This work examines the changes that have occurred in 12 homoeologous genomic regions from three rounds of polyploidy that contributed to the current soybean genome. It finds that genes in networks with a high level of connectivity are more strongly conserved than those with low connectivity.
Abstract
With the advent of high-throughput sequencing, the availability of genomic sequence for comparative genomics is increasing exponentially. Numerous completed plant genome sequences enable characterization of patterns of the retention and evolution of genes within gene families due to multiple polyploidy events, gene loss and fractionation, and differential evolutionary pressures over time and across different gene families. In this report, we trace the changes that have occurred in 12 surviving homoeologous genomic regions from three rounds of polyploidy that contributed to the current Glycine max genome: a genome triplication before the origin of the rosids (~130 to 240 million years ago), a genome duplication early in the legumes (~58 million years ago), and a duplication in the Glycine lineage (~13 million years ago). Patterns of gene retention following the genome triplication event generally support predictions of the Gene Balance Hypothesis. Finally, we find that genes in networks with a high level of connectivity are more strongly conserved than those with low connectivity and that the enrichment of these highly connected genes in the 12 highly conserved homoeologous segments may in part explain their retention over more than 100 million years and repeated polyploidy events.
INTRODUCTION
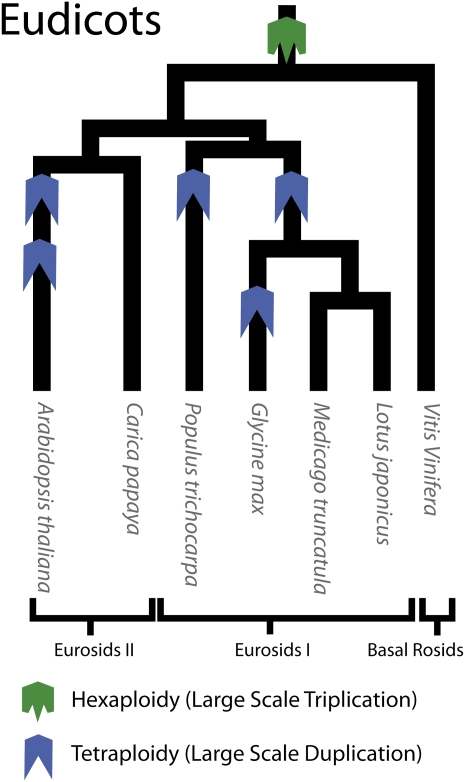
Literature on genome comparisons of homoeologous regions among eudicot species has focused primarily on polyploidy events (or whole-genome duplications [WGDs]) that have occurred in the last ~80 to 100 million years (Van de Peer et al., 2009). A recent report in soybean (Glycine max) describes a 1-Mb region, its homoeologous region generated from the ~13 million year WGD, and an orthologous region from Phaseolus vulgaris (Lin et al., 2010). A study in Arabidopsis thaliana describes four homoeologous segments deriving from two WGD (the alpha and beta events), estimated to have occurred in the last ~100 million years (Ziolkowski et al., 2003). The Arabidopsis study also describes some short, highly fragmented segments from an even earlier (Gamma) whole-genome event. It was in Arabidopsis that the presence of this earlier polyploidy event was first reported (Vision et al., 2000). This event was originally believed to be a tetraploidy or WGD event.
Analysis of the grape (Vitis vinifera) genome (Jaillon et al., 2007) revealed that the Gamma polyploidy event was a hexaploidy event or whole-genome triplication (WGT), which occurred in the common ancestor of grape, poplar (Populus spp), and Arabidopsis (Figure 1). This analysis also indicated that the Gamma WGT was not shared with rice (Oryza sativa) and therefore occurred sometime after the divergence of monocotyledonous and dicotyledonous plants, between ~130 and ~240 million years ago (Jaillon et al., 2007). Evidence of triplication in the papaya (Carica papaya) genome further supported the timing and triplicate nature of this event (Ming et al., 2008). A comparative genomics study of the rosids using the program CoGe (short for comparative genomics) and a study in Coffea both place the Gamma WGT before the asterid/rosid split in eudicots (Lyons et al., 2008; Soltis et al., 2009; Cenci et al., 2010). Possible paleogenomic models of extinct ancestors have also recently been published for angiosperms (Abrouk et al., 2010). When the whole-genome sequence from papaya, Arabidopsis, soybean, poplar, and grape are included in such a model, it suggests a shared ancestor containing seven chromosomes. Furthermore, this model supports a WGT rather than a WGD early in the evolution of these species. When this evidence of a Gamma WGT event is combined with the WGDs that occurred ~13 (Glycine) and 59 (legume) million years ago (Mya) in the evolutionary history of soybean (Schlueter et al., 2007; Schmutz et al., 2010), it is clear that soybean could contain as many as 12 copies of its ancestral genome with three sets of four homoeologous regions in soybean corresponding to a single ancestral region present before the Gamma event. We define these sets as being part of a Gamma hexaploidy lineage.
Figure 1.
Phylogenetic Tree of the Eudicots.
