This work provides evidence, using genetic perturbation of the MSH1 nuclear gene in five plant species, that MSH1 functions within the mitochondrion and plastid to influence organellar genome behavior and plant growth patterns.
Abstract
Mitochondrial-plastid interdependence within the plant cell is presumed to be essential, but measurable demonstration of this intimate interaction is difficult. At the level of cellular metabolism, several biosynthetic pathways involve both mitochondrial- and plastid-localized steps. However, at an environmental response level, it is not clear how the two organelles intersect in programmed cellular responses. Here, we provide evidence, using genetic perturbation of the MutS Homolog1 (MSH1) nuclear gene in five plant species, that MSH1 functions within the mitochondrion and plastid to influence organellar genome behavior and plant growth patterns. The mitochondrial form of the protein participates in DNA recombination surveillance, with disruption of the gene resulting in enhanced mitochondrial genome recombination at numerous repeated sequences. The plastid-localized form of the protein interacts with the plastid genome and influences genome stability and plastid development, with its disruption leading to variegation of the plant. These developmental changes include altered patterns of nuclear gene expression. Consistency of plastid and mitochondrial response across both monocot and dicot species indicate that the dual-functioning nature of MSH1 is well conserved. Variegated tissues show changes in redox status together with enhanced plant survival and reproduction under photooxidative light conditions, evidence that the plastid changes triggered in this study comprise an adaptive response to naturally occurring light stress.
INTRODUCTION
Several features link mitochondria and plastids within the plant cell. Both organelles maintain and express genetic information, conduct electron transport functions with the capacity to generate reactive oxygen species (ROS), and participate in organellar-nuclear signaling (Woodson and Chory, 2008). The relationship of these three processes is still not well defined. For example, whereas the plastid and mitochondrion are capable of supporting the replication, transcription, and translation of their own genetic information, much of the apparatus for doing so is nuclear encoded (Andersson et al., 2003; Richly and Leister, 2004; Woodson and Chory, 2008). Consequently, significant nuclear control exists over the synthesis and assembly of energy-transducing complexes within the organelles. It is assumed that organellar status is signaled to the nucleus, but the nature of these signals remains elusive. Studies of plastid dysfunction, conditioned by genetic mutation or chemical inhibitors, have implicated both ROS and chlorophyll biosynthetic intermediates in this signaling process, but these studies are not yet definitive (reviewed in Pfannschmidt, 2010). One difficulty in conducting these types of investigations is the inherent potential for secondary effects by chemical inhibitors and the relative paucity of genetic mutants influencing organellar function in a specific, well-defined manner.
In plants, the mitochondrial genome is unusually recombinogenic (Arrieta-Montiel and Mackenzie, 2010; Maréchal and Brisson, 2010). Asymmetric DNA exchange occurs at particular repeated sequences within the genome to influence the stoichiometry of subgenomic DNA molecules (known as substoichiometric shifting), allowing for differential copy number adjustments (Shedge et al., 2007). Over 47 recombination repeat pairs exist in the Arabidopsis thaliana mitochondrial genome, each of which becomes differentially active with disruption of the nuclear gene MSH1 (Arrieta-Montiel et al., 2009; Davila et al., 2011). MSH1 is a MutS homolog that suppresses homeologous mitochondrial DNA exchange in plants. First cloned in Arabidopsis (Abdelnoor et al., 2003), the gene appears to be well conserved in plants (Abdelnoor et al., 2006).
The substoichiometric shifting process that occurs in the msh1 mutant creates novel mitochondrial genotypes by altering relative copy number of various regions within the genome and influences the overall plant phenotype (for example, Shedge et al., 2010). We show here and in previous studies (Sandhu et al., 2007) that RNA interference (RNAi)–mediated suppression of MSH1 in tobacco (Nicotiana tabacum), tomato (Solanum lycopersicum), sorghum (Sorghum bicolor), and millet (Pennisetum glaucum) produces an array of phenotypes that are also observed in the msh1 mutant of Arabidopsis (previously designated chm1; Redei, 1973; Martínez-Zapater et al., 1992). These phenotypes include cytoplasmic male sterility (Sandhu et al., 2007), reduced growth rate with delayed flowering (Shedge et al., 2007), altered leaf morphology, and, under conditions for greatly enhanced recombination, increased thermotolerance (Shedge et al., 2010). Interestingly, these phenotypes are amenable to cytoplasmic sorting (Sandhu et al., 2007), implying their association with distinct organellar effects.
In this study, we examined another phenotype associated with MSH1 disruption: the emergence of green-white variegation. Leaf variegation arising by incomplete development or premature degeneration of plastids has been described in plants (Yu et al., 2007), and its induction by mitochondrial signaling in some cases has been implied (Roussell et al., 1991). It has not been feasible in previous studies to test for direct mitochondrial causation of the plastid effects. Here, we show that MSH1 has direct and distinct influences on plastid and mitochondrial behavior. Low frequency alterations of the plastid genome of an msh1 mutant are associated with plastid changes in the variegated tissue, altering the pattern of chloroplast development and redox status in these cells. This condition in msh1 provides tolerance to photooxidative light conditions, permitting plant survival and successful reproduction. Mitochondrial properties are also dramatically altered. We suggest, from these observations, that the dual targeting of MSH1 not only regulates mitochondrial recombination activity and plastid genome stability, but likewise enhances the plant’s repertoire for environmental response and interorganellar coordination.
RESULTS
MSH1 Disruption Can Result in Leaf Green-White Variegation
In a previous study (Sandhu et al., 2007), we implemented RNAi suppression of the nuclear gene MSH1 in the dicots tobacco and tomato. Here, an identical approach was used to downregulate MSH1 in the monocots millet and sorghum. Regenerated transgenic plants of all four species produced evidence of leaf variegation and plastid abnormalities within the T0 generation that were heritable (Figure 1). In tobacco, the effect was evidenced in a reticulated appearance of the leaf, whereas a more clearly defined green-white striping effect was seen in the other species. The observed variegation appeared to be random in pattern, with a small frequency (generally <5%) of an msh1 population showing near-albino levels of variegation and a small, variable frequency showing little or no evidence of variegation. Penetrance for the variegation phenotype in Arabidopsis ranged from 80 to 100%, depending on light quality (i.e., differences in penetrance were observed in plants grown under greenhouse versus controlled growth chamber conditions).
Figure 1.
RNAi Suppression of MSH1 Expression Results in a Variegation Phenotype in Multiple Plant Species.
Tomato (A), tobacco (B), pearl millet (C), and sorghum (D).
The MSH1 Gene Product Localizes within Mitochondria and Plastids
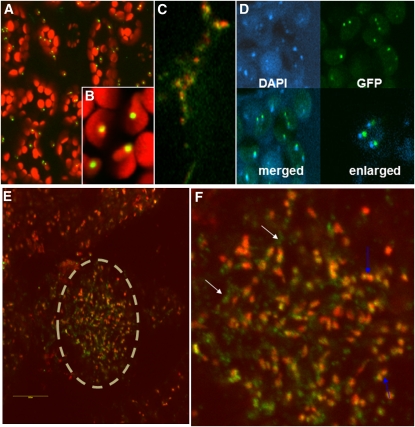
Earlier studies by our group to test MSH1 protein targeting within the cell were conducted using transient expression assays with the cauliflower mosaic virus (CaMV) 35S promoter. These studies resulted in ambiguous results, with some experiments showing mitochondrial targeting and others suggesting dual mitochondrial and plastid targeting (Abdelnoor et al., 2003, 2006). Consequently, protein targeting experiments were repeated in Arabidopsis stable transformants, with gene constructs comprising the full-length MSH1 genomic sequence under the control of its native promoter (758 bp upstream to the gene) and terminally fused to green fluorescent protein (GFP) at the carboxy end. GFP signal in these experiments was localized to both the mitochondrion and the plastid (Figures 2A to 2C). In both organelles, inclusion of the full-length gene sequence localized the protein to numerous punctate structures, shown in plastids by 4′,6–diamidino-2-phenylindole (DAPI) staining to be DNA-containing nucleoids (Figure 2D). Demonstration of MSH1 nucleoid localization, together with previous evidence of a canonical DNA binding domain within the gene (Abdelnoor et al., 2003) and participation in mitochondrial recombination surveillance (Shedge et al., 2007), suggest that the protein interacts with organellar genomes directly.
Figure 2.
MSH1 Localizes to Both Mitochondrial and Plastid Nucleoids in Arabidopsis (Col-0).
Images produced by confocal laser scanning microscopy.
(A) Young leaf cells showing GFP localization for a full-length MSH1-GFP fusion construct in a stable transformant. Red indicates plastid autofluorescence.
