This study finds that two reticulon proteins modulate the transport of an immune receptor to the cell membrane. The results highlight the role of receptor secretion in determining the receptor’s forward signaling efficacy and the cell’s response.
Abstract
Receptors localized at the plasma membrane are critical for the recognition of pathogens. The molecular determinants that regulate receptor transport to the plasma membrane are poorly understood. In a screen for proteins that interact with the FLAGELIN-SENSITIVE2 (FLS2) receptor using Arabidopsis thaliana protein microarrays, we identified the reticulon-like protein RTNLB1. We showed that FLS2 interacts in vivo with both RTNLB1 and its homolog RTNLB2 and that a Ser-rich region in the N-terminal tail of RTNLB1 is critical for the interaction with FLS2. Transgenic plants that lack RTNLB1 and RTNLB2 (rtnlb1 rtnlb2) or overexpress RTNLB1 (RTNLB1ox) exhibit reduced activation of FLS2-dependent signaling and increased susceptibility to pathogens. In both rtnlb1 rtnlb2 and RTNLB1ox, FLS2 accumulation at the plasma membrane was significantly affected compared with the wild type. Transient overexpression of RTNLB1 led to FLS2 retention in the endoplasmic reticulum (ER) and affected FLS2 glycosylation but not FLS2 stability. Removal of the critical N-terminal Ser-rich region or either of the two Tyr-dependent sorting motifs from RTNLB1 causes partial reversion of the negative effects of excess RTNLB1 on FLS2 transport out of the ER and accumulation at the membrane. The results are consistent with a model whereby RTNLB1 and RTNLB2 regulate the transport of newly synthesized FLS2 to the plasma membrane.
INTRODUCTION
Immune receptors associated with the plasma membrane (PM) called pattern recognition receptors (PRRs) are elements of cellular surveillance that alert the immune system to the presence of pathogens (Boller and Felix, 2009; Tör et al., 2009). PRRs bind to conserved bacterial molecules called pathogen-associated molecular patterns (PAMPs). Upon PAMP binding, plant PRRs trigger a defense response called PAMP-triggered immunity (PTI) (Jones and Dangl, 2006; Tör et al., 2009). A well-known element of PTI is FLAGELLIN SENSITIVE2 (FLS2), identified as a receptor for bacterial flagellin in Arabidopsis thaliana (Gómez-Gómez and Boller, 2000, Gómez-Gómez et al., 2001; Zipfel et al., 2004; Chinchilla et al., 2006). Multiple components essential for the signal transduction events downstream of FLS2 are known, including FLS2 heterodimerization partners and mitogen-activated protein kinase (MAPK) cascades that induce expression of transcription factors and other genes critical for PTI (Chinchilla et al., 2007; Lu et al., 2010). Much less understood are the mechanisms that dictate the intracellular transport of PRRs and their effect on receptor activity (Carter et al., 2004; Jurgens, 2004; Rojo and Denecke, 2008).
Membrane-associated proteins are transported via the secretory (or anterograde) pathway from the endoplasmic reticulum (ER), where they undergo folding and initial posttranslational modifications through the Golgi apparatus and the trans-Golgi network for further maturation and then are transported to the PM (Hanton and Brandizzi, 2006; Robinson et al., 2007). Studies in animal models established critical roles for trafficking pathways in rendering receptors signaling competent (Wiley, 2003; Mitchell et al., 2004). Recent studies underscore the importance of plant receptor trafficking as a regulatory mechanism of receptor activity. PRRs, including FLS2, were shown to be N-glycosylated during transport from the ER to the PM, in order to become competent (Häweker et al., 2010; von Numers et al., 2010). Also, ER-localized proteins were found to participate in the maturation and quality control of EFR, the receptor for bacterial EF-Tu (Li et al., 2009b; Lu et al., 2009; Nekrasov et al., 2009). Currently, the molecular determinants of FLS2 intracellular transport are unknown.
Genetic redundancy and low cellular abundance of signaling components are two of the factors that may preclude the identification of direct substrates of plant receptors (Tang et al., 2008). Protein microarrays circumvent these obstacles and have contributed to understanding protein function (Popescu et al., 2007, 2009; Hu et al., 2009; Mok et al., 2010; Ray et al., 2010). Here, we screened Arabidopsis protein microarrays and identified RTNLB1 (for RETICULON-LIKE PROTEIN NON-METAZOAN GROUP B; Oertle et al., 2003) as a FLS2-interacting protein. We found that manipulation of the expression levels of RTNLB1 and its homolog RTNLB2 affected the intracellular trafficking of FLS2 and FLS2-induced signaling and immunity. Furthermore, we identified in RTNLB1 an N-terminal structural element and two intracellular sorting signals that are critical for RTNLB1 functions. Our results indicate that RTNLB1 and RTNLB2 modulate FLS2 immune activity through a mechanism that involves the control of the anterograde transport of FLS2.
RESULTS
RTNLB1 Interacts with the Kinase Domain of FLS2 on Arabidopsis Protein Microarrays
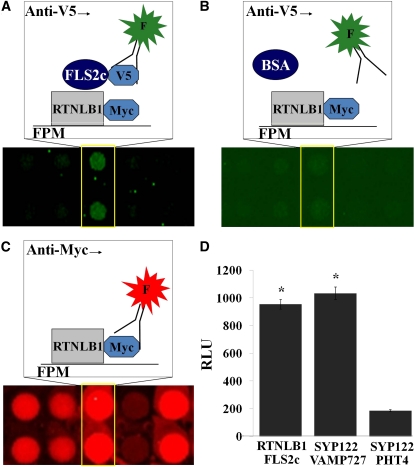
We performed a search for proteins that physically associate with FLS2 on functional protein microarrays with over 5000 Arabidopsis proteins (FPM-5000) (see Supplemental Figures 1A and 1B online). To generate recombinant proteins, we employed an extended Arabidopsis-tagged expression collection (Popescu et al., 2007) (see Supplemental Data Set 1 online). The protein microarrays were screened with the cytosolic portion of FLS2 (FLS2c) (Figures 1A to 1C) and over 50 FLS2c-interacting proteins were identified using a series of data processing steps (Popescu et al., 2009). The interactions between FLS2c and 10 candidate FLS2c-interacting proteins were further analyzed in Arabidopsis by split luciferase complementation using Renilla reniforms luciferase (Luc) vectors (Fujikawa and Kato, 2007). The confirmed FLS2c-interacting proteins included the reticulon-like protein RTNLB1 (Nziengui et al., 2007), also called BTI1 (VirB2-interacting protein 1) (Hwang and Gelvin, 2004). A high level of Luc activity was detected when RTNLB1-NLuc and FLS2c-CLuc were coexpressed, indicating a strong interaction (Figure 1D). FLS2c also interacted on the protein microarray with the receptor-like kinase PBS1, and the interaction was verified by split luciferase complementation (see Supplemental Figure 1C online). PBS1 was recently found to interact with FLS2 in the absence of flg22 (Zhang et al., 2010), confirming our results. The positive control, PM-localized SYP122 and endosomal VAMP727 (Kwon et al., 2008), showed high Luc activity. The negative control, SYP122 and PHT4 (Kato et al., 2010), had background levels of activity. The results indicate that RTNLB1 and FLS2 are part of the same protein complex and suggest a direct interaction.
Figure 1.
Identification of RTNLB1 as a Protein That Binds to the Cytosolic Kinase Domain of FLS2.
(A) to (C) Representative functional protein microarray (FPM) images showing interaction between the FLS2 cytosolic domain (FLS2c) and RTNLB1. The arrays were probed with recombinant FLS2c-V5 (A) or BSA (B) and an anti-V5 fluorophore-labeled antibody (F). The printed proteins were visualized with an anti-Myc fluorophore-labeled antibody (C).
(D) RTNLB1 interacts with FLS2c in Arabidopsis protoplasts. Average relative luminescence units (RLUs) and the associated standard errors are graphed (n = 12). The asterisks indicate significantly higher RLUs for RTNLB1/FLS2c and positive control SYP122/VAMP727 pairs relative to the negative control pair SYP122/PHT4 (Student’s t test).
[See online article for color version of this figure.]
RTNLB1 and Its Homolog RTNLB2 Contain Conserved Motifs for Protein–Protein Interaction and Intracellular Trafficking
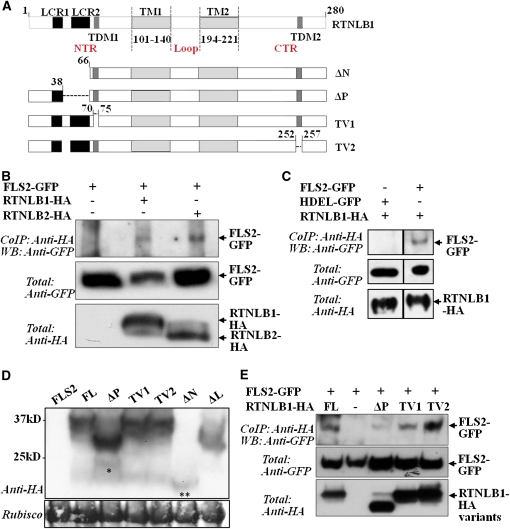
RTNL1 and RTNL2 contain two long, wedge-shaped transmembrane domains that divide the proteins into an N-terminal region, an intertransmembrane loop, and a C-terminal region (Nziengui et al., 2007; Sparkes et al., 2010). RTNLB1 has the highest sequence similarity to RTNLB2, followed by RTNL4/BTI3 and RTNL3. A search of motifs in the primary sequence of RTNLB1, using ELM (Puntervoll et al., 2003), identified two types of structural elements (Figure 2A; see Supplemental Figure 2 online). The first category is represented by two low-complexity regions (LCRs) (Coletta et al., 2010), LCR1 and LCR2. LCRs were found to have a tendency for establishing protein interactions (Tompa, 2002). Primary sequence alignment (Corpet, 1988) of RTNLB1 and related RTNLs revealed that the 17-residue-long LCR1 is present in the N-terminal regions of both RTNLB1 and RTNLB2 but not of RTNL3 or RTNL4. LCR2 is a Ser-rich region of 30 residues in RTNLB1. Notably, Ser-61 of LCR2 was found to be phosphorylated following flagellin elicitation (Benschop et al., 2007). LCR2 is four residues shorter in RTNLB2 and further truncated in RTNL4 and RTNL3. The second type of element identified in RTNLB1 and its homologs is represented by two putative Tyr-dependent trafficking motifs (TDMs), with TDM1 located close to LCR2 in the N-terminal region and TDM2 located in the C-terminal region. Thus, the putative protein interaction regions and trafficking motifs conserved in RTNLB1 and RTNLB2 suggest a potential model in which they function in intracellular trafficking through assembly of FLS2 protein complexes.