Depiction of the phylogenetic relationship between several eudicot species. Branch length and timing of polyploidies are not drawn to scale. Phylogenetic tree was approximated from phytozome.net and Van de Peer et al. (2009).
Previous comparative genomic studies of homoeologous segments have described patterns of nonrandom gene loss and retention. The gene balance hypothesis (GBH) for gene retention has been successful at describing gene loss and retention in many eukaryotic genomes (Papp et al., 2003; Maere et al., 2005; Aury et al., 2006; Blomme et al., 2006; Dopman and Hartl, 2007; Freeling, 2009). The GBH provides predictions for gene retention following different patterns of genome duplication (whole-genome, tandem, or segmental duplication). The premise behind the GBH is as follows. The biochemistry in a cell comprises networks of interactions, signaling cascades, and protein complexes. The hypothesis is that the more connected a gene product is within the cellular biochemistry, the more important its stoichiometry will be to the fitness of the organism and, therefore, the greater the impact a change in its dosage (number of copies of the gene) will have on the organism. Genes that encode for transcription factors or components of ribosomes and proteasomes are examples of highly connected genes. Examples of genes that work alone or are poorly connected include those involved in DNA repair, peptidases, nucleases, and small molecule biosynthesis (Freeling, 2009). After WGDs, the highly connected genes are in balance with their respective interactions, cascades, and complexes and are subject to purifying selection, whereas poorly connected genes may have redundant function and are subject to random loss. In a tandem duplication or isolated segmental duplication, highly connected genes will create a stoichiometric imbalance resulting in reduced fitness and are therefore likely to be selected against. This is not the case for genes that act alone or are poorly connected. In this report, we describe a genomic region that has been retained in 12 copies and has survived ~130 to 240 million years of evolution and three polyploidy events. Gene retention found in the 12 homoeologous regions is shown to be consistent with the GBH. These regions serve as a model to help us improve our understanding of both general evolutionary patterns and genomic evolution in a particular set of regions in soybean and other eudicots.
RESULTS
Identification of Homoeologous Segments
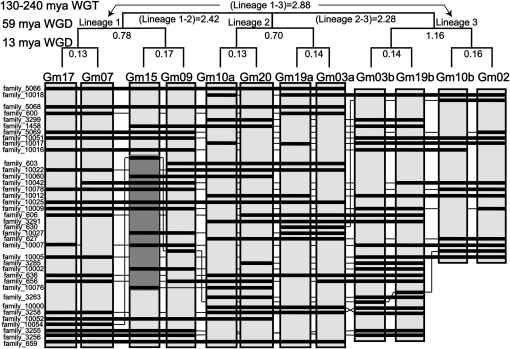
Twelve large genomic regions with extensive sequence similarity based on their relative gene content and gene order were identified using homology data processed with DAGchainer (Haas et al., 2004). These 12 homoeologous segments contain, in total, 753 gene models. Gene homologs identified using BLASTALL (Altschul et al., 1990) between the 12 homoeologous segments revealed 104 singleton genes and 649 genes with two or more homologs. These homologs fall into 174 gene families (see Supplemental Data Set 1A online), which span as many as nine of the 12 homoeologous segments. The 12 segments are found as single regions on chromosomes Gm02, Gm07, Gm09, Gm15, Gm17, and Gm20 and as two regions on each of chromosomes Gm3, Gm10, and Gm19. The families of homologous genes that span at least two lineages from the Gamma WGT show the colinearity of genes within the 12 homoeologous segments (Figure 2).
Figure 2.
Synteny and Phylogenetic Relationship between Homoeologous Segments.
The colinearity of gene families for 12 homoeologous segments that are the remnant of three polyploidy events. Phylogenetic relationships between syntenic blocks were determined from averaged median-block Ks values shown below each branch. Only gene families that contain genes in at least two of the three Gamma hexaploidy lineages are represented. A gray box on chromosome 15 represents an inversion event that occurred after the most recent duplication event and inverted for clarity.
Phylogenetic Origin of the Homoeologous Segments
Comparison of the shared gene complements and average rates of synonymous substitution (Ks) values between each region suggested that the 12 segments are remnants from the WGT and two WGD events, resulting in three Gamma hexaploidy lineages of four segments each (Figure 2). The range of Ks values between the homoeologous segments (0.13 to 0.17, 0.78 to 1.16, and 2.28 to 2.88) fall within the expected range of values previously reported for the Glycine WGD (~13 Mya, Ks ~0 to 0.3), the legume WGD (~58 Mya, Ks ~0.3 to 1.5), and the WGT events (~130 to 240 Mya, Ks > 1.5) (Schmutz et al., 2010). Organisms that experienced only the Gamma WGT event and not subject to an early release data usage policy were used to determine the phylogenetic origin of these segments. These include V. vinifera and C. papaya genomes that were reported to have experienced the Gamma WGT event, with no subsequent WGDs (Jaillon et al., 2007; Ming et al., 2008).
