This work examines the pollen defective in guidance1 (pod1) mutant and finds that it is defective in pollen tube guidance to the micropyle. Homozygous pod1 embryos also show lethality. POD1 is an endoplasmic reticulum (ER) luminal protein involved in ER protein retention.
Abstract
The pollen tube germinates from pollen and, during its migration, it perceives and responds to guidance cues from maternal tissue and from the female gametophyte. The putative female cues have recently been identified, but how the pollen tube responds to these signals remains to be unveiled. In a genetic screen for male determinants of the pollen tube response, we identified the pollen defective in guidance1 (pod1) mutant, in which the pollen tubes fail to target the female gametophyte. POD1 encodes a conserved protein of unknown function and is essential for positioning and orienting the cell division plane during early embryo development. Here, we demonstrate that POD1 is an endoplasmic reticulum (ER) luminal protein involved in ER protein retention. Further analysis shows that POD1 interacts with the Ca2+ binding ER chaperone CALRETICULIN3 (CRT3), a protein in charge of folding of membrane receptors. We propose that POD1 modulates the activity of CRT3 or other ER resident factors to control the folding of proteins, such as membrane proteins in the ER. By this mechanism, POD1 may regulate the pollen tube response to signals from the female tissues during pollen tube guidance and early embryo patterning in Arabidopsis thaliana.
INTRODUCTION
Fertilization completes the cycle from the haploid gametophyte generation to the diploid sporophyte generation in the life history of animals and plants. In single-celled organisms and animals, the egg cells may emit attractant molecules, which form a concentration gradient to guide the sperm cells by chemotaxis. The sperm cells perceive and respond to these attractant signals to target the egg cells and achieve fertilization (Higashiyama and Hamamura, 2008). However, the sperm cells in flowering plants have lost their mobility during evolution. To compensate for this, a new mechanism called siphonogamy evolved in which a pollen tube is produced by the male gametophyte to deliver the two sperm cells to the female gametophyte. Pollen tubes grow unidirectionally and are guided by multiple signals from the maternal tissues and the female gametophytes (embryo sacs). Indeed, pollen tube guidance is a precisely regulated process analogous to axon guidance in animals (Palanivelu and Preuss, 2000). Pollen tube guidance can be divided into sporophytic and gametophytic guidance (Shimizu and Okada, 2000; Higashiyama et al., 2003). Sporophytic guidance refers to pollen tube growth that is guided by female sporophytic signals within the transmitting tract. Gametophytic guidance is directed by signals from the embryo sac, and it is often divided into funicular guidance and micropylar guidance. Funicular guidance directs the tubes from the septum (placenta) surface to the funicular surface, and micropylar guidance directs the tubes from the funiculus into the micropylar opening of the ovule.
Genetic studies revealed that the female gametophyte plays an important role in pollen tube guidance (Hülskamp et al., 1995; Ray et al., 1997; Shimizu and Okada, 2000). For example, cell ablation experiments in Torenia fournieri showed that the synergid cells of the embryo sac are key to attracting pollen tubes (Higashiyama et al., 2001). Several proteins produced in the embryo sac, such as MYB98 in the synergid cells (Kasahara et al., 2005; Márton et al., 2005), CENTRAL CELL GUIDANCE in the central cell (Chen et al., 2007), and GAMETE-EXPRESSED3 in the egg cell (Alandete-Saez et al., 2008), have been shown to be involved in micropylar pollen tube guidance. Recently, the secreted defensin-like peptides LUREs have been shown to be able to guide pollen tube growth in T. fournieri (Okuda et al., 2009). LUREs are Cys-rich proteins that contain a motif conserved among antimicrobial peptides. In addition, maize (Zea mays) EA1, a small peptide secreted from the egg apparatus, is also essential for micropylar pollen tube guidance (Márton et al., 2005). These results suggest that the attracting cues from the embryo sac are small secreted peptides. Such peptides are encoded by a large family of genes in Arabidopsis thaliana, and more investigation is necessary to elucidate which are the attracting or repelling cues (Jones-Rhoades et al., 2007).
Although the pollen tube has long been considered a model system to study polar cell growth in plants (Yang, 1998; Hepler et al., 2001; Cheung and Wu, 2008; Kost, 2008; Cai and Cresti, 2009), very little is known about how it perceives and responds to the attractants to achieve directed growth. It was recently shown that Glu receptor-like proteins function as Ca2+ channels to regulate the cytosolic [Ca2+]cyt in pollen tubes, and, interestingly, the activities of these Glu receptor-like proteins are modulated by d-Ser in pistils (Michard et al., 2011). This is a conserved mechanism also used in neurotransmission in animal systems. Recently, it was reported that two endoplasmic reticulum (ER)–localized K+ transporters in pollen are involved in funicular guidance in vivo and micropylar guidance in a semi–in vivo assay (Lu et al., 2011). These findings suggest a role for cation and pH dynamics in pollen tube guidance. Mutation of the Rab GTPase RABA4D severely reduced the growth rate and efficiency of micropylar pollen tube targeting (Szumlanski and Nielsen, 2009). In addition, mutations of HAPLESS2/GENERATIVE CELL-SPECIFIC1, a conserved, sperm cell–specific gene mediating cell fusion during fertilization (Mori et al., 2006; Liu et al., 2010), also impairs pollen tube guidance (von Besser et al., 2006).
Guided cell growth is a common phenomenon in the plant and animal kingdoms, and much has been learned in yeast and animal systems (Madden and Snyder, 1998; Huber et al., 2003; O’Donnell et al., 2009), but very little is known about the signaling processes in plants (Chae and Lord, 2011; Cheung and Wu, 2008). Pollen tube guidance is a species-specific process (Swanson et al., 2004; Higashiyama et al., 2006), which serves as an ideal model system to investigate mechanisms involved in signaling and guided cell growth in plants. To identify male factors controlling pollen tube guidance, we performed a forward genetic screen and identified the mutant pollen defective in guidance1 (pod1), which was found to be specifically defective in the pollen tube response to female attractants during micropylar guidance. Molecular analysis shows that POD1 is a novel ER luminal protein involved in ER protein retention and interacts with CALRETICULIN3 (CRT3), a luminal chaperone involved in Ca2+ homeostasis and ER quality control. This work demonstrates that POD1 plays a specific role in the micropylar response and is also essential for cell patterning during early embryogenesis in Arabidopsis.
RESULTS
Genetic Screen to Identify Male Determinants in Pollen Tube Guidance
To investigate how pollen tubes perceive and respond to the female guidance signals during pollen tube growth, we used a series of tests to identify mutants that were defective in pollen tube entry into the gametophyte. As a first step, we screened Arabidopsis Ds and T-DNA insertion lines for reduced transmission efficiency of the mutation through the male gametophyte. In this broad screen, we selected mutations that affect many processes, including pollen development, pollen function, and pollen tube guidance. Second, we tested the candidate mutants to determine whether their pollen could target ovules in a limited pollination assay. A limited number of pollen grains (<40) from these candidate mutants were pollinated manually onto a wild-type pistil, which harbors ~50 to 60 ovules. This eliminates competition between pollen tubes and ensures that each pollen tube has the opportunity to target one ovule. To observe the entry of the pollen tubes into the ovules, 12 h after pollination the pistil was stained with aniline blue, which specifically labels the callose wall of the pollen tube. Mutants that displayed normal pollen tube growth but failed to enter the micropylar opening of the ovule were chosen for further investigation and designated as pod.
The pod1 mutant was isolated from our Arabidopsis Ds mutant pool (Sundaresan et al., 1995). The Ds element used for mutagenesis contains a kanamycin resistance gene (Kanr), so the transmission of the mutation can be tracked by the Kanr segregation of its progeny. Progeny from a self-pollinated pod1 plant showed a Kanr/Kans (kanamycin-sensitive) segregation ratio of 1:1 (550:554, n = 1104) (Table 1), and this ratio is stable over three consecutive generations, indicating that the mutant is heterozygous for the Ds insertion and its fertility is compromised. In addition, reciprocal crosses between the wild type and pod1 mutants were performed. When pod1/POD1 pistils were pollinated with wild-type pollen, the Kanr/Kans segregation ratio of the F1 progeny was 1:1 (500:498). This ratio was maintained in three independent crosses, indicating that the transmission of the Ds through the female gametophyte is not affected and the pod1 ovule is completely fertile. However, when wild-type pistils were pollinated with pollen from a pod1/POD1 plant, the Kanr/Kans segregation ratio of the F1 progeny was 0.04:1 (51:1215) with a transmission efficiency of 4.1%. This indicates that pollen development or/and function is severely affected in the pod1 mutant.
Table 1.
Segregation Analysis of pod1/POD1 Mutants
| Parental Genotype |
||||
| Kanr | Kans | Kanr/Kans | ||
| Male | Female | |||
| pod1-1/POD1-1 | pod1-1/POD1-1 | 550 | 554 | 1:1a |
| POD1-1/POD1-1 | pod1-1/POD1-1 | 500 | 498 | 1:1b |
| pod1-1/POD1-1 | POD1-1/POD1-1 | 51 | 1215 | 0.04:1b |
| pod1-2/POD1-2 | pod1-2/POD1-2 | 500 | 500 | 1:1b |
| pod1-3/POD1-3 | pod1-3/POD1-3 | 580 | 582 | 1:1b |
| pod1-1/POD1-1 | POD1-1/POD1-1 | 181 | 342 | 0.53:1LP, b |
| pod1-1/POD1-1 | pod1-1/POD1-1 | 269 | 254 | 1.06:1LP, b |
pod1-1, 1-2, and 1-3 refer to different Ds or T-DNA insertion alleles. LP, experiment carried out under limited pollination conditions. a, more than 10 replicates; b, three replicates.