Figure 2.
The N Terminus of RTNLB1 Is Critical for Its in Vivo Association with FLS2.
(A) Structural elements identified in the amino acid sequence of RTNLB1: transmembrane domains (TM1 and 2), low complexity regions (LCR1 and 2), and Tyr-dependent intracellular motifs (TDM1 and 2). The RTNLB1 deletions ΔN, ΔP, TV1, and TV2 are shown.
(B) Immunoblot (WB) of coimmunoprecipitation (Co-IP) reactions of RTNLB1- or RTNLB2-HA with FLS2-GFP.
(C) HDEL-GFP does not coimmunoprecipitate with FLS2-GFP. The Co-IPs were done in parallel and run on the same gel.
(D) RTNLB1 full-length (FL) and variants accumulate normally in plants. Immunoblot of RTNLB1 and variants transiently expressed in N. benthamiana. The single asterisk indicates a possible degradation product of ΔP. The double asterisk indicates the position of ΔN. The Ponceau-stained blot shows equal loading. Rubisco, ribulose-1,5-bis-phosphate carboxylase/oxygenase.
(E) Immunoblot of coimmunoprecipitates showing that deletion of the LCR2 from RTNLB1 negatively impacts its interaction with FLS2. The coimmunoprecipitates were repeated at least three times with consistent results. “Total” represents the input amount of proteins as shown by probing the total protein extract with the indicated antibodies.
[See online article for color version of this figure.]
Both RTNLB1 and RTNLB2 Interact with FLS2 in Planta
To investigate further the possible relationship between RTNLB1, RTNLB2, and FLS2, we performed coimmunoprecipitation assays of RTNLB1-HA or RTNLB2-HA and FLS2–green fluorescent protein (GFP) fusions coexpressed in Nicotiana benthamiana. C-terminal tagged FLS2, RTNLB1, and RTNLB2 were previously shown to remain functional (Robatzek et al., 2006; Tolley et al., 2008; Sparkes et al., 2010). RTNLB1- and RTNLB2-HA were purified from crude extracts using anti-HA antibody. Coimmunoprecipitated FLS2-GFP was detected using anti-GFP antibody. FLS2-GFP was detected in both RTNLB1- and the RTNLB2-HA complexes immunoprecipitated with anti-HA antibody (Figure 2B). The negative control HDEL-GFP failed to immunoprecipitate RTNLB1-HA (Figure 2C). Furthermore, we analyzed the interaction between RTNLB1 and EFR, a PRR structurally related to FLS2. RTNLB1-HA coimunoprecipitated with FLS2-FLAG but failed to coimmunoprecipitate with EFR-FLAG or GFP-FLAG, indicating that EFR is not a likely in vivo partner of RTNLB1 (see Supplemental Figure 3 online). We conclude that RTNLB1 and RTNLB2 physically associate with full-length FLS2 and suggest that the interaction is specific.
The Ser-Rich LCR2 Contributes to RTNLB1 Interaction with FLS2
RTNLB1 was shown to adopt a membrane spanning topology in which the N-terminal region, the loop region, and C-terminal region reside on the cytosolic side of the ER membrane (Sparkes et al., 2010). Thus, these regions are available for interaction with other cytosolic proteins. Among reticulon proteins, the N-terminal regions are most divergent and a source of functional diversification through interaction with substrates (Oertle et al., 2003). To test the requirement of the N-terminal region and its structural elements for the interaction with FLS2, we generated two variants, ΔN lacking LCR1 and LCR2, and ΔP lacking LCR2. To analyze the relative importance of the TDMs for RTNLB1 function, we generated TV1 and TV2, mutants lacking the first or the second putative TDM, respectively (Figure 2A). ΔP, TV1, and TV2 were expressed normally in plants; ΔN accumulated only in low amounts, suggesting that LCR1 contributes to RTNLB1 stability (Figure 2D). Coimmunoprecipitation of FLS2-GFP with full-length RTNLB1 or deletion variants revealed that a much smaller amount of FLS2 was precipitated by ΔP compared with RTNLB1 (Figure 2E). Both TV1 and TV2 coimmunoprecipitated FLS2 in similar amounts to RTNLB1. We conclude that LCR2 is necessary but not solely sufficient for the interaction of RTNLB1 with FLS2. Additional RTNLB1 regions may contribute to binding FLS2, similar to the interaction between the human RTN4 and NogoR receptor involving both the N-terminal region and the transmembrane loop (Hu et al., 2005). An RTNLB1 mutant without the loop (ΔL) did not express well and migrated aberrantly in SDS-PAGE (Figure 2D), precluding its analysis.
PAMP-Dependent Signaling Is Obstructed in rtnlb1 rtnlb2 Plants
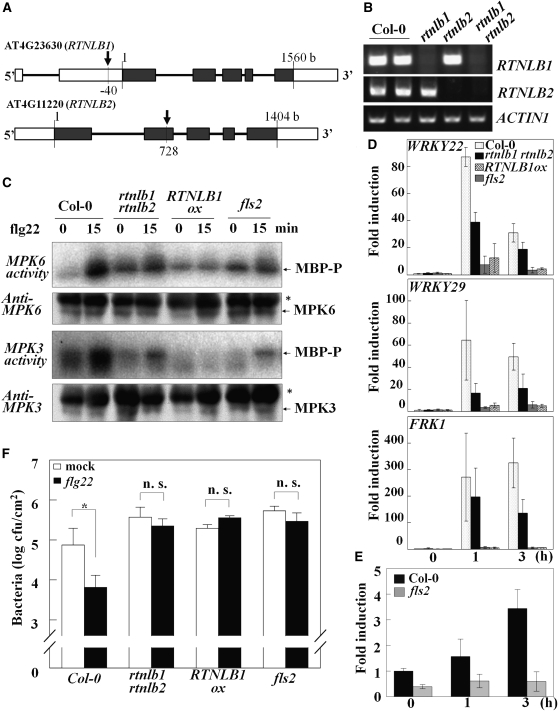
To test the putative roles of RTNLB1 and RTNLB2 in FLS2-mediated activation of signaling pathways, we developed transgenic plants to test their responses to the flagellin 22 peptide (flg22). The homozygous T-DNA insertion mutants rtnlb1 and rtnlb2 generated from heterozygous stocks were crossed to obtain rtnlb1 rtnlb2 lines (Figures 3A and 3B). No obvious growth defects were detected in rtnlb1 rtnlb2 (see Supplemental Figure 4 online). Analysis of the flg22-treated mutants revealed that the activity of endogenous MPK3 and MPK6 was reduced in rtnlb1 rtnlb2 (Figure 3C), and transcriptional induction of the early PTI markers was impaired compared with the wild type (Figure 3D) The rtnlb1 and rtnln2 homozygous lines also exhibited impaired FLS2-dependent signaling but to a lower extent than the rtnlb1 rtnlb2 lines (see Supplemental Figure 5 online). A null fls2 mutant was used as control (Navarro et al., 2004). We also tested EFR-dependent signaling in elf18-treated rtnlb1 rtnlb2. Interestingly, rtnlb1 rtnlb2 showed moderately impaired marker gene activation compared with the wild type, indicative of defective EFR signaling (see Supplemental Figure 6 online). Next, we measured RTNLB1 transcript accumulation and found a threefold increase in RTNLB1 transcript at 3 h after flg22 elicitation in control but not in fls2 (Figure 3E). This indicates that RTNLB1 is induced during PTI in an FLS2-dependent manner, supporting a role for RTNLB1 in PTI. We conclude that both RTNLB1 and RTNLB2 are necessary for the PAMP-triggered activation of MAPKs and PTI early marker genes.
Figure 3.
RTNLB1 Is a Positive Regulator of PTI.
(A) Schematic representation of RTNLB1 and RTNLB2 with exons shown as black boxes, 5′ and 3′ untranslated regions as white boxes, and the sites of T-DNA insertion indicated by arrows.
(B) RTNLB1 and RTNLB2 transcript levels in rtnlb1, rtnlb2, and rtnlb1 rtnlb2 mutants measured by RT-PCR. Columbia-0, Col-0.
(C) Defective activation of MPK3 and MPK6 in transgenic lines treated with flg22. The kinase activity of immunoprecipitated MAPKs was assayed using myelin basic protein (MBP). Phosphorylated MBP (MBP-P) was detected by radiography. The asterisk shows position of the IgG heavy chain.
(D) The induction of WRKY22, WRKY29, and FRK1 is impaired in rtnlb1 rtnlb2 and RTNLB1ox at 1 and 3 h after flg22 treatments but not in controls. Standard deviation bars are shown.
(E) RTNLB1 transcription is enhanced after flg22 treatment in the wild type (Columbia-0) but not in fls2. Average data from three real-time PCR replicates and the standard deviations are graphed.
(F) rtnlb1 rtnlb2 and RTNLB1ox overexpression lines pretreated with flg22 are more susceptible to P. syringae pv tomato DC3000 than the wild type. The asterisk indicates significant differences between mock- and flg22-treated samples (Student’s t test). cfu, colony-forming units; n. s., not statistically significant.
Excess RTNLB1 Suppresses FLS2-Triggered Signaling
To examine the effects of RTNLB1 gain of function on FLS2, we analyzed flg22 responses in mutants with ectopic RTNLB1 expression (RTNLB1ox) that were shown to accumulate higher-than-wild type levels of RTNLB1 (Hwang and Gelvin, 2004). RTNLB1ox plants exhibited a normal phenotype, indicating that excess RTNLB1 does not markedly alter development (see Supplemental Figure 4 online). However, RTNLB1ox displayed severe impairment in the activation of MAPKs and marker expression at similar levels to fls2 (Figures 3C and 3D). Overall, the results indicate that excess RTNLB1 is detrimental to the FLS2 immune activity.
Flg22-Induced Pathogen Resistance Is Reduced in rtnlb1 rtnlb2 and Abolished in RTNLB1ox
Defects in MAPK activation dynamics and gene expression following flg22 treatment of RTNLB1 and RTNLB2 loss- or gain-of-function mutants indicate lowered PTI. To test this, we pretreated plants with flg22, infected them with Pseudomonas syringae pv tomato DC3000 (Pst), and quantified the FLS2-dependent resistance by measuring bacterial multiplication (Göhre et al., 2008). Control flg22-treated plants showed higher resistance to Pst compared with mock plants. On the other hand, no significant differences were found in bacterial growth in mock-treated versus flg22-treated rtnlb1 rtnlb2, RTNLB1ox, or, as expected, fls2 (Figure 3F). Pst multiplied to similar levels in rtnl1 rtnl2, RTNLox, and fls2 lines, indicating pathogen susceptibility. Thus, RTNLB1 and RTNLB2 are positive regulators of flg22-activated PTI against Pst.
