This study identifies the last remaining unknown enzyme responsible for the chlorophyll cycle. It shows that 7-HYDROXYMETHYL CHLOROPHYLL A REDUCTASE is a flavoprotein capable of carrying out a difficult reaction involving the substitution of an OH group with an H.
Abstract
The interconversion of chlorophyll a and chlorophyll b, referred to as the chlorophyll cycle, plays a crucial role in the processes of greening, acclimation to light intensity, and senescence. The chlorophyll cycle consists of three reactions: the conversions of chlorophyll a to chlorophyll b by chlorophyllide a oxygenase, chlorophyll b to 7-hydroxymethyl chlorophyll a by chlorophyll b reductase, and 7-hydroxymethyl chlorophyll a to chlorophyll a by 7-hydroxymethyl chlorophyll a reductase. We identified 7-hydroxymethyl chlorophyll a reductase, which is the last remaining unidentified enzyme of the chlorophyll cycle, from Arabidopsis thaliana by genetic and biochemical methods. Recombinant 7-hydroxymethyl chlorophyll a reductase converted 7-hydroxymethyl chlorophyll a to chlorophyll a using ferredoxin. Both sequence and biochemical analyses showed that 7-hydroxymethyl chlorophyll a reductase contains flavin adenine dinucleotide and an iron-sulfur center. In addition, a phylogenetic analysis elucidated the evolution of 7-hydroxymethyl chlorophyll a reductase from divinyl chlorophyllide vinyl reductase. A mutant lacking 7-hydroxymethyl chlorophyll a reductase was found to accumulate 7-hydroxymethyl chlorophyll a and pheophorbide a. Furthermore, this accumulation of pheophorbide a in the mutant was rescued by the inactivation of the chlorophyll b reductase gene. The downregulation of pheophorbide a oxygenase activity is discussed in relation to 7-hydroxymethyl chlorophyll a accumulation.
INTRODUCTION
In land plants and cyanobacteria, chlorophyll is synthesized from Glu (Tanaka and Tanaka, 2006; Tanaka and Tanaka, 2007) and exists as chlorophyll-protein complexes (Barber et al., 2000). During greening, newly synthesized chlorophyll assembles with various proteins to form the chlorophyll-protein complexes of the photosystems (Shimada et al., 1990), which harvest and transfer light energy and drive the electron transport that is indispensable to photosynthesis (Green and Durnford, 1996; Fromme et al., 2003). Because chlorophyll is a potentially dangerous molecule that generates reactive oxygen species (op den Camp et al., 2003), the chlorophyll that is released from the complex during senescence is converted to safe molecules of nonfluorescent chlorophyll catabolites (Hörtensteiner, 2006). When chlorophyll metabolism is not well regulated, and intermediate molecules accumulate (Papenbrock et al., 2000; Meskauskiene et al., 2001), necrotic lesions appear on leaves (Pruzinská et al., 2003; Hirashima et al., 2009). By contrast, when the chlorophyll supply is limited, photosynthesis activity becomes low, resulting in the retardation of plant growth (Liu et al., 2004). Therefore, it is important that the synthesis and degradation of chlorophyll be strictly regulated during both greening and senescence.
In order for the plant to achieve a desirable chlorophyll a/b ratio, a portion of newly synthesized chlorophyll a or chlorophyllide a is converted to chlorophyll b or chlorophyllide b by chlorophyllide a oxygenase (CAO) (Tanaka et al., 1998; Espineda et al., 1999). When the plant needs to decrease the level of chlorophyll b, chlorophyll b is converted to chlorophyll a by chlorophyll b reductase (CBR) (Scheumann et al., 1996) and 7-hydroxymethyl chlorophyll a (HMChl) reductase (HCAR) (Ito et al., 1996; Scheumann et al., 1998). This interconversion pathway between chlorophyll a and chlorophyll b is referred to as the chlorophyll cycle (Rüdiger, 2002). However, the chlorophyll cycle entails more than just the interconversion of chlorophyll a and chlorophyll b; it also plays an essential role in the formation and degradation of light-harvesting chlorophyll a/b–protein complexes (LHC). The level of LHC has been shown to be greatly reduced in chlorophyll b–less mutants (Murray and Kohorn, 1991), which have a defect in CAO (Oster et al., 2000); by contrast, the formation of LHC of photosystem II (LHCII) has been reported to be accelerated in CAO-overexpressing plants (Tanaka et al., 1999, 2001; Tanaka and Tanaka, 2005). Two CBR genes, NOL and NYC1, have been identified using the stay-green mutant of rice (Oryza sativa) (Kusaba et al., 2007). The degradation of the LHC was found to be almost completely inhibited, but other chlorophyll a–protein complexes were degraded in cbr mutants (Kusaba et al., 2007). It has been reported that when trimeric LHCII was incubated with recombinant CBR (NOL), chlorophyll b in LHCII was converted to HMChl, and all of the chlorophyll, including chlorophyll a, was released from LHCII, indicating that CBR participates in the first step of LHC degradation (Horie et al., 2009). The chlorophyll cycle plays a crucial role in the processes of greening, acclimation to light intensity, seed formation, and senescence, where LHCII is actively synthesized or degraded. Therefore, the elucidation of the properties and the regulation of the enzymes for the chlorophyll cycle are indispensable for an understanding of various physiological processes.