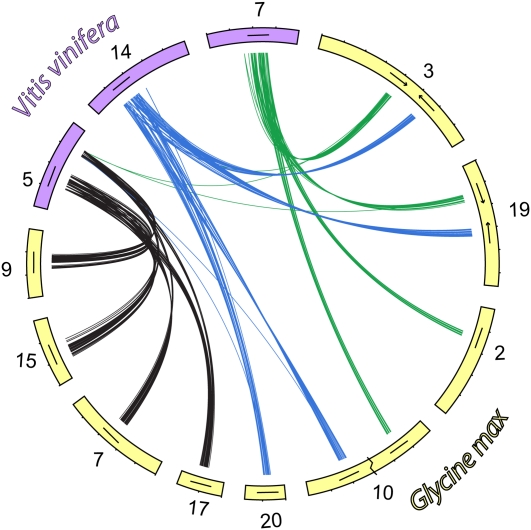
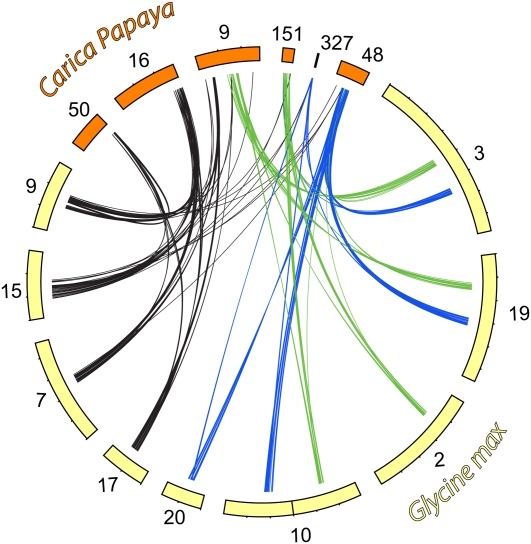
We reasoned, based on the shared evolutionary history between soybean, grape, and papaya, that if the genomic segment was equally preserved in grape and papaya, four homoeologous segments in soybean corresponding to the Gamma hexaploidy lineages should each map to a single distinct region in V. vinifera and C. papaya, respectively. Unidirectional top BLAST hits from G. max to V. vinifera or C. papaya, respectively, were consistent with this hypothesis. The majority of the top BLAST hits from soybean Gamma hexaploidy lineage 1 (Gm17, Gm07, Gm15, and Gm09) or soybean Gamma hexaploidy lineage 2 (Gm10a, Gm20, Gm19a, and Gm03a) or soybean Gamma hexaploidy lineage 3 (Gm3b, Gm19b, Gm10b, and Gm02) were to three different chromosomes in grape: Vv05, Vv14, and Vv07, respectively (Figure 3; see Supplemental Data Set 1B online). Similarly, top BLAST hits from soybean Gamma hexaploidy lineages to supercontigs in the draft sequence of papaya suggest a high level of conservation of these homoeologous regions to distinct regions in papaya (Figure 4; see Supplemental Data Set 1B online). The regions in papaya that correspond to the Gamma hexaploidy lineage 1 (black lines and supercontigs 50, 16, and 9) and Gamma hexaploidy lineage 3 (green lines and supercontigs 9 and 151) have had some rearrangement. However, the region in papaya that corresponds to Gamma hexaploidy lineage 2 (blue lines and supercontigs 327 and 48) appears to be largely intact. The region on supercontig 327 is either an assembly error or a small segmental rearrangement as a close inspection of Figure 4 reveals that the small gap on supercontig 48 corresponds perfectly to the region on supercontig 327. These results imply the origin of the 12 homoeologous segments in soybean arose from the three ancestral homoeologous segments from the Gamma WGT event and two WGD events rather than several independent large segmental duplications.
Figure 3.
Orthologous Segments between Soybean and Grape.
Circos plot of the segments conserved between soybean (yellow) and grape (purple). Curved lines represent top BLAST hits that mapped from soybean to grape. Segments of the genome that arose from the Gamma-hexaploid lineages are colored black, green, and blue. The black line within the chromosome represents 1 Mb, and directionality is indicated for regions on soybean chromosomes Gm03 and Gm19.
Figure 4.
Orthologous Segments between Soybean and Papaya.
Circos plot of the segments conserved between soybean (yellow) and papaya (orange). Curved lines represent top BLAST hits that mapped from soybean to papaya. Segments of the genome that arose from the Gamma-hexaploid lineages are colored black, green, and blue.
Comparison of Homoeologous Segments
Previous analyses of homoeologous segments in Arabidopsis and soybean found large variation in size between the segments (Ziolkowski et al., 2003; Lin et al., 2010). To obtain a better understanding of size variation in homoeologous segments in soybean, average sizes in base pairs and genes between and within each Gamma hexaploidy lineage were examined using our data set of homoeologous segments that have well-defined boundaries based on gene families (Figure 2). A gene family in this context is defined as a set of genes, with BLAST homology at 1e−10 or lower and contained within the 12 homoeologous segments. Gene family numbering is for reference within this article. Two gene families, 5066 and 659 (protein kinase), span eight of the 12 homoeologous segments and are found at the first and last positions for these segments. Families 10018 (DUF3511), 3255 (zinc-finger), and 10005 (epimerase) span at least six of the 12 homoeologous segments and serve as the first and last positions for the remainder of the segments. Using members of these families as boundaries, the length of the 12 homoeologous segments ranged from 252 to 1011 kb with a mean of 483 kb. The average lengths for each set of four segments that arose from the Gamma WGT event also varied markedly. Gamma hexaploidy lineages 1, 2, and 3 have average lengths of 697 ± 211 kb, 408 ± 59 kb, and 344 ± 90 kb and average number of genes of 341 ± 16, 214 ± 3, and 198 ± 11 genes, respectively. With a range of gene density between 5.8 kb per gene and 9.6 kb per gene, our data also suggest a variable rate of fractionation has occurred between homoeologous segments (see Supplemental Data Set 1C online).