FLS2 Transport to the PM Is Impaired in rtnlb1 rtnlb2
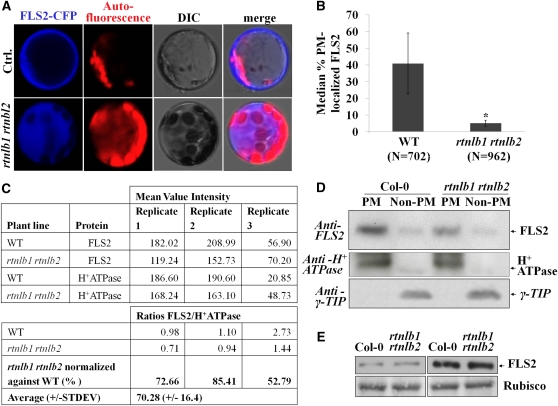
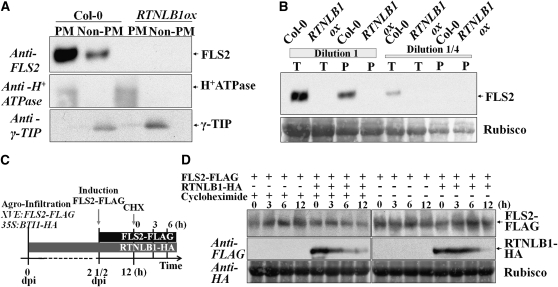
As FLS2 does not possess known trafficking signals (Geldner and Robatzek, 2008), interactions with components that link FLS2 to transport pathways may be critical. To test if RTNLB1 and RTNLB2 are able to regulate FLS2 transport to the PM, we first monitored the localization of an FLS2–cyan fluorescent protein (CFP) fusion in protoplasts. In the wild type, FLS2-CFP localized to PM as previously shown (Ali et al., 2007). In rtnlb1 rtnlb2 protoplasts, FLS2-CFP fluorescence was detected in internal structures reminiscent of the modified ER structures found in reticulon-depleted yeast cells (Voeltz et al., 2006) (Figures 4A and 4B), indicating defective FLS2 intracellular localization. Next, we examined the localization of endogenous FLS2 in rtnlb1 rtnlb2 lines. Total protein extracts were subjected to two-phase partitioning to separate the PM and non-PM fractions, followed by FLS2 detection with specific Ab. In rtnlb1 rtnlb2, consistently less FLS2 was detected in the PM fraction compared with the wild type (Figures 4C and 4D). FLS2 was not completely absent from the PM, suggesting a tight regulation of FLS2 secretion and functional redundancy. A PM-localized H+ATPase (Page et al., 2009) and the tonoplast marker γ-TIP (Fleurat-Lessard et al., 1997; Nelson et al., 2007) partitioned with the expected fractions and confirmed enrichment in membrane and endosomal proteins, respectively, and the lack of fraction cross-contamination. The amounts of markers separated in fractions from the wild type and mutants were similar, indicating that their transport was not obviously perturbed. We also measured the steady state amount of FLS2 in rtnlb1 rtnlb2. FLS2 accumulated in similar amounts in wild-type and rtnlb1 rtnlb2 lines (Figure 4E). This indicates that reduced FLS2 accumulation at the PM in the mutant is not a consequence of lower overall FLS2 levels. The data indicate that RTNLB1 and RTNLB2 specifically regulate FLS2 transport and accumulation at the PM.
Figure 4.
FLS2 Accumulation at the PM Is Impaired in rtnlb1 rtnlb2 Mutants.
(A) and (B) Aberrant localization of FLS2-CFP in rtnlb1 rtnlb2 protoplasts.
(A) FLS2-CFP localizes at the PM in the wild type (Ctrl.) but not rtnlb1 rtnlb2.
(B) For quantification, the number of protoplasts showing PM-localized FLS2-CFP was counted in electronic images. N is the number of total protoplasts counted for each line. The P value is shown above the column. The asterisk shows a statistically significant difference (Student’s t test). WT, wild type.
(C) Quantification of three biological replicates of phase partitioning. Total extracts from wild-type or mutant plants separated proteins in PM and non-PM fractions. Band intensities were measured using QuantityOne. The PM/non-PM ratios were calculated and normalized against the wild-type control.
(D) Representative blot showing reduced accumulation of FLS2 at the PM in rtnlb1 rtnlb2. The PM (H+-ATPase) and tonoplast γ-TIP markers showed the expected patterns.
(E) Immunoblot showing steady state amounts of FLS2 in total protein extracts from the wild type and mutants; the blot was stained with Ponceau, and Rubisco is shown as loading control.
RTNLB1ox Lines Do Not Accumulate FLS2
The degree of impairment in the signaling flux downstream of FLS2 in RTNLB1ox mutants was similar to the null fls2, suggesting a drastic effect on FLS2 trafficking and accumulation at the PM. We analyzed FLS2 levels in protein extracts subjected to two-phase partitioning from RTNLB1ox plants. We found that FLS2 amount was below the limit of detection by immunoblots in both PM and non-PM fractions (Figure 5A). Moreover, we could not detect FLS2 in the total protein extracts from RTNLB1ox (Figure 5B). Thus, RTNLB1ox contains undetectable amounts of FLS2, consistent with its fls2-like phenotype in immune assays. Accumulation of protein in the ER has been previously linked to activation of the ER protein degradation pathways (Liu and Howell, 2010). Our data argue that RTNLB1ox represents an extreme case of impaired FLS2 trafficking where FLS2 is degraded via an ER-dependent mechanism.
Figure 5.
Effects of Excess RTNLB1 on FLS2 Expression and Stability.
(A) Representative blot showing that FLS2 is undetectable in RTNLB1ox. The PM (H+-ATPase) and tonoplast γ-TIP separate into the expected fractions, the PM and non-PM fraction, respectively.
(B) Immunoblot to visualize the amount of FLS2 in total protein extracts from the wild type and RTNLB1ox mutants; undiluted (dilution 1 ≥ 2 μg/μL) and diluted (dilution 1/4) extracts were used. T represents the total protein fraction extracted using the same buffer used in the two-phase fractionation in (A); P represents the total protein extracted with the same buffer but containing 0.5% Triton X, from the pellet left after extraction of T. The blot was stained with Coomassie blue, and Rubisco is shown as a loading control.
(C) Schematics of the protein expression and CHX treatment of plants. dpi, days postinfiltration.
(D) FLS2 stability in the presence of RTNLB1. Samples were collected before CHX treatment and at 3 and 6 h after treatment from plants expressing RTNLB1-HA and/or FLS2-FLAG. The blot was stained with Coomassie blue; Rubisco is shown as an internal control.
RTNLB1 Does Not Directly Influence FLS2 Stability
To verify a possible direct effect of RTNLB1 on the stability of FLS2 we evaluated the accumulation of an inducible FLS2-FLAG, coexpressed with a constitutive RTNLB1-HA, at various time points after treatment with the protein synthesis inhibitor cycloheximide (CHX) (Figure 5C). In the CHX-treated samples, we could not detect major differences in FLS2-FLAG levels when RTNLB1-HA was present compared with samples expressing only FLS2-FLAG (Figure 5D). Irrespective of the presence or absence FLS2-FLAG, RTNLB1-HA levels remained constant at the 3- and 6-h time points. We also evaluated FLS2-FLAG levels in samples not treated with CHX (Figure 5D). Similarly, no considerable variations were found in FLS2-FLAG levels when expressed in the presence versus absence of RTNLB1-HA. Interestingly, evaluation of RTNLB1-HA accumulation in plants coinfiltrated with FLS2-FLAG revealed that RTNLB1-HA levels started decreasing very fast (3 h) and further decreased in a time-dependent manner after CHX treatment, pointing toward high RTNLB1 instability. In the presence of FLS2-FLAG and without CHX, RTNLB1-HA levels were high and remained constant at the equivalent time points, ruling out a possible negative effect of FLS2 on RTNLB1 stability. Taken together, the data show that RTNLB1-HA does not directly influence FLS2 stability. On the contrary, in this transient expression system, FLS2-FLAG seems to possess a lower degradation rate and/or higher protein stability compared with RTNLB1-HA.
Excess RTNLB1 Promotes FLS2 Sequestration in the ER
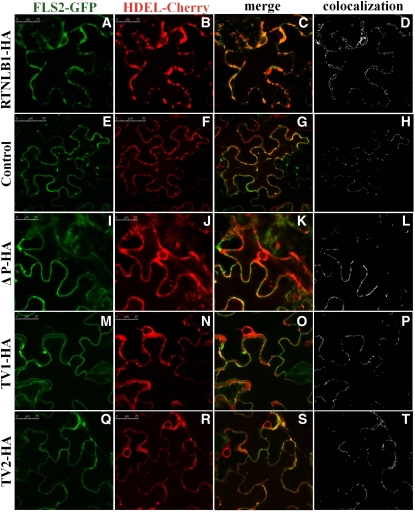
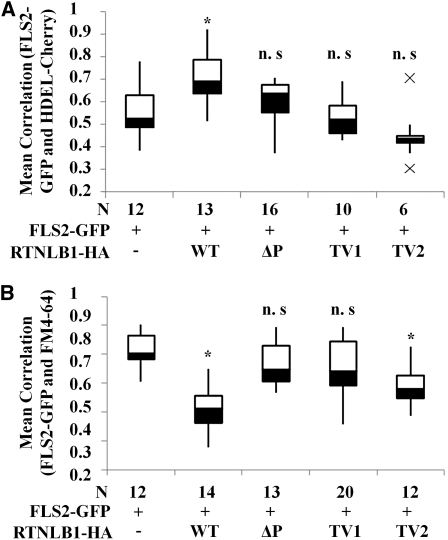
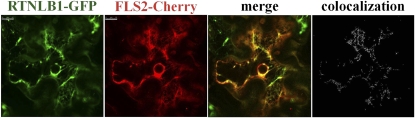
Our data strongly suggest that RTNLB1 and RTNLB2 regulate FLS2 secretion to the PM. Human reticulons are known to interact with and regulate the transport out of the ER of various PM-associated proteins (Wakana et al., 2005; Liu et al., 2008; Chang et al., 2009). To investigate further the role of RTNLB1 and RTNLB2 in FLS2 anterograde transport, we took advantage of the strong effects that excess RTNLB1 has on FLS2 activity. The intracellular distribution of an inducible FLS2-GFP was examined in the presence of RTNLB1- or RTNLB2-HA by transient expression in N. benthamiana. FLS2-GFP exhibited a predominantly PM localization, as previously shown (Robatzek et al., 2006), indicating that FLS2-GFP is rapidly transported to the PM. Coexpression of FLS2-GFP with RTNLB1- or RTNLB2-HA resulted in a dramatic shift in the distribution of fluorescence from the PM to intracellular organelles (see Supplemental Figures 7 and 8 online). To reveal the identity of these intracellular compartments, we coexpressed FLS2-GFP and RTNLB1-HA with the ER marker HDEL-Cherry. When RTNLB1 was present, most FLS2-GFP was trapped in enlarged ER bodies in close association with the PM, representing the PM-adjacent cortical ER domain (pmaER) (Figures 6A to 6D; see Supplemental Figure 9 online). The pmaER is tightly associated to the PM and was previously linked to the synthesis of membrane proteins and their release into the secretion pathway (Park and Blackstone, 2010). FLS2-GFP was also detected in the perinuclear (pn) ER domain. In the absence of RTNLB1, FLS2-GFP preferentially colocalized with HDEL-Cherry in the pmaER, visible as punctiform structures in the close vicinity or overlapping the PM (Figures 6E to 6H; see Supplemental Figure 10 online). Interestingly, SERK1 was also found to localize in the pmaER domain (Aker et al., 2006). We did not detect significant accumulation of FLS2-GFP in the ER reticulate network or in the pnER, where another leucin-rich repeat–receptor-like kinase, XA21, was found to localize (Park et al., 2010). Quantification of the degree of colocalization between FLS2-GFP and the ER marker indicated a significant difference in FLS2 intracellular distribution in plants coexpressing RTNLB1 versus controls (Figure 7A; see Supplemental Data Set 2 online). Thus, overexpression of RTNLB1 and RTNLB2 inhibits FLS2 transport out of the ER.
