This work presents the transcriptome profile of the meristemoid, a transient and low-density proliferating stomatal precursor cell with stem cell–like properties. The work identifies a new protein exhibiting polar localization during asymmetric division of stomatal cell lineages, reveals molecular characteristics of the meristemoid, and illuminates common themes in gene expression among plant stem cells.
Abstract
The balance between maintenance and differentiation of stem cells is a central question in developmental biology. Development of stomata in Arabidopsis thaliana begins with de novo asymmetric divisions producing meristemoids, proliferating precursor cells with stem cell–like properties. The transient and asynchronous nature of the meristemoid has made it difficult to study its molecular characteristics. Synthetic combination of stomatal differentiation mutants due to loss- or gain-of-function mutations in SPEECHLESS, MUTE, and SCREAM create seedlings with an epidermis overwhelmingly composed of pavement cells, meristemoids, or stomata, respectively. Through transcriptome analysis, we define and characterize the molecular signatures of meristemoids. The reporter localization studies of meristemoid-enriched proteins reveals pathways not previously associated with stomatal development. We identified a novel protein, POLAR, and demonstrate through time-lapse live imaging that it exhibits transient polar localization and segregates unevenly during meristemoid asymmetric divisions. The polar localization of POLAR requires BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE. Comparative bioinformatic analysis of the transcriptional profiles of a meristemoid with shoot and root apical meristems highlighted cytokinin signaling and the ERECTA family receptor-like kinases in the broad regulation of stem cell populations. Our work reveals molecular constituents of stomatal stem cells and illuminates a common theme among stem cell populations in plants.
INTRODUCTION
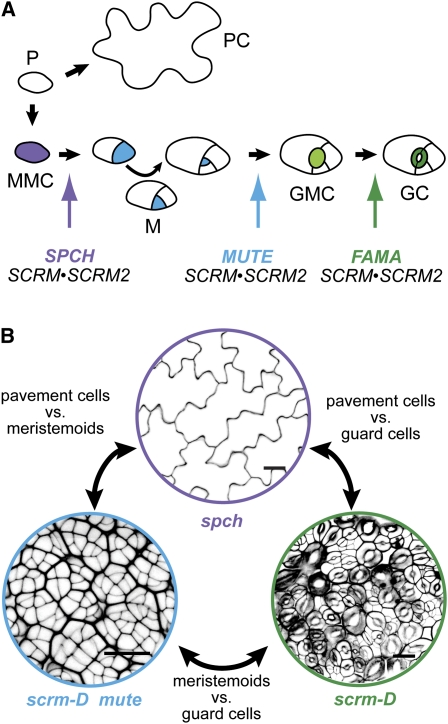
Plant stem cells continuously produce new organs to sustain indeterminate growth. To this end, these cell populations must ensure stem cell survival through self-renewal and produce daughter cells destined for specialization. The two major stem cell niches, the root and shoot apical meristems (RAMs and SAMs, respectively), have provided the majority of our current knowledge of plant stem cells (Clark et al., 1993; Di Laurenzio et al., 1996; Leibfried et al., 2005; Sarkar et al., 2007). However, other cell types outside the meristems, such as procambium cells and stomatal meristemoids, share distinct properties with meristematic stem cells and are likely to provide additional insight into cell self-renewal in plants (Fisher and Turner, 2007; Pillitteri et al., 2007a). Meristemoids represent a transitional state in the cell lineage producing stomata, valves on the plant epidermis (Figure 1A) (Sachs, 1991). Specifically, meristemoids have transient self-renewing capacity, and their differentiation can be blocked in the absence of a key regulator (Bergmann and Sack, 2007; Pillitteri and Torii, 2007).
Figure 1.
Stomatal Cell Lineage Transitions and Cell Enrichment Strategy.
(A) Diagram of the sequential cell state transitions during stomatal development specified by the combinatorial and sequential actions of the stomatal bHLH genes. A protodermal cell (P) can differentiate into a pavement cell (PC) or undergo a transition to become an MMC (purple). An MMC enters the stomatal lineage through an asymmetric division to create a meristemoid (M; blue). Meristemoids have transient stem cell–like properties that can undergo several rounds of amplifying divisions before differentiating into a GMC (light green). GMCs divide symmetrically to produce two GCs (dark green), which form a mature stoma. The point of action is indicated for each bHLH gene (colored arrows).
(B) Strategy for comparing molecular signatures associated with each epidermal cell state/type. Images are of abaxial cotyledon epidermis of spch, scrm-D mute, and scrm-D mutant seedlings at 5 d after germination. Colors coincide with those described in (A). Bars = 10 μm.
A central mechanism for stem cell maintenance and the generation of cellular diversity in both plants and animals is through asymmetric cell division, which ensures that the two daughter cells maintain separate identities (Abrash and Bergmann, 2009; Fichelson et al., 2009; Menke and Scheres, 2009). Asymmetric cell division during development can occur through signals from surrounding neighbors (extrinsic control) or, alternatively, intrinsic polarity within the cell can trigger partitioning of cell fate determinants (intrinsic control) (Abrash and Bergmann, 2009). Due to the tractability and accessibility of the epidermis, stomatal development has emerged as a model to study asymmetric division and cellular self-renewal. In Arabidopsis thaliana, stomatal development initiates from a subset of protodermal cells, termed meristemoid mother cells (MMCs) (Figure 1A). An MMC undergoes an asymmetric cell division that creates a stomatal precursor called a meristemoid. The meristemoid reiterates several rounds of asymmetric cell divisions, producing neighboring nonstomatal cells (stomatal lineage ground cells [SLGCs]) prior to differentiating into a guard mother cell (GMC). The GMC undergoes a single symmetric division and terminally differentiates into a set of paired guard cells (GCs) that constitute a stoma (Bergmann and Sack, 2007; Peterson et al., 2010) (Figure 1A).
Progression through the stomatal lineage is specified through the combinatorial activities and heterodimerization of the basic helix-loop-helix (bHLH) transcription factors SPEECHLESS (SPCH), MUTE, FAMA, SCREAM (SCRM), and SCRM2 (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007a; Kanaoka et al., 2008). SPCH, MUTE, and FAMA act as molecular switches for cell state transitions: SPCH drives the initial asymmetric division of MMCs and entry into the stomatal lineage, whereas MUTE triggers the differentiation of meristemoids into GMCs (MacAlister et al., 2007; Pillitteri et al., 2007a, 2008). FAMA is required for the proper differentiation of GCs (Ohashi-Ito and Bergmann, 2006). Knockout mutations in SPCH, MUTE, or FAMA result in a complete block of stomatal lineage progression at their respective points of action (Figure 1A). SCRM and SCRM2, two paralogous and partially redundant bHLH proteins, integrate the three cell state transitional events for stomatal differentiation by forming heterodimers with SPCH, MUTE, and FAMA (Kanaoka et al., 2008) (Figure 1A).
The unique ability of a meristemoid to reiterate asymmetric cell divisions makes this cell type an attractive system to understand the molecular and cellular basis of self-maintenance and differentiation in plants. To date, the molecular character of meristemoids remains unclear due to the transient nature and low density of meristemoids on the epidermal surface. Here, we report genome-wide transcriptional profiling of meristemoids through targeted enrichment of stomatal cell states enabled through combinatorial, synthetic mutants of stomatal differentiation. Pairwise comparisons of transcriptomes among mutant seedlings that develop an epidermis composed solely of pavement cells, meristemoids, and SLGCs or GCs identified new genes that are highly and specifically expressed in the stomatal lineage (Figure 1B). Meristemoid-enriched gene products include those involved in cell–cell signaling, hormone metabolism and signaling, and the cell cycle, as well as cell polarity components associated with the meristemoid state. A survey of commonly upregulated genes in meristemoids and other stem cell populations emphasizes a broad role for ERECTA family genes in cell proliferation and differentiation within the SAM and stomatal cell lineages. Through this transcriptome analysis, we identify a protein, POLAR, that is polarly localized and unequally segregated during asymmetric cell divisions of the stomatal lineage. Combined, our study enables the isolation of a unique gene set actively expressed during the transient meristemoid cell state and provides high-quality data for building a gene regulatory network. Furthermore, our study reveals the molecular character of the meristemoid and illuminates common themes in gene expression among plant stem cells.
RESULTS
Transcriptional Profiling of Stomatal Lineage Cells Enabled by Combinatorial Mutations in Stomatal Differentiation Genes
To obtain gene expression profiles reflecting cell state transitions in the stomatal lineage, we took advantage of mutant resources identified in our previous studies, which enable targeted enrichment of pavement cells, meristemoids, and GCs. To assess the transcriptional profile of meristemoids, we used a synthetic mutant carrying both scrm-D and mute. Meristemoids of mute mutants never differentiate into GMCs or transdifferentiate into pavement cells (Pillitteri et al., 2007a). By contrast, the gain-of-function scrm-D mutation triggers constitutive initiation and differentiation of the stomatal lineage, resulting in the production of stomata over the entire epidermis (Kanaoka et al., 2008) (Figure 1B). Therefore, the combined scrm-D mute mutations produce a synthetic phenotype of an epidermis composed almost entirely of meristemoids (Figure 1B). The scrm-D mutant was used to represent the guard cell state (Figure 1B). Finally, the spch mutant, characterized by an epidermis composed solely of pavement cells, was used as a genotype devoid of any stomatal cell lineage–derived transcripts (MacAlister et al., 2007; Pillitteri et al., 2007a) (Figure 1B). Pairwise comparisons of transcriptomes among three biological replicates of spch, scrm-D mute double mutants, scrm-D, and the wild type using Affymetrix ATH1 expression arrays were performed in an effort to maximize the signal-to-noise ratio between molecular signatures associated with each cell state in a way that was not previously possible (Figure 1B). To minimize secondary effects due to the absence or constitutive differentiation of stomata, very young seedlings 5 d after germination were used for profiling (see Supplemental Figure 1 online). To minimize diurnal effects on gene expression (Harmer et al., 2000), all seedlings were harvested in the middle of the light period.