In order to better understand why there might be large variation in sizes between the retained blocks from the Gamma hexaploidy lineages, deletions and tandem duplications were examined. There appears to have been a large deletion in two of the homoeologous segments in Gamma hexaploidy lineage 3 that likely occurred between the legume WGD and Glycine WGD and that explain the smaller than average size of this hexaploidy lineage (Figure 2). These two segments are found at the end of the chromosome and likely lost some of their size due to segmental reorganization. The two largest homoeologous segments are part of hexaploidy lineage 1 and are found on chromosomes Gm09 and Gm15. The larger size is in part due to an increase in the number of tandem duplications found in the homoeologous segment on chromosome Gm15. The number of tandem duplications was determined by the total number of genes found in tandem arrays. On chromosome Gm15, there were 26 tandem duplications, which is 2.8 standard deviations above the mean of eight tandem duplications for all 12 homoeologous segments. In the 12 homoeologous segments, a total of 90 TE were identified based on annotation from the SoyTE database (Du et al., 2010). The accumulation of TE in each segment ranged from two to 21 and counts of TE were similar for homoeologous segments that are related by the most recent Glycine WGD. TE density of the 12 homoeologous segments varies independently of segment size from 36K bases/TE to 264 bases/TE (see Supplemental Data Set 1D online). This suggests that sequence similarity and size of a genomic region are not the only factors that determine the density of TE in a genomic region and are not a large contributing factor in retained segment size for these 12 regions.
Putative ancestral gene complements were inferred in order to explore gene retention and loss following the three polyploidy events. There are six pairs of homoeologous segments that arose after the Glycine WGD (13 Mya). Comparisons of genes in these pairs show that on average 68.5% ± 3.3% of the genes were retained after the duplication. Three comparisons were made between the gene families contained within the six pairs of homoeologous segments (Gm17/Gm07-Gm15/Gm09, Gm10a/Gm20-Gm19a/Gm03a, and Gm03b/Gm19b-Gm10b/Gm02) that arose after the legume WGD (~58 Mya). This comparison reveals that 29.2% ± 1.7% of the genes were retained in duplicate (see Supplemental Data Set 1E online). These average values of gene retention for the homoeologous segments are similar to gene retention values of 71 and 24% determined on four different homoeologous regions in soybean described by Kim et al. (2009). Schmutz et al. (2010) reported a genome-wide estimate for gene retention after the Glycine and legume WGD events of 43.4 and 25.9%. No estimate for gene retention after the WGT in soybean has been reported. Examination of gene families contained within the three sets of four segments that arose after the Gamma WGT event (~130 to 240 Mya) indicate that 11.8% of the genes were retained in duplicate and 1.8% were retained in triplicate. These data suggest that the majority of the duplicated and triplicated genes in the legume WGD and Gamma WGT event have reverted to singleton status, were lost, or moved outside of these regions. However, for the Glycine WGD event, a little over two-thirds of the duplicated genes have been retained. This retention is more than twice the retention of the legume WGD and five times the retention of the Gamma WGT events.
GBH Explains Preferential Gene Loss and Retention
There are several patterns of gene retention within the 12 homoeologous regions. These patterns are specific to genes that were both retained after one or multiple polyploidy event and were not move out of the 12 homoeologous regions. In Figure 2, it is evident that the gene families that span more than one Gamma hexaploidy lineage were often retained in subsequent polyploidies to varying degrees. For example, the gene families 5066, 5068 (α-importin), 10025 (pectinesterase), and 659 (protein kinase) were retained after each WGD in two Gamma hexaploidy lineages, whereas the gene families 3263 (WD repeat containing), 10002 (transporter), and 1458 (Di19) were retained in two Gamma hexaploidy lineages and again during the Glycine WGD. The gene family 656 (copper transporter) shows variability in gene retention and loss as it was retained in triplicate after the Gamma WGT but to varying degrees in each hexaploidy lineage after the subsequent legume and Glycine WGD events.
To get a feel for the functional annotations of genes that were retained following the WGT event, the GOSlim Biological process categories (Berardini et al., 2004) were determined for the 38 gene families that span at least two Gamma hexaploidy lineages (Figure 2, Table 1). Most notable were the following GOSlim categories: signal transduction and response to biotic and abiotic stimulus, as these findings are consistent with genomic trends for retained genes after multiple polyploidy events in Arabidopsis (Blanc and Wolfe, 2004; Maere et al., 2005). Furthermore, the genes retained after the Gamma WGT are annotated primarily as transcription factors and proteins involved in signaling cascades or complexes, as predicted by the GBH (Birchler and Veitia, 2007; Veitia et al., 2008; Edger and Pires, 2009; Freeling, 2009).