Figure 6.
Effect of RTNLB1 and Its Deletion Variants on the Transport of FLS2 out of ER in N. benthamiana.
(A) to (D) In the presence of excess RTNLB1-HA, FLS2-GFP (green) colocalizes mostly with the ER marker HDEL-mCherry (red).
(E) to (H) In the absence of RTNLB1, FLS2-GFP partially colocalizes with HDEL-Cherry.
(I) to (T) Coexpression of FLS2-GFP with ΔP ([I] to [L]), TV1 ([M] to [P]), or TV2 ([Q] to [T]), decreases the accumulation of FLS2-GFP in ER. Bars = 25 μm for all, except the control image in which it is 50 μm. Colocalization images were generated using Image-Pro Plus.
Figure 7.
Quantification of the RTNLB1 Effect on FLS2 Anterograde Transport.
Box plots representing the quantification of the colocalization degree between FLS2-GFP and the PM dye FM4-64 (A) or the ER marker HDEL-Cherry (B). Pearson correlation coefficients were calculated using Image-Pro Plus. N is the number of images analyzed for each combination of markers. The box plots show in black the distance from quartile 1 to the median; the distance from the median to quartile 2 is shown in white. The whiskers extend to the smallest or largest nonoutlier, respectively, in the data set. The asterisk indicates statistically significant differences from control (Student’s t test). WT, wild type.
Excess RTNLB1 Blocks FLS2 Accumulation at the PM
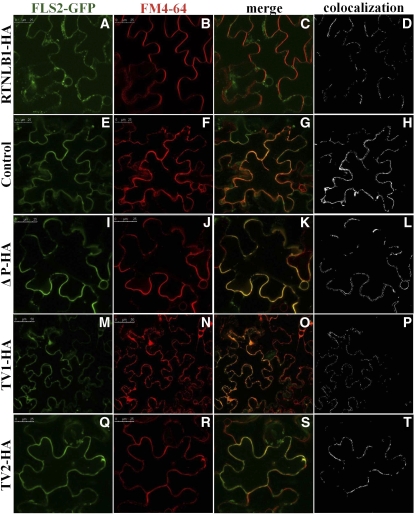
A block in the ER exit of FLS2-GFP in the presence of excess RTNLB1 would result in reduced accumulation of newly synthesized FLS2-GFP at the PM. To examine this possible consequence of excess RTNL1, we colocalized FLS2-GFP with the PM dye FM4-64. In the presence of RTNLB1, FLS2-GFP fluorescence at the PM was very low or absent as shown by colocalization with FM4-64 (Figures 8A to 8D). In the absence of RTNL1, as expected, the fluorescence detected at the PM was much higher and FLS2-GFP efficiently colocalized with FM4-64 (Figures 8E to 8H). Quantification of the degree of colocalization between FLS2-GFP and the PM marker in the presence or absence of RTNLB1 indicated statistically significant differences (Figure 7B). To test the specificity of RTNLB1 effects on the anterograde trafficking, we analyzed the intracellular localization of a known PM-anchored protein, CaM53 (GFP-BDCaM53) (Rodríguez-Concepcíon et al., 1999; Kato et al., 2008). GFP-BDCaM53 colocalized efficiently with FM4-64 both in the presence or absence of RTNLB1, indicating that RTNLB1 did not disrupt its transport to the PM (see Supplemental Figure 11 online). Taken together, our data clearly show that RTNLB1 regulates FLS2 anterograde transport.
Figure 8.
Effect of RTNLB1 and Its Deletion Variants on the Accumulation of FLS2 at the PM in N. benthamiana.
(A) to (D) FLS2-GFP (green) is absent from the PM when coexpressed with full-length RTNLB1-HA and the PM dye FM4-64 (red).
(E) to (H) In the absence of RTNLB1, FLS2-GFP colocalizes with FM4-64.
(I) to (T) Coexpression of FLS2-GFP with ΔP ([I] to [L]), TV1 ([M] to [P]), or TV2 ([Q] to [T]) partially increases the extent of FLS2-GFP colocalization with FM4-64. Bars = 25 μm for all except TV1-HA images, in which it is 50 μm. Colocalization images were generated with Image-Pro Plus.
The LCR2 Ser-Rich Element and the Tyr-Dependent Sorting Motifs Are Critical for the Negative Effects of Excess RTNLB1 on FLS2 Transport out of the ER
To understand further the functions of the RTNLB1 structural elements, we examined FLS2-GFP intracellular distribution in the presence of RTNLB1 variants ΔP, TV1, and TV2. The results showed that removal of LCR2, TDM1, or TDM2 suppressed to various degrees RTNLB1’s ability to block FLS2 transport and to induce accumulation of FLS2-GFP in the ER, as shown by colocalization with HDEL-Cherry. Among all the RTNL1 structural elements analyzed, LCR2, when removed, had the lowest impact on the negative effects of excess RTNLB1 on FLS2-GFP exit from the ER (Figures 6I to 6T; see Supplemental Figure 7 online). Notably, when coexpressed with individual RTNLB1 variants, FLS2-GFP was mostly found in the peripheral pmaER but not in the other ER domains, such as the pnER,where FLS2 was found to accumulate when coexpressed with full-length RTNLB1. We quantified the extent of FLS2-GFP colocalization with HDEL-Cherry in the presence versus the absence (control samples) of RTNLB1 mutants (see Supplemental Data Set 2 online). The differences in the ER accumulation of FLS2-GFP when coexpressed with ΔP, TV1, or TV2 versus the controls were not statistically significant (Figure 7A). These results indicate that LCR2 and both TDMs are required the inhibition of FLS2-GFP exit out of the ER by excess RTNLB1.
LCR2 and the TDM1 Sorting Motif Are Necessary for RTNLB1 Effects on FLS2 Accumulation at the PM
Lower FLS2-GFP accumulation in the ER in the presence of RTNLB1 variants suggests a partially restored FLS2-GFP functional anterograde transport. Indeed, FLS2-GFP accumulated at the PM in higher amounts when coexpressed with RTNLB1 variants than when coexpressed with full-length RTNLB1, as shown by colocalization with FM4-64 (Figures 8I to 8T). The degree of colocalization between FLS2-GFP and FM4-64 was quantified (see Supplemental Data Set 2 online). The data showed that in the presence of ΔP or TV1, FLS2-GFP accumulated at the PM in similar amounts to controls (Figure 7B). Interestingly, when TV2 was coexpressed with FLS2-GFP, although the plants showed control levels of FLS2-GFP fluorescence in the ER (Figure 7B), the accumulation of FLS2-GFP at the PM was significantly inhibited, suggesting that TDM1 and TDM2 perform distinct roles. These data suggest that LCR2 and TDM1 regions of RTNLB1 are important for regulating FLS2 transport to the PM.
RTNLB1 and FLS2 Colocalize in the ER
To investigate the cellular compartment where the interaction between RTNLB1 and FLS2 may occur, we coexpressed RTNLB1-GFP and an estradiol-inducible FLS2-Cherry. The fusion proteins colocalized in an intracellular compartment morphologically similar to the ER. Colocalization signals were high in the PM-adjacent pmaER and the pnER but reduced in the tubular ER structures (Figure 9; see Supplemental Figure 12 online). The average colocalization coefficient was 0.74 ± 0.04 (see Supplemental Data Set 3 online). RTNLB1-GFP coexpressed only with HDEL-Cherry was found localized in the ER reticulate network, as previously shown (Sparkes et al., 2010), and in the peripheral pmaER, similar to the yeast RTN1 (Schuck et al., 2009) (Figures 10A to 10D; see Supplemental Figure 13 online). These results indicate a direct association between RTNLB1 and FLS2 in the ER.
Figure 9.
RTNLB1-GFP and FLS2-Cherry Colocalize in the ER.
Coexpressed RTNLB-GFP and FLS2-Cherry colocalize in the ER as shown by the merged and colocalization images. The colocalization image was generated using Image-Pro Plus. Bars = 10 μm.
Figure 10.
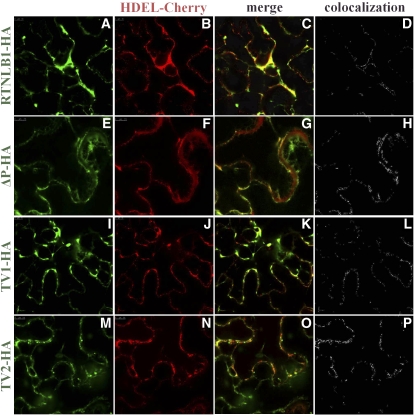
RTNLB1-GFP Variants Are Localized in the ER.
Full-length RTNLB1-GFP (A) and the RTNLB1 variants ΔP-GFP (E), TV1-GFP (I), and TV2-GFP (M) were coexpressed with HDEL-Cherry ([B], [F], [J], and [N]). All RTNLB1 fusions were localized in the ER, as shown in the merged images ([C], [G], [K], and [O]) and the colocalization analysis ([D], [H], [L], and [P]). Bars = 7.5 μm (ΔP and TV2) or 10 μm (RTNLB1-GFP and TV1). Colocalization images were generated with Image-Pro Plus.