Pollen Germination and Tube Growth Are Normal in pod1
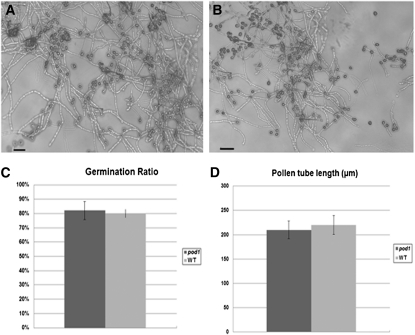
To investigate whether the reduced male transmission of pod1 is caused by a pollen developmental defect, we first checked the morphology of mature pod1 pollen grains by 4′,6-diamidino-2-phenylindole staining and Alexander staining for cell viability. The results showed that the pollen grains from pod1/POD1 plants are morphologically normal and contain two generative nuclei and one vegetative nucleus at maturity (n = 1000) (see Supplemental Figure 1 online); no difference in morphology or cell viability was observed between mutant and wild-type pollen. This indicates that pod1 pollen develop normally. We next used an in vitro pollen germination assay to test whether the reduced male transmission of pod1 is caused by a pollen germination defect. A mean value of 81% germination (n = 857, from six independent Kanr plants) is obtained for pollen grains from pod1/POD1 plants, which is comparable to that of the wild-type pollen grains (81%, n = 211, χ2 = 0.0057, P > 0.05) (Figure 1). In addition, no abnormality in pollen tube morphology or growth in vitro was observed (Figures 1A and 1B). To test pollen tube growth in vivo, 6 to 12 pollen grains from pod1/POD1 were pollinated on each wild-type pistil (24 h after emasculation). The pollinated pistils were collected 2 h later and stained with aniline blue. We found that 93.3% of the pollen grains (n = 453, χ2 = 0.06, P = 3.841) germinated on the stigma, their pollen tubes entered the style, and the tubes grew within the transmitting tract at the same growth rate (Figures 2A and 2B). These data indicate that the germination rate and tube growth of pod1 pollen in the sporophytic tissues are the same as that of POD1 pollen. Taken together, these data demonstrate that the pod1 mutation does not affect pollen morphology, germination, or pollen tube growth in vitro and in vivo.
Figure 1.
Germination Ratio and Pollen Tube Length in Vitro.
(A) DIC image of pollen grains from pod1/POD1 germinated in vitro.
(B) Pollen grains from the wild type germinated in vitro. Bars = 50 μm.
(C) Percentage of germination of pollen from pod1/POD1 and the wild type (WT).
(D) Pollen tube length of pod1/POD1 and the wild type 1 h after germination in vitro.
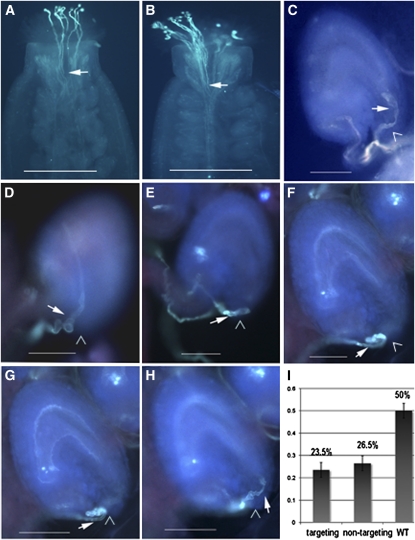
Figure 2.
Phenotype of Pollen Tubes Germinated in Planta.
Pollen tubes are stained with aniline blue and visualized by fluorescence microscopy.
(A) and (B) Similar pollen tube growth of pod1/POD1 and wild-type plants in wild-type stigma and style under limited pollination condition.
(A) Pollen grains of a pod1/POD1 plant germinated on the stigma and grown for 2 h.
(B) Pollen grains of a wild-type plant germinated on the stigma and grown for 2 h.
(C) Wild-type pollen tubes enter the micropyle directly.
(D) to (I) Pollen tubes of pod1/POD1 plant growing in wild-type pistils.
(E) to (H) Images of the same ovule in different focal planes showing the configuration of the pod1 pollen tube outside the micropyle. The pollen tube twisting outside the micropyle and growing on the integument (D) or twisting outside the micropyle and finally entering the ovule ([E] to [H]). Bar graph of the percentage of the pollen tubes showing each phenotype (n = 684). The nontargeting phenotype is shown in (D), and the targeting phenotype is shown in (E) to (H). Arrows indicate the pollen tubes. Arrowhead indicates the micropyle. WT, wild type.
Bars = 400 μm in (A) and (B) and 100 μm in (C) to (H).
The Micropylar Response of the pod1 Pollen Tube Is Compromised
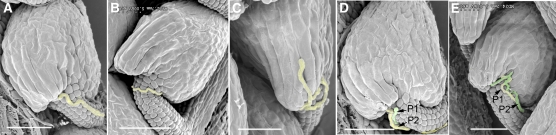
To further investigate the effect of the pod1 mutation on pollen function, we pollinated pollen grains from pod1/POD1 plants on wild-type or pod1/POD1 stigma under limited pollination conditions and then analyzed the Kanr/Kans ratio of the F1 progeny. When wild-type stigmas were pollinated with pollen grains from pod1/POD1 plants, the Kanr/Kans ratio of the F1 progeny was 0.53:1 (n = 523). The ratio in this limited-pollination test is higher than the 0.04:1 seen in the reciprocal crosses described above, but lower than the expected 1:1 ratio expected if pollen function is normal. This indicates that pod1 pollen transmission increases when there is no competition for ovules, suggesting that the competence of pod1 pollen is compromised. To determine whether pollen tube guidance was compromised, we tracked the path of pollen tube growth under limited pollination conditions. Some tubes approach the micropyle but fail to enter the micropylar opening of the ovule, and some pollen tubes entered the micropyle directly (Figure 2). Because the pollen used here comes from a heterozygous pod1/POD1 plant, 50% of the pollen grains should be pod1 and 50% should be POD1; if pod1 is gametophyte specific, then the POD1 pollen tubes should migrate normally, but the pod1 tubes may show defective migration. When we examined pollen tube migration, we found that the normal pollen tubes entered the micropyle directly (50%) and no twisting growth around the micropyle was observed (Figures 2C and 3A). The defective pollen tube phenotypes can be divided into two categories: (1) pollen tubes growing on the ovule surface or twisting around the micropyle or just turning back on the funicular surface (26.5%, n = 684) (Figures 2D, 3B, and 3C) or (2) a portion of pollen tubes twisting initially around the micropyle of the ovule and finally entering the micropyle (23.5%, n = 684) to complete fertilization (Figures 2E to 2H). Furthermore, scanning electron microscopy confirmed that the 50% of pollen tubes defective in micropylar entry also displayed abnormal growth behavior (n = 391) (Figure 3). For example, we often observed two pollen tubes targeting the same ovule (Figures 3D and 3E), with the first pollen tube failing to enter the micropyle and the second pollen tube entering the embryo sac. We speculate that the first one is a pod1 pollen tube and the second one is a POD1 tube. Pollen tubes defective in funicular guidance in the pod1/POD1 mutant were not observed in our assay. We also investigated the behavior of wild-type pollen tubes under limited pollination (pollen grains from the wild-type plants were pollinated on the wild-type stigma). Occasionally, wild-type tubes that did not target to the micropyle (0.26%, n = 1155) were observed. Together, these experiments under limited pollination indicate that pod1 pollen tubes are defective specifically in micropylar pollen tube guidance.
Figure 3.
Scanning Electron Microscopy Analysis of Pollen Tube Guidance.
Wild-type pistils were pollinated with a limited number of pollen grains from pod1/POD1 plants and processed for scanning electron microscopy analysis 24 h after pollination. Pollen tubes are highlighted in yellow or green where a second tube is seen. Bars = 100 μm.
(A) The normal pollen tube enters the micropyle after growth along the funiculus.
(B) The pollen tube bypasses the micropyle.
(C) The pollen tube grows on the integument.
(D) and (E) The first pollen tube (P1) fails to enter the micropyle, and the second pollen tube (P2) follows up and enters the micropyle.
POD1 Is Essential for Early Embryo Development
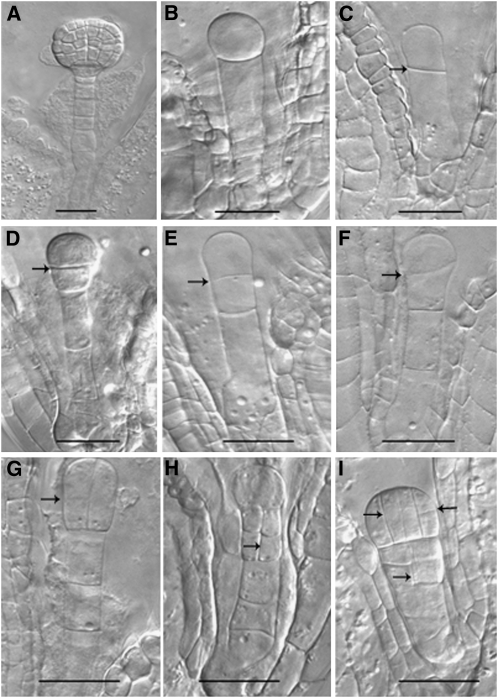
The low transmission rate of the pod1 mutation indicated that there could be additional defects in the pod1 mutants. For example, under limited pollination conditions, we found that 23.5% of the pod1 pollen tubes can enter the micropyle, but the Kanr/Kans ratio rises from 1:1 to 1.06:1 (n = 508), when the pollen of pod1/POD1 were pollinated on the stigma of pod1/POD1. However, under normal pollination conditions, where pollen is in excess, we seldom observe aborted seed due to the very low transmission efficiency of the pod1 mutation. The low transmission rate indicates that the pod1/pod1 embryos may be inviable. To confirm this, we self-pollinated pod1/POD1 pistils using limited pollen, then opened the siliques and examined the ovules (Ding et al., 2006). When the sibling embryos reached the globular embryo stage (88.5%) within the same silique (Figure 4A), 11.5% of the embryos were arrested at several early stages and also showed aberrant division planes (n = 578) (Figures 4B to 4I). The putative mutant zygotes underwent cell division with the aberrant cell plate usually at the wrong position or in the wrong orientation, yielding abnormal apical and basal cells (Figures 4B and 4C). The subsequent division plane of these cells was also aberrant. In the wild type, the division plane of apical cell was parallel to the apical-basal axis, but the apical cell of the putative pod1/pod1 often divided perpendicular to the apical-basal axis (Figures 4D to 4F). Rarely, the pre-embryo was able to divide further and formed an abnormal embryo, which gradually collapsed, and a suspensor with a likely reversed apical-basal axis (Figures 4G to 4I). These observations indicate that the cell plate does not form at the right position or orientation in the putative pod1/pod1 embryos. The percentage of embryo abortion is ~11.5%, which is half of the percentage of pod1 pollen tube targeting under limited pollination conditions, so the aborted embryos are most likely the pod1/pod1 homozygotes. Consistent with this, pod1/pod1 homozygous seedlings were never recovered in the progeny of plants fertilized by limited pollination. These results indicate that POD1 plays an essential role in cell plate orientation/positioning in early embryo patterning.