Table 1.
GOSlim Categories Identified for the 16 Singleton Genes with No Homology to Genes outside the 12 Homologous Regions and 38 Gene Families Retained after the WGT, Respectively
| GOSlim Category | Singletons | WGT Retained |
| DNA or RNA metabolism | 1 | 0 |
| Signal transduction | 0 | 3 |
| Response to abiotic or biotic stimulus | 0 | 6 |
| Cell organization and biogenesis | 0 | 4 |
| Nucleus | 0 | 1 |
| Other biological processes | 0 | 8 |
| Unknown biological processes | 5 | 11 |
| Developmental processes | 1 | 7 |
| Other cellular processes | 1 | 19 |
| Other metabolic processes | 1 | 13 |
| Protein metabolism | 1 | 3 |
| Response to stress | 1 | 7 |
| Transport | 1 | 5 |
Genes that were resistant to duplication and resistant to movement to regions outside the 12 homoeologous segments were also investigated. Approximately 14% of all genes (105/753) within the 12 homoeologous segments reverted to a singleton within the homoeologous segments after every polyploidy event. Singletons represented 38% (105/279) of the gene families in regions in this study that were involved in the three polyploidy events. Half of the singleton genes (53/105) had no annotation. A closer inspection of those genes without annotation revealed that the mean size for the singleton genes (503 bp) is considerably smaller than the mean gene size for singleton genes with annotations (949 bp) or genes genome wide (1241 bp). Genes of this size are also common within the low confidence gene annotations and may represent nonfunctional gene remnants that were misidentified during the automatic gene annotation (Schmutz et al., 2010; Woody et al., 2011). To test this, expression data from the publicly available RNA-Seq atlases (Libault et al., 2010; Severin et al., 2010b) for the unannotated singletons were examined. One-fourth of these genes (14/53) had total expression counts from 28 tissues and developmental stages of less than one read per kilobase per million normalized count. Over half of these genes (29/53) had a sum of <10 read per kilobase per million normalized counts. This suggests that many of the genes with no known function have little to no expression. This may support the conclusion that the singleton genes with unknown function are nonfunctional remnants of genes, although this conclusion is not definitive without expression data that encompasses a broader sampling of tissues and environmental conditions. It is possible that the unannotated (and often short) genes may have functions that are highly expressed under very specific developmental stages or environmental conditions.
The remaining 52 singletons were then filtered to eliminate genes with homology to genes elsewhere in the genome. This resulted in 16 singletons that have functional annotation and are not homologous to genes outside this region. All but one of the GOSlim categories for these genes (Table 1) are also represented in the genes that were retained following the WGT event. However, the GOSlim category that varies between singleton genes and genes retained after the WGT event is “DNA or RNA metabolism,” which was previously found to be less likely to be retained after a polyploidy event (Blanc and Wolfe, 2004; Maere et al., 2005) and is consistent with the GBH (Birchler and Veitia, 2007; Veitia et al., 2008; Edger and Pires, 2009; Freeling, 2009).
GBH and Syntenic Block Retention
To determine if the presence of highly connected genes has an effect on the retention of chromosomal segments, a gene list was created comprising the known transcription factors, genes that encode for ribosomal proteins, and soybean genes with high BLAST similarity to Arabidopsis genes that are known or predicted to have at least five protein–protein interactions (Brandão et al., 2009). A bootstrap method was then employed to test if any subset of these genes were clustered on the soybean chromosomes (Severin et al., 2010a). Genes on a chromosome were considered clustered if the number of genes in a given interval was at least three standard deviations above the mean of 1000 simulations of the same number of randomly chosen genes on the chromosome. Interestingly, significant intervals of clustering for these genes were identified that overlap with six of the 12 homoeologous regions (see Supplemental Data Set 1F online).
DISCUSSION
Homoeologous Segments Originated from Polyploidy, Not Segmental Duplications
Comparative genomic analysis between and within related genomes revealed the evolutionary origin of homoeologous segments. Upon first inspection, it was unclear whether these segments arose from polyploidy events or from a combination of polyploidy events and segmental duplications because two pairs of the homoeologous segments (3a/19a and 3b/19b) are located within 1.3 million bases of each other in a reversed orientation (Figure 3). Our original hypothesis was that sometime between the legume and Glycine WGD events there was a segmental duplication, which would explain the close proximity of the two homoeologous segments and their presence on the Gm03 and Gm19 chromosomes.
