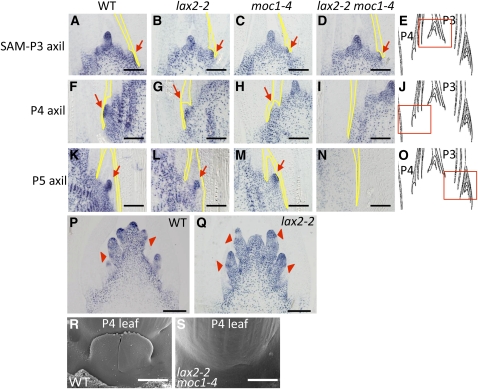
The reduced branching at both the vegetative and reproductive stages in lax2 single and double mutants was caused by a defect in the formation of the AM, suggesting that LAX2 is involved in the initiation or maintenance of the AM. To confirm this, we observed the expression of the OSH1 protein (a marker of the meristem) and the morphology of the AM in single and double mutants of lax2 and moc1.
At the vegetative stage in a wild-type plant, a population of cells that expresses OSH1 can be detected at the axil of the second and third youngest leaf primordia (P2-P3) as a bulge (Steeves and Sussex, 1989), depending on the sampling stage within a plastochron (Figures 3A and 3E). Similar structures were observed in both lax2-2 and moc1-4 single and double mutants (Figures 3B to 3D). These bulges possibly include cells that give rise to an incipient AM. At the P4 leaf axil, the AM is formed and grows in wild-type plants and in lax2 and moc1 single mutants (Figures 3F to 3H and 3J). At the P5 leaf axil, the AM grows continuously and is covered by prophyll in wild-type plants and in lax2 and moc1 single mutants (Figure 3K to 3M and 3O). However, in the lax2 moc1 double mutant, the bulge with OSH1-expressing cells, although seen initially at the P2-P3 leaf axil, did not proliferate at the P4 leaf axil (Figure 3I) and was absent at the P5 leaf axil (Figure 3N). The lack of the AM in the lax2 moc1 double mutant was also observed by scanning electron microscopy analysis (Figures 3R and 3S). This phenotype is quite consistent because there is never a tiller in the lax2 moc1 double mutant. This observation negates the possibility that the initiation of the SAM is stochastically arrested in the mutant and indicates that the double mutant is defective in the maintenance of the AM. Thus, at the vegetative stage, both LAX2 and MOC1 are involved in the maintenance of the AM.
Figure 3.
Anti-OSH1 Immunostaining of the Wild Type, lax2, moc1, and lax2 moc1 Double Mutant and Scanning Electron Microscopy Image of the Wild Type and lax2 moc1 Double Mutant.
(A), (F), and (K) Vegetative shoot of wild-type (WT) Nipponbare.
(B), (G), and (L) Vegetative shoot of lax2-2.
(C), (H), and (M) Vegetative shoot of moc1-4.
(D), (I), and (N) Vegetative shoot of the lax2-2 moc1-4 double mutant.
(E), (J), and (O) Position of photograph in shoot in each row is shown in red boxes.
(P) Developing wild-type Nipponbare inflorescence with SB primordia.
(Q) Developing inflorescence of lax2 at the same stage as in (P).
(R) and (S) Scanning electron microscopy images of the wild type (R) and double mutant (S) at the base of P4 leaves.
P3, P4, and P5 represent the 3rd, 4th, and 5th youngest leaf primordia, respectively (Steeves and Sussex, 1989). Red arrows indicate the position of AMs marked by morphology and OSH1 expression. Red arrowheads indicate the position of incipient AMs identified by the condensed OSH1 signals. Black and white bars indicate 100 μm and 1 mm, respectively.
At the reproductive stage, AM primordia that give rise to PBs formed normally in the lax2 mutant as in the wild type (Figures 3P and 3Q). The dense staining of OSH1 indicated by red arrowheads in Figures 3P and 3Q reside just above the place of OSH1 downregulation both in the wild type and lax2 mutant. These places are also marked by compact cells produced by frequent cell divisions and correspond to incipient meristems destined to become SBs or spikelets (Komatsu et al., 2003; McSteen et al., 2007). Considering that the number of lateral branches formed on the PBs, such as SBs or spikelets, was reduced in lax2, LAX2 is also involved in the maintenance of AMs at the reproductive stage.
LAX2 Encodes a Novel Nuclear Factor
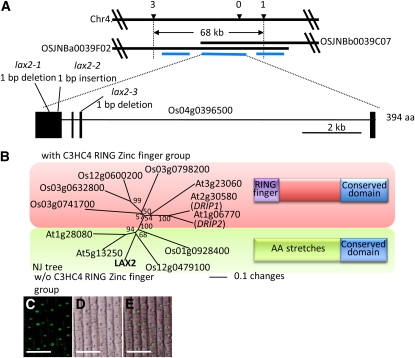
We cloned lax2 by a map-based method. lax2 mutations were roughly mapped on chromosome 4. We screened recombinants between two genetic markers that reside 68 kb apart on the physical map and identified four recombinants between these markers. Three genes were predicted to exist between two genetic markers, and among them we found mutations in gene Os04g0396500 for all three lax2 mutant alleles (Figure 4A). The putative LAX2 gene consisted of four exons and three introns and encoded a plant-specific conserved domain. The cDNA clone of the putative LAX2 driven by its own promoter was introduced into the lax2 mutant, and the characteristic lax2 phenotype (i.e., in which the SB does not form on the PB) was complemented (see Supplemental Figure 1 and Supplemental Table 1 online). Based on these results, we conclude that Os04g0396500 is LAX2.