In the chlorophyll cycle, CAO and CBR have been identified by mutant screening of Chlamydomonas reinhardtii (Tanaka et al., 1998) and rice (Kusaba et al., 2007), respectively, and their physiological functions, regulation, and distribution in photosynthetic organisms have been elucidated. However, HCAR has not yet been identified. HCAR catalyzes the reduction of a hydroxymethyl group to a methyl group, a process in which the substitution of a hydroxyl (OH) with a hydrogen (H) occurs. The reduction of an OH group is a chemically difficult reaction; therefore, chlorophyll b–to–chlorophyll a conversion had not been considered to occur prior to the discovery of chlorophyll b to chlorophyll a conversion within isolated plastids (Ito et al., 1993). Although enzymatic examples of this reaction are rare, a similar reaction has been reported and well studied with ribonucleotide reductase, which catalyzes the substitution of the 2′-OH group of a ribonucleotide with a hydrogen (Bollinger et al., 2008). This reaction occurs via a free radical mechanism. However, the mechanism of the substitution of the OH with a hydrogen by HCAR might be different from that by ribonucleotide reductase because the OH exists on the pyrrole ring of HMChl. Thus, enzymatic information is indispensable for an understanding of this difficult reaction.
In this study, we obtained an Arabidopsis thaliana mutant that accumulated HMChl, a substrate of HCAR. This mutant was impaired in a putative iron-sulfur flavoprotein, which has a high sequence similarity to divinyl chlorophyll vinyl reductase (DVR) of Synechocystis PCC6803. The recombinant protein expressed in Escherichia coli converted HMChl to chlorophyll a, a reaction that used ferredoxin as a reducing equivalent. We emphasize that this difficult reaction involving the substitution of an OH with an H was achieved by a single enzyme, the flavoprotein now referred to as HCAR. We also found accumulation of pheophorbide a, a degradation product of chlorophyll, in the hcar mutant, and further discuss the evolution of HCAR.
RESULTS
Arabidopsis Homolog of Cyanobacterial DVR
Figure 1 shows the chlorophyll metabolic pathway, including the later steps of chlorophyll a synthesis, the chlorophyll cycle, and the initial steps of chlorophyll degradation. Divinyl chlorophyllide a is converted to monovinyl chlorophyllide a and is then phytylated to chlorophyll a, which is converted to chlorophyll b by CAO. In the degradation processes, chlorophyll b is converted to chlorophyll a before its degradation and is subsequently dechelated and dephytylated to form pheophorbide a.
Figure 1.
Chlorophyll Metabolic Pathway in Land Plants.
Land plants possess two CBR isozymes, NYC1 and NOL. The metal-chelating substance (MCS) has not yet been identified. CS, chlorophyll synthase; PPH, pheophytinase; PAO, pheophorbide a oxygenase.
DVR is responsible for the reduction of a vinyl group to an ethyl group in the chlorophyll synthesis pathway, and the Arabidopsis DVR (At-DVR) is encoded by AT5G18660 (Nagata et al., 2005). Recently, a cyanobacterial DVR (Sy-DVR) has been identified from Synechocystis PCC6803 (Islam et al., 2008; Ito et al., 2008). The Sy-DVR protein encoded by slr1923 is the only protein responsible for the reduction of the 8-vinyl group. Interestingly, Sy-DVR demonstrates no sequence similarity to At-DVR, and Synechocystis PCC6803 has no genes showing homology to At-DVR. However, the Arabidopsis gene AT1G04620 was found to be homologous to Sy-DVR. Figure 2 shows the alignment of the amino acid sequences of Sy-DVR and AT1G04620. These two proteins have a high sequence similarity (54% identity), and both exhibit predicted binding motifs for a flavin and an iron-sulfur cluster. In spite of this high sequence similarity, it appears that AT1G04620 cannot substitute for AT5G18660 as mutants in AT5G18660 have no monovinyl chlorophyll (Nagata et al., 2005).