Figure 4.
Early Embryo Development in pod1/POD1.
(A) to (I) When the sibling embryos in the same silique reach the globular embryo stage (A), the putative homozygous mutant embryos are blocked at different stages ([B] to [I]).
(B) and (C) Mutant embryos are blocked after the first zygotic division.
(D) to (F) Mutant embryos development are blocked when the apical cell divides once with a abnormal horizontal or oblique division plane.
(G) to (I) Mutant embryos with abnormal division patterns in the apical cell and basal cell lineage.
Arrows indicate the presence of an aberrant cell plate. Bars = 20 μm.
POD1 Encodes a Novel Conserved Protein with Unknown Function
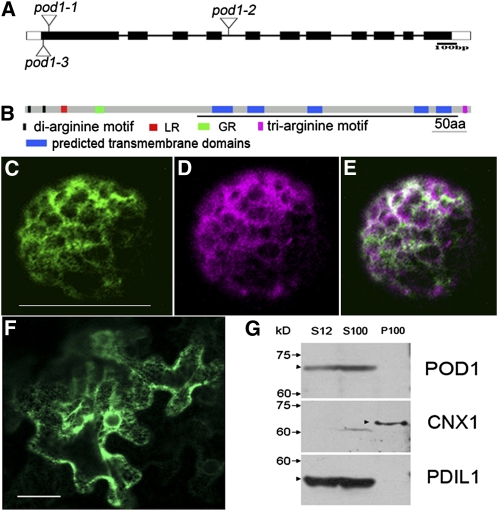
To identify the gene disrupted in pod1 mutant, the flanking sequence of the Ds element was isolated by thermal asymmetrical interlaced PCR and sequenced. Examination of the resulting sequence showed that the Ds was inserted into the first exon of At1G67960, 31 bp downstream of the ATG start codon, resulting in an 8-bp duplication (5′-GCCAAAAC-3′) typical for Ds insertions (Figure 5A). This Ds insertion allele is designated pod1-1. We also obtained two additional alleles, pod1-2 (SALK_049247) with a T-DNA insertion in the fourth intron and pod1-3 with a Ds insertion at 54 bp downstream of the ATG start codon in the first exon (Figure 5A). For both pod1-2/POD1 and pod1-3/POD1, when pollinated by wild-type pollen, the progeny display a Kanr/Kans ratio of 1:1 (500:500 for pod1-2 and 580:582 for pod1-3; Table 1), indicating 100% transmission through the female gametophyte. The efficiency of transmission through the male gametophyte of pod1-2 is 4% (n = 850) and the transmission of pod1-3 is 7% (n = 520). Moreover, pollen mutant for pod1-2 or pod1-3 showed defective micropylar pollen tube guidance (see Supplemental Figure 2 online). The analysis of these mutant alleles indicates that At1G67960 is the POD1 gene, and the disruption of this gene results in a defective pollen tube response to guidance cues from the female gametophyte.
Figure 5.
The Structure and Subcellular Localization of POD1.
(A) Three insertion alleles of pod1 designated as pod1-1 (Ds insertion line), pod1-2 (T-DNA insertion line), and pod1-3 (Ds insertion line).
(B) Domain structure of POD1 protein. The DUF747 domain is underlined. GR, Gly-rich motif; aa, amino acids.
(C) A confocal image showing POD1-GFP localization in an Arabidopsis protoplast. Bar = 50 μm.
(D) A confocal image of the same cell as (C) showing the localization of the ER marker mCherry-HDEL.
(E) Merged image of (C) and (D) shows the colocalization of POD1-GFP and mCherry-HDEL.
(F) A confocal image showing POD1-GFP localization in tobacco leaf cells. Bar = 50 μm.
(G) Immunoblot showing that POD1 protein is detected in total proteins (S12) and the soluble fraction (S100), but not the microsomal fraction (P100). CNX1 and PDIL1-1 are the controls as the membrane protein and soluble protein, respectively. Markers of molecular weight and POD1 protein are indicated on the left with arrows and arrowheads, respectively.
To further confirm that the phenotype is caused by mutation in At1G67960, a genetic complementation experiment was performed. A genomic fragment including a 1.628-kb promoter, the coding region including the exons and introns, and a 482-bp fragment downstream the stop codon was cloned and transformed into pod1/POD1 heterozygous plants. After double antibiotic selection for the Ds and the transgene, 30 T1 transgenic lines were obtained. The T2 generation seeds from six randomly chosen independent T1 lines were plated on Murashige and Skoog media supplemented with kanamycin for Kan segregation analysis. The Kanr/Kans ratio rose to 1.8–3:1, and plants homozygous for the Ds insertion were obtained in the T3 generation of these lines (Table 2). In addition, a construct containing the POD1 coding sequence (CDS) expressed in antisense orientation from the pollen-specific promoter LAT52 was transformed into wild-type plants. Six randomly selected lines out of 18 T1 transgenic plants were analyzed for pollen tube guidance, and all of them showed defective micropylar pollen tube guidance (see Supplemental Figure 3 online). Together, these data demonstrate that At1G67960 is indeed the POD1 gene. The CDS of POD1 driven by the LAT52 promoter can rescue the male defect of pod1 mutant and increase the Kanr/Kans of progeny from pod1-1/POD1 from 1:1 up to 2:1 (n = 2000). The LAT52 promoter is expressed in the vegetative cell but not the sperm cell (Twell, 1992; Eady et al., 1994). This demonstrates that POD1 functions in the vegetative cell of pollen tube to control the pollen tube response.
Table 2.
Complementation Analysis of the pod1/POD1 Mutant
| T2 Transgenic Plants | Kanr | Kans | Kanr/Kans |
| Genomic POD1-6 | 234 | 149 | 2.3:1 |
| Genomic POD1-10 | 204 | 109 | 1.87:1 |
| Genomic POD1-18 | 130 | 55 | 2.36:1 |
| Genomic POD1-20 | 314 | 98 | 3.2:1 |
| Genomic POD1-21 | 434 | 241 | 1.8:1 |
| Genomic POD1-26 | 220 | 101 | 2.2:1 |
| pLAT52:POD1 | 1324 | 676 | 1.96:1 |
| pPOD1:RXRRR#x003ERXAAA | 1186 | 600 | 1.97:1 |
| pPOD1:#x0394LR | 841 | 450 | 1.86:1 |
Constructs as indicated were transformed into pod1-1/POD1-1 background, and kanamycin selection was performed in T2 progeny. Genomic POD1 construct contains 1.628-kb promoter, coding region (including exons and introns), and 482-bp sequence downstream stop codon. The suffix numbers 6, 10, 18, 20, 21, and 26 indicate different transgenic lines. pLAT52:POD1 refers to POD1 CDS driven by LAT52 promoter. pPOD1:POD1RXRRR>RXAAA and pPOD1:POD1ΔLR refer to mutated POD1 CDS driven by the POD1 promoter, respectively. The Kanr/Kans ratio was obtained from at least five independent transgenic plants.
POD1 encodes a predicted protein of 624 amino acids belonging to the eukaryotic membrane protein superfamily pfam05346, the distinguishing feature of which is the presence of a domain of unknown function 747 (DUF747). POD1 homologs exist in many eukaryotic organisms, with the amino acid sequence similarity highest in the DUF747 domain. In addition to the DUF747 domain, POD1 contains a Lys-rich motif (LR; KRKRSKKKKKK) at the N terminus, followed by an Arabidopsis-specific Gly-rich motif (GGGGSGSSGGG) and five predicted hydrophobic transmembrane domains (Figure 5B). In general, the Lys/Arg-rich motif functions as a putative nuclear localization signal. However, it can also serve as a recognition site for molecular chaperones or in some cases it can serve as an interaction site for protein–protein or protein–RNA interaction (Wang et al., 2000; Boeddrich et al., 2006; Teft et al., 2009). The LR may also facilitate the interaction with diacylglycerol and/or acidic phospholipids for the full function of the enzyme activity (Rodríguez-Alfaro et al., 2004). In addition, two N-terminal di-arginine motifs (NRR1 and NRR2) and one C-terminal tri-arginine motif (CRRR) are present in POD1; these may serve as potential ER retention signals (Boulaflous et al., 2009). Sequence alignment shows that there is high similarity within the plant homologs and animal counterparts, respectively. Furthermore, plant POD1 proteins are ~150 amino acids longer at the N-terminal end and ~100 amino acids shorter at the C-terminal end than their homologs in other organisms (see Supplemental Figure 4 online). Phylogenetic analysis shows that Arabidopsis POD1 belongs to the plant clade, and the animal homologs form another clade (see Supplemental Figure 5 online).