The grape genome, having split early in the eudicot evolution, shortly after the shared triplication event, is ideal for discriminating these structural and phylogenetic relationships. Using unidirectional top BLAST hits (Berardini et al., 2004) from the soybean to the grape genome and the median-block Ks values, we determined that the two segments on chromosomes Gm03 and Gm19 are in fact homoeologous segments from two different lineages that arose from the Gamma WGT event (Figure 3, green and blue lines). Top BLAST hits from soybean to papaya also indicate these two regions arose from two different lineages (Figure 4, green and blue lines). This implies that sometime between the legume and Glycine WGD events, a chromosomal rearrangement occurred that brought these two homoeologous segments together within 1.3 million bases of each other and in a reversed orientation. The presence of two homoeologous segments on different arms of chromosome Gm10, and their most recent homoeologous segmental duplications on chromosomes Gm02 and Gm20, suggests a segmental rearrangement also occurred after the most recent WGD. These data show that genome shuffling of homoeologous segments has occurred after each major polyploidy event and is likely an ongoing process.
Why So Well Preserved?
If the homoeologous segments are well preserved due to a specific family of genes contained within the segments, then we might expect gene retention within the family to be localized to the homoeologous segments. To this end, the gene families with >50% of their genes localized to the 12 homoeologous segments in soybean were identified (Table 2). The resulting 22 families were enriched for housekeeping-like genes and in particular for ribosomal proteins and transcription factors that contained zinc-finger domains. Other gene families of possible interest include family 607 with an annotation relating to dormancy and family 602 with an annotation of exonuclease activity. These gene families have annotations with functions in line with the GBH and are orthologous to genes found in the corresponding regions of grape and papaya.
Table 2.
Analysis of Gene Families Localized to the 12 Homoeologous Segments
| Gene Family | 12 Regionsa | Genome-Wideb | Percentagec | Function | Present in Grape and Papayad |
| family_659 | 8 | 10 | 80.0% | Protein kinase | |
| family_5067 | 4 | 7 | 57.1% | DUF1218 | Yes |
| family_10018 | 7 | 12 | 58.3% | DUF3511 (flower specific) | |
| family_607 | 4 | 5 | 80.0% | Dormancy/auxin | Yes |
| family_1458 | 6 | 11 | 54.5% | Drought Induced Protein 19 | |
| family_602 | 4 | 4 | 100.0% | Exonuclease | Yes |
| family_5599 | 6 | 11 | 54.5% | Flowering promoter-like protein | |
| family_606 | 13 | 24 | 54.2% | Hydrolase (Alpha/Beta) | |
| family_10007 | 7 | 8 | 87.5% | Late embryogenesis abundant | |
| family_11004 | 4 | 4 | 100.0% | Unknown | |
| family_3258 | 8 | 13 | 61.5% | Unknown | |
| family_5066 | 8 | 11 | 72.7% | Unknown | |
| family_5106 | 4 | 6 | 66.7% | Unknown | Yes |
| family_5101 | 19 | 19 | 100.0% | Pathogenesis/BET-VI family | Yes |
| family_10014 | 6 | 10 | 60.0% | Ribosomal (60s/L19) | Yes |
| family_3291 | 6 | 7 | 85.7% | Ribosomal (L18\60s) | |
| family_627 | 6 | 10 | 60.0% | Ribosomal (L22\23s) | |
| family_5594 | 3 | 3 | 100.0% | Ribosomal (S21e\40s) | Yes |
| family_3299 | 4 | 4 | 100.0% | SNARE (Syntaxin) | |
| family_10033 | 4 | 5 | 80.0% | Zinc-finger (C2H2) | Yes |
| family_3255 | 6 | 6 | 100.0% | Zinc-finger (C3HC4) | |
| family_628 | 4 | 4 | 100.0% | Zinc-finger (CW) | Yes |
This column contains the number of genes in the gene family within the 12 homoeologous segments.
This column contains the number of genes in the gene family anywhere in the genome.
This column contains the percentage of genes within the 12 homoeologous segments.
This column indicates if the gene family was found in at least one orthologous region in grape or papaya.
The presence of multiple gene families in highly conserved regions that span such a wide taxonomic space (the Fabidae or eurosids I [soybean], the Malvidae or eurosids II [papaya], and an outgroup to the eurosids [grape]) led us to suspect that the conservation of these homoeologous segments may be due in part to the presence of the multiple families of highly connected genes, consistent with the GBH. This model predicts that following a polyploidy event, a gene is more likely to be retained if it has many connections in a network of genes, is part of a macromolecular structure, or is a transcription factor, since a deletion of such a gene would result in an imbalance leading to a reduced fitness. It follows that if a genomic region contains many highly connected genes, then the probability of a chromosomal rearrangement negatively affecting one of these highly connected genes resulting in reduced fitness is also higher. A list of highly connected genes was identified and a bootstrap clustering method determined that six of the 12 homoeologous regions contained at least one region with significant clustering. Six of the genes contained in family 10014 and three of the four genes contained in family 10033 fell within the significantly clustered intervals that were identified. These two families, encoding for a ribosomal protein and a zinc-finger, respectively, are also present in grape and papaya in the corresponding orthologous regions (Table 2). Further study will be required to determine if clustering of highly connected genes reduces the probability of a breakpoint occurring within a syntenic block during chromosomal rearrangement.