Gene Expression Clustering Reveals Dramatic Changes in Transcript Accumulation during Cell State Transition
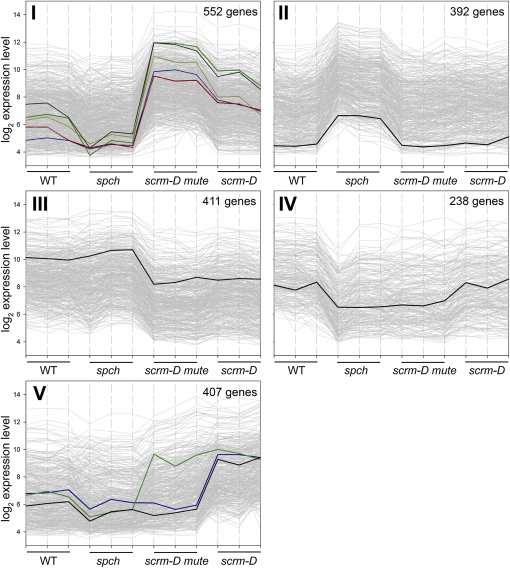
To assess genotype- or cell type–specific trends in gene expression, we performed a K-means cluster analysis using normalized expression values from each individual replicate of four different genotypes (Figure 2). Genes were prefiltered using the variance filter implemented in the MeV software to remove those that showed no differential expression across the genotypes (Saeed et al., 2003). This yielded 2000 genes with the highest variance among the different arrays. Five clusters (I to V; Figure 2) were built from a maximum of 200 iterations using a Pearson correlation distance matrix. A suite of genes specifically enriched in one or two genotypes was identified (see Supplemental Data Sets 1 to 3 online).
Figure 2.
K-Means Clustering of Genes Showing Differential Expression in Stomatal Differentiation Mutants.
The log2 expression levels are plotted against the three replicates of each genotype (the wild type, spch, scrm-D mute, and scrm-D). Genes were prefiltered using the variance filter implemented in the MeV software to yield the 2000 genes showing highest variance across the 12 arrays. Cluster I: genes upregulated in scrm-D mute and to a lesser extent in scrm-D (meristemoid and GMC/GC enriched, respectively). Black, TMM; green, EPF2; red, ARR16; blue, CLE9; yellow, POLAR (At4g31805). WT, wild type. Cluster II: genes upregulated in spch (pavement cell–only epidermis). Highlighted in black is a representative gene (Orp4C, At5g57240) of this cluster. Cluster III: genes with reduced expression in scrm-D mute and scrm-D. Highlighted in black is IAA7 (At3g23050), which shows the representative expression pattern in this cluster. Cluster IV: genes expressed most strongly in the wild type and scrm-D. Highlighted in black is a representative gene (integral membrane family protein, At5g44550) of this cluster. Cluster V: genes upregulated in scrm-D (GMC/GC-enriched). Black, CHX20, a cation/H+ exchanger of GCs; blue, FAMA; green, EPF1.
Gene cluster I showed robust change associated with the meristemoid state: 552 genes are represented in this cluster, with significant upregulation in the scrm-D mute background, but less in scrm-D (Figure 2; see Supplemental Data Set 1 online). Within this group are all genes known to be highly expressed in meristemoids, including TOO MANY MOUTHS (TMM), ERECTA-LIKE1 (ERL1), ERL2, STOMATAL DENSITY AND DISTRIBUTION1 (SDD1), SPCH, MUTE, SCRM, SCRM2, BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL), and EPIDERMAL PATTERNING FACTOR2 (EPF2).
Gene cluster V includes genes highly upregulated in GCs (scrm-D) and to a lesser extent in scrm-D mute: 407 genes are represented in this cluster (Figure 2; see Supplemental Data Set 2 online). These likely represent genes implicated in guard cell development or function, as well as GMC differentiation. For instance, KAT1 and KAT2, two genes encoding GC-specific potassium channel proteins (Schachtman et al., 1992; Nakamura et al., 1995), are represented in this cluster (Figure 2). FAMA and EPF1, a signaling peptide expressed in late meristemoids and GMCs to enforce stomatal spacing (Hara et al., 2007), are also in this cluster (Figure 2). No FAMA transcripts were detected in the scrm-D mute population, supporting the previous finding that in the absence of MUTE, stomatal precursor cells will not progress to GMC and express FAMA (Pillitteri et al., 2007a). By contrast, the presence of EPF1 transcripts in scrm-D mute implies that EPF1 is not directly controlled by MUTE. Genes in clusters I and V show lower expression in the wild type and fall under the detection limit in spch (Figure 2), consistent with the absence of stomatal lineage cells in spch. Combined, the results emphasize that our strategy enabled us to document robust changes in gene expression associated with specific stomatal precursor cell states, which are normally asynchronously transient and cannot be effectively captured in the wild-type background.
Gene Categories Overrepresented in Each Epidermal Cell State
To identify genes with statistically significant enrichment in each mutant background, we used a significance of microarrays analysis using a Q-value cutoff of <0.005 and a two- or threefold-change cutoff as a threshold for deregulation (see Methods). Using these stringent criteria, we identified gene signatures for each background compared with the wild type (see Supplemental Data Sets 4 to 6 online). We first performed overrepresentation analysis (Usadel et al., 2006) to determine the gene categories enriched in each background (see Supplemental Figure 2 online). No overrepresentation of genes involved in photosynthesis, carbohydrate, or central tricarboxylic acid metabolism was observed. A moderate overrepresentation of stress-related genes was found in spch and scrm-D, but not in scrm-D mute. This indicates that the alteration of epidermal phenotype did not significantly affect overall metabolism or stress level of the seedlings. Whereas cell wall biosynthesis genes were mainly upregulated in scrm-D, they were downregulated in scrm-D mute. This may reflect the increased cell wall deposition at the stomatal pore in scrm-D and the low degree of cell expansion in scrm-D mute.
We also found that signaling components, such as receptor kinases, were greatly enriched in the scrm-D mute background but not in spch or scrm-D. This is in accordance with the well-documented findings that cell–cell signals from meristemoids via secreted ligands, membrane receptors, and mitogen-activated kinase cascades are critical for proper stomatal patterning and spacing (Geisler et al., 2000; Bergmann et al., 2004; Shpak et al., 2005; Hara et al., 2007, 2009; Hunt and Gray, 2009).
Molecular Profiling Reveals Genes Highly Enriched in Meristemoids
The significance of microarray analysis identified 527 genes significantly deregulated (277 up/250 downregulated) in the scrm-D mute background. The 2-kb upstream promoter regions of this group of genes were enriched in the E-box (CANNTG) cis-regulatory sequence known to be bound by bHLH DNA binding domains (P = 10−10; Athena data mining tool; O’Connor et al., 2005). This is in contrast with the 74 genes deregulated (39 upregulated/35 downregulated) in the spch background, where no significant enrichment of the E-box element was detected. The finding is consistent with the notion that combinatorial actions of master regulatory bHLH proteins specify meristemoid gene expression.
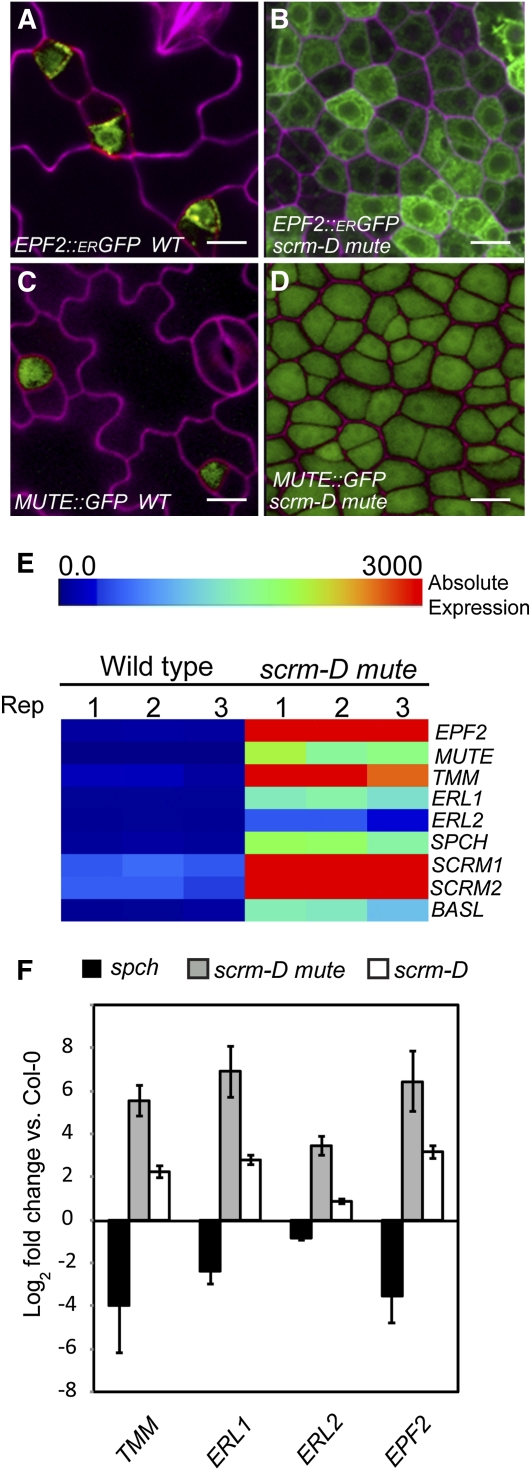
It is important to address whether the arrested meristemoid-like cells constituting the epidermis of scrm-D mute share molecular characteristics of meristemoids, as indicated by the cluster analysis (Figure 2). We first introduced representative markers of early and late meristemoids, EPF2pro:erGFP (for green fluorescent protein) and MUTEpro:GFP, respectively, to scrm-D mute (Figure 3). Indeed, both reporter constructs showed high and nearly constitutive expression in the epidermis of scrm-D mute (Figures 3B and 3D).