POD1 Is an ER Luminal Protein
To determine the subcellular localization of POD1, we transformed a POD1-GFP (green fluorescent protein) fusion construct driven by the constitutive 35S promoter of Cauliflower mosaic virus into Arabidopsis mesophyll protoplasts and tobacco (Nicotiana tabacum) leaf cells. To mark the ER, we cotransformed the cells with an ER marker, mCherry-HDEL (Batoko et al., 2000). POD1-GFP colocalized with mCherry-HDEL in Arabidopsis protoplasts (Figures 5C to 5E) and tobacco leaf cells (Figure 5F), showing a reticulate ER network. Surprisingly, no membrane localization of POD1-GFP was observed although five putative transmembrane domains were predicted by hydrophobicity analysis. To make sure that these constructs are functional, we tested whether they could complement the pod1 mutation. Two fusion constructs, POD1-GFP and GFP-POD1, driven by the native POD1 promoter, were introduced into the pod1 mutant and were able to rescue the phenotype completely. To visualize the cellular localization of POD1, the 10-d-old seedlings were stained with the ER-specific dye ER-Tracker Blue-White DPX and then observed by confocal microscopy (Yi et al., 2009). The signal of POD1-GFP or GFP-POD1 colocalized with the ER-Tracker dye staining (see Supplemental Figure 6 online), confirming POD1’s ER localization.
To further clarify whether POD1 is a membrane protein or a soluble protein in the ER, we performed cellular fractionation by ultracentrifugation of total proteins of wild-type inflorescences. POD1 protein was detected in the soluble fraction (S100) by a POD1-specific antibody but was not detected in the microsomal fraction (P100). As a control, the ER membrane protein CNX1 was detected in the P100, and the ER luminal protein Protein Disulfide Isomerase 1-1 (PDIL1-1) was only detected in the S100 fraction (Figure 5G). Together, these data confirm that POD1 is an ER luminal protein.
POD1 Is Involved in ER Protein Retention
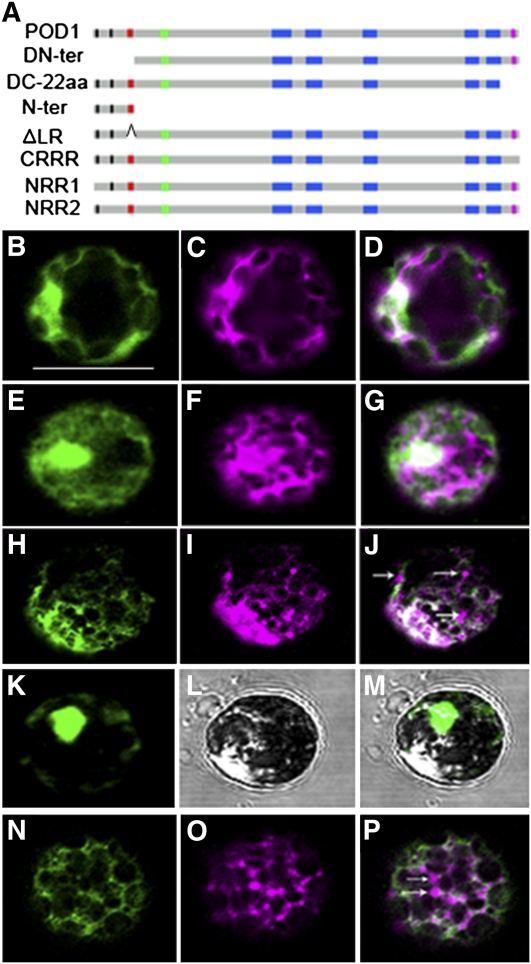
To dissect the functional domains of POD1, deletion and mutation experiments were performed. Sequence comparison of POD1 with its homologs in vertebrates, invertebrates, dicots, monocots, moss, ferns, and yeast indicates that, in addition to the DUF747 domain, the N- and C-terminal domains are conserved in plants (see Supplemental Figures 4 and 7 online). To determine whether the plant-specific domains are essential for POD1 function, we made two truncation constructs driven by the native POD1 promoter; the first construct lacked the N-terminal 60–amino acid residues, including the di-arginines (NRR1 and NRR2) and the LR motif (POD1DN-ter), and the second lacked the last C-terminal 22 amino acids, which is hydrophilic and basic, including the tri-arginine motif (CRRR) (POD1DC-22aa) (Figure 6A). The constructs were cotransformed with mCherry-HDEL into Arabidopsis protoplasts. The result shows that both deletions totally disrupted the ER localization of POD1 (Figures 6B to 6G). Consistent with the disrupted localization, these truncation constructs did not rescue the pod1 phenotype when introduced into the pod1 mutant. This indicates that both the N- and C-terminal domains are essential for the localization and function of POD1. In addition, no dominant-negative effect was observed when the two truncation constructs were transformed into wild-type plants.
Figure 6.
Mutation Analysis of POD1.
(A) A diagram showing different mutations and deletions of POD1 protein for POD1-GFP fusion constructs. aa, amino acids.
(B) to (P) Confocal images showing the localization of POD1m-GFP ([B], [E], [H], [K], and [N]), ER marker mCherry-HDEL ([C], [F], [I], and [O]) in respective channels, and the merged images ([D], [G], [J], [M], and [P]).
(B) to (D) Confocal images of the same protoplast showing that the N-terminal deletion (POD1DN-ter-GFP) disrupted the ER localization of POD1.
(E) to (G) Confocal images of the same protoplast showing C-22aa deletion (POD1DC-22aa-GFP) abolished the ER localization of POD1.
(H) to (J) Confocal images of the same protoplast showing that the CRRR mutation (POD1RXRRR>RXAAA-GFP) disrupted the ER localization of mCherry-HDEL. Arrows indicate the mislocalization of mCherry-HDEL.
(K) to (M) Confocal images of the same protoplast showing the nuclear localization of POD1Nter-GFP.
(N) to (P) Confocal images of the same protoplast showing POD1ΔLR-GFP disrupted the ER localization of mCherry-HDEL. Arrows indicate the mislocalization of mCherry-HDEL.
DN-ter, POD1 with the 60–amino acid N-terminal sequence deleted; DC-22aa, POD1 with the C-terminal 22 amino acids deleted; Nter, The N-terminal sequence including the LR; ΔLR, POD1 with the LR deleted. Bar = 50 μm.
To further dissect if the di-arginine motif in the N-terminal and the tri-arginine motif in the C-terminal are required for the ER localization of POD1, site-directed mutagenesis was performed. Sequence analysis of the N- and C-terminal sequence of the plant clade showed that the first (NRR1) is conserved in plants, but the second di-arginine (NRR2) is unique in Arabidopsis. The tri-arginine RRR motif (CRRR) in the C terminus is also conserved to some extent, and an Arg substitutes the Lys in monocots (see Supplemental Figure 7 online). So the conserved basic sequence may be important for the function of POD1. They could also be potential ER retention signals. To test these possibilities, mutations were introduced at NRR1 (POD1RXXXR>AXXXA), NRR2 (POD1RR>AA), and CRRR (POD1RXRRR>RXRAA, POD1RXRRR>RXAAR, and POD1RXRRR>RXAAA) (Figure 6A), respectively. Mutated POD1-GFP constructs (POD1m-GFP) were transiently expressed in Arabidopsis protoplasts. The result shows that mutations in neither NRR1 nor NRR2 disrupted the ER localization of POD1 (see Supplemental Figure 8 online). In the case of CRRR, the POD1RXRRR>RXAAA mutation, but not POD1RXRRR>RXRAA or POD1RXRRR>RXAAR, impaired the ER localization of mCherry-HDEL but not POD1 itself (Figures 6H to 6J). These data indicate that the tri-arginine motif of POD1 is essential for ER retention of HDEL proteins and this process requires at least one Arg in the tri-arginine motif.
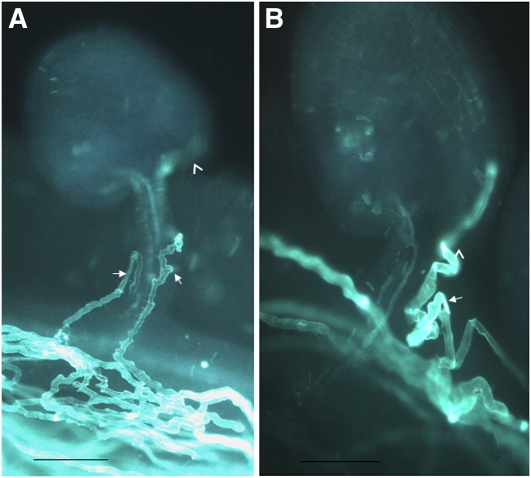
To investigate whether ER retention is essential for pollen tube guidance, POD1RXRRR>RXAAA driven by its native promoter was introduced into the wild-type and pod1/POD1 plants. Indeed, micropylar pollen tube targeting was affected in the transgenic plants under limited pollination conditions (Figure 7A). However, after twisting, ~80% of the pollen tubes (n = 800) finally enter the micropyle and finish fertilization. The POD1RXRRR>RXAAA construct can recover the Kanr/Kans ratio of pod1/POD1 from 1:1 to 2:1 in the T2 generation (n = 1786). In addition, pollen tubes carrying the POD1RXRRR>RXAAA construct displayed a zigzag morphology (Figure 7A), and this implies that the POD1RXRRR>RXAAA mutation also impairs pollen tube morphology.
Figure 7.
Dominant-Negative Effect of POD1RXRRR>RXAAA and POD1ΔLR.
(A) Micrograph showing defective pollen tube guidance and abnormal morphology in POD1RXRRR>RXAAA transgenic plants.
(B) Micrograph showing defective pollen tube guidance and abnormal morphology in POD1ΔLR transgenic plants.
Arrowhead, micropyle; arrow, pollen tube. Limited pollen grains from POD1RXRRR>RXAAA and POD1ΔLR transgenic plants were pollinated onto the wild-type stigma. Bars = 100 μm.