METHODS
Identification of Homoeologous Regions
Homoeologous regions were identified with DAGchainer (Haas et al., 2004), on homology data derived from protein–protein comparisons made with BLASTALL (Altschul et al., 1997), with an E-value cutoff of 1e−10 and BLAST output filtered to top reciprocal best BLAST hits per chromosome pair. DAGchainer settings were default, except for requiring a minimum of four aligned pairs (i.e., run_DAG_chainer.pl –A 4). Median-block Ks values per block were calculated for all gene pairs with Ks ≤ 2, using PAML (Zhang and Nei, 1997). The gene pairs and Ks values are in Supplemental Data Set 1G online. Boundaries for each homoeologous block were determined based on homologous genes. No single gene family was conserved in all 12 homoeologous segments. Therefore, five gene families define the boundaries for the 12 homoeologous segments. Family_5066 and family_659 serve as boundaries that span eight of the 12 homoeologous segments. Family_10018, family_3255, and family 10005 serve as boundaries for the remaining four homoeologous segments and span at least six of the 12 homoeologous segments (Figure 2).
Determination of Phylogenetic Relationships
Ks values between homoeologous segments represented in Figure 2 were determined by averaging median-block Ks values for all homologies found in Supplemental Data Set 1F online. BLASTp analysis using the BLASTALL program was performed between gene models found in the 12 homoeologous segments of soybean (Glycine max) and gene models from the grape (Vitis vinifera) genome (Jaillon et al., 2007) or papaya (Carica papaya) genome (Ming et al., 2008). The BLASTp analysis was unidirectional from soybean to grape or soybean to papaya, respectively, with an E-value cutoff of 1e−3. The E-value of 1e−3 was chosen in the analysis to identify as many potential orthologous genes as possible. An E-value of 1e−10 does not significantly reduce the number of genes identified in the orthologous regions of grape (177/195) and papaya (176/198). Top BLAST hits were analyzed and organized into groups based on Gamma hexapolidy lineages in soybean. Supplemental Data Set 1B online contains the top BLAST hits and hit counts between the soybean Gamma hexaploidy lineages and either the grape genome (phytozome.net ID 145) or the papaya draft genome (phytozome.net ID 113).
Gene Retention
Gene retention was determined based on the presence or absence of a gene family in a homoeologous segment. The homoeologous segments that are related by the Glycine WGD were compared first, and those families that contained genes in both segments were considered duplicated. Then, the homoeologous segments that are related by the legume WGD were compared, and those families that were contained in at least one homoeologous segment from each pair of segments generated by the Glycine WGD and related by the legume WGD were considered duplicated. Finally, the families in the four homoeologous segments in each of the three Gamma hexaploidy lineages were compared, and those families that were present in at least one of the four homoeologous segments in two of the three Gamma hexaploidy lineages were considered duplicated. Similarly, if a family was found in all three Gamma hexaploidy lineages, then it was considered triplicated (see Supplemental Data Set 1G online).
Gene Family Annotation
Annotation for gene families that includes GO, Pfam, KEGG, KOG, and Panther were taken from Soybase.org (Supplemental Data Set 1A online). GOSlim terms were determined by taking the longest cDNA for each soybean gene and blasting against Arabidopsis thaliana (TAIR10) cDNAs with an E-value cutoff of 1e−6. The Arabidopsis GOSlim categories were than associated with the best soybean blast hit.
Clustering of Highly Connected Genes
The list of genes chosen to represent some portion of the highly connected genes in soybean included the known transcription factors, genes that are components of ribosomes, and genes with high sequence similarity to Arabidopsis genes predicted to have at least five protein–protein interactions. The At-PIN database (Brandão et al., 2009) was downloaded and Arabidopsis genes with at least five or more known or predicted protein–protein interactions were retained. BLAST was performed from the cDNAs of soybean to the cDNAs of the highly connected genes taken from At-PIN. Only BLAST hits that had an E-value of zero were included in the list of highly connected genes for soybean. Clustering based on a bootstrap method was performed based on a variation of the method described for SNP clustering to identify introgressions between near-isogenic lines (Severin et al., 2010a).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Data Set 1A. Gene Families and Singletons in the 12 Homoeologous Segments along with the Predicted Annotation Obtained from Soybase.org.
Supplemental Data Set 1B. Top BLAST Hits Output between Soybean and Grape and Putative Orthologs.
Supplemental Data Set 1C. Analysis of Homoeologous Segments.
Supplemental Data Set 1D. Table of Transposable Elements Contained within the 12 Homoeologous Regions.
Supplemental Data Set 1E. Median-Block Ks Values Output from PAML and DAGchainer Used to Calculate Ks Values between Homoeologous Segments.
Supplemental Data Set 1F. Clustering of Highly Connected Genes.
Supplemental Data Set 1G. Gene Family Retention after the Paleo-Hexaploidy Event.
Acknowledgments
We thank Rex T. Nelson and Nathan Weeks for helpful discussions and IT support on this article. We acknowledge financial support from the USDA-Agricultural Research Service and National Science Foundation Grant 0822258-DBI.