Figure 2.
Multiple Amino Acid Sequence Alignment of AT1G04620, Slr1923, and FpoF
AT1G04620 homologous sequences were aligned using ClustalW, as implemented in BioEdit. Conserved residues are shaded with black. Filled circles represent the Cys residues that are predicted to coordinate the iron-sulfur cluster. Open circles represent the conserved sequence motif in FAD-containing proteins (KxxxxxGxG) (Dym and Eisenberg, 2001). FpoF does not contain this motif in the corresponding region. FpoF might contain an unidentified FAD binding motif.
The extended N-terminal region found in AT1G04620 was predicted to be a chloroplast transit peptide (Emanuelsson et al., 1999). Chloroplast localization of the protein was confirmed by the expression of AT1G04620 fused to green fluorescence protein (GFP) (see Supplemental Figure 1 online). GFP fluorescence colocalized with chlorophyll fluorescence, indicating chloroplast localization of AT1G04620.
Phenotype of the hcar Mutant
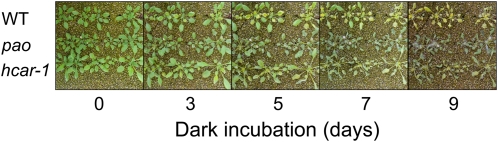
The high sequence similarity of AT1G04620 to Sy-DVR of the chlorophyll synthesis pathway suggested the participation of the AT1G04620 gene product in chlorophyll metabolism. To gain insight into the function of the AT1G04620 gene product, we analyzed two T-DNA insertion lines, SALK_018790C (hcar-1) and CS908281 (hcar-2). We referred to this mutant as the hcar mutant, as we found that the AT1G04620 gene product catalyzes the conversion of HMChl to chlorophyll a in this study as it is described hereafter. To confirm the absence of the AT1G04620 gene product in the hcar-1 mutant, we performed an immunoblot analysis using an antibody against the protein. No signal was obtained with protein extracts from the mutant (see Supplemental Figure 2 online), indicating that the mutant completely lacked the gene product. However, we could not detect any difference in the visible phenotype of green plants between the hcar-1 mutant and the wild type (Figure 3). Interestingly, the mutant exhibited the stay-green phenotype when the plants were transferred to the darkness: the mutant remained green during dark incubation, whereas in the wild type, the leaves turned yellow after 5 d of dark incubation. To quantify the changes in chlorophyll content, we measured chlorophyll levels during dark incubation (see Supplemental Figure 3 online). Decrease in chlorophyll content was suppressed in both hcar mutants.
Figure 3.
The Visible Phenotype of the Wild Type, pao, and hcar-1 during Dark Incubation.
Wild-type (WT), pao, and hcar-1 plants were grown for 4 weeks under continuous light and then incubated in complete darkness at 24°C for the indicated number of days.
A closer examination of the mutant revealed that some leaves were wilted after dark incubation. To quantify the cell death phenotype of the hcar-1 mutant, electrolyte leakage was measured in the hcar-1 mutant during dark incubation (see Supplemental Figure 4 online). A large increase in electrolyte leakage was observed in hcar-1 leaves, indicating that hcar-1 leaf cells lost membrane integrity during dark incubation. This phenotype was similar to the pao mutant, which has been shown to accumulate a large amount of pheophorbide a (Hirashima et al., 2009).