(B) Enlargement of plastids showing GFP localization to punctate structures.
(C) Enlargement of mitochondria (red indicates colocalization of a second mitochondrially targeted RFP fusion construct as control) showing MSH1-GFP localization (green) to punctate structures.
(D) DAPI staining and MSH1-GFP localization images from plastids merged and enlarged. Merged images were slightly offset to allow visualization of both signals.
(E) MSH1-GFP signal in ovule of stable transformant. Ovule is shown within dashed circle.
(F) Enlargement of ovule image from (C), with blue arrows showing plastids and white arrows showing mitochondrial GFP signals.
The tissues used to test MSH1 localization, young Arabidopsis leaves, show low levels of the native MSH1 transcript (Shedge et al., 2007), and previous genetic and expression studies have suggested that MSH1 (Shedge et al., 2007) and mitochondrial substoichiometric shifting (Johns et al., 1992) occur within reproductive tissues. Consequently, we confirmed that both mitochondrial and plastid localization of the protein occurred in ovules as well (Figures 2E and 2F).
MSH1 Functions within Plastids
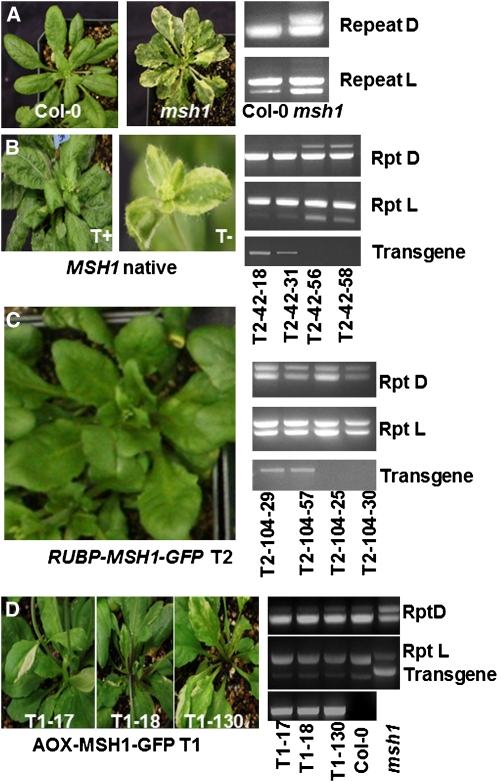
In planta localization of a transgenic protein to the plastid does not necessarily reflect in vivo activity. To test for MSH1 function within the plastid, we performed genetic hemicomplementation experiments. The complementation experiments involved development of three transgene constructs. The first included the full-length MSH1 gene, with the At-MSH1 native promoter and targeting presequence, fused to GFP at the C terminus. This construct was designed to confirm the feasibility of MSH1 genetic complementation and demonstrate its function in reproductive tissues of the T0 transformants. The second construct, testing hemicomplementation, substituted a plastid targeting sequence derived from a ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit gene (Lee et al., 2006) in place of the MSH1 presequence. The third substituted the alternative oxidase (AOXI) mitochondrial targeting presequence in place of the MSH1 presequence. The constructs were each introduced to Arabidopsis plants derived from the cross Columbia-0 (Col-0) × msh1 and confirmed heterozygous for the msh1 mutation. Transformation was followed by screening for msh1/msh1 transformed segregants in the first (T1) and second (T2) generations. Selected plants were tested for evidence of transgenic complementation of msh1 mitochondrial recombination and variegation phenotypes.
Successful complementation with the native MSH1 construct was assessed by complete absence of variegation on leaves of msh1/msh1 transformants and lack of mitochondrial DNA rearrangements using a diagnostic PCR-based assay (Figures 3A and 3B). Data from these experiments were consistent with full complementation of the msh1 phenotype (Table 1). In both T1 and T2 generations, we consistently observed functional complementation to cosegregate with presence of the transgene. We observed identical results in constructs with and without a GFP sequence fused to the C terminus (Table 1), indicating that the terminal GFP fusion did not interfere with MSH1 gene function.
Figure 3.
MSH1 Genetic Complementation.
(A) The msh1 mutant is distinguished from the wild type in its variegation and mitochondrial genome configuration. A PCR-based assay shows diagnostic polymorphisms at repeats D and L.
(B) Complementation of both the variegation and mitochondrial phenotypes occurs with the MSH1 native transgene. A PCR-based assay allows detection of the transgene in segregating T2 progeny (T+ indicates transgene present, T− transgene absent).
(C) A construct for chloroplast-targeted RUBP-MSH1 complements only the variegation, but not the mitochondrial, phenotype in a segregating T2 population.
(D) A construct for mitochondrially targeted AOX1-MSH1 complements the mitochondrial phenotype by suppressing recombination, but plants display evidence of variegation.
Table 1.
Genetic Complementation of the msh1 Phenotype with Native MSH1-GFP Gene Constructs
| Construct | Pop | MM | Mm | mm | Pχ2(1:2:1) | T+a | T− | Pχ2(3:1) |
| MSH1-GFP | T1 | 13 | 29 | 22 | 0.21 | |||
| MSH1 | T1 | 28 | 47 | 15 | 0.13 | |||
| MSH1 T1-42 (mm) | T2 | 0 | 0 | 67 | 50 | 17b | 0.94 | |
| MSH1-GFP T1-37 (mm) | T2 | 0 | 0 | 95 | 68 | 0.44 | 27b | |
| MSH1-GFP (mm background) | T1 | 0 | 0 | 9b | 6 | 3 |
Transgenes were introduced to MSH1/msh1 heterozygous plants, with normal (χ2 1:2:1, 3:1) Mendelian segregation (MM, Mm, mm) of the resulting populations. T2 populations display cosegregation for the transgene (T) and complemented phenotype.
T designates transgene.
All displayed variegation and mitochondrial genome rearrangements.
When the construct fusing MSH1 to a plastid targeting sequence was introduced, we observed hemicomplementation of the variegation phenotype (Figure 3C, Table 2). A small (<7%) proportion of the population still showed some evidence of variegation, suggesting that expression level of the construct may have been inadequate to fully compensate for the lack of endogenous activity. The first MSH1 intron resides within the native presequence and thus was removed to develop the plastid targeting construct. Deletion of the first intron may have altered transgene expression, since complete complementation of variegation was achieved with the native MSH1 presequence. All confirmed msh1/msh1 plants containing the plastid- targeted transgene displayed evidence of mitochondrial recombination (Figure 3C), indicating that the plastid form of the protein, while influencing chloroplast development, did not complement mitochondrial processes.
Table 2.
Genetic Complementation of the msh1 Phenotype with Plastid MSH1-GFP Gene Constructs
| Hemicomplementation with Chloroplast-Targeting MSH1 Gene Construct | ||||||||
| Construct | Pop | MM | Mm | mm/va | Pχ2(1:2:1) | Ta+/vb | T−/v | Pχ2(3:1) |
| RuBP-MSH1-GFP | T1 | 61 | 97 | 42/5 | 0.15 | |||
| MSH1-GFP T1-104(mm grn) | T2 | 87 | 60/7 | 27/23 | 0.19 | |||
| MSH1-GFP T1-117(mm var) | T2 | 78 | 62/2 | 16/14 | 0.36 | |||
Transgenes were introduced to MSH1/msh1 heterozygous plants, with normal (χ2 1:2:1, 3:1) Mendelian segregation (MM, Mm, mm) of the resulting populations. T2 populations display cosegregation for the transgene (T) and complemented phenotype.
Number of plants showing evidence of variegation.
T designates transgene.
Introduction of a mitochondrially targeted MSH1:GFP construct produced a variegated phenotype, but no evidence of mitochondrial recombination (Figure 3D). These observations are consistent with hemicomplementation of the mitochondrial phenotype without complementation of the plastid effects.
The transformation experiments involved introduction of the MSH1 transgene to heterozygous (MSH1/msh1) plants by the floral dip method (Clough and Bent, 1998). Confirmed homozygous msh1/msh1 segregants derived from self-pollination of the T0 plants were tested for evidence of complementation. Apparent full complementation by the native MSH1 construct within the first generation in this study supports earlier evidence from transcription studies that MSH1 functions during reproduction (Shedge et al., 2007).