As shown in Figures 6B to 6D, the deletion of the N-terminal 60 amino acids (POD1DN-ter-GFP) impairs the ER localization of POD1. This raises the possibility that an ER localization signal may exist in the N-terminal region. To our surprise, POD1N-ter-GFP was mainly targeted to the nucleus (Figures 6K to 6M). This indicated that the N-terminal sequence may be targeted into the nucleus by the LR motif, although we did not observe nuclear localization of the full-length POD1 protein. Usually the Lys-rich motif is predicted to be a nuclear localization signal. To analyze the function of this potential nuclear localization signal, POD1ΔLR with the KRKRSKKKKKK sequence deletion was fused with GFP reporter gene. Transient assay in protoplasts showed that the localization pattern of POD1ΔLR-GFP did not change compared with that of POD1, but a portion of mCherry-HDEL escaped from the ER (Figures 6N to 6P). This is nearly the same as that of the POD1RXRRR>RXAAA mutation. Interestingly, transformation of this mutated POD1 into wild-type plants also reduced the ratio of micropylar pollen tube targeting (78%, n = 496) and resulted in abnormal tube morphology in vivo, similar to that of POD1RXRRR>RXAAA (Figure 7B). Consistent with this, the POD1ΔLR mutation can also restore the Kanr/Kans ratio of pod1/POD1 plants to 2:1 (n = 1291). These results confirm the indispensible role of the conserved Lys/Arg-rich motifs for POD1. These studies suggest that the N-terminal Lys/Arg-rich motif and the C-terminal tri-arginine of POD1 are essential for HDEL protein retention, which is required for pollen tube response in Arabidopsis. An HDEL retrieval signal generally exists in some ER luminal chaperones. Therefore, POD1 may be involved in the ER retention of these chaperones.
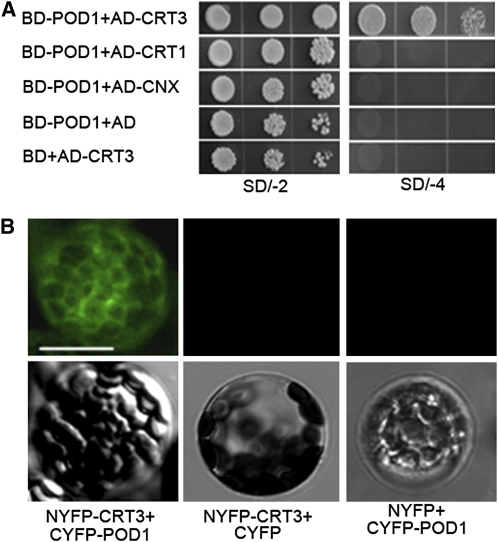
POD1 Interacts with CRT3
To investigate whether POD1 acts as an ER chaperone as it plays a role in ER protein retention, a yeast two-hybrid screen was performed. One candidate protein isolated was CRT3. To confirm the interaction, the full-length POD1 CDS and CRT3 CDS with the signal peptide for ER targeting deleted were cloned into pGBKT7 and pGADT7, respectively. The yeast cells cotransformed with these two plasmids grew well on the Trp-, Leu-, His-, Ade-dropout media (Figure 8A), indicating an interaction between these two proteins. To further test if POD1 also interacts with the other two closely related ER chaperones, CRT1 and CALNEXIN (CNX) with both the signal peptide and the transmembrane domain deletion were cloned into pGADT7. Yeast cells cotransformed with pGBKT7-POD1 and pGADT7-CRT1 or pGADT7-CNX did not grow on the Trp-, Leu-, His-, Ade-dropout media. This suggests that POD1 interacts specifically with CRT3 but not with its homologs CRT1 and CNX. The interaction between POD1 and CRT3 was further confirmed by bimolecular fluorescence complementation (BiFC). Protoplasts cotransformed with NYFP-CRT3 and CYFP-POD1 showed a yellow fluorescent protein (YFP) signal (Figure 8B), and no YFP signal was detected in the control experiment.
Figure 8.
Interaction between POD1 and CRT3.
(A) Yeast cotransformed with the plasmids as indicated on the left. The left picture shows yeast grown on SD/-2 (SD/-Trp-Leu) dropout media, and the right picture shows yeast grown on SD/-4 (SD/-Trp-Leu-His-Ade) dropout media. Cells grown on SD/-4 dropout media are indicative of physical interaction between POD1 and CRT3. Note that there is no interaction between POD1 and CRT1 or CNX.
(B) BiFC experiment showing YFP signal in protoplasts cotransformed with NYFP-CRT3 and CYFP-POD1, indicating physical interaction between CRT3 and POD1. No YFP signal is detected when cotransformed with NYFP-CRT3 and CYFP, or NYFP and CYFP-POD1 plasmids. Top row, confocal micrograph; bottom row, the corresponding differential interference contrast images. Bar = 25 μm.
Consistent with the interaction of CRT3 with POD1, CRT3 is expressed in pollen grains and pollen tubes, as revealed by Arabidopsis microarray analysis (Qin et al., 2009). Similarly, the transcripts of CRTs are detected in pollen grains, pollen tubes, and pistils in Haemanthus (Lenartowska et al., 2009). Together, these data suggest that POD1 most likely interacts with CRT3 and plays an important role in ER quality control.
Expression Pattern of POD1 in Planta
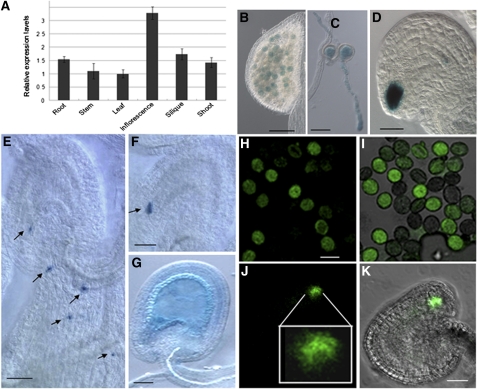
To characterize the temporal and spatial regulation of POD1, its expression pattern was analyzed. First quantitative RT-PCR (qRT-PCR) was performed with RNAs from root, stem, leaf, inflorescence, silique, and shoot. The data showed that POD1 mRNA is present in multiple tissues, with a relatively high level in inflorescence, silique, root, and shoot (Figure 9A). To provide a more precise expression pattern, a genomic sequence fusion with β-glucuronidase (GUS) or GFP driven by the POD1 native promoter was transformed into Arabidopsis, and four of five transgenic lines showed positive GUS staining. The GFP line is the same as in Supplemental Figure 6 online. GUS staining of transgenic lines and confocal microscopy of GFP-expressing transgenic lines showed that POD1 is expressed in mature pollen and pollen tubes (Figures 9B, 9C, 9H, and 9I). Although pod1 shows no female gametophytic defect, POD1 is expressed abundantly in synergid cells and weakly in the antipodal cells (Figures 9D, 9J, and 9K). When pollen from the GUS line was pollinated onto the wild-type stigma, the GUS signal was observed at the tip of the pollen tube (Figure 9E). When the tubes burst in the embryo sac, the POD1-GUS protein was released into the synergid cell (Figure 9F). Consistent with the phenotype of embryo abortion, POD1 is also expressed in the early embryo and endosperm (Figure 9G). The expression pattern of POD1 suggests that it functions in multiple developmental processes.
Figure 9.
Expression Pattern of POD1.
(A) Relative expression levels of POD1 in multiple tissues by real-time PCR. Bars represent the average ± se of normalized relative transcript levels of three replicates.
(B) to (G) GUS staining of POD1:POD1-GUS transgenic plants.
(H) to (K) POD1-GFP in pollen and synergid cells of POD1:POD1-GFP transgenic plants.
(B) POD1-GUS in pollen grains.
(C) POD1-GUS in pollen tubes germinated in vitro.
(D) POD1-GUS in synergid cells.
(E) POD1-GUS at pollen tube tips (arrows).
(F) POD1-GUS in wild-type ovules fertilized with POD1-GUS pollen (arrow).
(G) POD1-GUS in early embryo and endosperm.
(H) Confocal laser scanning microscopy images showing the POD1-GFP in pollen grains.
(I) Overlay of (H) and DIC image.
(J) Confocal laser scanning microscopy image showing the POD1-GFP in synergid cells. The image in the square is the enlarged view of the synergid cells.
(K) Overlay of (J) and DIC image. Bars = 50 μm.
DISCUSSION
We identified a male gametophytic mutant specifically defective in micropylar pollen tube guidance and cell plate patterning during early embryogenesis. Confocal microscopy and cellular fractionation showed that POD1 is an ER luminal protein. Mutagenesis indicates that the C-terminal tri-arginine motif and the N-terminal Lys/Arg-rich motif are essential for the function of POD1 in the ER retention of proteins with an HDEL motif. Interaction of POD1 with CRT3 indicates that POD1 might act as a cochaperone in the ER lumen. POD1 likely acts as a component of the CRT3 complex, which is involved in ER retention and perhaps also in ER-mediated protein secretory pathways, to regulate the pollen tube response to female gametophyte signals and cell plate patterning during early embryogenesis in plants.
POD1 Functions in ER Protein Retention
ER resident proteins are necessary for normal ER function. They usually contain specialized targeting signals (the signal peptide) consisting of specific amino acids, which is recognized by the signal recognition particle and translocated into the ER. The signal peptide is often located at the N or C terminus or sometimes internal to the proteins. In general, the signal peptides are cleaved by signal peptidase, but in some cases, they are retained in the mature protein sequence (Lingappa et al., 1979; Guo et al., 2011). Our data suggest that the N-terminal 60 amino acids and the C-terminal 22 amino acids are essential for the localization of POD1 in the ER. Therefore, the ER targeting signal of POD1 may reside in these two regions, but no typical ER retention or targeting signal peptide is predicted in these regions. The fact that both the N- and C-terminal GFP fusion proteins of POD1 showed ER localization suggests that neither the N nor C terminus is cleaved by signal peptidase. So, we propose two possible molecular mechanisms of the ER localization of POD1. First, POD1 does not possess the typical ER retention or targeting sequence, and its translocation into the ER may be signal recognition particle independent. Second, the N- or C-terminal domains required for POD1 ER retention may interact with ER resident proteins that possess the HDEL retrieval signals. Both scenarios require more studies on POD1 sequence and its interacting proteins. Further detailed mapping of the N- and C-terminal domains essential for the ER localization of POD1 will be helpful to identify new mechanism of the ER retention signal.