RTNLB1 Variants Are Localized in the ER
RTNLB1 and its homologs were previously localized in the ER tubules, edges of ER cisternae, and pnER (Sparkes et al., 2010). To test whether deletion of TDMs or LCR2 modified the localization of RTNLB1 variants, we coexpressed ΔP-, TV1-, or TV2-GFP with HDEL-Cherry. We found that all RTNLB1 variants colocalized with the ER marker (Figures 10E to 10P; see Supplemental Figure 13 online). The average colocalization coefficients were 0.52 ± 0.1, 0.75 ± 0.02, and 0.67 ± 0.1, respectively (see Supplemental Data Set 4 online). These data indicate that deletion of the structural domains analyzed did not interfere with RTNLB1 localization in the ER.
RTNLB1 Overexpression Inhibits FLS2 Glycosylation
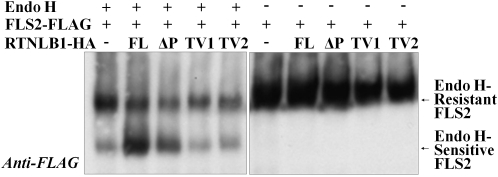
To investigate further the hypothesis that in the presence of excess RTNLB1 the transport of FLS2 is blocked at the ER-to-Golgi step, we tested the maturation status of FLS2. An FLS2-FLAG fusion expressed with or without RTNLB1 or its variants was purified and treated with Endoglycosidase H (Endo H) (Figure 11). Endo H treatment distinguishes between the Endo H–sensitive fraction from the ER and the Endo H–resistant fraction from the PM (Maley et al., 1989). If FLS2 traffic is inhibited upstream of the Golgi, the Endo H–sensitive FLS2-FLAG would be expected to accumulate at the expense of the Endo H–resistant form. When FLS2-FLAG was expressed alone, it was efficiently glycosylated, as shown by accumulation of Endo H–resistant FLS2-FLAG over the sensitive fraction. FLS2-FLAG coexpression with TV1 or TV2 also produced more Endo H–resistant than –sensitive FLS2-FLAG, indicative of control levels of FLS2-FLAG glycosylation. When RTNLB1 or ΔP was coexpressed with FLS2-FLAG, the sensitive fraction exceeded in abundance the resistant fraction, indicating defective glycosylation. The negative effects of RTNLB1 and its variants on FLS2-FLAG glycosylation correlated with their ability to block or partially interfere with FLS2 transport out of the ER (Figure 6), a strong indication that RTNL1 regulates FLS2 transport to the membrane. The data also suggest a strong relationship between the FLS2 transport pathway regulated by RTNLB1/2 and the FLS2 maturation process.
Figure 11.
Effect of RTNLB1 on the Maturation of FLS2.
Immunoblot of FLS2-FLAG precipitated with anti-FLAG and treated with Endo H. In controls (samples without full-length RTNLB1 or variants), Endo H–resistant FLS2-FLAG is more abundant than the Endo H–sensitive form. Removal of the Tyr signals but not LCR2 from RTNLB1 causes an increase in Endo H–resistant over Endo H–sensitive FLS2-FLAG.
DISCUSSION
RTNLB1 and RTNLB2 Are Components of the FLS2 Secretory Pathway
The main finding of this study is that the Arabidopsis reticulon-like RTNLB1 and RTNLB2 are previously unknown regulators of FLS2 immune activity and critical components in FLS2 transport to the PM. Several data strongly support this conclusion: (1) RTNLB1 and RTNLB2 interacted with FLS2 in vivo; (2) RTNLB1 and FLS2 have overlapping intracellular localization patterns and colocalize in the ER; (3) in the presence of excess RTNLB1, FLS2, but not another PM-associated protein, accumulated in the ER domain adjacent to the PM, indicating a block in the ER-to-Golgi anterograde transport specific to FLS2; (4) deletion of the TDMs or LCR2 from RTNLB1 relieved the block in ER export and had a positive effect on the PM accumulation of FLS2; (5) consistent with (4), in the presence of RTNLB1, FLS2 accumulated in the Endo H–sensitive form, strongly indicating that newly produced FLS2 did not reach the medial Golgi compartment, whereas higher levels of the FLS2 Endo H–resistant form accumulated in the presence of excess RTNLB1 variants; (6) RTNLB1 does not interfere directly with the stability of FLS2. Our data combined with previous observations by Sparkes et al. (2010) support a model in which RTNLB1 and RTNLB2 localized in the ER membranes interact with the cytosolic domain of newly synthesized FLS2 to facilitate its transport to the PM. RTNLB1 and RTNLB2 are known to interact through their reticulon domains (Hwang and Gelvin, 2004; Sparkes et al., 2010); thus, they may function as a heterodimer or as part of a larger protein complex to regulate FLS2 secretion. Furthermore, considering the role of the human RTNs in intracellular protein trafficking (Wakana et al., 2005; He et al., 2006; Liu et al., 2008; Chang et al., 2009; Shi et al., 2009; Yang et al., 2009), our findings indicate functional conservation between RTNs and plant RTNLBs.
We demonstrated that FLS2-activated signaling and immunity are dependent on efficient FLS2 transport to the PM. In rtnlb1 rtnlb2 lines, reduced FLS2 accumulation at the PM correlated with defective FLS2 signaling and lowered PTI. Interestingly, FLS2-dependent signaling was not abolished in rtnlb1 rtnlb2, indicating the presence of compensatory factors. In this regard, RTNLB1ox lines, with no detectable FLS2 and an fls2 immune phenotype, represent an extreme case. Previous studies showed that a lower concentration of receptors, including FLS2, at the PM was strongly correlated with weak outcomes of the downstream signaling (De Smet et al., 2009; Boutrot et al., 2010; Mersmann et al., 2010). Our findings are consistent with these studies and further indicate that FLS2 anterograde trafficking represents a regulatory mechanism for its immune activity.
Interestingly, either abrogation of RTNLB1 and RTNLB2 expression or upregulation of RTNLB1 compromised FLS2 immune activity. It is likely that the absence of RTNLB1 and RTNLB2 or excess of RTNLB1 causes perturbations in the stoichiometry of the transport pathway components and, consequently, impaired FLS2 activity. This opens the possibility that FLS2 abundance at the PM is regulated by RTNLB1 and RTNLB2 in a concentration-dependent manner so that strictly regulated levels are required for the optimal transport of FLS2.
The widespread expression of RTNLB1 and RTNLB2 shown by public expression data, along with the finding that rtnlb1 rtnlb2 plants have a strong impairment in FLS2-triggered PTI but only a modest inhibition in FLS2 accumulation at the PM, suggest that RTNLB1 and RTNLB2 may have roles that extend beyond FLS2. An indication that they may regulate other immune receptors or coreceptors comes from the observation that the EFR downstream signaling is deficient in rtnlb1 rtnlb2. Thus, it is possible that RTNLB1 and RTNLB2 influence common FLS2 and EFR regulatory components or other independent pathways that lead to PTI. Nevertheless, several of our data indicate that RTNLB1 and RTNLB2 are not critical for the general intracellular protein trafficking: (1) normal partitioning and accumulation of the PM-associated H+ATPase and the tonoplast-associated γ-TIP in both rtnlb1 rtnlb2 and RTNLB1ox lines, (2) unperturbed localization of the PM marker GFP-BDCaM53 in RTNLB1ox, and (3) the absence of obvious growth phenotypes in rtnlb1 rtnlb2 and RTNLB1ox plants. In support of our observations, the ectopic expression of RTNLB13, a protein with similar localization and morphology to RTNLB1 and RTNLB2 (Sparkes et al., 2010), did not cause major ER-to-Golgi trafficking defects (Tolley et al., 2008). Notably, mutants deficient in factors required for bulk protein transport usually exhibit drastic phenotypes, ranging from major growth defects to lethality (Niihama et al., 2009; Uemura et al., 2009; Hashiguchi et al., 2010). It remains to be investigated if the pathway for intracellular traffic dependent on RTNLB1/RTNLB2 is used by other PTI components.
RTNLB1 and RTNLB2 and Their Relationship with Two Processes Initiated in the ER: Protein Anterograde Transport and the ER Stress and Protein Degradation
Using various experimental approaches (transient expression, gene overexpression, and homozygous T-DNA insertion lines) we investigated multiple facets of the regulation of FLS2 secretion by RTNLBs. We showed that transient coexpression of FLS2 and RTNLB1 fused to fluorescent markers induced FLS2 accumulation in the ER, indicative of an ER-to-Golgi trafficking block. In a similar way, transient expression of a mutant Arabidopsis Rab1 triggered specific blockage of the ER to Golgi step, manifested by the ER accumulation of a secreted form of GFP (Batoko et al., 2000). Indeed, accumulation of protein cargo in intracellular organelles has been exploited as a way to identify components of the trafficking machinery (Rothman and Stevens, 1986; Lippincott-Schwartz et al., 1998). Whereas the immediate consequences of blocked FLS2 trafficking were tested by transient expression, the end-point effects of impaired FLS2 secretion on the plant were analyzed in the rtnlb1 rtnlb2 and RTNLB1ox lines. Interestingly, although the consequences of RTNLB1 and RTNLB2 depletion versus RTNLB1 overexpression on the FLS2 steady state levels were markedly different, the effects on FLS2 anterograde trafficking and immune function were similar in the respective mutants. In rtnlb1 rtnlb2 lines, accumulation of FLS2-GFP in the PM fraction was not completely inhibited compared with the wild type. One possible explanation is that an alternative RTNLB1/RTNLB2-independent pathway may account for the FLS2 transport to the PM; however, this potential second pathway is unlikely to be critical when considering the drastic effect that excess RTNLB1 had on FLS2-GFP export from the ER. Alternatively, other RTNLBs may be able to compensate partially for the loss of RTNLB1 and RTNLB2 to promote FLS2 anterograde transport in rtnlb1 rtnlb2. Lastly, it is possible that changes in the distribution and/or morphology of various ER domains following removal of both RTNLB1 and RTNLB2 may preclude an efficient two-phase partitioning. We showed that FLS2 preferentially accumulated in the pmaER when expressed in N. benthamiana. The pmaER was recently found to be particularly responsive to variations in RTN expression in yeast where the lack of RTNs considerably increased both the abundance of the pmaER relative to other domains and its degree of connectivity to the PM (West et al., 2011). Another important observation that needs further clarification is the apparent FLS2 depletion in the RTNLB1ox lines. For now, we can only speculate that defective FLS2 anterograde trafficking leads accumulation in the ER of nascent FLS2, which may trigger the activation of ER stress and the ER-associated degradation machinery (ERAD). FLS2 degradation in RTNLBox may be a consequence of indirect activation of ERAD following a severe block in the early FLS2 anterograde transport. Interestingly, in the presence of RTNLB1, FLS2-GFP accumulated in the pnER, whereas improved FLS2 trafficking to the PM in the presence of excess RTNLB1 variants correlated with its reduced accumulation in the pnER. The pnER was found to be the site of ERAD in yeast and mammals (Enenkel et al., 1998; Kamhi-Nesher et al., 2001; Kaganovich et al., 2008). In plants, SERK1 accumulation in the pnER was suggested to be linked to its degradation (Aker et al., 2006), and defective receptors accumulating in the ER were shown to be rapidly degraded via ERAD (Li et al., 2009b; Nekrasov et al., 2009; Su et al., 2011). The connection between RTNLBs and ER stress remains to be studied.