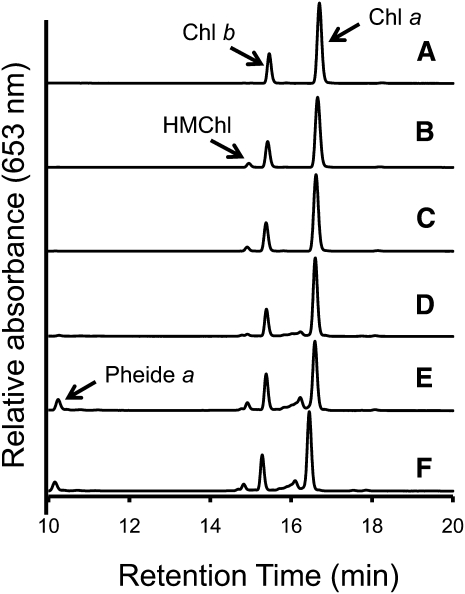
Mutants of the enzymes for chlorophyll metabolism are known to accumulate intermediate molecules of chlorophyll synthesis or degradation. To investigate the effect of the mutation in the AT1G04620 locus on chlorophyll metabolism, chlorophyll and its precursors were extracted from green leaves of the wild type and hcar mutants and subjected to HPLC (Figure 4). We found that the levels of chlorophyll a and chlorophyll b were similar between the wild type and the mutants. Although we could not find any visual differences between the green leaves, HMChl, an intermediate molecule between chlorophyll a and chlorophyll b, was found in hcar mutants (Figures 4B and 4C). When senescent leaves were examined by HPLC, an accumulation of pheophorbide a was found in addition to HMChl (Figures 4E and 4F). HMChl and pheophorbide a were either not found or found at low levels in wild-type leaves (Figures 4A and 4D).
Figure 4.
The HPLC Profile of Chlorophyll Accumulated in the Wild Type and hcar Mutants.
Seedlings were grown for 4 weeks under continuous light. Chlorophyll was extracted from the wild type ([A] and [D]), hcar-1 ([B] and [E]), and hcar-2 ([C] and [F]) before ([A] to [C]) and after ([D] to [F]) dark incubation for 5 d. Chl a, chlorophyll a; Chl b, chlorophyll b; Pheide a, pheophorbide a.
Next, the levels of HMChl (Figure 5A) and pheophorbide a (Figure 5B) in hcar-1 were quantified. The levels of HMChl were between 30 and 60 nmol/g fresh weight, which corresponds to ~2% of the total chlorophyll. Furthermore, the level of pheophorbide a in hcar-1 was comparable to the level that has been reported in an antisense pao mutant with an impaired PaO previously reported as As-ACD1 (Tanaka et al., 2003). In addition, we found that pheophorbide a did not accumulate before the dark incubation in the wild type, pao, or hcar-1 (Figure 5B); a low level of pheophorbide a accumulated after 3 d of dark incubation and drastically increased after 5 d in both of the mutants. The level of pheophorbide a in hcar-1 corresponded to ~60% of that in the pao mutant after 5 d of dark incubation. These results suggest that HCAR and PaO were both downregulated in hcar-1. Although HMChl did not accumulate in the wild type during the early phase of dark incubation, substantial amounts of HMChl accumulated after 5 d in the dark. The levels of HMChl in the wild type varied among experiments. Unidentified environmental or developmental changes might induce the accumulation of HMChl in the wild type.
Figure 5.
Chlorophyll Accumulation in the Wild Type and Mutants.
The wild type (WT), pao, and the hcar-1 and hcar-1 nyc1 nol mutants were grown for 4 weeks under continuous light and then incubated in complete darkness at 24°C for the indicated number of days. HMChl (A) and pheophorbide a (B) contents are indicated per gram fresh weight (FW) of leaf. The error bars represent the SD of three samples from a single harvest.
Because the accumulation of pheophorbide a indicated an impairment of PaO in the hcar-1 mutant, we examined the PaO protein level by immunoblotting (see Supplemental Figure 2 online). The PaO level was similar to that of the wild type, indicating that HCAR did not affect the expression of PaO. Next, we examined the effect of HMChl accumulation on PaO activity using the hcar-1 nyc1 nol triple mutant, in which HMChl is not produced due to a lack of CBRs (Figure 5A). HMChl accumulated in the hcar-1 mutant but not in the hcar-1 nyc1 nol triple mutant. A large amount of pheophorbide a accumulated in the hcar-1 mutant, and small amount of pheophorbide a accumulated in the wild type. Interestingly, the triple mutant did not accumulate pheophorbide a even after 5 d of dark incubation (Figure 5B), indicating a strong correlation between HMChl and pheophorbide a accumulation. HMChl is also an intermediate molecule of chlorophyll b synthesis that is catalyzed by CAO. However, HMChl was at a very low level or not detected in the hcar-1 nyc1 nol triple mutant, indicating that HMChl was produced from chlorophyll b by CBR in the hcar-1 mutant.