Figure 3.
Arrested Meristemoids Constituting the Epidermis of scrm-D mute Express Meristemoid Markers.
EPF2 and MUTE promoter activity (green) is detected only in early stomatal lineage cells and not in mature stomata. Bars = 10 μm.
(A) and (B) EPF2pro:erGFP (green) in the wild type (WT) (A) and scrm-D mute (B).
(C) and (D) MUTEpro:GFP in the wild type (C) and scrm-D mute (D). Cell peripheries were highlighted with either propidium iodide or FM4-64 (magenta).
(E) Known regulators of meristemoid development are highly expressed in scrm-D mute. Shown is a heat map showing the absolute expression levels in the wild type and scrm-D mute (meristemoid enriched). All three replicates are shown to demonstrate consistency.
(F) qRT-PCR verification of a subset of stomatal regulators known to be expressed in meristemoids. Error bars represent the se (n = 3).
Consistent with the reporter activity, among the top signatures (based on fold increase) in scrm-D mute are genes known to be highly expressed in meristemoids and regulate meristemoid development: MUTE (79-fold increase), EPF2 (39-fold increase), TMM (22-fold increase), ERL1 (22-fold increase), SPCH (20-fold increase), BASL (17-fold increase), and ERL2 (7.5-fold increase) (Figure 3E). A quantitative RT-PCR (qRT-PCR) analysis of these genes further verified the microarray results (Figure 3F). It is worth noting that mute displays the same phenotype as a transcriptional null allele, mute-2 (Pillitteri et al., 2008), but accumulates mis-spliced transcripts with premature termination codons (see Supplemental Figure 3 online) (Pillitteri et al., 2008). Overall, the scrm-D mute epidermal cell population has the molecular characteristics of meristemoids, which validates our approach and provides the basis for further characterization of genes enriched in this cell population.
Reporter Expression and Cellular Localization of Meristemoid-Enriched Genes
To gain insight into the roles of meristemoid-enriched genes, we chose 14 genes previously uncharacterized in relation to stomatal development and produced reporter constructs (Figure 4). We designed constructs containing up to 2.5 kb of the 5′ upstream regulatory sequence for each gene driving the expression of the full-length coding region fused to GFP. Alternatively, for those genes whose translational fusions did not result in detectable GFP signals, transcriptional fusion constructs were produced. They include the following: At1g26600 (CLE9; 30-fold), At4g31805 (misannotated as WRKY family transcription factor [see below]; 21-fold), At5g13220 (JAZ10; 15-fold), At2g40670 (ARR16; 14-fold), At3g17640 (leucine-rich repeat [LRR] protein; 9-fold), and At5g62210 (EMBRYO-SPECIFIC PROTEIN3 [ESP3]; >20-fold in scrm-D mute compared with spch) (see Supplemental Table 1 online).
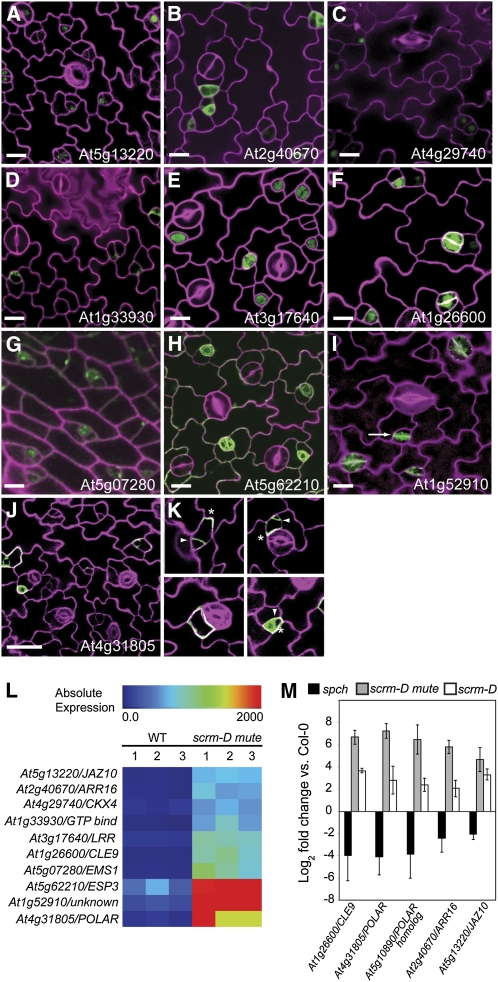
Figure 4.
Localization in Stomatal Cell Lineages and Expression Levels.
Genes represent those upregulated in the scrm-D mute background. All lines carry a C-terminal GFP protein fusion (green) of the indicated gene driven by its native promoter, except for CLE9 and EMS1, which carry the native promoter driving GFP.
(A) to (K) Confocal images of wild-type seedling leaf epidermis. Images are of the abaxial surface, except (E) (adaxial surface). Cell peripheries were highlighted with either propidium iodide or FM4-64 (magenta). Arrow in (I) indicates cell plate. Asterisks in (K) indicate asymmetrical localization of At4g31805-GFP; arrowheads indicate location of division. Bars = 10 μm in (A) to (I) and 25 μm in (J).
(L) Heat map representing the degree of upregulation of each indicated gene in scrm-D mute compared with the wild type. Three replicates are shown to demonstrate consistency. Scale represents absolute expression of each gene.
(M) qRT-PCR verification of selected meristemoid-enriched genes identified in this study. Values are relative to expression of corresponding genes in the wild type. Error bars represent the se (n = 3).
Of the 14 reporter constructs, 10 showed expression patterns in the stomatal lineage (Figures 4A to 4K). Some are components of signaling pathways that have not been assigned to stomatal development. JAZ10-GFP was found in the nucleus through all transitional states of the stomatal lineage but not in fully differentiated GCs (Figure 4A). JAZ10 acts as a downstream repressor of jasmonate signaling (Chung and Howe, 2009). ARR16-GFP showed strong and specific expression in meristemoids (Figure 4B). ARR16 belongs to the A-type Arabidopsis response regulator (ARR) family acting downstream in cytokinin signaling (Kiba et al., 2002). Interestingly, a gene coding for the cytokinin catabolic enzyme At4g29740 (CKX4) (Werner et al., 2006) is also enriched in scrm-D mute and its transcriptional GFP fusion exhibited expression in meristemoids (Figure 4C). GFP translational fusions of At1g33930, a protein with a GTP binding motif, and At3g17640 are also strongly detected in the cytoplasm of meristemoids (Figures 4D and 4E). CLE9, one of the CLE peptide genes, and At5g07280 (EMS1), an LRR–receptor-like kinase (RLK) regulating microsporogenesis (Zhao et al., 2002; Jun et al., 2010), showed strong expression in meristemoids and GMCs as GFP transcriptional fusions (Figures 4F and 4G).
Three proteins displayed unique expression patterns. ESP3, which is predicted to be membrane anchored via glycosylphosphatidylinositol (Borner et al., 2002), displayed patchy plasma membrane localization across the epidermis, including pavement cells, but showed strong stomatal lineage–specific expression on internal membranes (Figure 4H). At1g52910 (unknown protein) exhibited a characteristic, vesicular localization and strong accumulation at the cell plate of dividing meristemoids and GMCs (Figure 4I, arrow), suggesting that this protein may be involved in the cytokinesis of stomatal lineage cells. Lastly, At4g31805 exhibited an intriguing localization, with strong GFP signal detected in the cytoplasm of meristemoids and cell periphery in SLGCs (Figure 4J). Strikingly, At4g31805-GFP was localized polarly in SLGCs, distal to the newly divided meristemoid and parallel to the division plane (Figure 4K, asterisks and arrowheads). Based on the dynamic behavior of this protein during asymmetric divisions revealed by time-lapse imaging (see below), we named At4g31805 POLAR (for POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION).
Cellular Dynamics of POLAR Protein That Is Polarly Localized and Unequally Segregated during Asymmetric Cell Division of the Stomatal Lineage
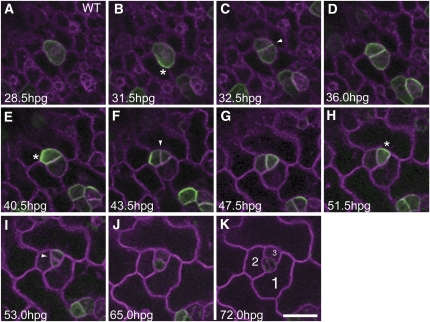
The polar localization of POLAR in SLGCs (Figures 4J and 4K) resembles that of BASL (Dong et al., 2009). Unlike GFP-BASL, which remains in the nucleus in meristemoids (Dong et al., 2009), however, POLAR-GFP displays no nuclear localization in any stage of stomatal development. Based on publicly available data sets, POLAR and BASL display little or no detectable expression in other asymmetrically dividing tissues, such as roots (see Supplemental Figure 4 online). To resolve the cellular dynamics of POLAR during asymmetric divisions of the stomatal lineage, we developed a time-lapse imaging technique (Figure 5; see Supplemental Movie 1 online). Mature embryos were dissected at germination, and cotyledons were subjected to time-lapse imaging at 30-min intervals over 72 h to capture the real-time movement of this protein in relation to cell division during stomatal development (see Methods). During the process of imaging, cotyledons developed normally and eventually formed stomata.
Figure 5.
Polar Localization of POLAR Precedes the Asymmetric Division of Stomatal Cell Lineages.