Figure 4.
Molecular Cloning of LAX2.
(A) Positional cloning of LAX2. Black horizontal bars represent chromosomal segments or BAC clones corresponding to the region of interest on chromosome 4. Numbers above the top bar represent the number of recombinants between markers and the lax2 mutation after screening an F2 population derived from crosses between the lax2 mutation and an indica cultivar. Blue lines represent the positions of predicted genes. The detailed gene structure of LAX2 is shown at the bottom. Boxes and lines connecting boxes represent exons and introns, respectively. The gene structure presented here is modified from that predicted in the Rice Annotation Project Database or The Institute for Genomic Research database based on our experiments. aa, amino acids.
(B) Phylogenetic analysis of amino acid sequences of conserved domains shared with LAX2 from rice and Arabidopsis. The alignment used to construct the phylogenetic tree was made using ClustalX (Jeanmougin et al., 1998) and is shown in Supplemental Figure 2 online. The phylogenetic tree was constructed by the NJ method (Saito and Nei, 1987) using PAUP* 4.0 software. Proteins sharing the conserved domain with LAX2 are divided into two groups, one with a zinc finger motif at the N-terminal side of the conserved domain (red box) and one without a zinc finger (green box). Numbers adjacent to each branch point of the tree indicate bootstrap support of that branch after 1000 replicates. The bar in the figure represents the degree of amino acids changes.
(C) to (E) Subcellular localization of LAX2:GFP fusion protein in transgenic rice plants. GFP fluorescence, differential interference contrast, and merged images are shown in (C), (D), and (E), respectively. Bars = 50 um.
Using the predicted amino acid sequence of LAX2, we identified six genes with homology to LAX2 in the rice genome and five genes from the Arabidopsis genome (Figure 4B; see Supplemental Data Set 1 and Supplemental Figures 2A and 2B online). We conducted a phylogenetic analysis of the amino acid sequences of conserved domains from rice and Arabidopsis that are shared with LAX2. Proteins containing the conserved domains shared with LAX2 were divided into two groups based on their conservation of amino acid residues. Members of one group had a C3HC4-type RING zinc finger motif at the N-terminal side of the conserved domain, while members of the other group did not have this motif; instead, the other group had numerous stretches of the same amino acid residues, including stretches of Arg, Ala, Glu, His, Pro, Gly, Ser, and Thr, at the N-terminal region (Figure 4B; see Supplemental Figure 2B online).
To identify the molecular function of LAX2, we investigated the cellular localization of LAX2 by expressing LAX2:green fluorescent protein (GFP) fusion proteins driven by a 35S constitutive promoter of Cauliflower mosaic virus in the root cells of stable rice transformants (Figures 4C to 4E). GFP fluorescence was localized exclusively in the nucleus, suggesting that LAX2 is a nuclear protein, although we could not find the typical nuclear localization signal in the predicted LAX2 amino acid sequence.
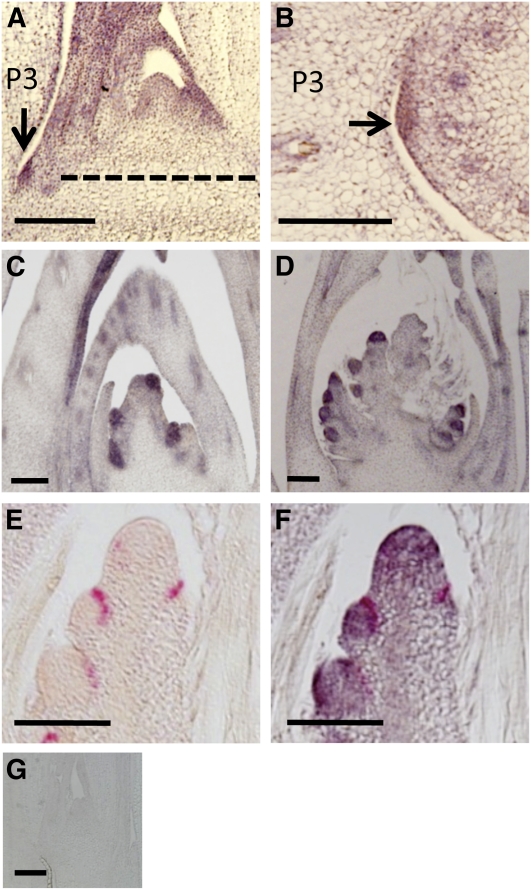
We then examined the in situ expression of LAX2. In the vegetative shoots, LAX2 expression was observed at the axil of the P3 leaf, at the location where the AM is formed, as well as in young leaves (Figures 5A, 5B, and 5G). In the reproductive stage, LAX2 expression was observed at all the AMs that give rise to PBs, SBs, and spikelets (Figures 5C and 5D). The lax1 and lax2 mutants had similar phenotypes during the vegetative and reproductive stages. Furthermore, the lax1 lax2 double mutant resulted in a more extreme lax panicle phenotype than either single mutant, without markedly affecting the number of PBs (Figures 2H, 2I, and K). Based on these findings, we hypothesized that LAX1 and LAX2 act at a related process in AM formation. To test this, we observed whether the spatiotemporal expression domains of LAX1 and LAX2 overlapped using two-color in situ hybridization (Figures 5E and 5F). The domain of LAX1 mRNA localization, visualized in red in Figures 5E and 5F, resides at the margin of the LAX2 expression domain, visualized in purple in Figure 5F. At the tip of the primary branch meristem, we also observed LAX2 expression, and this is consistent with the fact that PBs are also branches made at the inflorescence. LAX1 is directionally trafficked to the body of the AM, from its mRNA localization domain, which is the boundary between the AM and SAM (Oikawa and Kyozuka, 2009). Thus, it is possible that the functional domains of LAX1 and LAX2 proteins largely overlap.