Effects of the Mutation in HCAR on the Conversion of Chlorophyll b to Chlorophyll a in Greening Cotyledons
The accumulation of HMChl in hcar suggested the involvement of HCAR in the conversion of chlorophyll b to chlorophyll a. It has been reported that the conversion of chlorophyll b to chlorophyll a is high during both greening (Ito and Tanaka, 1996) and senescence (Scheumann et al., 1999). Therefore, we next examined the effect of the mutation in HCAR on the conversion of chlorophyll b to chlorophyll a in greening tissues. Previously, we reported that when greening cucumber (Cucumis sativus) cotyledons were exposed to light for several hours and then incubated in the dark, chlorophyll a increased with a concomitant decrease in chlorophyll b without a significant change in the total chlorophyll content (Tanaka et al., 1991). Considering that chlorophyll is not newly synthesized in the dark in angiosperm species, it was concluded that the increase in chlorophyll a and decrease in chlorophyll b was caused by the conversion of chlorophyll b to chlorophyll a during the dark incubation. When we performed the same experiments with greening Arabidopsis seedlings (Figure 6), both wild-type and hcar-1 mutant etiolated seedlings that were exposed to light for 4 h accumulated chlorophyll a and chlorophyll b. When the wild-type seedlings were incubated in the dark for 24 h, chlorophyll a increased with a decrease in chlorophyll b (Figure 6), which is consistent with our results obtained with cucumber cotyledons. This clearly shows that the conversion of chlorophyll b to chlorophyll a actively occurred in the Arabidopsis seedlings. When the same experiment was performed with the mutant, chlorophyll b decreased as in wild type, but there was no concomitant chlorophyll a increase (Figure 6). Interestingly, a large amount of HMChl accumulated after the dark incubation, indicating that the mutation did not seriously affect the conversion of chlorophyll b to HMChl but almost completely inhibited the conversion of HMChl to chlorophyll a. This clearly shows that HCAR is involved in the conversion of HMChl to chlorophyll a.
Figure 6.
Changes in the Chlorophyll Content during the Dark Incubation of Greening Seedlings.
Wild-type and hcar-1 seeds were germinated for 3 d under dark conditions and transferred to light conditions (100 μmol m−2 s−1) at 24°C for 4 h (0 hD). The seedlings were then transferred to complete dark conditions for 24 h (24 hD). Chlorophylls were extracted before and after the dark incubation from the seedlings and were subjected to HPLC analysis. The error bars represent the sd for three samples. Black, chlorophyll a; gray, chlorophyll b; white, HMChl.
Measurement of HCAR Activity of the AT1G04620 Recombinant Protein Expressed in E. coli
The accumulation of HMChl in greening cotyledons and green leaves of the hcar mutant impaired in AT1G04620 suggested that AT1G04620 is responsible for the conversion of HMChl to chlorophyll a. To examine this possibility, we expressed the recombinant AT1G04620 protein in E. coli and determined whether it could convert HMChl to chlorophyll a. The His-tagged protein lacking the transit peptide was purified from E. coli as a single band (see Supplemental Figure 5 online), as revealed by SDS-PAGE analysis, which showed 48 kD that corresponded to the expected size of 50.9 kD (AT1G04620 lacking the transit sequence). The substrate, 7-hydroxymethyl chlorophyllide a (HMChlide), was incubated with the recombinant protein in the presence of ferredoxin, ferredoxin NADPH oxidoreductase (FNR), and NADPH, and the reaction products were examined by HPLC. After incubation, we found that chlorophyllide a was formed with a concomitant decrease in HMChlide (Figure 7), indicating that AT1G04620 had catalytic activity. Furthermore, the requirement for ferredoxin in this reaction is consistent with the previously reported results obtained with isolated plastids (Scheumann et al., 1998). The natural substrate of HCAR should be HMChl instead of HMChlide because NOL, which produces the substrate of HCAR, converts chlorophyll b to HMChl and because HMChl, not HMChlide, accumulated in the hcar mutant. Accordingly, we used HMChl as the substrate instead of HMChlide for the enzymatic assay. An E. coli cell extract was used for this experiment because the cell extract exhibited a higher activity than purified HCAR probably because HCAR was partly inactivated during the purification procedures. Although the level of chlorophyll a was low, HMChl was converted to chlorophyll a by the recombinant protein (Figure 7). These results indicate that the AT1G04620 product converted a hydroxymethyl group on the pyrrole ring to a methyl group using reduced ferredoxin. Based on these experiments, AT1G04620 was named HCAR.
Figure 7.
The Conversion of HMChl(ide) to Chlorophyll(ide) a by HCAR.