Whereas MSH1 protein localized to both mitochondria and plastids and appeared to function differentially in the two organelles, we could detect no definitive evidence of plastid genome changes in response to MSH1 depletion when we pooled white sectors from a variegated msh1 plant for analysis. Consequently, we used a genetic approach. If no heritable plastid genome alterations accompany loss of MSH1, introduction of wild-type MSH1 to a variegated msh1/msh1 mutant by crossing or transgene complementation should permit complementation of the variegation phenotype in the resulting F1 or T1 plants. Crossing results (msh1 × MSH1) and transgene complementation experiments (Table 3, MSH1-GFP mm background) to introduce the native MSH1 construct to an msh1/msh1 mutant produced 80 to 100% variegated progeny. These results suggest that variegation cannot be complemented in the first generation, when the wild-type MSH1 allele is introduced to the msh1 mutant, and imply that organellar genomic changes, perhaps at unusually low frequency, accompany the plastid phenotype. These results differ from the hemicomplementation experiments described in Figure 3, where transformations were conducted prior to formation of the msh1/msh1 homozygous condition; thus, transgene expression preceded organellar genomic changes.
Table 3.
Genetic Complementation of the msh1 Phenotype with Mitochondrially Targeted MSH1-GFP Gene Constructs
| Construct | Pop | MM | Mm | mm | Pχ2(1:2:1) | Ta + | T− | Pχ2(3:1) |
| AOX-MSH1-GFP | T1 | 40 | 52 | 41b | 0.04 | |||
| MSH1-GFP T1-17 (mm var) | T2 | 70 | 57b | 23 | 0.15 |
Transgenes were introduced to MSH1/msh1 heterozygous plants, with normal (χ2 1:2:1, 3:1) Mendelian segregation (MM, Mm, mm) of the resulting populations. T2 populations display cosegregation for the transgene (T) and complemented phenotype.
T designates transgene.
100% negative for mitochondrial recombination, and 80 to 90% positive for variegation.
MSH1 Influences Plastid Genome Stability
To further test for evidence of plastid genome rearrangement in msh1, we sequenced the Arabidopsis msh1 plastid genome using Illumina paired-end sequencing and looked for evidence of repeat-mediated recombination. DNA sequence analysis produced no evidence of plastid genome rearrangement or sequence changes relative to the wild type, suggesting that any genomic changes may be localized exclusively to white sectors and represent low frequency events. Likewise, tests of DNA exchange at putative repeat sites identified by sequence analysis or shown by others to recombine (Maréchal et al., 2009) produced no evidence of alterations in the msh1 mutant at high frequencies, as were observed in the mitochondrial genome (see Supplemental Figure 1 online).
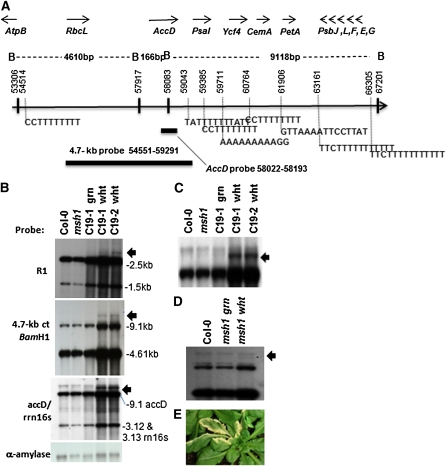
We speculated that the lack of evident DNA changes was due to plastid genomic heterogeneity in the variegated tissues. In fact, variegated sectors in msh1 mutants are visibly stippled and mosaic in their patterns, not purely white stripes (Figure 3B). We noticed that the hemicomplementation lines expressing the chloroplast-targeted form of MSH1 and displaying very low frequency variegation (Figure 3C) produced a purely white striping pattern with no evident mosaic or green stippling (Figure 4). We sectioned these white sectors and tested them for evidence of plastid genome changes, probing a plastid site previously shown to undergo rearrangement in other mutants (Maréchal et al., 2009). These experiments produced clear evidence of plastid genomic change localized to the white sectors (Figure 4).
Figure 4.
Evidence of Plastid Genome Change in the msh1 Mutant.
(A) Map of the chloroplast region encompassing one site of rearrangement. Genes encoded in this region are shown as well as the BamHI restriction map of the interval. Numerous short repeats associated with micro-homology-mediated recombination are present, and two probes used in (B) to (D) are shown.
(B) DNA gel blot analysis shows recombination polymorphisms (arrows) in dissected white tissues from two chloroplast hemicomplementation lines (C19-1 and 19-2). R1 is a small repeat located at coordinates 38655 to 38777 for copy 1 and 40879 to 41001 for copy 2. Copy 1 is part of the psaB gene, and copy 2 is part of the psaA gene. The bottom panel shows nuclear α-amylase probe as DNA loading control for each lane.
(C) Enlargement of DNA gel blot in (B) probed with accD and rrn16s.
(D) The wild type and msh1 mutant dissected to white and green sector tissues. DNA samples were probed with the 4.7-kb chloroplast fragment.
(E) Chloroplast hemicomplementation line showing the white sectors used for analysis.
These results may be the consequence of a more uniformly altered plastid population within the sectors of the hemicomplementation line. The probes used to detect the rearrangement overlap the rearranged region by only ~100 nucleotides, so the apparent low abundance of the recombinant form in msh1 white sectors may be due to both heterogeneity of plastid DNA molecules in these sectors, but also to insufficient probe overlap of the chloroplast DNA region in question.
The observed plastid DNA rearrangement in msh1 occurred within a segment of the genome that is similarly unstable in the why1 why3 double mutant, although the rearrangement and variegation frequencies are much lower in why1 why3 (Maréchal et al., 2009). The segment contains a number of very small repeats (Figure 4A). DNA gel blot analyses of msh1 plastid hemicomplementation lines showed the plastid genomic change predominantly within the white sectors and, although extremely difficult to detect in pooled msh1 white sectors (Figure 4D), the rearrangements were likewise restricted to white tissue samples. The low frequency of the plastid genomic change, coupled with heteroplasmy of the tissues, has presumably obscured efforts in the past by our group and others to detect these msh1 genomic effects.
The msh1 Variegation Involves Altered Mitochondrial and Plastid Phenotypes
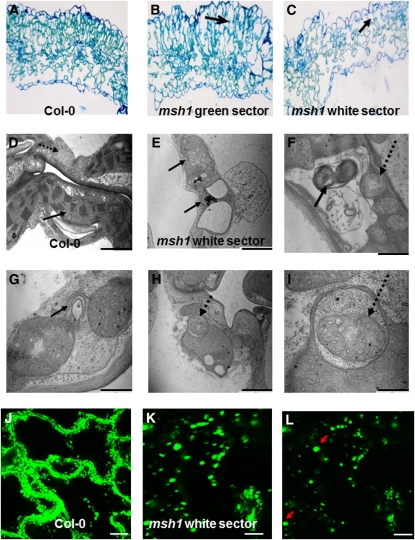
Cytological evaluation of the variegated tissues showed disorganized cellular arrangements in white tissue, with collapse of the palisade layer (Figures 5A to 5C). Organelle morphology was altered in both plastids and mitochondria within the white tissues. Plastids, greatly reduced in number per cell, demonstrated rudimentary and dramatically altered thylakoid structures in organelles (Figures 5D and 5E). Many plastids appeared completely devoid of organized thylakoids.
Figure 5.
Cellular and Organelle Morphology and Physiological Changes Associated with MSH1 Mutation.
(A) to (C) Cytological effects in the msh1 mutant are shown by light microscopy of a leaf cross section from wild-type (A) and green (B) or white (C) msh1 sectors. Arrows indicate loss of palisade cell organization.
(D) and (E) Transmission electron micrographs of wild-type and msh1 mutant organelle morphology in the white sector. Solid arrows indicate plastids; dashed arrows indicate mitochondria.
(F) to (I) Transmission electron micrographs show evidence of mitophagy and autophagy-like activity within the msh1 mutant.
(J) to (L) Confocal laser scanning microscopy of mitochondrial morphology and movement, visualized in wild-type and msh1 mutant plants transformed with a mitochondrial-targeting GFP construct. Time projection of mitochondrial movement over 5 min allows visualization of relative changes in mitochondrial motility ([J] and [K]). Mitochondria in msh1 white sectors are nearly stationary and enlarged. Arrows indicate unusually large-sized mitochondrial forms (single image, no time lapse) in (L). Bar = 10 μm.
Mitochondria within white sectors showed poorly defined cristae and appeared to undergo morphological changes characteristic of mitophagy, with changes in membrane organization, altered electron density within the organelles, and possible vesicle associations (Figures 5F to 5I). Col-0 and the msh1 mutant lines were both stably transformed with a mitochondrially targeted GFP gene construct to allow live-cell visualization of mitochondrial behavior in the mutant versus the wild type. Whereas mitochondrial movement appeared rapid, consistent, and well distributed in the Col-0 line (Figure 5J), cells within the mutant displayed mitochondria that were variable and reduced in movement in green sectors and enlarged and virtually stationary in the white sectors (Figures 5K and 5L). These observations indicate a physiologically altered mitochondrial state and likely reflect a condition of ATP depletion (see below).