The Importance of the Tyr-Dependent Motifs and LCR2 for RTNLB1 and RTNLB2 Functions
Our data indicate that the N termini and the Tyr sorting motifs have significant contributions to the functions of RTNLB1 and RTNLB2. We identified two Tyr-dependent sorting motifs in RTNLB1 and showed that removal of either one of them interferes with the effects of excess RTNLB1 on FLS2 trafficking. Removal of LCR2 from RTNLB1 negatively affects its interaction with FLS2, lessens the negative impact of excess RTNLB1 on FLS2 trafficking, and interferes with the accumulation of complex-glycosylated FLS2. Both TDMs and LCRs have a known propensity for establishing protein–protein interactions. TDMs are known to recruit components of the general trafficking machinery and to perform multiple roles in the transport of membrane proteins (Bonifacino and Traub, 2003; Happel et al., 2004; Jurgens, 2004; daSilva et al., 2006; Derby et al., 2007; Saint-Jean et al., 2010). LCR-containing proteins were shown to interact with numerous protein partners within protein interaction networks and were more often associated with hubs (Coletta et al., 2010). Particularly, mammalian RTNs were shown to bind to components of the general transport pathways, such as SNAREs (Steiner et al., 2004), and the adaptor protein complex AP2 (Iwahashi and Hamada, 2003) with roles in transport vesicle formation and recycling. It is possible that the TDMs and LCRs present in RTNLB1 and RTNLB2 offer interfaces for the interaction with protein partners necessary for regulating aspects of FLS2 transport.
Multiple residues within LCR2 were found to be phosphorylated in both RTNLB1 and RTNLB2 (Benschop et al., 2007; Li et al., 2009a; Reiland et al., 2009), suggesting interactions with kinases through their N termini. LCR2 phosphorylation may affect the binding properties of the adjacent Tyr signals to trafficking components as in AQP4 aquaporin, where phosphorylation of residues preceding a Tyr motif enhanced the interaction with AP2 (Madrid et al., 2001), or it may modulate binding to FLS2 and other substrates. Our data cannot distinguish between direct roles of the TDM motifs in the FLS2 export out of the ER and an indirect effect on FLS2 traffic resulting from other potential roles of TDMs outside the ER. Further work is necessary to investigate the functions of the LCRs and TDMs in RTNLB1 and RTNLB2.
Current models postulate that animal receptor trafficking is controlled by ER membrane–localized receptor accessory proteins that bind to their cytoplasmic tails and recruit transport complexes (McLatchie et al., 1998; Petaja-Repo et al., 2000; Bermak et al., 2001; Dupré and Hébert, 2006; Cooray et al., 2009; Díaz, 2010). Interestingly, RTNLB1 and RTNLB2 were also identified outside the ER compartment, namely, at the PM and in intracellular punctuate structures (Marmagne et al., 2004; Benschop et al., 2007; Nziengui et al., 2007; Mitra et al., 2009), suggesting their transport between various cellular compartments. Thus, an intriguing possibility that remains to be investigated is that RTNLB1 and RTNLB2 are part of a complex pathway of receptor accessory proteins that regulate the transport of transmembrane receptors to the PM.
METHODS
Protein Microarray Screening
Protein microarrays printed on PATH slides (Gentel Biosciences) using a contact printer (Genomic Solutions) were screened with FLS2c. To prepare FLS2c probe, the cytosolic domain of FLS2 (starting from the residue F870) was cloned from genomic DNA (Forw_FLS2_2604 5′-TTCAACAGTGCCAACATCATTGGC-3′ and Rev_FLS2 5′-AACTTCTCGATCCTCGTTACGATC-3′) into a Gateway-compatible plant expression vector carrying a V5 C-terminal peptide tag. Recombinant FLS2c was produced in Nicotiana benthamiana (Popescu et al., 2007). Protein microarrays were blocked using Pierce blocking solution (Pierce) at 4°C, followed by a 1.5-h incubation with 250 μL mix of probing buffer (1× PBS, 5 mM MgCl2, 0.5 mM DTT, 0.05% Triton, 2.5% glycerol, and 1% BSA) and purified FLS2c at 4°C and under a cover slip. The slides were washed three times for 1 min in cold PBS supplemented with 150 mM NaCl followed by 30 min incubation with Alexa-labeled anti-V5 Ab (Sigma-Aldrich), a brief wash in PBS-Tween 0.1%, drying, and processing as described (Popescu et al., 2009).
Split-Luciferase Complementation
The C-terminal half of Renilla reniformis luciferase was fused to the C-terminal end of RTNLB1 or PBS1 and the N-terminal half of luciferase was fused to the C- terminal end of FLS2c. The protein pairs were coexpressed in Arabidopsis thaliana protoplasts, and the luminescence was measured using a Veritas microplate luminometer (Turner BioSystems). The cloning, protoplast preparation, and split-luciferase complementation assays were performed as described by Fujikawa and Kato (2007).
DNA Constructs
For subcellular localization, FLS2 cDNA was cloned into the AscI sites of the binary vector pER8-GFP or pER-Cherry containing the estrogen-inducible XVE promoter (PXVE); to create FLS2-CFP, FLS2 cDNA was cloned into pEarlyGate102 using Gateway reactions (Zuo et al., 2000). The ORFs for RTNLB1 and variants were cloned using Gateway reactions into pDonor 221 and then into the plant binary transformation vector pYY63, containing a 35S cauliflower mosaic virus promoter, P35S. RTNLB1 variants were generated using a PCR-based method. Plasmids were incorporated into the Agrobacterium tumefaciens strain GV2260 for the transient expression analyses performed as described by Voinnet et al. (2003). Two-week-old N. benthamiana leaves were infiltrated with GV2260 carrying PXVE:FLS2-GFP (or Cherry), P35S:RTNLB1-HA (or GFP), or P35S:RTNLB1variants-HA (or GFP) for the subcellular localization experiments or with GV2260 carrying P35S:FLS2-FLAG, PCaMV35s:BTI-HA, or P35S: RTNLB1variants-HA for the Endo H (NEB) treatments. Primer sequences are listed in Supplemental Table 1 online.
Plant Materials
Arabidopsis plants were grown at 22°C with 60% relative humidity, a 16-h photoperiod, and a photon flux density of 120 μmol m−2 s−1. Mature leaves of 3- to 4-week-old plants were used for the majority of the experiments. The RTNLB1 and RTNLB2 T-DNA insertion lines (SALK_016408 and SALK_124414; Alonso et al., 2003) were obtained from the ABRC. fls2 line was generated by Alonso et al. (2003). Protoplasts were produced using published protocols (Sheen, 2001).
Fluorescence Microscopy and Quantification of Colocalization
Images were collected on a Leica TCS-SP5 confocal microscope (Leica Microsystems) after samples were treated with β-estradiol (Sigma-Aldrich) for 6 h, when inducible constructs were used, using a ×63 water immersion objective (numerical aperture 1.2), zoom 1.6. Images were processed using Leica LAS-AF software version 1.8.2. FM4-64 (Sigma-Aldrich) was used according to the manufacturer’s protocol. The colocalization was calculated by importing the colored digitized images to Image-Pro Plus version 4.5 (Media Cybernetics). Pearson’s correlation coefficients were calculated for multiple images per sample, and statistical parameters were calculated using Excel.
CHX Treatment
N. benthamiana plants expressing XVE:FLS2-FLAG in the presence or absence of 35S:RTNLB1-HA at 2 d after infiltration were induced with estradiol for FLS2-FLAG expression. Twelve hours after FLS2-FLAG induction, the estradiol was removed from the leaves by repeated washing with buffer (1 mM MES) and then the leaves were treated with CHX at 10 μM final concentration to terminate protein synthesis. Equal numbers of leaf discs per sample were collected at the indicated time points to prepare the protein extracts as described above. The extracts were analyzed by SDS-PAGE and immunoblotting. The bands corresponding to fusion proteins were detected using chemiluminescence.
Endo H Treatment
The FLS2-GFP coexpressed with RTNLB1 or RTNLB1 variants was immunoprecipitated using Anti-Flag M2 magnetic beads (Sigma-Aldrich). Purified FLS2-FLAG was treated with Endo H following the manufacturer’s protocol and then visualized by immunoblot analysis using an anti-FLAG antibody (Sigma-Aldrich).
Coimmunoprecipitation
Ground plant tissue was mixed with EB buffer (40 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Triton X-100, and 1× Roche protease inhibitor cocktail) and centrifuged at 15,000 rcf for 10 min at 4°C. The supernatant was incubated with prewashed anti-HA-agarose beads for 12 h at 4°C with gentle agitation and the beads washed with EB. The coimmunoprecipitated proteins were visualized on immunoblots using anti-GFP Ab (Santa Cruz Biotechnology). The proteins in total extracts were detected using anti-HA Ab (Pierce).
Treatment with flg22 and Quantification of Bacterial Growth
Leaves were infiltrated with 10 μM flg22 (Genscript) in 1 mM MgCl2 followed at 6 h by spray inoculation with a Pseudomonas syringae DC3000 bacterial suspension (1 × 108 colony-forming units mL−1 in 0.01% [v/v] Silwet L-77). Bacterial titers were assayed at 3 d after inoculation as described (Zipfel et al., 2004).
Analysis of MPK3 and MPK6 Activation by flg22 or elf18
Total extracts were prepared from frozen leaf tissue in EB buffer supplemented with 25 mM NaF, 1 mM Na3VO4, and 50 mM β-glycerophosphate and incubated for 3 h at 4°C with beads prebound to anti-MPK6 or -MPK3 Ab (obtained from S.B. Ryu). The beads were washed three times in EB and one time in KB buffer (40 mM HEPES, pH 8.0, 1 mM DTT, 12 mM MgCl2, 1 mM EGTA, and 0.1 mM Na3VO4). Myelin basic protein (Sigma-Aldrich) phosphorylation by immunoprecipitated MPKs was assayed in KB supplemented with 50 μCi [γ-P32], followed by boiling in SDS sample buffer, SDS-PAGE, and radiography.