Purified recombinant HCAR was incubated with HMChlide without any reductant (A) or with NADPH, FNR, and ferredoxin (B). After incubation, the pigments were extracted from the mixture and analyzed by HPLC.
HMChl was incubated with lysed E. coli harboring the empty vector (C) or expressing recombinant HCAR (D), in the presence of NADPH, FNR, and ferredoxin. After incubation, the pigments were extracted from the mixture and analyzed by HPLC. Chlide a, chlorophyllide a; Chl a, chlorophyll a.
The Redox Cofactors of HCAR
The amino acid alignment predicted binding motifs for both a flavin (Dym and Eisenberg, 2001) and an iron-sulfur cluster (Figure 2). As these two redox cofactors were expected to play an important role in the enzymatic reaction, we examined the presence of these factors by biochemical methods. Spectral analysis of purified HCAR showed a peak at 450 nm, which is typical of an oxidized flavin (Figure 8). The absorption peak at 450 nm disappeared when the flavin was reduced with dithionite, indicating that HCAR contained a flavin molecule that may function in catalysis. To identify which flavin species was present, we employed HPLC analysis. Figure 9 shows the HPLC profile of authentic flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN), which were clearly separated into two peaks. Next, the flavin molecule was isolated from purified HCAR and subjected to HPLC. Only one peak corresponding to FAD was detected, indicating that the HCAR enzyme contains FAD.
Figure 8.
The Absorbance Spectra of the Oxidized and Reduced HCAR.
Purified HCAR (1 mg/mL, solid line) expressed in E. coli was reduced by the addition of dithionite (10 mM, dashed line).
Figure 9.
HPLC Analysis of the Released Flavin from HCAR.
Authentic FAD and FMN (A) and flavin extracted from purified HCAR (B) were analyzed by HPLC. The flavin was released from recombinant HCAR by heat treatment.
We found that the four conserved Cys residues for the iron-sulfur cluster binding motif were conserved in all of the HCARs of green plants and in Sy-DVR (Figure 2). However, we could not determine the presence of an iron-sulfur cluster merely by absorbance spectra. Thus, we measured the iron content of HCAR by a colorimetric method and found that the iron content was 0.85 atoms per HCAR protein. It is well known that cofactors are often lost during heterologous expression. Indeed, the expression of iron-sulfur cluster-containing proteins has been reported to be difficult, and a significant amount of expressed proteins have been found to lack their iron-sulfur cluster (Oppenheimer et al., 2010). Assuming that some portion of recombinant HCAR was denatured and had lost the iron, holo-HCAR might potentially have more iron per protein moiety. Both sequence analysis and iron determination suggested the presence of an iron-sulfur cluster in HCAR, though we have no information concerning the type or stoichiometry of iron binding. Further information would be obtained by x-ray crystallographic analysis.
Phylogenetic Analysis of HCAR
As described in Figure 2, HCAR has a high sequence similarity to DVR of Synechocystis PCC 6803. These types of DVRs were found in other cyanobacteria and in photosynthetic bacteria, including purple bacteria and green sulfur bacteria. Figure 10 shows the phylogenetic tree of HCAR and DVR using F420H2 dehydrogenase as an outgroup. HCARs were found in both green plants and green algae. In this tree, HCAR branched off within the DVR cluster and was most closely related to diatom DVR phylogenetically, suggesting that HCAR branched off from DVR at the eukaryotic stage. These results indicate that HCAR of the chlorophyll cycle evolved from DVR of the bacterial chlorophyll synthesis pathway.
Figure 10.
Phylogenetic Tree of HCAR.
Neighbor-joining tree constructed with the translated sequence of HCAR and DVR. Bootstrap values for each clade are indicated on each node, and the scale bar indicates the number of amino acid substitutions per site. The F subunit of Methanosaeta F420H2 dehydrogenase was used as an outgroup. The alignment used for this analysis is available as Supplemental Data Set 1 online.