POD1 functions in the retention of ER proteins with HDEL retrieval signals by an unknown mechanism. It is known that ER luminal resident proteins are constitutively secreted to the Golgi. To be retained in the ER, soluble ER-resident proteins have to escape from the bulk flow of secreted proteins. Many ER luminal proteins, such as CRT, PDIL, CNX, and BiP, have a C-terminal H/KDEL motif that acts as an ER retrieval signal (Munro and Pelham, 1987). H/KDEL is recognized and retrieved by the integral membrane H/KDEL receptors in the Golgi complex (Pagny et al., 1999; Capitani and Sallese, 2009). However, the amount of secretion of resident proteins, such as CRT3 and BiP, is very low, and one possible reason for this is that these chaperones may associate with newly folding proteins (Pfeffer, 2007). So H/KDEL acts by retrieval rather than retention and other mechanisms yet to be unveiled (Capitani and Sallese, 2009). This view is also supported by the fact that some ER chaperones, like SDF2 and ERdj3b, lack an H/KDEL motif at the C terminus. Our data show that POD1 acts as an ER luminal protein without the K/HDEL motif and plays a role in the ER retention of HDEL proteins. The basic motifs in the N and C terminus are likely involved in this process. Two dominant-negative mutations, POD1RXRRR>RXAAA and POD1ΔLR, cause migration of mCherry-HDEL protein out of the ER and impair pollen tube guidance and morphology. Therefore, ER retention of specific ER resident proteins by POD1 plays an important role in pollen tube guidance. Similarly, mutations in YER140W, the yeast homolog of POD1, result in the escape of a well-characterized ER chaperone (BiP) from the ER (Copic et al., 2009) and consequently inhibit filamentous growth (Jin et al., 2008) and induce unfolded protein response (Jonikas et al., 2009). So HDEL protein retention may be a general function of POD1 and its homologs in eukaryotic organisms.
The Role of POD1 in Pollen Tube Guidance and Early Embryogenesis
Pollen tube growth in the pistil is a continuous process of guidance and reception. Previous genetic and molecular analysis suggests that different mechanisms function during sporophytic and gametophytic pollen tube guidance (Ray et al., 1997; Higashiyama et al., 1998; Shimizu and Okada, 2000; Chae and Lord, 2011). The ER luminal POD1 acts specifically in the micropylar response to female cues, but obviously through an indirect mechanism. The interaction with CRT3 suggests that POD1 may be involved ER quality control, in which ER chaperones assist and monitor the proper maturation and formation of the nascent proteins in the secretory pathway. In Arabidopsis, ~30% of total proteins enter the secretory pathway through the ER, and among them 33% are transmembrane proteins.
ER-dependent maturation of membrane receptors during host–pathogen interactions in plants has been reported. CRTs are multifunctional ER chaperones involved in protein folding and Ca2+ homeostasis. Three CRTs are found in the Arabidopsis genome, and CRT1 and CRT2 function as general ER chaperones, while CRT3 is specifically associated with the formation of some membrane receptors, such as elongation factor Tu receptor (EFR) (Christensen et al., 2010). The maturation of EFR is dependent on the ER quality control components STT3A, SDF2, BiP, and CRT3. These proteins are essential for the N-glycosylation, accumulation, folding, and maturation of EFR. The processing of EFR is abolished and the immunity response to the pathogen is lost when the genes encoding the chaperones are knocked out (Nekrasov et al., 2009; Häweker et al., 2010). In rice (Oryza sativa), the ER chaperone BiP3 interacts with the pattern recognition receptor XA21, which is involved specifically in resistance to pathogen Xoo (Park et al., 2010). It has also been shown that SHEPHERD, an ER-resident HSP90-like protein in Arabidopsis, controls the formation and/or folding of CLAVATA, a receptor controlling meristem activity (Ishiguro et al., 2002). Similarly, the folding of the bradykinin receptor in ES cells and the Toll receptor in mice is also dependent on ER chaperones (Nakamura et al., 2001; Randow and Seed, 2001). All these findings suggest that the ER quality control system is essential for formation and accumulation of receptors. In this context, it is plausible to speculate that POD1, via modulating CRT3 function, plays a key role in the ER quality control of putative membrane receptors perceiving the female signal molecules, such as LUREs, during pollen tube guidance in plants.
In addition to protein folding and vesicle trafficking, ion homeostasis, which is fundamental to the axis of polarity in yeast, animals, and plants, requires the proper function of the ER. So another possibility is that POD1 affects the folding of ER-localized cation transporters, such as the putative K+ transporters CHX21 and 23 (Lu et al., 2011) and cyclic nucleotide-gated channels (Frietsch et al., 2007), and subsequently disrupts the Ca2+ and K+ homeostasis in the ER and Ca2+ signaling. Another possibility is that POD1 may regulate cytosolic Ca2+ homeostasis and its regional concentration by tethering CRT3 activity to mediate the precise micropylar guidance. Finally, POD1 may have a general effect on proteins that are folded or modified in the ER. To clarify these possibilities, more factors involved in the pollen tube response need to be identified. It would be interesting to compare the proteome of pod1 with that of the wild-type pollen tubes, which may facilitate the identification of POD1-interacting proteins that may be the male factors directly involved in pollen tube guidance.
In addition to its role in pollen tube guidance, POD1 is also important for early embryogenesis. Occasionally, the pod1 egg cells can be fertilized by the pod1 sperm cells under limited pollination to form arrested zygotes or embryos that are defective in positioning and orientation of the cell plate. In mouse, mutation of TAPT1, the putative homolog of POD1, results in a reversed posterior-anterior axis polarity of the axial skeleton, suggesting a role for TAPT1 in body axis formation (Howell et al., 2007). Similar reversion of apical-basal polarity likely takes place in pod1 embryos in Arabidopsis, as shown earlier. The knockdown of POD1 homolog NP507972 in Caenorhabditis elegans causes embryo lethality, but no detailed analysis of embryo polarity has been reported (http://www.wormbase.org/).
The mechanism of the pod1 embryo defect is not clear, but it is plausible to speculate that the ER quality control and subsequently the secretory systems that require the function of POD1 are needed for positioning and orientation of the cell plate in zygotes and embryos. In plant cells, studies on cell plate orientation have revealed roles of microtubule and associated proteins on preprophase band and phragmoplast formation (Müller et al., 2009) but have not yet provided clues on the role of the ER in these processes. It is believed that cell plate formation requires the deposition of newly synthesized materials from Golgi apparatus–derived secretory vesicles. Recently, it was also shown that protein secretion and endocytic pathways are essential for cell plate formation (Dhonukshe et al., 2006; Reichardt et al., 2007; Boutté et al., 2010). The inhibition of ER-Golgi trafficking causes the retention of newly synthesized proteins, such as the cytokinesis-specific syntaxin KNOLLE in the ER, therefore preventing cell plate formation (Reichardt et al., 2007). In conclusion, POD1 might function in ER quality control or the protein secretory system that is required for proper positioning and orientation of the cell plate during embryogenesis.
METHODS
Plant Materials and Growth Conditions
Seeds of Arabidopsis thaliana were surface-sterilized with 20% bleach for 10 min, washed four times with sterile water, and plated onto Murashige and Skoog media (Sigma-Aldrich) supplemented with 50 mg/L kanamycin for Ds and T-DNA insertion lines or 40 mg/L hygromycin for pCAMBIA-based constructs. After 7 d of growth on a Murashige and Skoog plate in the greenhouse (16 h light/8 h dark, 22°C), seedlings are transferred to soil for further growth. SALK_049247 was obtained from The Arabidopsis Information Resource (TAIR) seed stock center. pod1-3 is a Ds insertion line obtained from De Ye’s lab at China Agricultural University.
Phenotypic Analysis
To investigate pollen development, flowers were dipped several times in the 4′,6-diamidino-2-phenylindole or Alexander solution droplets directly on microscope slides and observed under a fluorescence microscope. For the pollen in vitro germination assay, mature pollen grains were spread onto the pollen germination medium containing 10 mM CaCl2, 10 mM Ca(NO3)2, 10 mM MgSO4, 10 mM H3BO4, 1.8% Suc, and 0.1% agarose and cultured for 5 to 12 h at 25°C. The aniline blue staining, GUS staining, and scanning electron microscopy were performed as described previously (Chen et al., 2007). For embryo observation, the ovules were dissected out with a fine needle, cleared according to Li et al. (2010), and observed using a Zeiss microscope equipped with differential interference contrast (DIC) optics.
Protein Extraction and Immunoblot Analysis
Flowers from Arabidopsis were collected and ground in liquid nitrogen to a fine powder, and then total protein was extracted with ice-cold extraction buffer (50 mM Tris, pH 7.5, 1 mM EDTA, and 0.25 M Suc supplemented with PMSF and protease inhibitor cocktail [Roche]). The mixture was centrifuged at 1,000g for 25 min at 4°C. Supernatant (S1) was filtered through four layers of miracloth (Calbiochem). The filtered supernatant was centrifuged at 12,000g for 30 min. Then the supernatant (S12) was transferred to a new ultracentrifuge tube (Beckman) and subsequently centrifuged at 100,000g for 2 h at 4°C. The supernatant (S100) was collected and the pellet (P100) was rinsed several times with cold extraction buffer. The pellet was then incubated with detergent buffer (5 mM HEPES-KOH, pH 7.0, 20% Triton X-100, and 1% Suc) on ice for 30 min. All the samples were mixed with SDS-PAGE loading buffer and boiled before gel fractionation. Immunoblots were performed using specific antibody, which was generated with the POD1 peptide INRCRRRNSSHLHND as antigen to immune rabbit and affinity purified. Antibody specificity of POD1, CNX1, and PDIL1-1 was verified using protein extracts from the wild type and corresponding mutant or transgenic plants carrying 35S: FLAG-POD1.