Analysis of Gene Expression
Total plant RNA was isolated using an RNA extraction kit (Qiagen). First-strand cDNAs were synthesized from 4 ug total RNA using random primers and the ThermoScript RT-PCR system (Invirtogen) according to the manufacturer’s instructions. PCR amplification was conducted using 3 μL of the cDNA reaction mixture and the primers listed in Supplemental Table 1 online. Quantitative real-time PCR was performed using the CFX96 real-time PCR detection system (Bio-Rad) and 2× SYBR Green qPCR Master Mix (Takara). Actin1 served as internal control for both semiquantitative and quantitative real-time PCR. All primers are listed in Supplemental Table 1 online.
Aqueous Two-Phase Separation
Arabidopsis membranes were extracted and fractionated as previously described (Wirthmueller et al., 2007) with the following modifications: ~2 g of Arabidopsis leaves were homogenized in 10 mL of the described buffer at 4°C without a freeze-thaw and using a T10 Ultra-Turrax homogenizer (IKA). Anti-FLS2 antibody was obtained from A. Heese (University of Missouri).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers At5g46330, At4g23630, At4g11220, and At5g41600.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Example of an Arabidopsis thaliana FPM-5000.
Supplemental Figure 2. Sequence Alignment of RTNLB1 and Its Closest Homologs.
Supplemental Figure 3. RTNLB1 Coimmunoprecipitates with FLS2-FLAG but Not with EFR-FLAG.
Supplemental Figure 4. Phenotype of rtnlb1 rtnlb2 and RTNLB1ox Lines.
Supplemental Figure 5. FLS2-Dependent Signaling in rtnlb1, rtnlb2, and rtnlb1 rtnlb2 Mutants.
Supplemental Figure 6. EFR-Dependent Signaling in rtnlb1 rtnlb2 Mutants.
Supplemental Figure 7. Expression of Full-Length RTNLB1 and Variants in N. benthamiana for Confocal Analysis.
Supplemental Figure 8. Aberrant Localization of FLS2 in the Presence of RTNLB1 or RTNLB2.
Supplemental Figure 9. Colocalization of FLS2-GFP and HDEL-Cherry in the Presence of RTNLB1-HA.
Supplemental Figure 10. Colocalization of FLS2-GFP and HDEL-Cherry in the ER.
Supplemental Figure 11. RTNLB1 Does Not Interfere with the Localization of a PM Marker.
Supplemental Figure 12. Colocalization of RTNLB1-GFP and FLS2-Cherry.
Supplemental Figure 13. RTNLB1-GFP Variants Are Localized in the ER.
Supplemental Table 1. Oligonucleotides Used in This Work.
Supplemental Data Set 1. FPM-5000 Layout.
Supplemental Data Set 2. Quantification of FLS2-GFP Colocalization with PM and ER Markers.
Supplemental Data Set 3. Quantification of the Colocalization Degree of RTNLB1-GFP and FLS2-Cherry.
Supplemental Data Set 4. Quantification of the Colocalization Degree of RTNLB1-GFP Variants and FLS2-Cherry.
Acknowledgments
We thank Stanton Gelvin, Antje Heese, Maria Harrison, Nate Pumplin, and Stephen B. Ryu for generous gifts of reagents. We also thank Mamta Srivastava for technical assistance with the microscopy experiments and Giulio Zampogna for editing the manuscript. This work was supported by start-up funds to S.C.P. from the Boyce Thompson Institute and by National Science Foundation Grants DBI-0723722 and DBI-1042344 to S.D.-K and M.S.
AUTHOR CONTRIBUTIONS
H.Y.L., C.H.B., H.-G.K., N.K., G.V.P., and S.C.P. performed research. G.V.P. and S.C.P analyzed data. S.M., S.D.-K., and M.S. contributed new reagents. S.C.P. and H.Y.L. designed the research. S.C.P. wrote the article.
References
- Aker J., Borst J.W., Karlova R., de Vries S. (2006). The Arabidopsis thaliana AAA protein CDC48A interacts in vivo with the somatic embryogenesis receptor-like kinase 1 receptor at the plasma membrane. J. Struct. Biol. 156: 62–71 [DOI] [PubMed] [Google Scholar]
- Ali G.S., Prasad K.V.S.K., Day I., Reddy A.S. (2007). Ligand-dependent reduction in the membrane mobility of FLAGELLIN SENSITIVE2, an arabidopsis receptor-like kinase. Plant Cell Physiol. 48: 1601–1611 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Batoko H., Zheng H.-Q., Hawes C., Moore I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop J.J., Mohammed S., O’Flaherty M., Heck A.J., Slijper M., Menke F.L. (2007). Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6: 1198–1214 [DOI] [PubMed] [Google Scholar]
- Bermak J.C., Li M., Bullock C., Zhou Q.Y. (2001). Regulation of transport of the dopamine D1 receptor by a new membrane-associated ER protein. Nat. Cell Biol. 3: 492–498 [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., Traub L.M. (2003). Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72: 395–447 [DOI] [PubMed] [Google Scholar]
- Boutrot F., Segonzac C., Chang K.N., Qiao H., Ecker J.R., Zipfel C., Rathjen J.P. (2010). Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc. Natl. Acad. Sci. USA 107: 14502–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C.J., Bednarek S.Y., Raikhel N.V. (2004). Membrane trafficking in plants: New discoveries and approaches. Curr. Opin. Plant Biol. 7: 701–707 [DOI] [PubMed] [Google Scholar]
- Chang K., Seabold G.K., Wang C.-Y., Wenthold R.J. (2009). Reticulon 3 is an interacting partner of the SALM family of adhesion molecules. J. Neurosci. Res. 88: 266–274 [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Coletta A., Pinney J.W., Solís D.Y., Marsh J., Pettifer S.R., Attwood T.K. (2010). Low-complexity regions within protein sequences have position-dependent roles. BMC Syst. Biol. 4: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooray S.N., Chan L., Webb T.R., Metherell L., Clark A.J. (2009). Accessory proteins are vital for the functional expression of certain G protein-coupled receptors. Mol. Cell. Endocrinol. 300: 17–24 [DOI] [PubMed] [Google Scholar]
- Corpet F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- daSilva L.L.P., Foresti O., Denecke J. (2006). Targeting of the plant vacuolar sorting receptor BP80 is dependent on multiple sorting signals in the cytosolic tail. Plant Cell 18: 1477–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derby M.C., Gleeson P.A., Kwang W.J. (2007). New insights into membrane trafficking and protein sorting. Int. Rev. Cytol. 261: 47–116 [DOI] [PubMed] [Google Scholar]
- De Smet I., Voss U., Jürgens G., Beeckman T. (2009). Receptor-like kinases shape the plant. Nat. Cell Biol. 11: 1166–1173 [DOI] [PubMed] [Google Scholar]
- Díaz E. (2010). Regulation of AMPA receptors by transmembrane accessory proteins. Eur. J. Neurosci. 32: 261–268 [DOI] [PubMed] [Google Scholar]
- Dupré D.J., Hébert T.E. (2006). Biosynthesis and trafficking of seven transmembrane receptor signalling complexes. Cell. Signal. 18: 1549–1559 [DOI] [PubMed] [Google Scholar]
- Enenkel C., Lehmann A., Kloetzel P.-M. (1998). Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 17: 6144–6154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurat-Lessard P., Frangne N., Maeshima M., Ratajczak R., Bonnemain J.L., Martinoia E. (1997). Increased expression of vacuolar aquaporin and H+-ATPase related to motor cell function in Mimosa pudica L. Plant Physiol. 114: 827–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa Y., Kato N. (2007). Split luciferase complementation assay to study protein-protein interactions in Arabidopsis protoplasts. Plant J. 52: 185–195 [DOI] [PubMed] [Google Scholar]
- Geldner N., Robatzek S. (2008). Plant receptors go endosomal: A moving view on signal transduction. Plant Physiol. 147: 1565–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre V., Spallek T., Häweker H., Mersmann S., Mentzel T., Boller T., de Torres M., Mansfield J.W., Robatzek S. (2008). Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18: 1824–1832 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Bauer Z., Boller T. (2001). Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13: 1155–1163 [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Hanton S.L., Brandizzi F. (2006). Protein transport in the plant secretory pathway. Can. J. Bot. 84: 523–530 [Google Scholar]
- Happel N., Höning S., Neuhaus J.M., Paris N., Robinson D.G., Holstein S.E.H. (2004). Arabidopsis mu A-adaptin interacts with the tyrosine motif of the vacuolar sorting receptor VSR-PS1. Plant J. 37: 678–693 [DOI] [PubMed] [Google Scholar]
- Hashiguchi Y., Niihama M., Takahashi T., Saito C., Nakano A., Tasaka M., Morita M.T. (2010). Loss-of-function mutations of retromer large subunit genes suppress the phenotype of an Arabidopsis zig mutant that lacks Qb-SNARE VTI11. Plant Cell 22: 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häweker H., Rips S., Koiwa H., Salomon S., Saijo Y., Chinchilla D., Robatzek S., von Schaewen A. (2010). Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 285: 4629–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Hu X., Shi Q., Zhou X., Lu Y., Fisher C., Yan R. (2006). Mapping of interaction domains mediating binding between BACE1 and RTN/Nogo proteins. J. Mol. Biol. 363: 625–634 [DOI] [PubMed] [Google Scholar]
- Hu F., Liu B.P., Budel S., Liao J., Chin J., Fournier A., Strittmatter S.M. (2005). Nogo-A interacts with the Nogo-66 receptor through multiple sites to create an isoform-selective subnanomolar agonist. J. Neurosci. 25: 5298–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., et al. (2009). Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 139: 610–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H.-H., Gelvin S.B. (2004). Plant proteins that interact with VirB2, the Agrobacterium tumefaciens pilin protein, mediate plant transformation. Plant Cell 16: 3148–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahashi J., Hamada N. (2003). Human reticulon 1-A and 1-B interact with a medium chain of the AP-2 adaptor complex. Cell. Mol. Biol. (Noisy-le-grand) 49: OL467–OL471 [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jurgens G. (2004). Membrane trafficking in plants. Annu. Rev. Cell Dev. Biol. 20: 481–504 [DOI] [PubMed] [Google Scholar]
- Kaganovich D., Kopito R., Frydman J. (2008). Misfolded proteins partition between two distinct quality control compartments. Nature 454: 1088–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamhi-Nesher S., Shenkman M., Tolchinsky S., Fromm S.V., Ehrlich R., Lederkremer G.Z. (2001). A novel quality control compartment derived from the endoplasmic reticulum. Mol. Biol. Cell 12: 1711–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N., Fujikawa Y., Fuselier T., Adamou-Dodo R., Nishitani A., Sato M.H. (2010). Luminescence detection of SNARE-SNARE interaction in Arabidopsis protoplasts. Plant Mol. Biol. 72: 433–444 [DOI] [PubMed] [Google Scholar]
- Kato N., Reynolds D., Brown M.L., Boisdore M., Fujikawa Y., Morales A., Meisel L.A. (2008). Multidimensional fluorescence microscopy of multiple organelles in Arabidopsis seedlings. Plant Methods 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature 451: 835–840 [DOI] [PubMed] [Google Scholar]