Figure 5.
In Situ mRNA Accumulation Pattern of LAX2.
(A) In situ hybridization with a LAX2-specific probe on a longitudinal section of a shoot during vegetative development. The dashed line indicates the approximate position of the cross section shown in (B).
(B) In situ hybridization with a LAX2-specific probe on a cross section of a shoot during vegetative development.
(C) In situ hybridization with a LAX2-specific probe on a longitudinal section of an inflorescence meristem with PB primordia.
(D) In situ hybridization with a LAX2-specific probe on a longitudinal section of an inflorescence meristem with secondary PB primordia.
(E) In situ hybridization with a LAX1-specific probe on a longitudinal section of a primary branch meristem with SB primordia.
(F) Two-color in situ hybridization with LAX1- and LAX2-specific probes on a longitudinal section of a primary branch meristem with SB primordia. LAX2 expression is detected by a purple color and is visualized on the same section in (E). LAX1 mRNA localization is visualized by red color.
(G) In situ hybridization with a LAX2 sense probe on a longitudinal section of the vegetative shoot.
Bars = 100 μm. Arrows indicate LAX2 expression at the vegetative AM.
Because the LAX1 protein and LAX2 mRNA localization domains largely overlap, we investigated the possibility that LAX1 and LAX2 regulate the expression of LAX2 and LAX1, respectively, by in situ hybridization (see Supplemental Figure 3 online) and RT-PCR analysis (see Supplemental Figure 4 online). We found that expression of LAX1 was not abolished in the lax2 mutant, indicating that the expression of LAX1 does not depend on the expression of LAX2 (see Supplemental Figure 3 online). We also found that LAX2 mRNA was not abolished in lax1 mutant (see Supplemental Figure 4 online). This indicates that LAX2 expression does not require LAX1, although there is a possibility that LAX1 affects LAX2 expression at a very subtle level, which cannot be distinguished by RT-PCR.
LAX2 Physically Interacts with LAX1, and the Interaction Requires the Conserved Domain
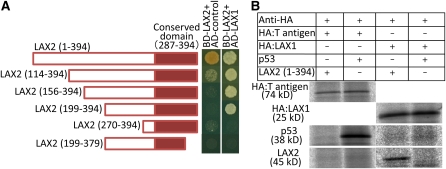
The sites where LAX1 and LAX2 function in the AM overlap, and both proteins localize in the nucleus; therefore, it is possible that LAX1 and LAX2 physically interact. To test this, we conducted a yeast two-hybrid assay (Figure 6A). Yeast cells carrying bait with the full-length LAX2 cDNA and the empty prey vector grew on the selective medium, suggesting the presence of a transcriptional activation domain in LAX2, at least in yeast cells. Consequently, we used a bait construct with a partial LAX2 cDNA lacking the region corresponding to the 113 amino acids at the N terminus of LAX2. We confirmed that yeast cells carrying this bait and empty prey vector barely grew on the selective medium. We then transformed yeast cells with this bait and the prey with LAX1 cDNA. The transformants grew on the selective medium, suggesting an interaction between LAX1 and LAX2 in yeast cells.
Figure 6.
Molecular Interaction between LAX1 and LAX2.
(A) Interaction between LAX1 and LAX2 was tested by a yeast two-hybrid assay. The full-size LAX2 cDNA and a deletion series of LAX2 cDNAs were fused with a DNA binding domain construct (BD), and the full-size cDNA of LAX1 was fused with an activation domain construct (AD). Yeast cells were grown on −Leu, Trp, His medium, and the interactions were monitored by growth on the selective medium. Numbers in parentheses are the positions of amino acid residues included in each construct. Red boxes indicate the position of the conserved domain.
(B) In vitro binding of LAX1 and LAX2. In vitro–synthesized HA-tagged T antigen and HA-tagged LAX1 along with LAX2 (1 to 394) and p53 proteins in the presence of 35S-Met were used as bait and prey, respectively. After incubation of bait and prey proteins, bait proteins were immunoprecipitated using anti-HA antibody. LAX2 and p53 proteins were coprecipitated with HA-LAX1 and HA-T antigen, respectively.