Programmed Nuclear Gene Expression Changes Alter Plastid Development
The variegation phenotype of msh1 shows a range of severity from near undetectable levels to an almost albino phenotype. Therefore, transcript profiling of the msh1 mutant was conducted in two experiments: one focused on moderately variegated plants and the second using more extremely variegated plants from the msh1 mutant population.
Nuclear transcript analysis results were consistent between the two transcript profiling experiments, but with greater magnitude changes observed in the most highly variegated. Arabidopsis variegated msh1 mutants were altered in expression of several nuclear genes for photosystem I and II assembly and plastid development (Table 4; see Supplemental Table 1 online). Transcript profiling experiments were likewise performed with MSH1 RNAi suppression lines of soybean (Glycine max) displaying the variegated leaf phenotype. A similar pattern of transcript changes involving nuclear encoded photosynthesis components was observed (Table 4). In fact, this altered pattern of gene expression is shared by two other Arabidopsis variegation mutants, var2 (Miura et al., 2010) and immutans (Aluru et al., 2009) (included in Table 4), and by ROS-mediated plastid nucleus signaling (Surpin et al., 2002), suggesting a programmed response. At least two transcription factors that participate in this nuclear-plastid interaction pathway, GLK1 and GLK2 (Waters et al., 2009), were also downregulated in the msh1 mutant. A subset of these gene expression changes was confirmed by quantitative RT-PCR analysis (see Supplemental Figure 2 online).
Table 4.
Downregulated Photosynthesis Genes in Different Variegation Mutants
| AGI | Gene | msh1 | msh1_alb | var2 | immutans | Soybean msh1 |
| AT3G54890 | LHCA1 | −1.3 | −1.8 | −1.4 | −1.7 | NS |
| AT1G61520 | LHCA3 | −1.2 | −1.9 | −1.5 | −1.5 | −1.3 |
| AT4G02770 | PSAD-1 | −1.6 | −1.9 | −1.6 | −1.6 | NA |
| AT3G27690 | LHCB2.4 | −1.7 | −4.4 | −5.7 | −3.1 | NS |
| AT3G08940 | LHCB4.2 | −1.2 | −2.1 | −2.2 | −2.3 | −1.2 |
| AT1G15820 | LHCB6 | −1.3 | −1.7 | −1.4 | −2.3 | −1.6 |
| AT5G66570 | PSBO-1 | −1.3 | −1.5 | NS | −1.4 | −1.4 |
| AT3G50820 | PSBO-2 | −1.4 | −1.6 | NS | −1.2 | −1.2 |
| AT2G30790 | PSBP-2 | −2.0 | −2.2 | −2.9 | −1.8 | NA |
| AT5G54190 | PORA | −2.4 | −2.4 | −2.8 | −1.6 | NS |
| AT4G27440 | PORB | −1.8 | −1.9 | −2.2 | −6.3 | −1.4 |
| AT2G20570 | GLK1 | −1.9 | −3.1 | −5.6 | −1.4 | −1.6 |
| AT5G44190 | GLK2 | −1.3 | −1.8 | −2.6 | −1.5 | NA |
Data are shown as fold changes in gene expression of various mutants relative to wild-type Arabidopsis (Col-0) or soybean (Thorne) (significant at false discovery rate < 0.1). Soybean msh1 refers to an RNAi suppression line, and msh1 and msh1-alb designate moderately and highly variegated Arabidopsis, respectively. Data shown for Arabidopsis var2 and immutans mutants were obtained from Miura et al. (2010) and Aluru et al. (2009), respectively. AGI, Arabidopsis Genome Initiative; NS, not significant; NA, no annotation on the soybean Affymetrix array.
Variegation Is Associated with Altered Plastid Redox State in the White Tissues
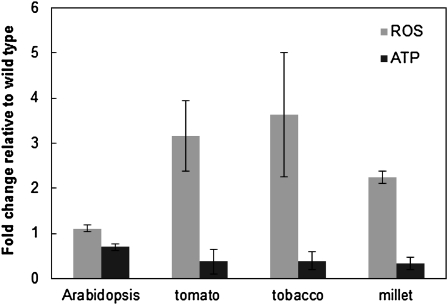
Initial investigation of physiological status in the msh1 mutant and RNAi suppression lines indicated that leaf tissues have reduced ATP and elevated ROS (H2O2) levels relative to the wild type (Figure 6), consistent with altered mitochondrial functions. The dramatic changes in plastid morphology of the white sectors prompted us to investigate the level and redox status of plastoquinone and phylloquinone in Arabidopsis leaf tissues. Because the photoactive pool of these two quinones is reduced during photosynthesis, tissue sampling and extractions were performed in the dark.
Figure 6.
Changes in ROS and ATP Levels Are Evident in the msh1 Mutant and RNAi Suppression Lines.
Arabidopsis msh1 mutant, and tomato, tobacco, and millet MSH1 RNAi suppression lines were assayed for changes in ROS and ATP levels relative to wild-type controls. Data shown are means ± se from three experiments.
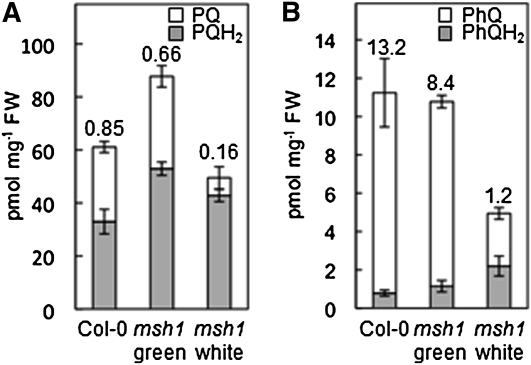
Plastoquinone and phylloquinone levels were lower in the msh1 white sectors compared with those of the green sectors or Col-0 tissues (Figure 7). More important, the quinone/quinol (oxidized/reduced) ratios were markedly lower in the white sectors (Figure 7), indicating a more reduced status of both quinones. White sectors were also found to accumulate the phylloquinone biosynthetic intermediate demethylphylloquinone (see Supplemental Figure 3 online), a diagnostic feature of impaired methylation of naphthoquinone conjugates in plants and bacteria (Lee et al., 1997; Lohmann et al., 2006).
Figure 7.
Chloroplast Redox Status Is Altered in the msh1 Mutant.
Levels and redox status of plastoquinone (A) and phylloquinone (B) in wild-type (Col-0) Arabidopsis leaf tissues and dissected green and white leaf sectors of the corresponding msh1 mutant. Tissues were sampled and extracted in the dark to avoid photoreduction and photodegradation of quinone species. Values above the bars indicate the quinone/quinol ratios. Data are means ± se of three replicates. FW, fresh weight.
Altered Plastid Properties in the msh1 Mutant Protect the Plant from High Light Damage
A more highly reduced state of the phylloquinone and plastoquinone pools within white sectors of the msh1 mutant led us to investigate whether variegation is adaptive for the plant under high light conditions causing photooxidative damage. Three approaches were taken to assess this: plant growth and reproduction, measurement of ROS levels, and MSH1 transcription under high light stress conditions.
Mature wild-type and msh1 mutant plants grown under high light conditions for 5 d were monitored for growth response and reproduction potential. Wild-type and, to a lesser extent, msh1 green plants produced visible levels of anthocyanin pigment in the leaves and, by day 5, showed signs of severe stress, leaf senescence, floral abortion, and little or no seed set. By contrast, highly variegated msh1 plants produced little visible anthocyanin in the same time period and were able to successfully flower and set seed (Figure 8A). In our initial experiments, 15 wild-type, 18 nonvariegated msh1 plants, three slightly variegated msh1 plants, and five highly variegated msh1 plants were prepared in individual pots and subjected to the high light regimen over 5 d. The five highly variegated plants remained viable, flowered, and set fully developed seeds; all others showed significant but variable stress responses to high light, with severity of phenotype greatest in the wild type. Subsequent experiments were performed in flats, where the most highly variegated plants remained viable, and evidence of plant stress was progressively more severe with decreasing variegation to most intense effects in wild-type plants. Appearance and extent of variegation of each plant did not change under exposure to high light. Seed collected from the surviving variegated plants gave rise to a range of green to variegated phenotypes.
Figure 8.
High Light Treatment in the msh1 Mutant.
(A) Wild-type Col-0 and an msh1 variegated plant following high light treatment for 5 d.