Real-time imaging was performed using wild-type cotyledons expressing a C-terminal GFP protein fusion of POLAR driven by the native promoter (POLARpro:POLAR-GFP). Individual fields from imaging are shown over time as indicated ([A] to [K]). Cell membranes (magenta) are visualized with the mCherry plasma membrane marker pm-RB. POLAR-GFP displays localized, transient accumulation at the cell periphery distal to the plane of division prior to asymmetric divisions of stomatal cell lineage (MMC and meristemoids), then is redistributed following asymmetric division. Asterisk indicates polar localization. Arrowhead indicates the plane of division. SLGCs resulting from the asymmetric division are numbered (K). hpg, hours post germination. Bar = 20 μm. See also Supplemental Movie 1 online.
The time-lapse imaging revealed that during germination, POLAR-GFP initially appears in a subset of protodermal cells, which are likely MMCs, both in the cytosol and at the cell periphery (Figure 5A). Approximately 2 h before each asymmetric division occurs, POLAR-GFP becomes dynamically localized at the cell cortex distal to the division plane (Figures 5B, 5E, and 5H). Clear polar localization of POLAR distal to the division plane was observed in all asymmetrically dividing cells of the stomatal lineage. After asymmetric cell division, POLAR-GFP is upregulated in only one of the daughter cells, which is predictive of the cell that will continue to divide asymmetrically. When the larger daughter cell (SLGC) retains POLAR expression, localization is directed to the periphery of the cell adjoining the adjacent meristemoid or GMC, correlated with a new satellite meristemoid being placed away from the existing one (see Supplemental Movie 1 online). Expression of POLAR appears to mark stomatal lineage cells with asymmetric division potential. Those protodermal cells that did not undergo asymmetric entry division but underwent pavement cell differentiation or, alternatively, differentiated into GMCs without asymmetric divisions immediately lost POLAR-GFP expression (see Supplemental Figure 5 online). Likewise, once a meristemoid terminated asymmetric division and differentiated into a GMC, POLAR-GFP expression rapidly diminished (Figures 5I to 5K).
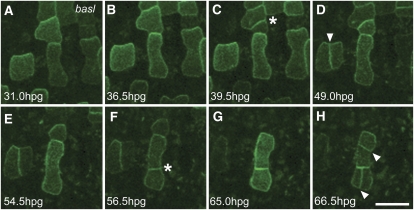
Asymmetric Localization of POLAR Is Disrupted by Loss of Function in BASL
To understand whether a known regulator of asymmetric cell divisions in the stomatal cell lineage influences the polar localization of POLAR, we next investigated cellular dynamics of POLAR-GFP in basl using time-lapse imaging (Figure 6; see Supplemental Movie 2 online). The basl mutant is characterized by a loss of physical asymmetry during division and concomitant loss of cell fate asymmetries, resulting in clustered stomata (Dong et al., 2009). Asymmetric localization of POLAR-GFP was disrupted in the basl mutant background, which correlated with a nonasymmetric division of the GFP-expressing cell (Figures 6D and 6H). During the division of stomatal lineage cells in basl, both daughters retain expression of POLAR-GFP and both continue to divide (Figures 6E to 6H). This is in sharp contrast with the wild type, where only one cell from a division retains expression of POLAR-GFP and continues asymmetric division (Figures 5E to 5G). This supports the previous finding that cell fate asymmetries after division are absent in the basl background (Dong et al., 2009). The observed perturbation of POLAR-GFP localization in the basl background suggests that a functional BASL protein is required for POLAR-GFP asymmetry. Some cells in the basl background may also express POLAR-GFP without dividing for over 24 h, a much longer duration than we observed in the wild type, which may indicate that the regulation of this protein is perturbed by the loss of BASL. The asymmetric division defects in basl are not fully penetrant; when meristemoids divide normally in basl, POLAR-GFP exhibits normal expression dynamics and polar localization (see Supplemental Movie 2 online).
Figure 6.
Localization of POLAR in the basl Mutant Defective in Asymmetric Division.
Real-time imaging was performed using basl cotyledons expressing POLARpro:POLAR-GFP. Individual fields from imaging are shown over time as indicated ([A] to [H]). Loss-of-function basl mutants have diffuse peripheral POLAR-GFP expression that is not strongly polarized. Arrowheads indicate symmetrical division plane ([D] and [H]) relative to the parent cell. Asterisks indicate asymmetric division. hpg, hours postgermination. Bar = 20 μm. See also Supplemental Movie 2 online.
[See online article for color version of this figure.]
Protein Structure and Loss-of-Function Allele of POLAR
POLAR codes for a 344–amino acid protein of unknown function. Although it is annotated as WRKY transcription factor (The Arabidopsis Information Resource: http://www.arabidopsis.org/), sequence analysis does not reveal any similarity to the WRKY domain or any other recognizable structural motifs or domains aside from its moderate similarity to an Arabidopsis myosin-heavy chain-like protein (At5g10890: 29% identity/49% similarity). Secondary structure analysis predicts a predominantly alpha-helical structure with a coiled-coil region at the C terminus (Lupas et al., 1991). We did not identify homologs of POLAR outside of plants. To unravel the function of POLAR, we analyzed two T-DNA insertion alleles (SALK_112914 and SALK 142820). The RT-PCR analysis detected no POLAR mRNA accumulation (see Supplemental Figure 6 online), indicating that the insertions lead to the loss/reduction of POLAR transcripts. These insertion lines did not reveal discernable growth or developmental phenotypes, suggesting that POLAR may be functionally redundant.
Meristemoid-Enriched Transcriptome Includes Specific Members of Core Cell Cycle Regulators
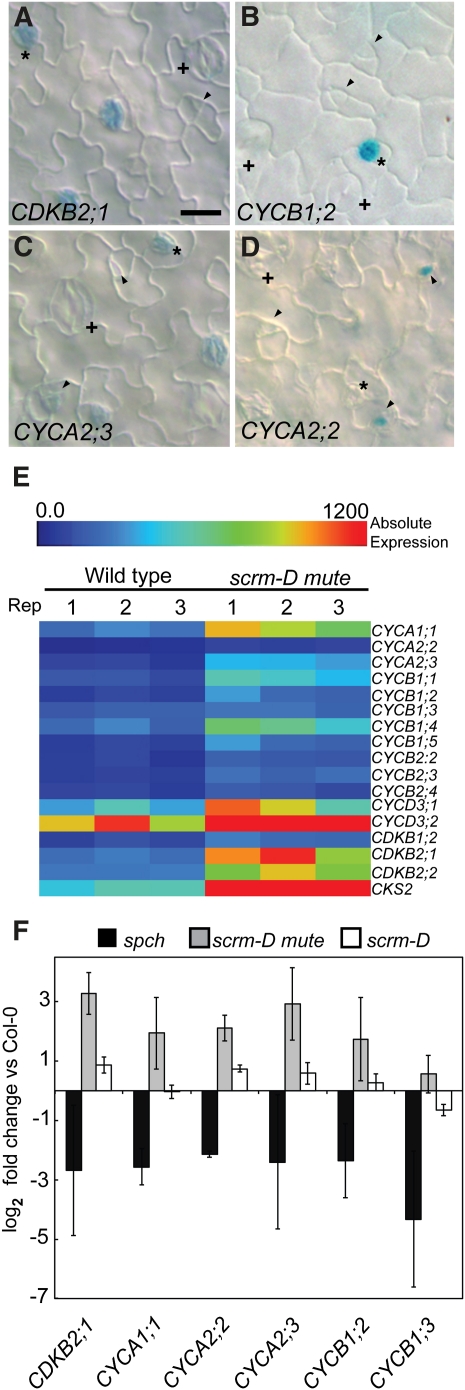
Meristemoids exhibit a rare capacity for reiterating asymmetric cell divisions, which is lost with the cell state transition to GMCs: GMCs instead undergo a symmetric division and differentiation. We therefore investigated whether cell cycle regulatory genes were enriched in meristemoids. Our analysis revealed that 17 core cell cycle genes were represented in gene cluster I (Figures 2 and 7). Among them, we observed the reporter β-glucuronidase (GUS) expression of CYCLIN DEPENDENT KINASE B2;1 (CDKB2;1), CYCB1;2, and CYCA2;3 (Figures 7A to 7C), all of which showed high promoter activity in stomatal lineage cells and have higher levels of transcript accumulation in the scrm-D mute background compared with spch or scrm-D (Figures 7E and 7F). It has been reported that ERECTA family receptor kinases, which regulate organ growth and stomatal patterning, affect expression of cyclin A2 genes (Pillitteri et al., 2007b). We further examined the reporter activity of the remaining cyclin A2 genes and found that CYCA2;2 is upregulated in scrm-D mute (Figure 7F) and CYCA2;2pro:GUS is expressed in a subset of meristemoids (Figure 7D), potentially those making a transition from asymmetric to symmetric division. Consistent with this result, CYCA2;2 transcript is slightly enriched in scrm-D mute (Figure 7F).
Figure 7.
Analysis of Cell Cycle Regulatory Gene Expression in the Stomatal Lineage.
(A) to (D) All lines carry the GUS coding sequence driven by the native promoter of each indicated gene. Shown are differential interference contrast microscope images of abaxial leaf epidermis from wild-type seedlings expressing the following reporter lines: CDKB2;1, At1g76540 (A); CYCB1;2, At5g06150 (B); CYCA2;3, At1g15570 (C); and CYCA2;2, At5g11300 (D). Arrowheads indicate meristemoids; asterisks indicate GMCs; (+) indicates stomata. Bar = 10 μm.
(E) Heat map representing the degree of upregulation of each indicated gene in the scrm-D mute background compared with the wild type. All three replicates of both genotypes are shown to demonstrate consistency. Scale represents absolute expression of each gene.
(F) qRT-PCR of selected core cell cycle regulators in spch, scrm-D mute, and scrm-D. Gene names are indicated. Values are relative to the Col wild-type background. Error bars represent the se (n = 3).