We then examined the LAX2 domain that is responsible for the interaction with LAX1. A series of truncated LAX2 cDNAs was cloned into the bait constructs and assayed for interaction with LAX1 by observing growth on selective medium. Constructs lacking the region corresponding to the 270 amino acids at the N terminus or the 15 amino acids at the C terminus did not interact with LAX1, indicating that the interaction of LAX2 and LAX1 requires regions 199 to 269 and 380 to 394 of LAX2. This suggests that the C-terminal region of LAX2, which includes the conserved domain, is involved in the interaction. Because LAX1 also shows strong transcriptional activation in yeast when LAX1 cDNA is used in the bait construct (see Supplemental Figure 5 online), we could not switch LAX2 and LAX1 cDNA inserts in the bait and prey construct, respectively, to confirm the interaction. Instead, we confirmed the interaction by an in vitro binding assay (Figure 6B). HA-tagged LAX1 protein and LAX2 were synthesized in vitro in the presence of 35S-labeled Met. These proteins were incubated and immunoprecipitated using anti-HA antibody. LAX2 coprecipitated with HA-LAX1, while it did not coprecipitate with the HA-T antigen. This assay confirmed that LAX1 and LAX2 interact in vitro.
DISCUSSION
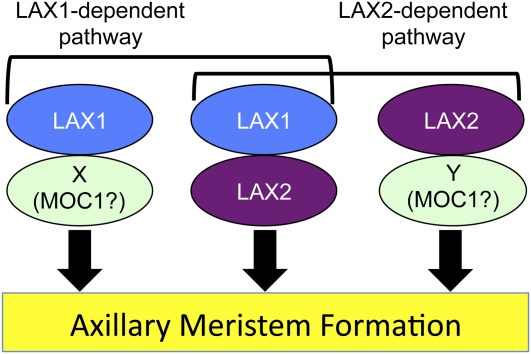
LAX2 functions in the aboveground branching of the rice plant throughout development. The phenotype of lax2 mutants is similar to that of lax1 mutants during both the vegetative and reproductive stages. The synergistic genetic interaction of lax2 with lax1 and moc1 in branching (as illustrated by the lax1 lax2 and lax2 moc1 double mutant phenotypes) indicates that there are multiple pathways in which LAX1, LAX2, and MOC1 function for branching (Figure 7). LAX2 encodes a nuclear protein with a plant-specific conserved domain, and it physically interacts with LAX1 through a region containing this domain. So far, many factors that regulate AM formation have been identified, but the biochemical relation between these factors is not well characterized. Thus, we propose that LAX2 is a novel factor that acts synergistically with LAX1 in rice to regulate AM formation.
Figure 7.
A Model for the Functions of LAX1, LAX2, and MOC1.
LAX2 is a novel nuclear factor that is involved in branching events throughout rice development. There are multiple pathways for AM formation where LAX1 and LAX2 are involved. LAX2 acts in two pathways, LAX1 dependent and LAX1 independent. In the LAX1-dependent pathway, LAX1 and LAX2 form a dimer. Both LAX1 and LAX2 can also function independently. Factors X and Y could be redundant factors of LAX2 and LAX1, respectively, and they could be partners of LAX1 and LAX2, respectively. Alternatively, it is also possible that factor X and factor Y could correspond to MOC1 because the phenotype of lax1 moc1 and lax2 moc1 double mutants are similar. In this case, MOC1 may or may not form dimers with LAX1 or LAX2. Once the activity of two proteins from the group consisting of LAX1, LAX2, and MOC1 is removed by mutations in the corresponding genes, the formation of the AM is almost completely suppressed at all developmental stages, except PB formation. Thus, there is expected be another layer of regulation for the formation of PB.
LAX2 Plays a Role in the Maintenance of AM
lax2 mutants have a sparse panicle that results in a reduced number of SBs and spikelets. lax2 also affects vegetative development in that lax2 mutants have fewer tillers. This is consistent with the double mutant phenotype of lax2 combined with either lax1 or moc1, in which a monoculm phenotype is observed. The reduction or absence of branching in lax2 plants could be due to either loss or arrest of the AM. Morphological, histological, and scanning electron microscopy observations did not provide evidence of arrested axillary shoots that give rise to tillers, SBs, or spikelets in lax2 single or double mutants. Interestingly, immunohistochemical analysis of the lax2 mutant inflorescence shows that AMs that give rise to SBs or spikelets are marked by anti-OSH1 antibody on PBs, similar to the wild type. Furthermore, in the lax2 moc1 double mutant, a population of OSH1-positive cells at the flank of the SAM is observed by both histological and immunohistochemical analysis. Thus, we propose that LAX2 is involved in the maintenance of the AM.
lax1 mutants are reported to be defective in the maintenance of the AM during the vegetative and reproductive stages (Komatsu et al., 2003; Oikawa and Kyozuka, 2009). During the vegetative stage, the phenotype of the lax1 and lax2 single mutants is relatively weak, as tillers are occasionally present. However, in the lax1 lax2 double mutant, no tillers form during the vegetative stage. In the reproductive stage, either the lax1 or lax2 mutation partially affects the formation of SBs and spikelets on PBs. However, in the double mutant, the formation of SBs and spikelets on PBs is completely suppressed. The similar phenotypes of single lax1 and lax2 mutants and the strong synergistic genetic interaction between the lax1 and lax2 mutation in the double mutant suggest that both LAX1 and LAX2 are involved in the maintenance of the AM throughout rice development.