DISCUSSION
The Reduction of HMChl to Chlorophyll a by HCAR
It has been reported that the F subunit of F420H2 dehydrogenase of Methanolobus tindarius (Fpo) is an iron-sulfur flavoprotein (Deppenmeier, 2004) and has sequence similarity to HCAR. In this study, we succeeded in detecting FAD in recombinant HCAR that was reduced by sodium dithionite. The binding motif for an iron-sulfur center was conserved at the N-terminal region of F420H2 dehydrogenase (Johnson and Mukhopadhyay, 2005) and HCAR, and iron was detected in recombinant HCAR. From these results, we concluded that HCAR contains a flavin and an iron-sulfur cluster as redox cofactors. However, the route of electron transfer from ferredoxin to HMChl in HCAR is uncertain at present. In the F subunit of the Fpo complex, the two-electron carrier F420H2 reduces FAD, from which an electron is transferred to an iron-sulfur cluster (Kulkarni et al., 2009); the iron-sulfur cluster of the F subunit then transfers an electron to the BCDI subunit of Fpo. The FrhB subunit of the F420-reducing hydrogenase complex also has weak homology to HCAR, and it has been reported to contain FAD but lack the iron-sulfur cluster (Johnson and Mukhopadhyay, 2005). FrhB passes the electron of F420H2 to the iron-sulfur cluster of the FrhG subunit in the complex. Accordingly, the sequence similarity between HCAR, FpoF, and FrhB suggests an electron transfer from FAD to the iron-sulfur cluster in HCAR. However, this hypothesis is not necessarily accepted because HCAR receives an electron from a one-electron carrier (ferredoxin), yet FpoF and FrhB receive electrons from a two-electron carrier (F420H2). Another proposal is that ferredoxin first reduces the iron-sulfur cluster and then an electron is transferred to FAD, which stores two electrons and reduces HMChl to chlorophyll a. This idea is consistent with the reported electron transfer in ferredoxin-dependent Glu synthase, an iron-sulfur flavoprotein, in which flavin is reduced by ferredoxin via the iron-sulfur cluster (Vanoni et al., 2005). Further study will be required to elucidate the electron transfer in HCAR.
The reductive elimination of a hydroxyl group is a difficult reaction due to the strong carbon-oxygen bond. This is the reason why enzymatic examples for this reaction are rarely found in the literature. A well-studied example is ribonucleotide reductase, which catalyzes the replacement of the ribose 2′-OH in ribonucleotides with hydrogen (Bollinger et al., 2008); a free radical is an essential component for this replacement. However, a catalytic mechanism for the replacement of an OH by a hydrogen on a pyrrole ring has not yet been elucidated. Based on the finding that the conversion of chlorophyll b to chlorophyll a occurs in vivo in the presence of reduced ferredoxin and ATP, Folly and Engel (1999) have proposed a reaction mechanism for the conversion of HMChl to chlorophyll a. These authors hypothesized that the unique electron arrangement of the cyclic 18-π electron porphyrin system facilitates the elimination of water and the stabilization of the carbocation, but this process demands an activator, such as ATP. In this report, we showed that ATP was not required for the substitution of the OH with hydrogen in the reaction catalyzed by HCAR, which has a high sequence similarity to the DVR enzyme that catalyzes the reduction of –CH=CH2 to –CH2–CH3. Thus, a portion of the reaction mechanism of HCAR may be similar to the DVR reaction. Based on these findings, we propose the following reaction mechanism for HCAR, as described below (see Supplemental Figure 6 online). The first step is the production of a hydroxymethyl cation. The loss of a water molecule and the addition of an electron yields a methylene radical, which is converted to a methylene anion or a methyl radical and then to a methyl group.
Clearly, the exact reaction mechanism may be elucidated by an electron transfer route and the crystal structure of HCAR.
The Regulation of PaO Activity by HCAR
The hcar-1 mutant carries a T-DNA in an exon, and HCAR was below the level detectable by immunoblotting, indicating that the mutant completely lacked HCAR activity; however, the level of HMChl was also low. One possible reason for this may be the conversion of HMChl to other molecules, such as 7-hydroxymehyl pheophorbide a and HMChlide, although these molecules were not detected by HPLC. The same phenomenon has been reported with the Arabidopsis pheophytinase mutant (Schelbert et al., 2009). In this mutant, the conversion from pheophytin to pheophorbide was reported to be completely blocked, yet the level of pheophytin was low. Therefore, it might be reasonable to speculate that the chlorophyll degradation pathway has another route, in addition to the major route, due to the wide substrate specificity.