(B) ROS levels measured over time in wild-type, green msh1, and variegated msh1 plants. Each data point represents the results of five experimental samples. Relative fluorescence was plotted by setting the initial time point at 1 unit for each sample and plotting relative change. Data shown are means ± se for three experiments.
(C) Changes in MSH1 transcript levels in Col-0 leaves during high light treatment for a period of 16 h. Data shown are means ± se for three biological replicates.
The wild type (Col-0) and msh1 moderately and highly variegated plants were grown in a walk-in growth chamber under controlled light conditions approaching 1000 μE/m2/s. ROS (H2O2) levels were monitored over time under the high light conditions, revealing marked differences among lines in response over time (Figure 8B). Observations suggested that msh1 plants displaying highest levels of variegation are altered in their capacity to regulate ROS levels under stress conditions. The msh1 plants categorized visually as nonvariegated green were comparable to the wild type in their ROS levels. Increased variegation appeared to alter ROS response as measured over time, particularly early in the light exposure period.
Whereas stress tolerance of the msh1 mutant was observable, it is unlikely that msh1 mutations are prevalent in nature. Consequently, we investigated whether MSH1 transcriptional modulation occurs in the wild-type Col-0 line under high light stress conditions. In young leaf tissues tested by quantitative RT-PCR, MSH1 transcript levels were reduced by over 50% during high light treatments (Figure 8C). These observations were consistent with public transcript profiling data for MSH1 (Hruz et al., 2008), indicating that steady state transcript levels are markedly lower under stress conditions in the plant and suggesting that MSH1 transcriptional downregulation can occur naturally in stress environments. Exposure to stress during flowering, when MSH1 transcripts are at highest levels, could result in sufficient suppression of MSH1 to influence the status of organelle populations in the forming embryo. Studies of transgenerational effects are ongoing.
DISCUSSION
The variegation phenotype of msh1 mutants appears to represent a plastid-deficient phenotype in response to MSH1 depletion in the plastid nucleoid. Arabidopsis MSH1 protein has the capacity to target both mitochondria and plastids, although this dual targeting potential might vary in some plant species (Abdelnoor et al., 2006). Here, we show functionally, by gene complementation, that the variegation phenotype is associated with loss of the plastid-targeted form of MSH1.
The plastid genome of plants does not undergo the extensive recombination characteristic of mitochondrial genomes (Kwon et al., 2010). Yet, it is possible that MSH1 could influence the fate of double-strand breaks, contributing to recombination-mediated replication and/or DNA repair. If MSH1 affects DNA repair, each white sector could represent a collection of discrete plastid sequence variants. The region of plastid DNA rearrangement that we detected was previously shown to be unstable with dual disruption of At-WHY1 and At-WHY3 loci that encode plastid-localized single-stranded DNA binding proteins (Maréchal et al., 2009). This earlier study showed plastid genome instability to be characterized by concatemerization or subcircularization of plastid genomic regions that included small (10 to 18mer) repeated sequences. This particular region of the plastid genome is highly populated by such repeats as well as essential genes (Figure 4A). Whereas the why1 why3 double mutations led to only low (~4%) frequency variegation, we presume a similar phenomenon is involved in the high frequency (80 to 100%) variegation observed in msh1. Kwon et al. (2010) recently showed that small (20 to 60 bp) repeats within the chloroplast genome participate in the repair of double-strand breaks. Based on these observations, and previous studies of mitochondrial genome effects in the msh1 mutant, we suggest that MSH1 functions in both mitochondria and chloroplasts to influence the frequency or fate of double-strand breaks within each genome. Whereas depletion of MSH1 in the mitochondrion results in high frequency asymmetric recombination at 50- to 550-bp repeats (Arrieta-Montiel et al., 2009), in the chloroplast, loss of MSH1 apparently leads to similar asymmetric exchange at much smaller repeats. Whether MSH1 stabilizes each genome to prevent double-strand breaks, or facilitates strand invasion once breaks occur, is not clear.
It was fortuitous that we discovered the unusual appearance of rare sectors in the plastid hemicomplementation lines. We presume these rare sectors to reflect leakiness in genetic complementation, permitting a few altered plastids to remain in the presence of the MSH1 transgene for transmission to the next generation. These rare sectors may originate from a minute number of altered plastids, permitting rapid sorting to a more homogenous population. By contrast, the variegated sectors of the msh1 mutant appear to be composed of a heterogeneous population of altered and unaltered plastid genomes, so that detecting evidence of a single genomic rearrangement is more difficult. In Arabidopsis, white sectors are too small to allow complete plastid sequence analysis from a single sector, so studies of these details must be pursued in variegated MSH1 suppression lines of other species, such as sorghum, where individual white sectors are much larger.
In the hemicomplementation experiments with a plastid-targeted MSH1, where we detected the low frequency occurrence of variegation, the leakiness of this complementation may have arisen by inadequate or inappropriately timed transgene expression. The manner in which the transgene was assembled might have influenced its transcription levels. We used confocal laser scanning microscopy to confirm GFP-tagged MSH1 protein localization in these lines, allowing us to observe considerable, albeit nonquantified, variation in signal strength among transformed plants. Several of the derived plants initially classified as variegated showed later initiating leaves to be fully green, suggesting that the few sectors were rare escapes, so that expression of the transgene in later stages of development might result in a regreening of the plant.
Features of the variegated msh1 mutant, including incompletely assembled plastids, altered cellular redox status of white tissues, apparent mitophagy activity, and reduced mitochondrial mobility, suggest significant physiological effects on the cell in association with organellar genome rearrangements. Our ability to deduce mitophagy activity was based on striking resemblance to events described previously by others (Bernales et al., 2006).
Although it is not clear how loss of a plastid DNA-associating protein like MSH1 would directly influence plastid development, there is evidence of a physical association between the thylakoid membrane, the nucleoid, and the plastid DNA directly (Liu and Rose, 1992; Jeong et al. 2003). This association may be the means of directly linking changes in plastid genome status to assembly and redox status of the photosynthetic apparatus (Allen and Pfannschmidt, 2000), distinct from events occurring within the mitochondrion.
Some of the physiological effects observed in the msh1 mutant, characteristic of ATP depletion (Brough et al., 2005), are also associated with senescence or early stages of programmed cell death (Azad et al., 2008). Similarly, the low phylloquinone content and quinone/quinol ratio observed in the white sectors of the msh1 mutant are typical of senescing tissues (Oostende et al., 2008). Therefore, the observation of increased plant viability and reproductive success in these plants under photooxidative damaging light conditions was unexpected, bearing certain resemblance to proposed mitochondrial hormesis effects in fungal and animal systems (Schulz et al., 2007; Mesquita et al., 2010). Apart from the photoactive plastoquinone pool in chloroplasts, nonphotoactive plastoquinone is present in chloroplast envelopes and plastoglobuli. Under heat stress, a redistribution of plastoquinone between photoactive and nonphotoactive pools occurs (Pshybytko et al., 2008). Enhanced stress tolerance as a consequence of altered mitochondrial functions has been reported in the tobacco CMSII mutant lacking complex I (Dutilleul et al., 2003), again supporting the influence of organelle status on cellular stress responses.
The reduced state of the conjugated quinone species in plastids may be unique to msh1-derived variegation, since other variegated Arabidopsis mutants appear to show light sensitivity of the variegation phenotype and/or enhanced susceptibility to photooxidative damage (Rosso et al., 2009). Of course, the msh1 mutant contains a null mutation and displays a more extreme phenotype than would be expected with suppressed MSH1 transcription during stress. Whereas the msh1 mutant undergoes extensive mitochondrial genomic rearrangements (Arrieta-Montiel et al., 2009) and plastid genomic and developmental alterations, transcriptional downregulation of MSH1 is likely to result in more subtle, largely reversible genomic and physiological effects and perhaps altered plastid redox status without observable variegation.
Our data suggest that the suppression of MSH1 might constitute an adaptive response that enhances progeny success. Galloway and Etterson (2007) described the phenomenon of transgenerational plasticity in the forest herb Campanulastrum americanum, where plants demonstrated enhanced fitness for growth in the same light environment (understory versus light gap) as the maternal parent. They reasoned that plants have the capacity to transmit environmental cues to progeny that increase progeny fitness for growth under similar conditions. These cues were shown to be maternal. We postulate that MSH1 represents one component of such an environmental signal transmission mechanism, operating not only in response to light conditions but likely other environmental factors as well, to condition mitochondria and plastids within the ovule, where MSH1 expression is most pronounced (Shedge et al., 2007).