By contrast, the formation of PBs in the inflorescence does not depend on LAX1 or LAX2, although both LAX1 and LAX2 are expressed at the site of PB formation (Figure 5C) (Komatsu et al., 2003). It is possible that there is a mechanism that forms AMs destined to become PBs in the inflorescence that is independent of LAX genes. Alternatively, PB formation might not require the full activity of the LAX genes, which promote the maintenance of lateral shoots because immediately after their formation, PBs start to elongate as independent main shoots.
Genetic Pathways That Regulate the Maintenance of AMs in Rice
The striking similarity of lax1 and lax2 mutants suggests an epistatic relationship between lax1 and lax2. However, the phenotype of the double mutant is synergistically enhanced compared with the single mutant, suggesting that LAX1 and LAX2 act in independent pathways in AM formation rather than a direct or indirect regulation of one gene by the other. In support of this, LAX1 or LAX2 expression was not abolished in the lax2 or lax1 mutant, respectively (see Supplemental Figures 3 and 4 online). Thus, we propose that there are multiple pathways that regulate the formation of the AM through the action of LAX1 and/or LAX2 (Figure 7).
MOC1 is another factor that regulates the formation of AM in rice (Li et al., 2003). In the moc1 mutant, the number of branches is reduced during both the vegetative and reproductive stages. The combination of the moc1 and lax1 mutations results in the complete loss of branching (Komatsu et al., 2003). This phenotype is similar to that of moc1 lax2 double mutants. We propose that there are multiple pathways for the formation of the AM, in which three factors (MOC1, LAX1, and LAX2) have overlapping functions (Figure 7). Once two of the three factors are removed by mutation of the corresponding genes, the process of AM formation is strongly suppressed at all developmental stages except PB formation. In the formation of PBs, MOC1 seems to have a more critical role than LAX1 or LAX2. It is interesting that the number of PBs is not reduced in lax1 and lax2 single mutants and in their double mutants. However, both LAX1 and LAX2 are also involved in the formation of PBs because lax1 moc1 or lax2 moc1 double mutants completely lose the PB. Thus, for the formation of PBs, there should be an additional layer of the regulation as discussed before.
LAX2 Encodes a Novel Nuclear Protein That Acts Together with LAX1 in Branching
The molecular cloning of lax2 revealed that it encodes a nuclear protein with a plant-specific conserved domain. Proteins containing this domain either have a C3HC4-type RING zinc finger motif or have stretches of the same amino acids, such as Arg, Ala, Glu, His, Pro, Ser, and Thr (see Supplemental Figure 2B online). The C3HC4-type RING zinc finger is often found in the single-subunit E3 ubiquitin ligase family (Kuroda et al., 2002; Stone et al., 2005). In Arabidopsis, there are ~400 RING domain–containing proteins, which may function as single-subunit E3 ligases (Stone et al., 2005), and some of these proteins are known to target transcription factors (Hardtke et al., 2000; Osterlund et al., 2000; Xie et al., 2002; Seo et al., 2003; Yang et al., 2005; Zhang et al., 2005; Dong et al., 2006). Among the proteins that share a conserved domain with LAX2, the function of two genes, DRIP1 and DRIP2, which encode the C3HC4 RING zinc finger, has been reported in Arabidopsis (Qin et al., 2008). These two genes target DREB2A, the key transcriptional factor for the response to water stress, to mediate DREB2A degradation by a mechanism that tightly modulates its protein level and thereby regulates gene expression in response to water stress.
LAX2 does not have a C3HC4-type RING zinc finger; thus, it is unlikely that LAX2 has E3 ligase activity. It has been shown that DRIP1 and DRIP2 interact with DREB2A through a region that includes a LAX2-conserved domain (Qin et al., 2008). We have also shown that LAX2 localizes to the nucleus and interacts with LAX1 through a region that includes this domain. Thus, it is possible that this domain is responsible for protein–protein interactions. The in situ mRNA accumulation of LAX2 partially overlaps with that of LAX1, although the mRNA expression domain of LAX2 is broader than that of LAX1 (Komatsu et al., 2003). It is noteworthy that LAX1 moves directionally to the newly formed AM primordium where LAX2 mRNA resides. Thus, it is possible that LAX1 and LAX2 protein localizations largely overlap. These observations suggest that LAX1 and LAX2 form a dimer and that the dimer is responsible for AM formation, at least in part.
Because LAX2 does not have a typical DNA binding domain, it is unlikely that LAX2 acts as a sequence-specific DNA binding protein. It is also unlikely that LAX2 regulates LAX1 expression. Rather, we propose that LAX2 acts as a cofactor of LAX1. LAX1 is a bHLH-type transcription factor, and its function is regulated at both the transcriptional and posttranscriptional levels (Komatsu et al., 2003; Oikawa and Kyozuka, 2009). Furthermore, BA1, a LAX1 ortholog in maize, is a substrate of the PINOID-like kinase BIF2 in vitro (Skirpan et al., 2008), suggesting that BA1 is regulated by BIF2 at the posttranscriptional level. Although LAX1 and LAX2 have a unique role in the maintenance of the AM, it is possible that LAX2 modulates the activity of LAX1 at the posttranscriptional level.