Constructs and Plant Transformation
All primers are listed in Supplemental Table 1 online. For the complementation construct, a 5479-bp genomic fragment containing 1.628-kb promoter region and the transcribed region (including intron and exon) plus a 482-bp fragment downstream stop codon was amplified with primers POD1g-F and POD1g-R from Arabidopsis genomic DNA and ligated to pCAMBIA1300 to generate p1300-gPOD1-Ter, which was transformed by floral dip method into pod1 mutants (Clough and Bent, 1998). Similarly, the pPOD1:gPOD1-GUS fusion with GUS CDS inserted before the stop codon was made and transformed into Arabidopsis plants for POD1 gene expression analysis. The POD1 CDS (1875 bp in length) amplified with POD1-F and POD1-R was cloned into p1300 flanked by the LAT52 promoter and the 498 bp POD1 3′ untranslated region amplified with primers T-F and T-R to generate LAT52:POD1. The LAT52:POD1AS construct was cloned in the same way as the LAT52:POD1 but with the POD1 CDS in a reverse direction.
For subcellular localization of POD1, pBSK-35S:POD1-EGFP-polyA was made by subcloning POD1 CDS into pBSK-35S:EGFP-polyA at the SalI-BamHI site. All the deletion and mutation constructs were cloned into pBSK-35S-EGFP-polyA to produce a POD1-GFP fusion. For the BiFC constructs, the full-length CDS of POD1 and CRT3 was cloned into the XbaI and KpnI sites of pSPYNE and pSPYCE plasmid (Walter et al., 2004). The protoplast transformation was performed by the method described by Asai et al. (2002). The protoplasts were transformed with NYFP-CRT3 and CYFP-POD1, and protoplasts were transformed with NYFP-CRT3 plus CYFP and NYFP plus CYFP-POD1 as the negative control. The protoplasts were then viewed with a Zeiss LSM510 META laser scanning microscope with a 488- and 543-nm laser. For ER staining, seedlings were stained in 2 μM ER Tracker Blue-White DPX (Invitrogen) solution diluted in W5 buffer (154 mM NaCl, 125 mM KCl, 5 mM Glc, and 0.03% MES, pH 5.8) for 15 min. The signal was excited with a 405-nm laser and detected using the band-pass 420- to 480-nm filter.
Yeast Two-Hybrid Assay
The POD1 CDS and CRT3 with its ER targeting signal deleted were amplified from floral cDNAs with primers POD1y-F/POD1y-R and CRT3y-F/CRT3y-R (primer sequences in Supplemental Table 1 online) and subcloned into pGBKT7 and pGADT7 (Clontech), respectively. The CRT1 cDNA was amplified with CRT1-F and CRT1-R, and CALNEXIN was amplified with CNX-F and CNX-R from plasmid, which was a kind gift from Jianming Li (Michigan State University).
qRT-PCR
The total RNA was extracted from different Arabidopsis tissues using TRIzol reagent (Invitrogen). First-strand cDNA synthesis was performed in a 20-μL reaction mixture with 1 μg of DNase-digested (Roche) total RNA using the superscript III reverse transcriptase (Invitrogen) with random primers (TaKaRa) according to the manufacturer’s instructions. The cDNA product was diluted and 2 ng was used in each qRT-PCR. Real-time PCR reactions containing SYBR Green PCR master mix were performed on the ABI PRISM 7900 HT Real-Time PCR system (Applied Biosystems). The following thermal reaction was used: 50°C for 2 min, 95°C for 5 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. Three technical replicates were performed with each cDNA sample. Primers for 18s rRNA were used to normalize the qRT-PCR data. The analysis of the qRT-PCR data was performed according to the geNORM manual (http://medgen.ugent.be/~jvdesomp/genorm/). The primers used were POD1q-F and POD1q-R. Primer sequences are provided in Supplemental Table 1 online.
Bioinformatics and Phylogenetic Analysis
We used the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and TAIR (http://www.Arabidopsis.org) to analyze the cDNA and genomic sequence. The phylogenetic tree is processed with MEGA 5 software. The alignment in Supplemental Figure 4 was generated using the alignment program ClustalW (network protein sequence analysis). The alignment used for phylogeny is produced with ClustalW2 program at the European Bioinformatics Institute (http://www.ebi.ac.uk/clustalw/index.html). The weight matrix used was Gonnet with the penalty of the gap opening 10 and gap extension 0.1 for slow pairwise alignment and gap opening 10, gap extension 0.2, and gap distance 5 for multiple sequence alignment. The alignment was manually fine-tuned with DNAMAN software (http://www.lynnon.com/pc/framepc.html) and has been made available in FASTA format (see Supplemental Data Set 1 online). The phylogeny of the aligned sequences was generated with neighbor-joining algorithm (Saitou and Nei, 1987) by the MEGA 5 software (Tamura et al., 2011), and bootstrap analysis was conducted with 1000 replications.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: POD1, At1g67960; CNX1, AT5G61790; CRT3, AT1G08450; and PDIL1-1, AT1G21750.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Development of pod1 Pollen Is Normal.
Supplemental Figure 2. pod1-2 and pod1-3 Pollen Tubes Showing the Micropylar Targeting Defect.
Supplemental Figure 3. Defective Pollen Tube Guidance in POD1 Antisense Transgenic Plants.
Supplemental Figure 4. Sequence Alignment of POD1 and Its Homologs.
Supplemental Figure 5. Phylogenetic Tree of Arabidopsis POD1 and Its Homologs.
Supplemental Figure 6. POD1-GFP Is Localized in the ER.
Supplemental Figure 7. Sequence Alignment of the N Terminus and C Terminus Showing NRR1, NRR2, CRRR, LR, and C-22aa Sequences.
Supplemental Figure 8. POD1RXXXR>AXXXA and POD1RR>AA Localization in the ER.
Supplemental Table 1. List of Primers Used in This Study.
Supplemental Data Set 1. Sequence Alignment Used for the Phylogenetic Analysis.
Acknowledgments
We thank Jianmin Li (University of Michigan) for kindly providing the CNX plasmid and Yihua Zhou (Institute of Genetics and Developmental Biology, Beijing, China) for the mCherry-HDEL plasmid. This work was supported by the Ministry of Science and Technology of China (2007CB947600) and the National Natural Science Foundation of China (30830063 and 30921003) to W.-C.Y.
AUTHOR CONTRIBUTIONS
H.-J.L. and W.-C.Y. designed the research and wrote the article. H.-J.L., Y.X., and T.W. performed the research and analyzed the data. F.C. and Q.X. provided the antibody of CNX and PDIL2-1. D.-J.J. and D.Y. provided the plant material of pod1-3. D.-Q.S. and J.L. provided technical assistance.
References
- Alandete-Saez M., Ron M., McCormick S. (2008). GEX3, expressed in the male gametophyte and in the egg cell of Arabidopsis thaliana, is essential for micropylar pollen tube guidance and plays a role during early embryogenesis. Mol. Plant 1: 586–598 [DOI] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Batoko H., Zheng H.Q., Hawes C., Moore I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeddrich A., et al. (2006). An arginine/lysine-rich motif is crucial for VCP/p97-mediated modulation of ataxin-3 fibrillogenesis. EMBO J. 25: 1547–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulaflous A., Saint-Jore-Dupas C., Herranz-Gordo M.C., Pagny-Salehabadi S., Plasson C., Garidou F., Kiefer-Meyer M.C., Ritzenthaler C., Faye L., Gomord V. (2009). Cytosolic N-terminal arginine-based signals together with a luminal signal target a type II membrane protein to the plant ER. BMC Plant Biol. 9: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutté Y., Frescatada-Rosa M., Men S., Chow C.M., Ebine K., Gustavsson A., Johansson L., Ueda T., Moore I., Jürgens G., Grebe M. (2010). Endocytosis restricts Arabidopsis KNOLLE syntaxin to the cell division plane during late cytokinesis. EMBO J. 29: 546–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G., Cresti M. (2009). Organelle motility in the pollen tube: a tale of 20 years. J. Exp. Bot. 60: 495–508 [DOI] [PubMed] [Google Scholar]
- Capitani M., Sallese M. (2009). The KDEL receptor: new functions for an old protein. FEBS Lett. 583: 3863–3871 [DOI] [PubMed] [Google Scholar]
- Chae K., Lord E.M. (2011). Pollen tube growth and guidance: Roles of small, secreted proteins. Ann Bot. 108: 627–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.H., Li H.J., Shi D.Q., Yuan L., Liu J., Sreenivasan R., Baskar R., Grossniklaus U., Yang W.C. (2007). The central cell plays a critical role in pollen tube guidance in Arabidopsis. Plant Cell 19: 3563–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.Y., Wu H.M. (2008). Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu. Rev. Plant Biol. 59: 547–572 [DOI] [PubMed] [Google Scholar]
- Christensen A., et al. (2010). Higher plant calreticulins have acquired specialized functions in Arabidopsis. PLoS ONE 5: e11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Copic A., Dorrington M., Pagant S., Barry J., Lee M.C., Singh I., Hartman J.L.T., IV, Miller E.A. (2009). Genomewide analysis reveals novel pathways affecting endoplasmic reticulum homeostasis, protein modification and quality control. Genetics 182: 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., Baluska F., Schlicht M., Hlavacka A., Samaj J., Friml J., Gadella T.W., Jr (2006). Endocytosis of cell surface material mediates cell plate formation during plant cytokinesis. Dev. Cell 10: 137–150 [DOI] [PubMed] [Google Scholar]
- Ding Y.H., Liu N.Y., Tang Z.S., Liu J., Yang W.C. (2006). Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 18: 815–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eady C., Lindsey K., Twell D. (1994). Differential activation and conserved vegetative cell specific activity of a late pollen promoter in species with bicellular and tricellular pollen. Plant J. 5: 543–550 [Google Scholar]