Molecular Signatures of Meristemoids and Meristems: Commonalities and Distinctions
Comparison of transcriptome profiles among different microarray experiments can identify unexpected molecular commonalities among specific cell types. Our meristemoid-enriched transcriptome provides the opportunity to compare the meristemoid state with the true stem cell population in the SAM and RAM. For this purpose, we obtained publicly available transcriptome data derived from isolated cells expressing the SAM-specific markers CLAVATA3 (CLV3; stem cell domain), WUSCHEL (WUS; rib meristem), and FILAMENTOUS FLOWER (FIL; peripheral zone of the floral meristem) (Yadav et al., 2009) as well as data from two independent root tip microarrays, termed root tips zone1 (Dinneny et al., 2008) and root tips (Sena et al., 2009). These data sets were compared with our meristemoid-enriched transcript (scrm-D mute) profile (Figure 8). Overrepresentation analysis showed that genes classified as transcription factors (RNA regulation of transcription) and receptor kinases are equally overrepresented in all stem cell populations included in this meta-analysis (see Supplemental Figure 7 online), highlighting the importance of transcriptional regulation and cell–cell signaling in maintenance and function of meristematic cells. Alternatively, these trends may reflect general characteristics of actively dividing cells.
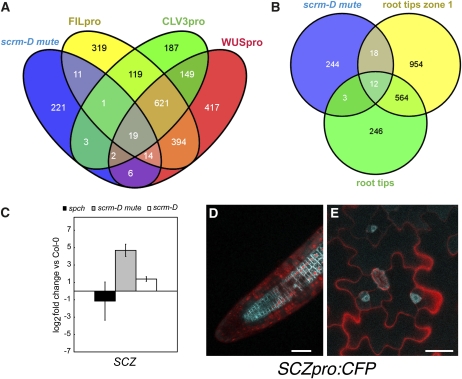
Figure 8.
Integrative Analysis of Transient and Permanent Stem Cell Populations.
(A) and (B) Venn diagrams showing the distribution of unique and shared genes upregulated among scrm-D mute, CLV3, FIL, or WUS expressing cells (A) as well as among scrm-D mute and root tips zone1 and root tips (B).
(C) qRT-PCR analysis showing that SCZ, a known regulator of asymmetric cell division within a root meristem, is highly upreguated in scrm-D mute and downregulated in spch. Error bars represent the se (n = 3).
(D) and (E) Expression of SCZ promoter driving cyan fluorescent protein (CFP). SCZ promoter is active in stelar tissue within the root meristem (D) as well as in meristemoids (E). Bars = 10 μm in (D) and 20 μm in (E).
We identified relatively little overlap in upregulated genes between scrm-D mute and individual SAM populations: 11, 3, and 6 genes for the FILpro, CLV3pro, and WUSpro arrays, respectively. This is far less than the overlap between the CLV3pro array and the WUSpro (149 genes) and FILpro arrays (119 genes) (Figure 8A). However, we identified 19 genes that are commonly upregulated in the all three SAM populations and scrm-D mute (see Supplemental Table 2 online). As expected, several cell division–related proteins were commonly upregulated: At3g19590 (BUDDING UNINHIBITED BY BENZYMIDAZOL [BUB3.1]), At2g06510 (REPLICATION PROTEIN A [RPA1A]), and At1g20930 (CDKB2;2). The most notable among the commonly upregulated genes are two members of the ERECTA family of genes, ERL1 and ERL2, which encode closely related LRR-RLKs (Shpak et al., 2004). Among all arrays, ERL1 (At5g62230) shows eightfold to 22-fold upregulation and ERL2 (At5g07180) a sevenfold to 35-fold increase. The finding underscores the known roles of the ERECTA family in promoting cell proliferation during shoot and flower development as well as restricting stomatal differentiation (Torii et al., 1996; Shpak et al., 2004, 2005; Pillitteri et al., 2007b; Hord et al., 2008). The LRR-RLK gene EMS1 was also transcriptionally enriched in scrm-D mute, WUSpro, and FILpro (19-, 14-, and 24-fold, respectively). Enrichment in CLV3pro was statistically insignificant (Q = 0.017) but likely a false negative due to our stringent cutoffs. EMS1 regulates anther cell differentiation (Zhao et al., 2002) by physical interaction with the small peptide TAPETUM DETERMINANT1 (TPD1) (Jia et al., 2008). A role for the TPD1/EMS1 ligand receptor system in the SAM has not been documented.
Comparison of scrm-D mute to transcriptional profiles from root tips revealed 12 genes commonly enriched in both root tip experiments and scrm-D mute (Figure 8B). Among them is SCHIZORIZA (SCZ), a heat shock family transcription factor, which regulates asymmetric cell divisions in the root stem cell niche (ten Hove et al., 2010). SCZ is also transcriptionally enriched in scrm-D mute and specifically expressed in meristemoids during stomatal development (Figures 8C to 8E), suggesting an exciting potential role for SCZ in asymmetric cell division in the stomatal lineage.
DISCUSSION
Here, we present the transcriptome profile of the meristemoid, a transient and low-density proliferating stomatal precursor cell with stem cell–like properties. Producing plants highly enriched in specific epidermal cell types (pavement cells, meristemoids, or GCs forming stomata) is possible using loss- and gain-of-function mutations in transcription factor genes (SPCH, MUTE, and SCRM) that act in a combinatorial and sequential manner to direct key cell state transitional events within the stomatal cell lineages (MacAlister et al., 2007; Pillitteri et al., 2007a; Kanaoka et al., 2008). Cell type–specific profiling has been widely performed using physical isolation of cells via laser capture microdissection (Kerk et al., 2003; Nakazono et al., 2003; Day et al., 2007) or protoplasting followed by fluorescence-activated cell sorting (FACS) (Iyer-Pascuzzi and Benfey, 2010). Protoplasting and FACS were used for the cell type–specific expression profiling of the SAM used in our meta-analysis (Yadav et al., 2009). These approaches, while accurately isolating specific cell types of interest from wild-type plants, are expensive and include experimental procedures, such as tissue sectioning and protoplast isolation, that could affect gene expression. In addition, the FACS process relies on the availability of GFP markers that specifically and exclusively mark the cell types of interest.
Using spch, scrm-D mute, and scrm-D mutants, we were able to isolate RNA directly without further manipulations to perform pairwise transcriptome comparisons. Using mutant backgrounds invites two major questions: whether the identity of the cells enriched in these mutants is truly normal and whether physiological consequences due to the mutations deregulate transcripts unrelated to the enriched cell types. We did not find significant alterations in stress- or photosynthesis-related transcripts in the scrm-D mute population. Top signatures from this population include genes known to be highly and/or specifically expressed in meristemoids and regulate stomatal development, including TMM, SPCH, MUTE, EPF2, BASL, ERL1, ERL2, SCRM, and SCRM2 (Figure 3E). Furthermore, a new gene associated with cellular asymmetry of meristemoids was identified (Figures 4 and 5). Combined, we conclude that our approach is a simple yet powerful alternative to unravel molecular signatures associated with this transient cell state.
Coexpression programs (ATTED-II: http://atted.jp/) (Obayashi et al., 2011) cluster only two of our 10 more closely analyzed genes, POLAR and CLE9, with known stomatal regulatory networks; the other eight genes identified in this analysis with stomatal-specific expression (Figure 4) were not predicted to be coexpressed with stomatal regulators. This inconsistency demonstrates the importance of continued transcriptome profiling in identifying coregulated genes and ultimately regulatory networks controlling developmental processes. Such discoveries provide a framework of how the molecular mechanisms driving cell division maintenance may be partially conserved among stem cell populations.
Cytokinin Signaling May Play a Universal Role in Stem Cell Regulation
We found that the cytokinin response genes ARR16, CKX4, and CLE9 are strongly upregulated in scrm-D mute, and their expression in stomatal lineage cells was confirmed by reporter analysis (Figure 4). CKX4 encodes a cytokinin oxidase, which degrades and thus inactivates cytokinin (Schmülling et al., 2003), and ARR16 is a member of the A-type ARR family, which function as negative regulators of cytokinin signaling (Ren et al., 2009). ARR proteins act as downstream components of cytokinin signaling via a two-component signaling pathway, where cytokinin receptors, ARABIDOPSIS HISTIDINE KINASE (AHK2, 3, and 4) (Inoue et al., 2001; Yamada et al., 2001), relay a phosphate group to ARR proteins. AHK3 has been shown to display stomatal lineage–specific epidermal expression (Stolz et al., 2011). Although T-DNA insertion lines of ARR16 and CKX4 did not confer discernible phenotypes, our finding implies that downregulation of cytokinin signaling and/or metabolism may play a role in the transition from proliferation to differentiation states of stomatal cell lineages.
Recently, a molecular link between cytokinin signaling and CLE peptide-mediated stem cell maintenance has been reported. In the SAM, the CLV3-WUS–mediated signaling pathway directly represses the transcription of Type-A ARRs (ARR5, ARR6, ARR7, and ARR15), which are required for proper meristem function (Leibfried et al., 2005). Similarly, during root vascular development, one of the CLE peptides, CLE10, inhibits protoxylem differentiation by repressing Type-A ARRs (ARR5 and ARR6) (Kondo et al., 2011). Based on the known roles of the CLE peptides in stem cell maintenance and high expression of CLE9 in meristemoids (Figure 4F) (Jun et al., 2010), it is attractive to hypothesize that CLE9 may act as a signaling molecule for meristemoid maintenance. Our results, in combination with the expanding evidence of crosstalk between cytokinin and CLE signaling in other stem cell populations, suggest that integration of these two pathways might be a general mechanism to mediate broad hormone signals into local changes of gene expression and, ultimately, cell behavior.