METHODS
Plant Material and Growing Conditions
The rice single-gene recessive mutants lax2-1, lax2-2, lax2-3, lax1-1, lax1-6, and moc1-4 and wild-type rice (all Oryza sativa subsp japonica) were used in this study. lax2-1 was obtained from an ethyl methanesulfonate–mutagenized population of Zhonghua 11. lax2-1 was provided by Qian Qian of the China National Rice Research Institute. lax2-2 and moc1-4 were screened from a population generated by tissue culture of Nipponbare followed by regeneration. lax2-3 and lax1-6 were generated by chronic γ-ray irradiation of Norin 8 and Nihonmasari, respectively, with a dose rate of 0.4 Gy/d from transplanting until the mature stage. Plants were grown in a paddy field or in pots in a greenhouse under standard growth conditions. Zhonghua 11, Nipponbare, or Norin 8 was used as the wild type for the observation of phenotypes.
Immunohistochemical Staining
Vegetative and reproductive shoots were fixed in FAA (1:1:18 [formaldehyde to glacial acetic acid to 70% ethanol]) for 24 h at 4°C and then dehydrated in a graded ethanol series. Dehydrated samples were embedded in Paraplast Plus (Oxford Labware), sectioned into 5- to 8-mm-thick sections with a rotary microtome, and used for immunostaining. Immunostaining was conducted based on the method described by Donlin et al. (1995), except that anti-OSH1 antibody and anti-rabbit IgG linked with alkaline phosphatase (ZYMED Laboratories) were used at 1:150 and 1:600 dilutions, respectively.
Scanning Electron Microscopy Analysis
For scanning electron microscopy, seedlings were harvested and young leaves were excised to expose P5 leaf axils. These samples were observed directly under an N-SEM system (S-3000N; Hitachi).
Map-Based Cloning of lax2
Map-based cloning was conducted using a mapping population made by crosses between the lax2 mutant and an indica cultivar; the lax2 mutation was mapped on chromosome 4. Using more than 1000 mutant individuals from the mapping population, we screened recombinants between two genetic markers that reside 68 kb apart on the physical map and identified four recombinants between these markers. This region corresponds to BAC clones OSJNBa0039F02 and OSJNBb0039C07. Mutations in lax2 were determined by PCR amplification and sequencing of genes predicted within these regions. We confirmed that LAX2 cDNA driven by the LAX2 promoter complements the mutant phenotype. LAX2 cDNA was amplified with primers LAX2F1 (5′-GCGTCGACGAGGGTGAGAGG-3′) and LAX2R2 (5′-TACCGGCAAAAGTTGTTAGAAG-3′), cloned into the pCRII Blunt TOPO vector (Invitrogen), and verified by sequencing. This plasmid was designated as pLAX2full. The promoter region of LAX2 was amplified using the primers LPM1-pm-3108+Asc-F (5′-AGGCGCGCCTATGGCACAGGGTGGTTAGTG-3′) and the 3′ primer LPM1-pm+531-R (5′-CAGCATGTGCGTGGGTATC-3′). The fragment was cloned into the pCRII Blunt TOPO vector and verified by sequencing. A BamHI-EcoRI fragment of this vector was cloned into pBluescript SK+ (Agilent Technologies). The resulting vector was digested with BamHI-XbaI and ligated with a BamHI-SpeI fragment of pLAX2full. The resulting vector and the pZH2Bik (Kuroda et al., 2010) were digested with AscI-SacI and ligated to produce vector pL2C. The pZH2Bik and pL2C vectors were introduced into scutellum-derived calli of lax2 by Agrobacterium tumefaciens–mediated transformation under selection with hygromycin at 50 mg/L (Toki, 1997). Transformants were used to analyze the panicle phenotype.
Phylogenetic Analysis
The alignment used to construct the phylogenetic tree was made using ClustalX (Jeanmougin et al., 1998), and the sequences were manually adjusted to optimize the alignment as shown in Supplemental Figure 2A online. The parameters used in the construction of the alignment are as follows. For the pairwise alignment parameters, we used a GAP extension penalty of 0.75 and a GAP opening penalty of 35.00. For the multiple alignment parameters, we used a GAP extension penalty of 0.3 and a GAP opening penalty 15.00. We also used the BLOSUM 30 protein weight matrix. The phylogenetic tree was constructed by the NJ method (Saitou and Nei, 1987) using PAUP* 4.0 software. Statistical support for nodes is shown as bootstrap values obtained by 1000 trials.
Subcellular Localization of LAX2
The pEGFP plasmid (Clontech) was digested with NcoI and XbaI, and pCAMBIA1305 was partially digested with NcoI and NheI. The EGFP fragment was then cloned into pCAMBIA1305 to produce the pCAMBIA1305-EGFP vector. To examine the subcellular localization of LAX2, the LAX2 coding sequence driven by the 35S promoter of Cauliflower mosaic virus was cloned into the pCAMBIA1305-EGFP vector in which LAX2 was fused in-frame to EGFP. The plasmid was introduced into wild-type callus, using the method described previously. The roots of transgenic plants were examined using an Olympus BX51 fluorescence microscope.