Other phenotypes arising in msh1 mutant or RNAi suppression lines also appear to influence plant fitness under particular environmental conditions. For example, cytoplasmic male sterility (Sandhu et al., 2007) represents a phenotype that, in natural populations, may enhance seed set and plant vigor by encouraging effective outcrossing (Charlesworth, 2002). The ability of a plant to spontaneously reverse the cytoplasmic male sterility trait to fertility is associated with mitochondrial genomic substoichiometric shifting (Janska et al., 1998); one could envision this shifting as a response to carbon flux signals upon unsuccessful pollination or carbon sink formation. Likewise, an msh1 recA3 double mutant in Arabidopsis shows greatly enhanced thermotolerance and reproduction under conditions of heat stress (Shedge et al., 2010).
Mechanisms by which mitochondria and chloroplasts alter plant growth are not well defined, but several plant responses are linked to elevated levels of ROS. It has been suggested that ROS might affect an array of distinct plant processes through the broad spectrum of modified peptides produced as a consequence of ROS damage within the cell (Pfannschmidt, 2010). Our observation of elevated ROS in the msh1 mutant, and of differences in ROS accumulation in wild-type and variegated lines under high light conditions, appear consistent with these models. However, organelle-associated changes in metabolism, electron transport, or redox status and ATP levels may also participate in developmental reprogramming. Others have shown that plastids and mitochondria generate synergistic signals to influence nuclear gene expression (Pesaresi et al., 2006). Cellular signals triggering the variegated phenotype, with implications for enhanced progeny survival in natural conditions of environmental stress, are especially intriguing concepts. MSH1 suppression variants, presenting a broad array of phenotypes in response to organellar perturbation, reveal the intersection of distinct developmental programs and suggest their direct or indirect reliance on organellar signals.
METHODS
Plant Materials
Arabidopsis thaliana Col-0 and msh1 (CS 3372) mutant lines were obtained from the Arabidopsis stock center. RNAi suppression lines of tomato (Solanum lycopersicum) and tobacco (Nicotiana tabacum) were reported previously (Sandhu et al., 2007), and millet (Pennisetum glaucum) and sorghum (Sorghum bicolor) RNAi lines were derived by similar procedures, with construct details below, and with transformations and plant regeneration performed according to the procedures of Howe et al. (2006) in the Center for Biotechnology Plant Transformation Facility at the University of Nebraska.
Molecular Biology Procedures
Total plant genomic DNA was extracted from mature Arabidopsis leaves with the DNeasy plant mini kit (Qiagen). One-leaf DNA extraction for PCR genotyping was performed according to published procedures (Martínez-Zapater and Salinas, 1997). Five micrograms of total genomic DNA was digested with restriction enzyme BamHI, fractionated by agarose gel electrophoresis (0.8% agarose in 0.5× TBE buffer at 70 V/cm overnight), and transferred to Hybond-N (GE Healthcare) for DNA gel blot analysis. PCR-amplified mitochondrial fragments from the repeated regions of the mitochondrial or chloroplast genome were used as probes. Probes were 32P-labeled (Prime-It II random primer labeling kit; Stratagene) for autoradiographic exposure. Primer pairs that amplify these probes as well as PCR primers used in this study are listed in Supplemental Table 2 online.
Plasmids and Gene Constructs for Complementation and Transformation Experiments
For development of RNAi transgene constructions, segments encoding 157 amino acids from the MSH1 C terminus were amplified from total cDNA of pearl millet and sorghum using primers zm-msf8 (5′-GGTTGAGGAGCCTGAATCTCTGAAGAAC-3′) and zm-msr8 (5′-CTCGCCAGAGATTCGAGATATACCGAAG-3′). PCR products were cloned in forward and reverse orientation, separated by an intron sequence. The base vector, pUCRNAi- intron, which harbors the second intron of the Arabidopsis small nuclear riboprotein (At4g02840), was graciously provided by H. Cerutti (University of Nebraska, Lincoln, NE). The vector pPTN290, a derivative of pPZP212 (Hajdukiewicz et al., 1994), was used to introduce the MSH1-RNAi cassettes under the control of the maize (Zea mays) Ubiquitin1 promoter coupled with its first intron, with transcription terminated by the CaMV 35S terminator. Agrobacterium tumefaciens strain NTL4 (Luo et al., 2001) was used for inoculating embryos from pearl millet Tift23 DBE1 and sorghum Tx430 lines.
All plasmid constructs for complementation experiments were prepared with the At-MSH1 native promoter (758 bp upstream to the 5′ untranslated region). To achieve this, the CaMV 35S promoter adjacent to mGFP in a plant binary vector pCAMBIA1302c (CAMBIA Labs) was removed by BglII and EcoRI double digestion. The At-MSH1 native gene construct, 7.1 kb in size, was PCR amplified from Col-0 genomic DNA with Pfu ultra (Stratagene) and directly ligated to the SpeI site of modified pCAMBIA1302c (Wamboldt et al., 2009). For the plastid-targeting At-MSH1 construct, the At-MSH1 native promoter, genomic DNA gene fragment minus targeting presequence, and ribulose-1,5-bisphosphate small subunit targeting presequence were PCR amplified separately and ligated sequentially to KpnI and SalI sites of an intermediate pBluescript SK+ vector. The final chimeric fragments were then released from pBluescript SK+ and ligated to the SpeI site of modified pCAMBIA1302c. Final constructs were also prepared without GFP terminal fusion by adding a stop codon 5′ to the GFP sequence. Orientation and in-frame fusion of chimeric clones were confirmed by DNA sequencing (Eurofins MWG Operon). Primers used in this study are listed in Supplemental Table 2 online. For Arabidopsis stable transformation, constructs were introduced into Agrobacterium strain C58C1. All oligomers were synthesized from Integrated DNA Technologies and restriction enzymes purchased from New England Biolabs and Fermentas.
RNA Isolation and Real-Time PCR Analysis
Total RNA was extracted from leaf tissues of wild-type and mutant plants using the TRIzol (Invitrogen) extraction procedure followed by purification on RNeasy columns (Qiagen). cDNA was synthesized using SuperScriptIII first-strand synthesis SuperMix for quantitative RT-PCR (Invitrogen) following the manufacturer’s instructions. Quantitative PCR was performed on an iCycler iQ system (Bio-Rad) using SYBR GreenER Supermix (Invitrogen). Primers used for PCR are listed in Supplemental Table 2 online. The transcript level of each gene was normalized to that of UBIQUITIN10.
ROS and ATP Assays
Mature tomato, tobacco, and millet leaves or 8-week-old Arabidopsis whole plants were used for ROS (Joo et al., 2005) and ATP assays. For ROS measurements, frozen tissue was ground, taken up in 10 mM Tris-HCl buffer, pH 7.3, and centrifuged twice at 15,000 rpm for 5 min. ROS production was assayed by adding 100 mM H2DCFDA in DMSO to a final concentration of 10 μM and measuring fluorescence (Cary Eclipse fluorometer; excitation, 485 nm; emission, 535 nm) (Varian). Average fluorescence values obtained from three replicates for each sample were normalized by protein content (Bio-Rad protein assay kit). For ATP assays, ground tissue was homogenized in extraction buffer (25 mM Tris-HCl, pH 7.8, 2 mM DTT, 2 mM EDTA, 10% glycerol, and 1% Triton X-100), homogenates were centrifuged 5 min at 13,000g, and supernatants were used for analysis. Each assay used 20 μL supernatant, background luminescence counts were determined, and 100 μL ATP assay mix (Sigma-Aldrich) was added. Luminescence counts were measured as 5-s integrations after a delay of 5 s using a Veritas microplate luminometer (Turner BioSystems). The average fluorescence values obtained from three replicates for each sample were normalized by protein content (Bio-Rad protein assay kit).
Confocal Laser Scanning Microscopy
Young leaves and ovules from stably transformed plants were isolated for evaluation of GFP expression. For DAPI staining in chloroplasts, leaves from stably transformed plants containing the At-MSH1 native construct expressing GFP were infiltrated with half-strength Murashige and Skoog medium, 10 mM MES, and 1 μg/mL DAPI (Invitrogen). Plants were placed in the dark at least 2 h prior to confocal imaging by sequential acquisition. All imaging was performed on the Olympus FluoView 500 confocal laser scanning microscope mounted on the BX51 upright compound microscope, with software FluoView 4.3. DAPI was excited at 405 nm, and fluorescence was detected at 430 to 460 nm, GFP was excited with 488 nm and detected at 505 to 525 nm, plastid autofluorescence with a long-pass filter at 660 nm. Images were acquired sequentially.