- Frietsch S., Wang Y.F., Sladek C., Poulsen L.R., Romanowsky S.M., Schroeder J.I., Harper J.F. (2007). A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. Proc. Natl. Acad. Sci. USA 104: 14531–14536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo K.K., Tang Q.H., Zhang Y.M., Kang K., He L. (2011). Identification of two internal signal peptide sequences: Critical for classical swine fever virus non-structural protein 2 to trans-localize to the endoplasmic reticulum. Virol. J. 8: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häweker H., Rips S., Koiwa H., Salomon S., Saijo Y., Chinchilla D., Robatzek S., von Schaewen A. (2010). Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 285: 4629–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler P.K., Vidali L., Cheung A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17: 159–187 [DOI] [PubMed] [Google Scholar]
- Higashiyama T., Hamamura Y. (2008). Gametophytic pollen tube guidance. Sex. Plant Reprod. 21: 17–26 [Google Scholar]
- Higashiyama T., Inatsugi R., Sakamoto S., Sasaki N., Mori T., Kuroiwa H., Nakada T., Nozaki H., Kuroiwa T., Nakano A. (2006). Species preferentiality of the pollen tube attractant derived from the synergid cell of Torenia fournieri. Plant Physiol. 142: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama T., Kuroiwa H., Kawano S., Kuroiwa T. (1998). Guidance in vitro of the pollen tube to the naked embryo sac of torenia fournieri. Plant Cell 10: 2019–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama T., Kuroiwa H., Kuroiwa T. (2003). Pollen-tube guidance: Beacons from the female gametophyte. Curr. Opin. Plant Biol. 6: 36–41 [DOI] [PubMed] [Google Scholar]
- Higashiyama T., Yabe S., Sasaki N., Nishimura Y., Miyagishima S, Kuroiwa H., Kuroiwa T. (2001). Pollen tube attraction by the synergid cell. Science 293: 1480–1483 [DOI] [PubMed] [Google Scholar]
- Howell G.R., Shindo M., Murray S., Gridley T., Wilson L.A., Schimenti J.C. (2007). Mutation of a ubiquitously expressed mouse transmembrane protein (Tapt1) causes specific skeletal homeotic transformations. Genetics 175: 699–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A.B., Kolodkin A.L., Ginty D.D., Cloutier J.F. (2003). Signaling at the growth cone: Ligand-receptor complexes and the control of axon growth and guidance. Annu. Rev. Neurosci. 26: 509–563 [DOI] [PubMed] [Google Scholar]
- Hülskamp M., Schneitz K., Pruitt R.E. (1995). Genetic evidence for a long-range activity that directs pollen tube guidance in Arabidopsis. Plant Cell 7: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S., Watanabe Y., Ito N., Nonaka H., Takeda N., Sakai T., Kanaya H., Okada K. (2002). SHEPHERD is the Arabidopsis GRP94 responsible for the formation of functional CLAVATA proteins. EMBO J. 21: 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R., Dobry C.J., McCown P.J., Kumar A. (2008). Large-scale analysis of yeast filamentous growth by systematic gene disruption and overexpression. Mol. Biol. Cell 19: 284–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Borevitz J.O., Preuss D. (2007). Genome-wide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genet. 3: 1848–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas M.C., Collins S.R., Denic V., Oh E., Quan E.M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J.S., Schuldiner M. (2009). Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323: 1693–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara R.D., Portereiko M.F., Sandaklie-Nikolova L., Rabiger D.S., Drews G.N. (2005). MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17: 2981–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B. (2008). Spatial control of Rho (Rac-Rop) signaling in tip-growing plant cells. Trends Cell Biol. 18: 119–127 [DOI] [PubMed] [Google Scholar]
- Lenartowska M., Lenartowski R., Smoliński D.J., Wróbel B., Niedojadło J., Jaworski K., Bednarska E. (2009). Calreticulin expression and localization in plant cells during pollen-pistil interactions. Planta 231: 67–77 [DOI] [PubMed] [Google Scholar]
- Li H.J., Liu N.Y., Shi D.Q., Liu J., Yang W.C. (2010). YAO is a nucleolar WD40-repeat protein critical for embryogenesis and gametogenesis in Arabidopsis. BMC Plant Biol. 10: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingappa V.R., Lingappa J.R., Blobel G. (1979). Chicken ovalbumin contains an internal signal sequence. Nature 281: 117–121 [DOI] [PubMed] [Google Scholar]
- Liu Y., Misamore M.J., Snell W.J. (2010). Membrane fusion triggers rapid degradation of two gamete-specific, fusion-essential proteins in a membrane block to polygamy in Chlamydomonas. Development 137: 1473–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Chanroj S., Zulkifli L., Johnson M.A., Uozumi N., Cheung A., Sze H. (2011). Pollen tubes lacking a pair of K+ transporters fail to target ovules in Arabidopsis. Plant Cell 23: 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden K., Snyder M. (1998). Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol. 52: 687–744 [DOI] [PubMed] [Google Scholar]
- Márton M.L., Cordts S., Broadhvest J., Dresselhaus T. (2005). Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307: 573–576 [DOI] [PubMed] [Google Scholar]
- Michard E., Lima P.T., Borges F., Silva A.C., Portes M.T., Carvalho J.E., Gilliham M., Liu L.H., Obermeyer G., Feijó J.A. (2011). Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 332: 434–437 [DOI] [PubMed] [Google Scholar]
- Mori T., Kuroiwa H., Higashiyama T., Kuroiwa T. (2006). GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8: 64–71 [DOI] [PubMed] [Google Scholar]
- Müller S., Wright A.J., Smith L.G. (2009). Division plane control in plants: New players in the band. Trends Cell Biol. 19: 180–188 [DOI] [PubMed] [Google Scholar]
- Munro S., Pelham H.R. (1987). A C-terminal signal prevents secretion of luminal ER proteins. Cell 48: 899–907 [DOI] [PubMed] [Google Scholar]
- Nakamura K., et al. (2001). Functional specialization of calreticulin domains. J. Cell Biol. 154: 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V., et al. (2009). Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28: 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell M., Chance R.K., Bashaw G.J. (2009). Axon growth and guidance: Receptor regulation and signal transduction. Annu. Rev. Neurosci. 32: 383–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S., et al. (2009). Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361 [DOI] [PubMed] [Google Scholar]
- Pagny S., Lerouge P., Faye L., Gomord V. (1999). Signals and mechanisms for protein retention in the endoplasmic reticulum. J. Exp. Bot. 50: 157–164 [Google Scholar]
- Palanivelu R., Preuss D. (2000). Pollen tube targeting and axon guidance: Parallels in tip growth mechanisms. Trends Cell Biol. 10: 517–524 [DOI] [PubMed] [Google Scholar]
- Park C.J., Bart R., Chern M., Canlas P.E., Bai W., Ronald P.C. (2010). Overexpression of the endoplasmic reticulum chaperone BiP3 regulates XA21-mediated innate immunity in rice. PLoS ONE 5: e9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S.R. (2007). Unsolved mysteries in membrane traffic. Annu. Rev. Biochem. 76: 629–645 [DOI] [PubMed] [Google Scholar]
- Qin Y., Leydon A.R., Manziello A., Pandey R., Mount D., Denic S., Vasic B., Johnson M.A., Palanivelu R. (2009). Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet. 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F., Seed B. (2001). Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat. Cell Biol. 3: 891–896 [DOI] [PubMed] [Google Scholar]
- Ray S.M., Park S.S., Ray A. (1997). Pollen tube guidance by the female gametophyte. Development 124: 2489–2498 [DOI] [PubMed] [Google Scholar]
- Reichardt I., Stierhof Y.D., Mayer U., Richter S., Schwarz H., Schumacher K., Jürgens G. (2007). Plant cytokinesis requires de novo secretory trafficking but not endocytosis. Curr. Biol. 17: 2047–2053 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Alfaro J.A., Gomez-Fernandez J.C., Corbalan-Garcia S. (2004). Role of the lysine-rich cluster of the C2 domain in the phosphatidylserine-dependent activation of PKCalpha. J. Mol. Biol. 335: 1117–1129 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Shimizu K.K., Okada K. (2000). Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development 127: 4511–4518 [DOI] [PubMed] [Google Scholar]
- Sundaresan V., Springer P., Volpe T., Haward S., Jones J.D., Dean C., Ma H., Martienssen R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Swanson R., Edlund A.F., Preuss D. (2004). Species specificity in pollen-pistil interactions. Annu. Rev. Genet. 38: 793–818 [DOI] [PubMed] [Google Scholar]
- Szumlanski A.L., Nielsen E. (2009). The Rab GTPase RabA4d regulates pollen tube tip growth in Arabidopsis thaliana. Plant Cell 21: 526–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (May 4, 2011). MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. http://dx.doi.org/10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teft W.A., Chau T.A., Madrenas J. (2009). Structure-function analysis of the CTLA-4 interaction with PP2A. BMC Immunol. 10: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D. (1992). Use of a nuclear-targeted β-glucuronidase fusion protein to demonstrate vegetative cell-specific gene expression in developing pollen. Plant J. 2: 887–892 [Google Scholar]
- von Besser K., Frank A.C., Johnson M.A., Preuss D. (2006). Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133: 4761–4769 [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang C.C., Morales A.J., Schimmel P. (2000). Functional redundancy in the nonspecific RNA binding domain of a class I tRNA synthetase. J. Biol. Chem. 275: 17180–17186 [DOI] [PubMed] [Google Scholar]
- Yang Z.B. (1998). Signaling tip growth in plants. Curr. Opin. Plant Biol. 1: 525–530 [DOI] [PubMed] [Google Scholar]
- Yi M., Chi M.H., Khang C.H., Park S.Y., Kang S., Valent B., Lee Y.H. (2009). The ER chaperone LHS1 is involved in asexual development and rice infection by the blast fungus Magnaporthe oryzae. Plant Cell 21: 681–695 [DOI] [PMC free article] [PubMed] [Google Scholar]