In Situ Hybridization
In situ hybridization was performed as previously described by Kouchi and Hata (1993). For the LAX2 probe, the full-length cDNA clone was used as a template for in vitro transcription. Hybridizations were conducted at 53°C overnight; slides were then washed four times at 50°C for 10 min each. An excess amount of sense transcript was used as the negative control. For two-color in situ hybridization, a DIG-labeled full-length LAX1 antisense RNA probe and a fluorescein-labeled LAX2 antisense probe were made and used for hybridization. Conditions of hybridization and the procedure for the two-color in situ hybridization were the same as those of the single-color method except that after the development of the first color using Fast Red TR (Sigma-Aldrich), slides were heat-treated at 70°C for 8 h to inactivate the enzyme and then used to detect the second color using the standard NBT/BCIP method.
Yeast Two-Hybrid Assay
We used the Matchmaker yeast two-hybrid system (Takara). The selection of transformants with bait and prey plasmids was performed on SD plates lacking Trp and Leu. Interactions were monitored by growth on SD plates lacking His, Trp, and Leu.
The bait and prey plasmids used in this experiment were constructed as follows. Regions of LAX1 and LAX2 cDNA spanning the entire open reading frame and a deletion series of the LAX2 cDNA were amplified using the primers listed in Supplemental Table 2 online and cloned into pGBKT7 with EcoRI and XhoI, and into pGADT7 with EcoRI and SalI. All of the PCR-derived fragments were sequenced after cloning, and an absence of errors in PCR amplifications was confirmed.
In Vitro Binding Assay
HA-LAX1, LAX2, HA-T antigen, and p53 proteins for coimmunoprecipitations were expressed from T7 promoters in the TnT in vitro transcription/translation system (Promega) in the presence of 35S-Met. After incubation of bait and prey proteins, bait proteins were immunoprecipitated using the anti-HA antibody derived from the 3F10 clone (Roche Diagnostics). The binding buffer was as follows: 25 mM Tris-Cl, pH 7.5, 100 mM NaCl, 10% glycerol, 0.2% Nonidet P-40, 5 mg/mL BSA, 2 mM EDTA, and 1× complete protease inhibitor cocktail [Roche Diagnostics]). After washing six times with binding buffer, samples were boiled in the presence of SDS-PAGE sample buffer and loaded onto SDS-PAGE gels. After electrophoresis, gels were fixed, dried, and developed with contacting imaging plates for BAS2000 (Fuji Film).
Detection of LAX2 Transcripts in lax1-6, lax2-2, moc1-4, and lax2-2 moc1-4 Mutants
Total RNA was extracted using TRIzol reagent (Invitrogen) from 3-week-old seedlings of the wild type and mutants. RT-PCR was performed using Omniscript (Qiagen). Primers used to amplify LAX2, MOC1, Os ACT1, and Os TB1 are shown in Supplemental Table 2 online. We performed RT-PCR at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, for 30 (LAX2 and Os TB1), 32 (MOC1), and 26 (Os ACT1) cycles, respectively. PCR products were run on agarose gels, stained with ethidium bromide, and visualized on a UV transilluminater. Data were obtained using RNA extracted from more than five biologically independent seedlings for the wild type and all mutants.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database under accession numbers AB669025 (LAX2), AB115668 (LAX1), AB088343 (OsTB1), AY242058 (MOC1), and AK058421 (OsACT1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Complementation of lax2 Mutants by LAX2 cDNA Driven by Its Own Promoter.
Supplemental Figure 2. Multiple Alignments of Amino Acid Sequences of Conserved Domains Shared with LAX2 from Rice and Arabidopsis.
Supplemental Figure 3. In Situ Expression of LAX1 in Mutants and the Wild Type.
Supplemental Figure 4. RT-PCR Analysis of Genes Expressed at AM in Mutants and the Wild Type.
Supplemental Figure 5. LAX1 and LAX2 Have Transactivation Activity in Yeast.
Supplemental Table 1. Quantification Data of the Complementation of the lax2 Mutant.
Supplemental Table 2. Primers Used in This Experiment.
Supplemental Data Set 1. Alignment Used to Generate the Phylogeny Presented in Figure 4B.
Acknowledgments
We thank Tomoko Atsumi for assistance in plant maintenance. This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science and Culture in Japan (Grant 21027017), Precursory Research for Embryonic Science and Technology program from the Japan Science and Technology Agency to Y.S., by grants from the Ministry of Agriculture, Forestry and Fisheries of Japan (Green Technology Project MP-2129) to Y.S. and H.K., and by the International Cooperation Program of the National Natural Science Foundation of China (Grant 30710103903) to Q.Q. and F.C.
AUTHOR CONTRIBUTIONS
H.T., Y.Z., S.H., M.O., S.S.-S., T.O., H.X., X.F., H.Y., F.C., J.K., and Y.S. designed and conducted the experiments. Q.Q., M.N., and H.K. generated the plant materials. Y.S., H.Y., and F.C. wrote the article.