AUTHOR CONTRIBUTIONS
A.J.S. and R.C.S. designed the research and wrote the article. A.J.S. and S.B.C. performed the research. A.J.S., M.M.G., and D.G. analyzed the data and contributed tools.
References
- Abrouk M., Murat F., Pont C., Messing J., Jackson S., Faraut T., Tannier E., Plomion C., Cooke R., Feuillet C., Salse J. (2010). Palaeogenomics of plants: Synteny-based modelling of extinct ancestors. Trends Plant Sci. 15: 479–487 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury J.-M., et al. (2006). Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444: 171–178 [DOI] [PubMed] [Google Scholar]
- Berardini T.Z., et al. (2004). Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 135: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J., Veitia R.A. (2007). The gene balance hypothesis: From classical genetics to modern genomics. Plant Cell 19: 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme T., Vandepoele K., De Bodt S., Simillion C., Maere S., Van de Peer Y. (2006). The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 7: R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão M.M., Dantas L.L., Silva-Filho M.C. (2009). AtPIN: Arabidopsis thaliana protein interaction network. BMC Bioinformatics 10: 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci A., Combes M.C., Lashermes P. (2010). Comparative sequence analyses indicate that Coffea (Asterids) and Vitis (Rosids) derive from the same paleo-hexaploid ancestral genome. Mol. Genet. Genomics 283: 493–501 [DOI] [PubMed] [Google Scholar]
- Dopman E.B., Hartl D.L. (2007). A portrait of copy-number polymorphism in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 104: 19920–19925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Grant D., Tian Z., Nelson R.T., Zhu L., Shoemaker R.C., Ma J. (2010). SoyTEdb: A comprehensive database of transposable elements in the soybean genome. BMC Genomics 11: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger P.P., Pires J.C. (2009). Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 17: 699–717 [DOI] [PubMed] [Google Scholar]
- Freeling M. (2009). Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60: 433–453 [DOI] [PubMed] [Google Scholar]
- Haas B.J., Delcher A.L., Wortman J.R., Salzberg S.L. (2004). DAGchainer: A tool for mining segmental genome duplications and synteny. Bioinformatics 20: 3643–3646 [DOI] [PubMed] [Google Scholar]
- Jaillon O., et al. ; French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Kim K.D., Shin J.H., Van K., Kim D.H., Lee S.-H. (2009). Dynamic rearrangements determine genome organization and useful traits in soybean. Plant Physiol. 151: 1066–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M., Farmer A., Joshi T., Takahashi K., Langley R.J., Franklin L.D., He J., Xu D., May G.D., Stacey G. (2010). An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 63: 86–99 [DOI] [PubMed] [Google Scholar]
- Lin J.-Y., Stupar R.M., Hans C., Hyten D.L., Jackson S.A. (2010). Structural and functional divergence of a 1-Mb duplicated region in the soybean (Glycine max) genome and comparison to an orthologous region from Phaseolus vulgaris. Plant Cell 22: 2545–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E., Pedersen B., Kane J., Alam M., Ming R., Tang H., Wang X., Bowers J., Paterson A., Lisch D., Freeling M. (2008). Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 148: 1772–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., De Bodt S., Raes J., Casneuf T., Van Montagu M., Kuiper M., Van de Peer Y. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 102: 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R., et al. (2008). The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452: 991–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B., Pál C., Hurst L.D. (2003). Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197 [DOI] [PubMed] [Google Scholar]
- Schlueter J.A., Lin J.-Y., Schlueter S.D., Vasylenko-Sanders I.F., Deshpande S., Yi J., O’Bleness M., Roe B.A., Nelson R.T., Scheffler B.E., Jackson S.A., Shoemaker R.C. (2007). Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genomics 8: 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Severin A.J., et al. (2010a). An integrative approach to genomic introgression mapping. Plant Physiol. 154: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin A.J., et al. (2010b). RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 10: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D.E., Albert V.A., Leebens-Mack J., Bell C.D., Paterson A.H., Zheng C., Sankoff D., dePamphillis C.W., Wall P.K., Soltis P.S. (2009). Polypoidy and angiosperm diversification. Am. J. Bot. 96: 336–348 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., Fawcett J.A., Proost S., Sterck L., Vandepoele K. (2009). The flowering world: A tale of duplications. Trends Plant Sci. 14: 680–688 [DOI] [PubMed] [Google Scholar]
- Veitia R.A., Bottani S., Birchler J.A. (2008). Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 24: 390–397 [DOI] [PubMed] [Google Scholar]
- Vision T.J., Brown D.G., Tanksley S.D. (2000). The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117 [DOI] [PubMed] [Google Scholar]
- Woody J.L., et al. (2011). Gene expression patterns are correlated with genomic and genic structure in soybean. Genome 54: 10–18 [DOI] [PubMed] [Google Scholar]
- Zhang J., Nei M. (1997). Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J. Mol. Evol. 44(Suppl. 1): S139–S146 [DOI] [PubMed] [Google Scholar]
- Ziolkowski P.A., Blanc G., Sadowski J. (2003). Structural divergence of chromosomal segments that arose from successive duplication events in the Arabidopsis genome. Nucleic Acids Res. 31: 1339–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]