Transmission and Light Electron Microscopy
Leaf samples for transmission electron microscopy were prepared as follows. Green and white leaf sectors of 2-week-old variegated and wild-type plants were dissected and fixed in 2.5% glutaraldehyde and 0.05 M sodium cacodylate, pH 7.4, under vacuum for 3 h, and postfixed in 1% osmium tetroxide in 0.05 M sodium cacodylate, pH 7.4, for 2 h. Samples were dehydrated in a graduated ethanol series and embedded in Epon 812 (Electron Microscopic Sciences). Thin sections (80 nm) were stained with uranyl acetate and lead citrate and observed under a transmission electron microscope (Hitachi H7500-I) in the University of Nebraska Lincoln Center for Biotechnology Microscopy Facility. For toluidine blue staining, samples were semithin sectioned at a 1-um thickness and examined under a light microscope (Olympus AX70).
Quantification of Quinones
Phylloquinone and phylloquinol were extracted and quantified as previously described (Oostende et al., 2008). All steps, including leaf tissue sampling from Col-0 and dissected msh1 green and white leaf sectors, were performed in the dark to avoid photoreduction and photodegradation of quinone species. Demethylphylloquinone (retention time 11.7 min) was analyzed in the same run as that of phylloquinone and phylloquinol and was quantified according to an external calibration standard purified from an extract of Synechocystis mutant knockout for demethylphylloquinone methyltransferase (Lohmann et al., 2006). Plastoquinone and plastoquinol analyses were modified from Kruk and Karpinski (2006). Briefly, fresh leaf tissues (10 to 30 mg) were homogenized in 1 mL of 100% methanol, and extracts were centrifuged (14,000g; 5 min) to pellet debris. Supernatant aliquots (100 μL) were immediately analyzed by HPLC using a 5 μM Discovery C-18 column (250 × 4.6 mm; Supelco) thermostated at 30°C. Samples were eluted in isocratic mode with 100% methanol at a flow rate of 1.5 mL·min−1. Plastoquinone was detected spectrophotometrically at 255 nm, and plastoquinol was detected fluorimetrically (290 and 330 nm for excitation and emission, respectively). External calibration standards were used for quantification. The external standard of plastoquinone was purified from Arabidopsis leaves and quantified spectrophotometrically using ε255 = 1940 M−1 cm−1. The external standard of plastoquinol was prepared by quantitative reduction of plastoquinone with sodium borohydride. Retention times for plastoquinol and plastoquinone were 13.5 and 38.5 min, respectively.
Microarray Experiments
For microarray experiments, total RNA was extracted from 8-week-old Col-0 plants and msh1 mutant Arabidopsis, and Thorne and MSH1 RNAi transgenic soybean (Glycine max) plants using TRIzol (Invitrogen) extraction procedures followed by purification on RNeasy columns (Qiagen). For highly variegated samples, Arabidopsis msh1 mutant and Col-0 plants were grown on Murashige and Skoog medium, and total RNA was extracted from 4-week-old plants. Three hybridizations were performed per genotype with RNA extractions from single plants for each microarray chip. Samples were assayed on the Affymetrix GeneChip oligonucleotide 22K ATH1 array for Arabidopsis and GeneChip Soybean microarray for soybean (Affymetrix) according to the manufacturer's instructions. Microarray data from each experiment were analyzed separately. Expression data from Affymetrix GeneChips were normalized using the Robust Multichip Average method (Bolstad et al., 2003). A separate mixed linear model analysis was conducted using the normalized data with SAS software (Wolfinger et al., 2001). Each mixed model includes a fixed effect for genotype and a random effect for experiment. Tests for differential expression across genotypes were performed as part of our mixed linear model analyses. The P values generated from tests of interest were converted to q-values to obtain approximate control of the false discovery rate at a specified value (Storey and Tibshirani, 2003). We obtained estimates of fold change for each probe by converting the mean treatment difference estimated as part of our mixed linear model analyses.
High Light Treatment Experiments
Arabidopsis Col-0 and msh1 plants were individually grown in 4 × 3-cm pots in a growth chamber with fluorescent light intensity set at 100 μmol photons/m2/s with a 12-h-light/12-h-dark photoperiod, at 24°C and 70% of air humidity. For high light exposure treatments, 4-week-old plants were transferred to a high light intensity chamber immediately following the dark cycle and exposed to 1000 μmol photons/m2/s for 5 d with continuous light. The plants were returned to the initial growth conditions after the high light exposure for recovery, and the survival ratio was analyzed after 7 to 10 d at these initial conditions.
ROS analysis of plants under high light generally followed procedures described by Joo et al. (2005) as described above. Four-week-old Arabidopsis Col-0, green msh1, and variegated msh1 plants were transferred following a dark cycle to a high light chamber and exposed to 1000 μmol photons/m2/s light intensity for 0, 60, 120, 150, 180, and 210 min. Three biological replicates of Col-0, green msh1, and variegated msh1 tissue were collected at each time point and ground in liquid nitrogen. Average fluorescence values obtained from three successive measurements were divided by protein content and expressed as relative fluorescence units per milligram of protein. These values were expressed as a ratio of relative fluorescence units obtained under high light versus control plants.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MSH1 (AT3G24320), PORA (AT5G54190), PORB (AT4G27440), GLK1 (AT2G20570), GLK2 (AT5G44190), and ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (AT1G67090). The microarray array data have been deposited at ArrayExpress (E-MEXP-1410) and the Gene Expression Omnibus (GSE30281 and GSE30112).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. DNA Gel Blot Assessment of Chloroplast Repeat-Mediated Recombination in the msh1 Mutant.
Supplemental Figure 2. Arabidopsis Genes PSBO1/2, PPD3, GLK1/2, and PORA/B Are Downregulated in the msh1 Mutant.
Supplemental Figure 3. Phylloquinone Biosynthesis Is Altered in the Arabidopsis msh1 Mutant.
Supplemental Table 1. Enriched Gene Ontology Categories Related to Chloroplast and Photosynthesis in the Highly Variegated msh1 Mutant.
Supplemental Table 2. Primers Used in This Work.
Acknowledgments
We thank Xuehui Feng and Natalya Nersesian for development of the transgenic millet and sorghum lines, Hardik Kundariya for assistance with plant genotyping, Han Chen for assistance with electron microscopy, Yuannan Xia for assistance with microarray experiments, Concetta DiRusso for use of the microplate reader for our analysis, and Markus Schwarzlander and Alan Christensen for invaluable discussions during these studies. Funding for these studies was provided by the Department of Energy (DE-FG02-07ER15564) and the National Science Foundation (IOS 0820668) to S.A.M.
AUTHOR CONTRIBUTIONS
Y.-Z.X. performed gene expression, physiological, redox, and phenotype analysis research and analyzed data. M.P.A.-M. performed genetic analysis, electron microscopy, and chloroplast recombination research and analyzed data. K.S.V. performed hemicomplementation research. W.B.M.D. performed light response research. J.R.W. performed redox research. G.J.B. designed redox research and analyzed data. J.I.D. performed DNA sequencing research. T.E.E. contributed physiological analysis tools. C.G.E. performed confocal microscopy research. S.J.S. and T.E.C. performed transformation research. S.A.M. designed research, analyzed data. and wrote the article.