Asymmetric Division and Segregation of Cell Fate in the Stomatal Lineage
Stomatal development serves as a model to study de novo asymmetric cell divisions in plants. BASL was the only known protein to be distributed unevenly during the asymmetric division of stomatal lineages (Dong et al., 2009). Our result adds a potential component to the BASL-mediated pathway. POLAR exhibits a striking, transient polar distribution during asymmetric cell division, and this polar localization requires functional BASL. POLAR possesses a coiled-coil motif, which is known to function in protein–protein interactions (Burkhard et al., 2001) and shows sequence similarity to myosin heavy chain–like protein (see Supplemental Figure 6 online). Based on the protein structure and localization patterns, it is tempting to hypothesize that POLAR associates with BASL, components of a complex that includes BASL, or cytoskeletal components to mark the site for rapid cell expansion, which facilitates the cell fate separation of SLGC from meristemoid. POLAR is evenly distributed at the cell periphery in meristemoids where BASL is known to be exclusively localized in the nucleus (Dong et al., 2009). This supports the idea that sequestering BASL in the nucleus is a mechanism preventing polar cell expansion, a process involving the dynamic relocation of POLAR to the cell periphery. The null allele of POLAR does not exhibit stomatal patterning defects like basl, implying the presence of functional redundancy. Indeed, the closest homolog of POLAR, At5g10890, which is not on the ATH1 expression array, is also highly expressed in scrm-D mute (88-fold upregulation), while being downregulated in spch (14-fold downregulation) (Figure 4M). The extent of up- and downregulation of At5g10890 in the meristemoid-enriched populations is similar to that of POLAR (149-fold upregulation in scrm-D mute; 17-fold downregulation in spch) (Figure 4M).
It is known that the bHLH transcription factor SPCH initiates the meristemoid state together with SCRM/SCRM2 (MacAlister et al., 2007; Pillitteri et al., 2007a; Kanaoka et al., 2008). Thus, genes driving asymmetric division of meristemoids may be downstream targets of these bHLH proteins. A coexpression gene network places both BASL and POLAR directly connected to SPCH and closely connected to SCRM (see Supplemental Figure 8 online). Further experiments should address the possibility of direct induction.
Cell Cycle Regulators and Stomatal Cell Divisions
Stomatal cell lineages undergo stereotypical cell divisions, from entry and amplifying divisions of meristemoids to the eventual symmetric divisions of GMCs. Our analysis revealed that promoter activity of CDKB2;1, CYCB1;2, CYCA2;2, and CYCA2;3 was highly specific to stomatal lineage cells (Figure 7). Consistent with a possible direct role in stomatal development, all three cyclin genes examined have been shown to interact directly with CDKB1;2 and CDKB2;1 in yeast and in planta (Van Leene et al., 2011). A direct link between CYCA2;3 and epidermal patterning was recently reported using ectopic overexpression analysis where the ectopic overexpression of both CYCA2;3 and CYCB1;1 resulted in increased epidermal cell division similar to a SPCH overexpression phenotype (Boudolf et al., 2009). Single overexpression of either regulator failed to confer a clear phenotype, suggesting that they function together as a complex (Boudolf et al., 2009). However, CYCD4, which is known to regulate stomatal cell lineage divisions specifically in the hypocotyl (Kono et al., 2007), was not represented in our scrm-D mute transcriptome. Plants encode significantly more cell cycle regulatory proteins than other eukaryotic organisms, presumably due to a sessile existence and subsequent need for plasticity during development. Our profiling and expression analysis add to the growing evidence supporting the idea that overlapping expression of plant core cell cycle regulators is a key component driving cell division and differentiation (Engler et al., 2009).
Molecular Commonalities among Stem Cell Populations in Plants
A notable feature of plant development is indeterminate growth, where organogenesis persists throughout the life cycle via continual activity of stem cells in the SAM and RAM. Our meristemoid-enriched transcriptome provides the opportunity to investigate the similarities and differences of molecular constituents between transient and permanent stem cell populations. The comparison of meristemoid versus SAM signatures reemphasizes the diverse roles of ERECTA family RLKs as regulators coordinating cell proliferation and differentiation during shoot and flower development as well as stomatal patterning (Torii et al., 1996; Shpak et al., 2003, 2004, 2005; Pillitteri et al., 2007b; Hord et al., 2008). In the epidermis, ERECTA restricts asymmetric cell divisions, while ERL1 and ERL2 inhibit the differentiation of meristemoids to GMC and enforce stomatal spacing. Genetic studies suggest that ERECTA family RLKs act together with TMM to perceive secreted signaling ligands EPF1 and EPF2 (Shpak et al., 2005; Hara et al., 2007, 2009; Hunt and Gray, 2009). Because TMM, EPF1, and EPF2 are not expressed in the SAM, an exciting possibility is that the ERECTA family perceives other EPF-LIKE (EPFL) peptides via dimerization with other receptor partner(s). There are 11 EPFL genes in Arabidopsis (Hara et al., 2009; Rowe and Bergmann, 2010; Rychel et al., 2010), for seven of which the expression patterns and functions are yet unknown. The highlighted roles of LRR-RLKs, ERECTA family, and EMS1 as commonly upregulated SAM meristemoid signatures underscore the importance of cell–cell signaling in orchestrating stem cell proliferation and differentiation.
In contrast with the SAM, where stem cell divisions are less organized, Arabidopsis RAM cells undergo stereotypical asymmetric divisions. Based on their positions relative to the quiescent center, each stem cell in the RAM gives rise to a defined cell file (Scheres, 2002). Our RAM versus meristemoid comparison revealed SCZ as a commonly upregulated gene. SCZ executes proper separation of cell fate in stem cells generating differentiated root tissues, including epidermis, cortex, endodermis, and root cap (ten Hove et al., 2010). The molecular mechanism by which SCZ mediates cell fate separation is unknown, but it appears to involve both cell-autonomous and non-cell-autonomous effects (ten Hove et al., 2010). Our study highlights specific components involved in asymmetric division within the stomatal lineage, BASL and POLAR, as well as a potential common component, SCZ. Future mechanistic studies of these proteins may molecularly define the commonalities and uniqueness of asymmetric divisions in plants.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana Columbia (Col) accession was used as the wild type. Mutants used in the study were in the Col background. Identification numbers for T-DNA insertional mutants of genes identified in this study are given in Supplemental Table 3 online. All insertion lines and the plasma membrane mCherry reporter, pm-RB (CD3-1008) (Nelson et al., 2007), were obtained from the ABRC. The following mutants and reporter lines were described previously: spch and mute (Pillitteri et al., 2007a), scrm-D and scrm-D mute (Kanaoka et al., 2008), basl-2 (Dong et al., 2009), MUTEpro:GFP (Pillitteri et al., 2008), EPF2pro:erGFP (Hara et al., 2009), CDKB2;1pro:GUS, CycB1;2pro:GUS, CycA2;2pro:GUS, and CycA2;3pro:GUS (gifts from Lieven de Veylder and Steffen Vanneste; Vanneste et al., 2011). Selected reporter lines were crossed with stomatal differentiation mutants. For microarray preparation, seeds of the wild type, scrm-D, scrm-D mute, and spch were sterilized using 33% bleach solution (bleach and 0.05% Tween 20) for 12 min and washed five times with sterile water. Seeds were plated on 1× Murashige and Skoog media and placed at 4°C for 5 d. Plates were then placed under standard growth conditions (16-h day/8-h night, 21°C), and whole seedlings were collected 5 d after germination.
Microarray Material Preparation
Five-day-old seedlings of the wild type, scrm-D, scrm-D mute, and spch were collected and immediately placed in liquid nitrogen. To avoid any complications due to circadian gene regulation, all genotypes were harvested at the same time (1 pm) within 1 h of each other for each replicate. spch plants were from a segregating population and were identified by the smooth appearance of the cotyledons compared with wild-type plants. Total RNA was extracted using the Qiagen RNeasy kit (Qiagen) following manufacturer's instructions. RNA purity and yield were confirmed using the Agilent 2100 Bioanalyzer (Brolingen). Probe preparation and hybridization to the ATH1 GeneChip (Affymetrix) were performed according to the manufacturer's instructions by the University of Washington Center for Array Technologies. Signal intensities from each array were converted to raw expression .CEL data using GeneChip operating software (Affymetrix).
Statistical Analysis of Microarrays
Our own and publicly available ATH1 microarray data were normalized using Robin software (Lohse et al., 2010). Quality control was performed with standard settings (Limma package using RMA normalized data with Benjamini Hochberg P value correction) (Benjamini and Hochberg, 1995). Further statistical analysis was performed using the MultiExperiment Viewer (MeV) software (Saeed et al., 2003). Genes with statistically significant deregulation compared with the wild type were identified using the significance of microarrays module with a Q-value cutoff of <0.005 and a twofold- or threefold-change cutoff. K-means clustering was performed on our own microarrays using log2-transformed normalized expression values as input. Genes were prefiltered using the variance filter implemented in the MeV software to remove genes that showed little variance in expression across the genotypes, to yield 2000 genes with the highest variance among the different arrays. Five clusters were built from maximal 200 iterations using a Pearson correlation distance matrix. Annotation of genes and classification into functional pathways (BINs) was performed with the Mapman software (Thimm et al., 2004). Pageman software (Usadel et al., 2006) was used for overrepresentation analysis.
Verification of Microarray Results by RT-PCR and qRT-PCR
The RNA isolated from Col-0 wild-type, spch, scrm-D mute, and scrm-D seedlings used for the microarray experiment was treated with DNase I (Invitrogen). RNA was converted to cDNA using iScript reverse transcriptase (Bio-Rad) and random hexamers. First-strand cDNA was used directly or diluted 1:10 in double distilled water and used as template for PCR. Real-time PCR was performed using a CFX96 real-time PCR detection system (Bio-Rad) using iTaq SYBR Green Supermix with ROX (Bio-Rad) and standard qPCR conditions in three technical and three biological replicates. Amplification of the ACT2 gene was used to verify equal loading of cDNA (RT-PCR) or as an internal control in reactions with relative expression calculated using the cycle threshold (Ct) 2−ΔΔCt method (qRT-PCR) (Livak and Schmittgen, 2001). For RT-PCR, all genes were amplified in 31 cycles, except ACT2 (30 cycles) and POLAR (35 cycles) using standard PCR conditions. DNA sequences of primer pairs used for RT-PCR and qRT-PCR can be found in Supplemental Table 4 online.