- Li H., Wong W.S., Zhu L., Guo H.W., Ecker J., Li N. (2009a). Phosphoproteomic analysis of ethylene-regulated protein phosphorylation in etiolated seedlings of Arabidopsis mutant ein2 using two-dimensional separations coupled with a hybrid quadrupole time-of-flight mass spectrometer. Proteomics 9: 1646–1661 [DOI] [PubMed] [Google Scholar]
- Li J., Zhao-Hui C., Batoux M., Nekrasov V., Roux M., Chinchilla D., Zipfel C., Jones J.D. (2009b). Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl. Acad. Sci. USA 106: 15973–15978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Cole N., Presley J. (1998). Unravelling Golgi membrane traffic with green fluorescent protein chimeras. Trends Cell Biol. 8: 16–20 [DOI] [PubMed] [Google Scholar]
- Liu J.-X., Howell S.H. (2010). Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22: 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Vidensky S., Ruggiero A.M., Maier S., Sitte H.H., Rothstein J.D. (2008). Reticulon RTN2B regulates trafficking and function of neuronal glutamate transporter EAAC1. J. Biol. Chem. 283: 6561–6571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L., He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Tintor N., Mentzel T., Kombrink E., Boller T., Robatzek S., Schulze-Lefert P., Saijo Y. (2009). Uncoupling of sustained MAMP receptor signaling from early outputs in an Arabidopsis endoplasmic reticulum glucosidase II allele. Proc. Natl. Acad. Sci. USA 106: 22522–22527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid R., Le Maout S., Barrault M.B., Janvier K., Benichou S., Mérot J. (2001). Polarized trafficking and surface expression of the AQP4 water channel are coordinated by serial and regulated interactions with different clathrin-adaptor complexes. EMBO J. 20: 7008–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley F., Trimble R.B., Tarentino A.L., Plummer T.H., Jr. (1989). Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180: 195–204 [DOI] [PubMed] [Google Scholar]
- Marmagne A., Rouet M.A., Ferro M., Rolland N., Alcon C., Joyard J., Garin J., Barbier-Brygoo H., Ephritikhine G. (2004). Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol. Cell. Proteomics 3: 675–691 [DOI] [PubMed] [Google Scholar]
- McLatchie L.M., Fraser N.J., Main M.J., Wise A., Brown J., Thompson N., Solari R., Lee M.G., Foord S.M. (1998). RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature 393: 333–339 [DOI] [PubMed] [Google Scholar]
- Mersmann S., Bourdais G., Rietz S., Robatzek S. (2010). Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 154: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell H., Choudhury A., Pagano R.E., Leof E.B. (2004). Ligand-dependent and -independent transforming growth factor-beta receptor recycling regulated by clathrin-mediated endocytosis and Rab11. Mol. Biol. Cell 15: 4166–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S.K., Walters B.T., Clouse S.D., Goshe M.B. (2009). An efficient organic solvent based extraction method for the proteomic analysis of Arabidopsis plasma membranes. J. Proteome Res. 8: 2752–2767 [DOI] [PubMed] [Google Scholar]
- Mok J., et al. (2010). Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 3: ra12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L., Zipfel C., Rowland O., Keller I., Robatzek S., Boller T., Jones J.D. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135: 1113–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V., et al. (2009). Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28: 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Niihama M., Takemoto N., Hashiguchi Y., Tasaka M., Morita M.T. (2009). ZIP genes encode proteins involved in membrane trafficking of the TGN-PVC/vacuoles. Plant Cell Physiol. 50: 2057–2068 [DOI] [PubMed] [Google Scholar]
- Nziengui H., Bouhidel K., Pillon D., Der C., Marty F., Schoefs B. (2007). Reticulon-like proteins in Arabidopsis thaliana: Structural organization and ER localization. FEBS Lett. 581: 3356–3362 [DOI] [PubMed] [Google Scholar]
- Oertle T., Klinger M., Stuermer C.A., Schwab M.E. (2003). A reticular rhapsody: Phylogenic evolution and nomenclature of the RTN/Nogo gene family. FASEB J. 17: 1238–1247 [DOI] [PubMed] [Google Scholar]
- Page M.D., Kropat J., Hamel P.P., Merchant S.S. (2009). Two Chlamydomonas CTR copper transporters with a novel cys-met motif are localized to the plasma membrane and function in copper assimilation. Plant Cell 21: 928–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.-J., Bart R., Chern M., Canlas P.E., Bai W., Ronald P.C. (2010). Overexpression of the endoplasmic reticulum chaperone BiP3 regulates XA21-mediated innate immunity in rice. PLoS ONE 5: e9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.H., Blackstone C. (2010). Further assembly required: Construction and dynamics of the endoplasmic reticulum network. EMBO Rep. 11: 515–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petaja-Repo U.E., Hogue M., Laperriere A., Walker P., Bouvier M. (2000). Export from the endoplasmic reticulum represents the limiting step in the maturation and cell surface expression of the human delta opioid receptor. J. Biol. Chem. 275: 13727–13736 [DOI] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Seay M., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2007). Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. USA 104: 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntervoll P., et al. (2003). ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 31: 3625–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S., Mehta G., Srivastava S. (2010). Label-free detection techniques for protein microarrays: Prospects, merits and challenges. Proteomics 10: 731–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S., Messerli G., Baerenfaller K., Gerrits B., Endler A., Grossmann J., Gruissem W., Baginsky S. (2009). Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 150: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Herranz M.-C., Bubeck J., Pepperkok R., Ritzenthaler C. (2007). Membrane dynamics in the early secretory pathw\ay. Crit. Rev. Plant Sci. 26: 199–225 [Google Scholar]
- Rodríguez-Concepción M., Yalovsky S., Zik M., Fromm H., Gruissem W. (1999). The prenylation status of a novel plant calmodulin directs plasma membrane or nuclear localization of the protein. EMBO J. 18: 1996–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E., Denecke J. (2008). What is moving in the secretory pathway of plants? Plant Physiol. 147: 1493–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J.H., Stevens T.H. (1986). Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell 47: 1041–1051 [DOI] [PubMed] [Google Scholar]
- Saint-Jean B., Seveno-Carpentier E., Alcon C., Neuhaus J.M., Paris N. (2010). The cytosolic tail dipeptide Ile-Met of the pea receptor BP80 is required for recycling from the prevacuole and for endocytosis. Plant Cell 22: 2825–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S., Prinz W.A., Thorn K.S., Voss C., Walter P. (2009). Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J. Cell Biol. 187: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Shi Q., Prior M., He W., Tang X., Hu X., Yan R. (2009). Reduced amyloid deposition in mice overexpressing RTN3 is adversely affected by preformed dystrophic neurites. J. Neurosci. 29: 9163–9173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I., Tolley N., Aller I., Svozil J., Osterrieder A., Botchway S., Mueller C., Frigerio L., Hawes C. (2010). Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell 22: 1333–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner P., Kulangara K., Sarria J.C., Glauser L., Regazzi R., Hirling H. (2004). Reticulon 1-C/neuroendocrine-specific protein-C interacts with SNARE proteins. J. Neurochem. 89: 569–580 [DOI] [PubMed] [Google Scholar]
- Su W., Liu Y., Xia Y., Hong Z., Li J. (2011). Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 870–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Deng Z., Oses-Prieto J.A., Suzuki N., Zhu S., Zhang X., Burlingame A.L., Wang Z.-Y. (2008). Proteomics studies of brassinosteroid signal transduction using prefractionation and two-dimensional DIGE. Mol. Cell. Proteomics 7: 728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolley N., Sparkes I.A., Hunter P.R., Craddock C.P., Nuttall J., Roberts L.M., Hawes C., Pedrazzini E., Frigerio L. (2008). Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic 9: 94–102 [DOI] [PubMed] [Google Scholar]
- Tompa P. (2002). Intrinsically unstructured proteins. Trends Biochem. Sci. 27: 527–533 [DOI] [PubMed] [Google Scholar]
- Tör M., Lotze M.T., Holton N. (2009). Receptor-mediated signalling in plants: Molecular patterns and programmes. J. Exp. Bot. 60: 3645–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Sato T., Aoki T., Yamamoto A., Okada T., Hirai R., Harada R., Mori K., Tagaya M., Harada A. (2009). p31 deficiency influences endoplasmic reticulum tubular morphology and cell survival. Mol. Cell. Biol. 29: 1869–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G.K., Prinz W.A., Shibata Y., Rist J.M., Rapoport T.A. (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124: 573–586 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- von Numers N., Survila M., Aalto M., Batoux M., Heino P., Palva E.T., Li J. (2010). Requirement of a homolog of glucosidase II beta-subunit for EFR-mediated defense signaling in Arabidopsis thaliana. Mol. Plant 3: 740–750 [DOI] [PubMed] [Google Scholar]
- Wakana Y., Koyama S., Nakajima K., Hatsuzawa K., Nagahama M., Tani K., Hauri H.P., Melançon P., Tagaya M. (2005). Reticulon 3 is involved in membrane trafficking between the endoplasmic reticulum and Golgi. Biochem. Biophys. Res. Commun. 334: 1198–1205 [DOI] [PubMed] [Google Scholar]
- West M., Zurek N., Hoenger A., Voeltz G.K. (2011). A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 193: 333–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley H.S. (2003). Trafficking of the ErbB receptors and its influence on signaling. Exp. Cell Res. 284: 78–88 [DOI] [PubMed] [Google Scholar]
- Wirthmueller L., Zhang Y., Jones J.D.G., Parker J.E. (2007). Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17: 2023–2029 [DOI] [PubMed] [Google Scholar]
- Yang Y.S., Harel N.Y., Strittmatter S.M. (2009). Reticulon-4A (Nogo-A) redistributes protein disulfide isomerase to protect mice from SOD1-dependent amyotrophic lateral sclerosis. J. Neurosci. 29: 13850–13859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.-W., Chua N.-H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273 [DOI] [PubMed] [Google Scholar]