References
- Abdelnoor R.V., Christensen A.C., Mohammed S., Munoz-Castillo B., Moriyama H., Mackenzie S.A. (2006). Mitochondrial genome dynamics in plants and animals: Convergent gene fusions of a MutS homologue. J. Mol. Evol. 63: 165–173 [DOI] [PubMed] [Google Scholar]
- Abdelnoor R.V., Yule R., Elo A., Christensen A.C., Meyer-Gauen G., Mackenzie S.A. (2003). Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl. Acad. Sci. USA 100: 5968–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.F., Pfannschmidt T. (2000). Balancing the two photosystems: photosynthetic electron transfer governs transcription of reaction centre genes in chloroplasts. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru M.R., Zola J., Foudree A., Rodermel S.R. (2009). Chloroplast photooxidation-induced transcriptome reprogramming in Arabidopsis immutans white leaf sectors. Plant Physiol. 150: 904–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson S.G., Karlberg O., Canbäck B., Kurland C.G. (2003). On the origin of mitochondria: A genomics perspective. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 165–177, discussion 177–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta-Montiel M., Mackenzie S.A. (2010). Plant mitochondrial genomes and recombination. In Advances in Plant Biology: Plant Mitochondria, Kempken F., (New York: Springer; ), pp. 65–84 [Google Scholar]
- Arrieta-Montiel M.P., Shedge V., Davila J., Christensen A.C., Mackenzie S.A. (2009). Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics 183: 1261–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad A.K., Ishikawa T., Ishikawa T., Sawa Y., Shibata H. (2008). Intracellular energy depletion triggers programmed cell death during petal senescence in tulip. J. Exp. Bot. 59: 2085–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S., McDonald K.L., Walter P. (2006). Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 4: e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Brough D., Schell M.J., Irvine R.F. (2005). Agonist-induced regulation of mitochondrial and endoplasmic reticulum motility. Biochem. J. 392: 291–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. (2002). What maintains male-sterility factors in plant populations? Heredity 89: 408–409 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Davila J.I., Arrieta-Montiel M.P., Wamboldt Y., Cao J., Hagmann J., Shedge V., Xu Y.-Z., Weigel D., Mackenzie S.A. (2011). Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 9: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C., Garmier M., Noctor G., Mathieu C., Chétrit P., Foyer C.H., de Paepe R. (2003). Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15: 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway L.F., Etterson J.R. (2007). Transgenerational plasticity is adaptive in the wild. Science 318: 1134–1136 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Howe A., Sato S., Dweikat I., Fromm M., Clemente T. (2006). Rapid and reproducible Agrobacterium-mediated transformation of sorghum. Plant Cell Rep. 25: 784–791 [DOI] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janska H., Sarria R., Woloszynska M., Arrieta-Montiel M., Mackenzie S.A. (1998). Stoichiometric shifts in the common bean mitochondrial genome leading to male sterility and spontaneous reversion to fertility. Plant Cell 10: 1163–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S.Y., Rose A., Meier I. (2003). MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res. 31: 5175–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns C., Lu M., Lyznik A., Mackenzie S. (1992). A mitochondrial DNA sequence is associated with abnormal pollen development in cytoplasmic male sterile bean plants. Plant Cell 4: 435–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo J.H., Wang S., Chen J.G., Jones A.M., Fedoroff N.V. (2005). Different signaling and cell death roles of heterotrimeric G protein α and β subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell 17: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruk J., Karpinski S. (2006). An HPLC-based method of estimation of the total redox state of plastoquinone in chloroplasts, the size of the photochemically active plastoquinone-pool and its redox state in thylakoids of Arabidopsis. Biochim. Biophys. Acta 1757: 1669–1675 [DOI] [PubMed] [Google Scholar]
- Kwon T., Huq E., Herrin D.L. (2010). Microhomology-mediated and nonhomologous repair of a double-strand break in the chloroplast genome of Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 13954–13959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Lee S., Lee G.J., Lee K.H., Kim S., Cheong G.W., Hwang I. (2006). Functional characterization of sequence motifs in the transit peptide of Arabidopsis small subunit of rubisco. Plant Physiol. 140: 466–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.T., Hsu A.Y., Ha H.T., Clarke C.F. (1997). A C-methyltransferase involved in both ubiquinone and menaquinone biosynthesis: isolation and identification of the Escherichia coli ubiE gene. J. Bacteriol. 179: 1748–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.W., Rose R.J. (1992). The spinach chloroplast chromosome is bound to the thylakoid membrane in the region of the inverted repeat. Biochem. Biophys. Res. Commun. 184: 993–1000 [DOI] [PubMed] [Google Scholar]
- Lohmann A., Schöttler M.A., Bréhélin C., Kessler F., Bock R., Cahoon E.B., Dörmann P. (2006). Deficiency in phylloquinone (vitamin K1) methylation affects prenyl quinone distribution, photosystem I abundance, and anthocyanin accumulation in the Arabidopsis AtmenG mutant. J. Biol. Chem. 281: 40461–40472 [DOI] [PubMed] [Google Scholar]
- Luo Z.-Q., Clemente T.E., Farrand S.K. (2001). Construction of a derivative of Agrobacterium tumefaciens C58 that does not mutate to tetracycline resistance. Mol. Plant Microbe Interact. 14: 98–103 [DOI] [PubMed] [Google Scholar]
- Maréchal A., Brisson N. (2010). Recombination and the maintenance of plant organelle genome stability. New Phytol. 186: 299–317 [DOI] [PubMed] [Google Scholar]
- Maréchal A., Parent J.S., Véronneau-Lafortune F., Joyeux A., Lang B.F., Brisson N. (2009). Whirly proteins maintain plastid genome stability in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 14693–14698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Zapater J., Salinas J., eds (1997). Methods in Molecular Biology: Arabidopsis Protocols. (Totowa, NJ: Humana Press Inc.). [Google Scholar]
- Martínez-Zapater J.M., Gil P., Capel J., Somerville C.R. (1992). Mutations at the Arabidopsis CHM locus promote rearrangements of the mitochondrial genome. Plant Cell 4: 889–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita A., Weinberger M., Silva A., Sampaio-Marques B., Almeida B., Leão C., Costa V., Rodrigues F., Burhans W.C., Ludovico P. (2010). Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H2O2 and superoxide dismutase activity. Proc. Natl. Acad. Sci. USA 107: 15123–15128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura E., Kato Y., Sakamoto W. (2010). Comparative transcriptome analysis of green/white variegated sectors in Arabidopsis yellow variegated2: Responses to oxidative and other stresses in white sectors. J. Exp. Bot. 61: 2433–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostende C., Widhalm J.R., Basset G.J. (2008). Detection and quantification of vitamin K(1) quinol in leaf tissues. Phytochemistry 69: 2457–2462 [DOI] [PubMed] [Google Scholar]
- Pesaresi P., Masiero S., Eubel H., Braun H.P., Bhushan S., Glaser E., Salamini F., Leister D. (2006). Nuclear photosynthetic gene expression is synergistically modulated by rates of protein synthesis in chloroplasts and mitochondria. Plant Cell 18: 970–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T. (2010). Plastidial retrograde signalling—A true “plastid factor” or just metabolite signatures? Trends Plant Sci. 15: 427–435 [DOI] [PubMed] [Google Scholar]
- Pshybytko N.L., Kruk J., Kabashnikova L.F., Strzalka K. (2008). Function of plastoquinone in heat stress reactions of plants. Biochim. Biophys. Acta 1777: 1393–1399 [DOI] [PubMed] [Google Scholar]
- Redei G.P. (1973). Extra-chromosomal mutability determined by a nuclear gene locus in Arabidopsis. Mutat. Res. 18: 149–162 [Google Scholar]
- Richly E., Leister D. (2004). An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene 329: 11–16 [DOI] [PubMed] [Google Scholar]
- Rosso D., Bode R., Li W., Krol M., Saccon D., Wang S., Schillaci L.A., Rodermel S.R., Maxwell D.P., Hüner N.P. (2009). Photosynthetic redox imbalance governs leaf sectoring in the Arabidopsis thaliana variegation mutants immutans, spotty, var1, and var2. Plant Cell 21: 3473–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussell D.L., Thompson D.L., Pallardy S.G., Miles D., Newton K.J. (1991). Chloroplast structure and function is altered in the NCS2 maize mitochondrial mutant. Plant Physiol. 96: 232–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu A.P., Abdelnoor R.V., Mackenzie S.A. (2007). Transgenic induction of mitochondrial rearrangements for cytoplasmic male sterility in crop plants. Proc. Natl. Acad. Sci. USA 104: 1766–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz T.J., Zarse K., Voigt A., Urban N., Birringer M., Ristow M. (2007). Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 6: 280–293 [DOI] [PubMed] [Google Scholar]
- Shedge V., Arrieta-Montiel M., Christensen A.C., Mackenzie S.A. (2007). Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell 19: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedge V., Davila J., Arrieta-Montiel M.P., Mohammed S., Mackenzie S.A. (2010). Extensive rearrangement of the Arabidopsis mitochondrial genome elicits cellular conditions for thermotolerance. Plant Physiol. 152: 1960–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M., Larkin R.M., Chory J. (2002). Signal transduction between the chloroplast and the nucleus. Plant Cell 14(Suppl): S327–S338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamboldt Y., Mohammed S., Elowsky C., Wittgren C., de Paula W.B., Mackenzie S.A. (2009). Participation of leaky ribosome scanning in protein dual targeting by alternative translation initiation in higher plants. Plant Cell 21: 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.T., Wang P., Korkaric M., Capper R.G., Saunders N.J., Langdale J.A. (2009). GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfinger R.D., Gibson G., Wolfinger E.D., Bennett L., Hamadeh H., Bushel P., Afshari C., Paules R.S. (2001). Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8: 625–637 [DOI] [PubMed] [Google Scholar]
- Woodson J.D., Chory J. (2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9: 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Fu A., Aluru M., Park S., Xu Y., Liu H., Liu X., Foudree A., Nambogga M., Rodermel S. (2007). Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ. 30: 350–365 [DOI] [PubMed] [Google Scholar]