Molecular Cloning and Generation of Transgenic Plants
See Supplemental Table 5 online for a list of plasmid constructs generated in this study and Supplemental Table 6 online for a list of primer DNA sequences used for molecular cloning. Briefly, the promoter region (up to ~2.5 kb) upstream of the translation start site was cloned into the pENTR 5′ TOPO (Invitrogen) vector according to the manufacturer's instructions. The full-length genomic coding region was amplified and cloned into the pENTR D-TOPO vector (Invitrogen). These vectors were combined in a three-way Gateway cloning reaction using LR Clonase II Plus (Invitrogen) to produce a C-terminal GFP fusion driven by the endogenous promoter. Each genomic plasmid was used in a two-way Gateway reaction to produce an overexpression construct driven by the cauliflower mosaic virus 35S promoter. The Gateway-based destination vectors were provided by Tsuyoshi Nakagawa (Shimane University) (Nakagawa et al., 2007, 2008). Generation and selection of transgenic plants and their phenotypic analyses were performed as described by Pillitteri et al. (2007a).
Microscopy
Confocal microscopy images were taken on either the Zeiss LSM700 or the Olympus FV1000 microscope. Cell borders were visualized with either propidium iodide (Molecular Probes) or FM4-64 (Invitrogen). The confocal images were false colored, and their brightness/contrast settings were adjusted using Photoshop CS4 (Adobe). Histochemical staining for GUS activity was performed as previously described (Sessions et al., 1999).
Time-Lapse Cotyledon Imaging
Seeds expressing GFP markers in the desired backgrounds were washed as described above and held at 4°C for 2 d. All seeds were imbibed and placed under germinating conditions within 1 h of the same time (10 am). Seeds were moved into room temperature and immediately dissected: the seed coat was removed and the hypocotyl severed. Removing the hypocotyl shows no noticeable effect on cotyledon development for the first 4 d under our conditions. A solution of 0.5% Bacto Agar was prepoured into chamber slides (Lab Tek II chambered 1.5 German cover glass system; Fisher Scientific) at room temperature, and dissected cotyledons were placed beneath it for immobilization and protection from desiccation. The Zeiss LSM700 (inverted, ×20 objective, ×1 zoom) was used to image the GFP reporter with excitation at 488 nm and a band-pass emission filter of 470 to 500 nm at intervals of 30 min. Z-stacks of 50 to 70 slices at 0.96 μm were captured for each time point and projected for maximum intensity. For POLAR-GFP, stable lines were crossed with pm-RB and the F1 generation imaged as above, with additional excitation at 555 nm and collection with a short-pass 555-nm filter.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers in Supplemental Table 1 online and the following: POLAR (At4g31805), CDKB2;2 (At1g20930), RPA1A (AT2G06510), BUB3.1 (AT3G19590), ER (AT2G26330), ERL1 (AT5G62230), ERL2 (AT5g07180), TPD1 (AT4g24977), CDKB2;1 (AT1G76540), CYCB1;2 (AT5G06150), CYCA2;3 (AT1G15570), CYCA2;2 (AT5G11300), IAA7 (AT3G23050), Integral membrane family protein (AT5g44550), Myosin heavy chain protein (AT5G10890), Orp4C (AT5G57240), MUTE (AT3G06120), SPCH (AT5G53210), FAMA (AT3G24140), ICE1/SCRM (AT3G26744), SCRM2 (AT1G12860), SDD1 (AT1G04110), TMM (AT1G80080), YODA (AT1G63700), BASL (AT5G60880), EPF1 (AT2G20875), EPF2 (AT1G34245), KAT1 (AT5G46240), KAT2 (AT4G18290), FIL (AT2G45190), WUS (AT2G17950), CLV3 (AT2G27250), AHK2 (AT5G35750), AHK3 (AT1G27320), AHK4 (AT2G08130), ARR5 (AT3G48100), ARR6 (AT5G62920), ARR7 (AT1G19050), ARR15 (AT1G74890), CLE10 (AT1G69320), and SCZ (AT1G46264). The GenBank accession number for POLAR cDNA is JN663804. The complete expression data set is available in the National Center for Biotechnology Information Gene Expression Omnibus (Edgar et al., 2002) under accession number GSE29814 (http://www.ncbi.nlm.nih.gov/geo).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Images of Representative 5–Days After Germination Seedlings Used in This Study.
Supplemental Figure 2. Summary of Overrepresented Functional Gene Categories Associated with Cell-Enriched Genotypes.
Supplemental Figure 3. RT-PCR Data for Selected Genes.
Supplemental Figure 4. Heat Map of Absolute Expression of Selected Stomatal Regulatory Genes in Rosettes and Roots.
Supplemental Figure 5. Expression of POLAR in a GMC Differentiating without Asymmetric Divisions.
Supplemental Figure 6. T-DNA Insertion Lines and Sequence Comparison Coiled-Coil Domain of POLAR.
Supplemental Figure 7. Overrepresentation Analysis of Transient and Permanent Stem Cell Populations.
Supplemental Figure 8. POLAR Coexpression Network.
Supplemental Table 1. Genes Used for Production of GFP Reporter Constructs.
Supplemental Table 2. Genes Common to Transient and Permanent Stem Cell Populations.
Supplemental Table 3. T-DNA Insertion Lines Examined in This Study.
Supplemental Table 4. Primers Used for Transcript Amplification in RT-PCR and qRT-PCR.
Supplemental Table 5. Reporter and Overexpression Constructs Produced for This Study.
Supplemental Table 6. Primers Used for Cloning in This Study.
Supplemental Data Set 1. Genes Showing High Expression in scrm-D mute.
Supplemental Data Set 2. Genes Showing High Expression in scrm-D.
Supplemental Data Set 3. Genes Showing High Expression in spch.
Supplemental Data Set 4. Genes Significantly Deregulated in the Meristemoid-Enriched Background, scrm-D mute.
Supplemental Data Set 5. Genes Significantly Deregulated in the Stomata-Enriched Background, scrm-D.
Supplemental Data Set 6. Genes Significantly Deregulated in the Pavement Cell-Enriched Background, spch.
Supplemental Movie 1. Time-Lapse Imaging of POLAR Localization.
Supplemental Movie 2. Time-Lapse Imaging of POLAR Localization in basl.
Acknowledgments
We thank the ABRC and SIGnAL for providing clones and insertion lines used in this study; S. Vanneste, P. Doerner, J. Murray, D. Inzé, and L. de Vylder for kindly providing transgenic plants expressing various cell cycle GUS reporter lines; R. Heidstra for the SCZpro:CFP line; J. Nemhauser for advice on designing the microarray experiments; D. Bergmann for discussion about stomatal cell lineage profiling; and T. Kuroha and A. Rychel for assisting construction of reporter plasmids. The work was supported by the University of Washington Royalty Research Fund (RRF-4098), the National Science Foundation (MCB-0855659), and the Precursory Research for Embryonic Science and Technology award from the Japan Science and Technology Agency to K.U.T. L.J.P.’s research was in part supported by the startup funds from Western Washington University. K.M.P. is supported by the National Science Foundation Graduate Research Fellowship (DGE-0718124), and R.J.H. is supported by the Deutsche Forschungssgemeinschaft research fellowship. K.U.T. is a Howard Hughes Medical Institute–Gordon and Betty Moore Foundation investigator.
AUTHOR CONTRIBUTIONS
K.U.T. conceived and directed the project. L.J.P. conducted microarray experiments and analyzed the results with R.J.H. R.J.H. performed bioinformatic analysis. L.J.P. and K.M.P. generated expression constructs and Arabidopsis reporter lines and performed analysis with R.J.H. K.M.P. developed and performed time-lapse imaging. Thus, L.J.P., K.M.P., and R.J.H. made uniquely equal contributions. All authors contributed to writing the article.