References
- Donlin M.J., Lisch D., Freeling M. (1995). Tissue-specific accumulation of MURB, a protein encoded by MuDR, the autonomous regulator of the Mutator transposable element family. Plant Cell 7: 1989–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.H., Agarwal M., Zhang Y., Xie Q., Zhu J.K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A., Zhao Q., Kyozuka J., Meeley R.B., Ritter M.K., Doebley J.F., Pè M.E., Schmidt R.J. (2004). The role of barren stalk1 in the architecture of maize. Nature 432: 630–635 [DOI] [PubMed] [Google Scholar]
- Greb T., Clarenz O., Schäfer E., Müller D., Herrero R., Schmitz G., Theres K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17: 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S., Gohda K., Osterlund M.T., Oyama T., Okada K., Deng X.W. (2000). HY5 stability and activity in arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19: 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara K.-i., Karim M.R., Takada S., Taoka K.-i., Furutani M., Aida M., Tasaka M. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18: 2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshikawa K. (1989). The Growing Rice Plant. (Tokyo: Nosan Gyoson Bunka Kyokai). [Google Scholar]
- Jeanmougin F., Thompson J.D., Gouy M., Higgins D.G., Gibson T.J. (1998). Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23: 403–405 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Maekawa M., Shimamoto K., Kyozuka J. (2001). The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev. Biol. 231: 364–373 [DOI] [PubMed] [Google Scholar]
- Komatsu K., Maekawa M., Ujiie S., Satake Y., Furutani I., Okamoto H., Shimamoto K., Kyozuka J. (2003). LAX and SPA: Major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA 100: 11765–11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H., Hata S. (1993). Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 238: 106–119 [DOI] [PubMed] [Google Scholar]
- Kuroda H., Takahashi N., Shimada H., Seki M., Shinozaki K., Matsui M. (2002). Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol. 43: 1073–1085 [DOI] [PubMed] [Google Scholar]
- Kuroda M., Kimizu M., Mikami C. (2010). A simple set of plasmids for the production of transgenic plants. Biosci. Biotechnol. Biochem. 74: 2348–2351 [DOI] [PubMed] [Google Scholar]
- Li S., Qian Q., Fu Z., Zeng D., Meng X., Kyozuka J., Maekawa M., Zhu X., Zhang J., Li J., Wang Y. (2009). Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 58: 592–605 [DOI] [PubMed] [Google Scholar]
- Li X., et al. (2003). Control of tillering in rice. Nature 422: 618–621 [DOI] [PubMed] [Google Scholar]
- McSteen P. (2009). Hormonal regulation of branching in grasses. Plant Physiol. 149: 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P., Hake S. (2001). barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 128: 2881–2891 [DOI] [PubMed] [Google Scholar]
- McSteen P., Leyser O. (2005). Shoot branching. Annu. Rev. Plant Biol. 56: 353–374 [DOI] [PubMed] [Google Scholar]
- McSteen P., Malcomber S., Skirpan A., Lunde C., Wu X., Kellogg E., Hake S. (2007). barren inflorescence2 Encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol. 144: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Müller D., Schmitz G., Theres K. (2006). Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18: 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T., Kyozuka J. (2009). Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 21: 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Otsuga D., DeGuzman B., Prigge M.J., Drews G.N., Clark S.E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25: 223–236 [DOI] [PubMed] [Google Scholar]
- Qin F., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S., Greb T., Peaucelle A., Blein T., Laufs P., Theres K. (2008). Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 55: 65–76 [DOI] [PubMed] [Google Scholar]
- Ritter M.K., Padilla C.M., Schmidt R.J. (2002). The maize mutant barren stalk1 is defective in axillary meristem development. Am. J. Bot. 89: 203–210 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Schmitz G., Tillmann E., Carriero F., Fiore C., Cellini F., Theres K. (2002). The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc. Natl. Acad. Sci. USA 99: 1064–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K., Schmitt T., Rossberg M., Schmitz G., Theres K. (1999). The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA 96: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Yang J.Y., Ishikawa M., Bolle C., Ballesteros M.L., Chua N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Skirpan A., Wu X., McSteen P. (2008). Genetic and physical interaction suggest that BARREN STALK 1 is a target of BARREN INFLORESCENCE2 in maize inflorescence development. Plant J. 55: 787–797 [DOI] [PubMed] [Google Scholar]
- Steeves T.A., Sussex I.M. (1989). Patterns in Plant Development, 2nd ed. (Cambridge, UK: Cambridge University Press). [Google Scholar]
- Stone S.L., Hauksdóttir H., Troy A., Herschleb J., Kraft E., Callis J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P.B., Adler H.T., Parks D.W., Comai L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121: 2723–2735 [DOI] [PubMed] [Google Scholar]
- Toki S. (1997). Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol. Biol. Rep. 15: 16–21 [Google Scholar]
- Xie Q., Guo H.S., Dallman G., Fang S., Weissman A.M., Chua N.H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170 [DOI] [PubMed] [Google Scholar]
- Yang J., Lin R., Sullivan J., Hoecker U., Liu B., Xu L., Deng X.W., Wang H. (2005). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Garreton V., Chua N.H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19: 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K., Tang D., Yan C., Chi Z., Yu H., Chen J., Liang J., Gu M., Cheng Z. (2010). Erect panicle2 encodes a novel protein that regulates panicle erectness in indica rice. Genetics 184: 343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]