References
- Abrash E.B., Bergmann D.C. (2009). Asymmetric cell divisions: A view from plant development. Dev. Cell 16: 783–796 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300 [Google Scholar]
- Bergmann D.C., Lukowitz W., Somerville C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Bergmann D.C., Sack F.D. (2007). Stomatal development. Annu. Rev. Plant Biol. 58: 163–181 [DOI] [PubMed] [Google Scholar]
- Borner G.H., Sherrier D.J., Stevens T.J., Arkin I.T., Dupree P. (2002). Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A genomic analysis. Plant Physiol. 129: 486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V., et al. (2009). CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol. 150: 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard P., Stetefeld J., Strelkov S.V. (2001). Coiled coils: A highly versatile protein folding motif. Trends Cell Biol. 11: 82–88 [DOI] [PubMed] [Google Scholar]
- Chung H.S., Howe G.A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Running M.P., Meyerowitz E.M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119: 397–418 [DOI] [PubMed] [Google Scholar]
- Day R.C., McNoe L., Macknight R.C. (2007). Evaluation of global RNA amplification and its use for high-throughput transcript analysis of laser-microdissected endosperm. Int. J. Plant Genomics 2007: 61028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Dinneny J.R., Long T.A., Wang J.Y., Jung J.W., Mace D., Pointer S., Barron C., Brady S.M., Schiefelbein J., Benfey P.N. (2008). Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Dong J., MacAlister C.A., Bergmann D.C. (2009). BASL controls asymmetric cell division in Arabidopsis. Cell 137: 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler Jde. A., et al. (2009). Systematic analysis of cell-cycle gene expression during Arabidopsis development. Plant J. 59: 645–660 [DOI] [PubMed] [Google Scholar]
- Fichelson P., Moch C., Ivanovitch K., Martin C., Sidor C.M., Lepesant J.A., Bellaiche Y., Huynh J.R. (2009). Live-imaging of single stem cells within their niche reveals that a U3snoRNP component segregates asymmetrically and is required for self-renewal in Drosophila. Nat. Cell Biol. 11: 685–693 [DOI] [PubMed] [Google Scholar]
- Fisher K., Turner S. (2007). PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr. Biol. 17: 1061–1066 [DOI] [PubMed] [Google Scholar]
- Geisler M., Nadeau J., Sack F.D. (2000). Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Kajita R., Torii K.U., Bergmann D.C., Kakimoto T. (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Yokoo T., Kajita R., Onishi T., Yahata S., Peterson K.M., Torii K.U., Kakimoto T. (2009). Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Harmer S.L., Hogenesch J.B., Straume M., Chang H.S., Han B., Zhu T., Wang X., Kreps J.A., Kay S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hord C.L., Sun Y.J., Pillitteri L.J., Torii K.U., Wang H., Zhang S., Ma H. (2008). Regulation of Arabidopsis early anther development by the mitogen-activated protein kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol. Plant 1: 645–658 [DOI] [PubMed] [Google Scholar]
- Hunt L., Gray J.E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19: 864–869 [DOI] [PubMed] [Google Scholar]
- Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., Tabata S., Shinozaki K., Kakimoto T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Iyer-Pascuzzi A.S., Benfey P.N. (2010). Fluorescence-activated cell sorting in plant developmental biology. Methods Mol. Biol. 655: 313–319 [DOI] [PubMed] [Google Scholar]
- Jia G., Liu X., Owen H.A., Zhao D. (2008). Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proc. Natl. Acad. Sci. USA 105: 2220–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J., Fiume E., Roeder A.H., Meng L., Sharma V.K., Osmont K.S., Baker C., Ha C.M., Meyerowitz E.M., Feldman L.J., Fletcher J.C. (2010). Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154: 1721–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka M.M., Pillitteri L.J., Fujii H., Yoshida Y., Bogenschutz N.L., Takabayashi J., Zhu J.K., Torii K.U. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell 20: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk N.M., Ceserani T., Tausta S.L., Sussex I.M., Nelson T.M. (2003). Laser capture microdissection of cells from plant tissues. Plant Physiol. 132: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., Yamada H., Mizuno T. (2002). Characterization of the ARR15 and ARR16 response regulators with special reference to the cytokinin signaling pathway mediated by the AHK4 histidine kinase in roots of Arabidopsis thaliana. Plant Cell Physiol. 43: 1059–1066 [DOI] [PubMed] [Google Scholar]
- Kondo Y., Hirakawa Y., Kieber J.J., Fukuda H. (2011). CLE peptides can negatively regulate protoxylem vessel formation via cytokinin signaling. Plant Cell Physiol. 52: 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono A., Umeda-Hara C., Adachi S., Nagata N., Konomi M., Nakagawa T., Uchimiya H., Umeda M. (2007). The Arabidopsis D-type cyclin CYCD4 controls cell division in the stomatal lineage of the hypocotyl epidermis. Plant Cell 19: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A., To J.P., Busch W., Stehling S., Kehle A., Demar M., Kieber J.J., Lohmann J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lohse M., et al. (2010). Robin: an intuitive wizard application for R-based expression microarray quality assessment and analysis. Plant Physiol. 153: 642–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. (1991). Predicting coiled coils from protein sequences. Science 252: 1162–1164 [DOI] [PubMed] [Google Scholar]
- MacAlister C.A., Ohashi-Ito K., Bergmann D.C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445: 537–540 [DOI] [PubMed] [Google Scholar]
- Menke F.L., Scheres B. (2009). Plant asymmetric cell division, vive la différence! Cell 137: 1189–1192 [DOI] [PubMed] [Google Scholar]
- Nakamura R.L., McKendree W.L., Jr., Hirsch R.E., Sedbrook J.C., Gaber R.F., Sussman M.R. (1995). Expression of an Arabidopsis potassium channel gene in guard cells. Plant Physiol. 109: 371–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Nakamura S., Tanaka K., Kawamukai M., Suzuki T., Nakamura K., Kimura T., Ishiguro S. (2008). Development of R4 gateway binary vectors (R4pGWB) enabling high-throughput promoter swapping for plant research. Biosci. Biotechnol. Biochem. 72: 624–629 [DOI] [PubMed] [Google Scholar]
- Nakazono M., Qiu F., Borsuk L.A., Schnable P.S. (2003). Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: Identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell 15: 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Obayashi T., Nishida K., Kasahara K., Kinoshita K. (2011). ATTED-II updates: Condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol. 52: 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor T.R., Dyreson C., Wyrick J.J. (2005). Athena: A resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Bergmann D.C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson K.M., Rychel A.L., Torii K.U. (2010). Out of the mouths of plants: The molecular basis of the evolution and diversity of stomatal development. Plant Cell 22: 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L.J., Bemis S.M., Shpak E.D., Torii K.U. (2007b). Haploinsufficiency after successive loss of signaling reveals a role for ERECTA-family genes in Arabidopsis ovule development. Development 134: 3099–3109 [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Bogenschutz N.L., Torii K.U. (2008). The bHLH protein, MUTE, controls differentiation of stomata and the hydathode pore in Arabidopsis. Plant Cell Physiol. 49: 934–943 [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Sloan D.B., Bogenschutz N.L., Torii K.U. (2007a). Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505 [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Torii K.U. (2007). Breaking the silence: Three bHLH proteins direct cell-fate decisions during stomatal development. Bioessays 29: 861–870 [DOI] [PubMed] [Google Scholar]
- Ren B., Liang Y., Deng Y., Chen Q., Zhang J., Yang X., Zuo J. (2009). Genome-wide comparative analysis of type-A Arabidopsis response regulator genes by overexpression studies reveals their diverse roles and regulatory mechanisms in cytokinin signaling. Cell Res. 19: 1178–1190 [DOI] [PubMed] [Google Scholar]
- Rowe M.H., Bergmann D.C. (2010). Complex signals for simple cells: The expanding ranks of signals and receptors guiding stomatal development. Curr. Opin. Plant Biol. 13: 548–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychel A.L., Peterson K.M., Torii K.U. (2010). Plant twitter: Ligands under 140 amino acids enforcing stomatal patterning. J. Plant Res. 123: 275–280 [DOI] [PubMed] [Google Scholar]
- Sachs T. (1991). Pattern Formation in Plant Tissues. (Cambridge, UK: Cambridge University Press) [Google Scholar]
- Saeed A.I., et al. (2003). TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sarkar A.K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Schachtman D.P., Schroeder J.I., Lucas W.J., Anderson J.A., Gaber R.F. (1992). Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science 258: 1654–1658 [DOI] [PubMed] [Google Scholar]
- Scheres B. (2002). Plant patterning: TRY to inhibit your neighbors. Curr. Biol. 12: R804–R806 [DOI] [PubMed] [Google Scholar]
- Schmülling T., Werner T., Riefler M., Krupková E., Bartrina y Manns I. (2003). Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J. Plant Res. 116: 241–252 [DOI] [PubMed] [Google Scholar]
- Sena G., Wang X., Liu H.Y., Hofhuis H., Birnbaum K.D. (2009). Organ regeneration does not require a functional stem cell niche in plants. Nature 457: 1150–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., Weigel D., Yanofsky M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20: 259–263 [DOI] [PubMed] [Google Scholar]
- Shpak E.D., Berthiaume C.T., Hill E.J., Torii K.U. (2004). Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131: 1491–1501 [DOI] [PubMed] [Google Scholar]
- Shpak E.D., Lakeman M.B., Torii K.U. (2003). Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA Leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 15: 1095–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E.D., McAbee J.M., Pillitteri L.J., Torii K.U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293 [DOI] [PubMed] [Google Scholar]
- Stolz A., Riefler M., Lomin S.N., Achazi K., Romanov G.A., Schmülling T. (2011). The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J. 67: 157–168 [DOI] [PubMed] [Google Scholar]
- ten Hove C.A., Willemsen V., de Vries W.J., van Dijken A., Scheres B., Heidstra R. (2010). SCHIZORIZA encodes a nuclear factor regulating asymmetry of stem cell divisions in the Arabidopsis root. Curr. Biol. 20: 452–457 [DOI] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Torii K.U., Mitsukawa N., Oosumi T., Matsuura Y., Yokoyama R., Whittier R.F., Komeda Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Steinhauser D., Gibon Y., Bläsing O.E., Redestig H., Sreenivasulu N., Krall L., Hannah M.A., Poree F., Fernie A.R., Stitt M. (2006). PageMan: An interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S., et al. (2011). Developmental regulation of CYCA2s contributes to tissue-specific proliferation in Arabidopsis. EMBO J. 30: 3430–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., Boruc J., De Jaeger G., Russinova E., De Veylder L. (2011). A kaleidoscopic view of the Arabidopsis core cell cycle interactome. Trends Plant Sci. 16: 141–150 [DOI] [PubMed] [Google Scholar]
- Werner T., Köllmer I., Bartrina I., Holst K., Schmülling T. (2006). New insights into the biology of cytokinin degradation. Plant Biol. 8: 371–381 [DOI] [PubMed] [Google Scholar]
- Yadav R., Girke T., Pasala S., Xie M., Reddy G.V. (2009). Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl. Acad. Sci. USA 106: 4941–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H., Suzuki T., Terada K., Takei K., Ishikawa K., Miwa K., Yamashino T., Mizuno T. (2001). The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 42: 1017–1023 [DOI] [PubMed] [Google Scholar]
- Zhao D.Z., Wang G.F., Speal B., Ma H. (2002). The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16: 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]