In the chlorophyll cycle, the first reaction, the conversion of chlorophyll b to HMChl, is catalyzed by CBR; the second reaction, the conversion of HMChl to chlorophyll a, is catalyzed by HCAR (Figure 1). In Arabidopsis, there are two CBR genes, NOL and NYC1 (Horie et al., 2009). These two CBRs have a high sequence similarity but are differentially expressed during development in Arabidopsis. HCAR is encoded by a single gene and is constitutively expressed. It is expected that HCAR and NOL should be regulated to convert chlorophyll b to chlorophyll a efficiently during greening; by contrast, HCAR and NYC1 should be coordinated during senescence. However, HCAR does not affect the activity of CBR because the conversion of chlorophyll b to HMChl occurred in the greening cotyledons of the hcar mutant, as well as in the wild type. The proposed independent regulation of these two enzymes would be different from PaO and red chlorophyll catabolite reductase (Pruzinská et al., 2003).
By contrast, the mutation in HCAR induced the accumulation of pheophorbide a, a degradation product of chlorophyll. The level of accumulated pheophorbide a in the hcar-1 mutant was almost 60% of what has been reported for the pao mutant (Tanaka et al., 2003), indicating that PAO activity was severely downregulated in the hcar mutant. Immunoblot analysis clearly showed that the PaO protein level in the hcar-1 mutant was almost the same as in the wild type, indicating that PaO catalytic activity was downregulated in the hcar mutant. One possible hypothesis is that PaO is active only when HCAR is present, for example, due to a direct interaction between the two enzymes. However, this idea is not consistent with the report that PaO is active without HCAR in in vitro experiments (Pruzinská et al., 2003). A second hypothesis is the inhibition of PaO activity via HMChl, which accumulated in the hcar mutant. This hypothesis is supported by the results that pheophorbide a did not accumulate in the hcar-1 nyc1 nol triple mutant, which also did not accumulate HMChl; it is also consistent with in vitro experiments that suggested that pheophorbide b was a competitor of PaO, and FCC production by PaO was inhibited by pheophorbide b (Hörtensteiner et al., 1995). However, this hypothesis cannot be fully accepted because PaO activity was not completely inhibited by pheophorbide b in in vitro experiments; by contrast, PaO seemed to be severely inhibited in the hcar mutant. We suggest that it would be difficult for HMChl to be a competitive inhibitor of PaO because HMChl retains the phytyl chain, making it difficult to access PaO, which resides in the chloroplast envelope. Although HMChl might not directly regulate PaO activity, it is possible to speculate that HMChl or its degradation products is involved in modulation of PaO activity through an unidentified mechanism. This idea is supported by the finding that both HMChl and pheophorbide a accumulated in the Arabidopsis hmc1 mutant, which has a defect in the Nap1 gene (Nagane et al., 2010).
The Distribution and Evolution of HCAR
DVR has been identified from Synechocystis PCC6803 by a bioinformatics method (Ito et al., 2008). This DVR is found to be present in other cyanobacteria, except for marine types of Synechococcus and Prochlorococcus, some photosynthetic bacterial groups, and diatoms. Interestingly, the phylogenetic tree of HCAR and DVR clearly showed that HCAR appeared within the DVR clade, indicating that HCAR evolved from DVR (Figure 10). This phylogenetic tree strongly suggests that the ancestor of chlorophytes and chromophytes should have both types of DVRs (At-DVR and Sy-DVR) and that Sy-DVR evolved to HCAR in the green lineage. Thus, a chlorophyll biosynthesis enzyme may have evolved to catalyze other steps of chlorophyll metabolism. Although the phylogenetic relationship is not clear, sequence similarities are found between the different enzymes for tetrapyrrole metabolism. PaO and CAO belong to the same Rieske-non heme iron oxygenase family (Gray et al., 2002) and have sequence similarity. The amino acid sequences of red chlorophyll catabolite reductase, which reduces the linear tetrapyrrole of the red chlorophyll catabolite, were shown to have a low degree of sequence similarity to ferredoxin-dependent bilin reductases (Sugishima et al., 2009). This phenomenon is reasonable because the substrates (linear or open tetrapyrroles) have common structure, and the reaction mechanisms are partly shared. In addition to chlorophyll a, there are many chlorophyll species, such as chlorophyll b, chlorophyll c, chlorophyll d, and chlorophyll f (Chen et al., 2010). It might be possible that changes in the catalytic properties of chlorophyll biosynthetic enzymes contribute to the diversity of chlorophyll species.
HCAR has been found only in green algae and land plants, such as Arabidopsis, mosses, and prasinophytes. Therefore, it may be concluded that HCAR was acquired in the green lineage. The distribution of CBR is consistent with that of HCAR, which suggests a coevolution of HCAR and CBR. However, not all of the chlorophyll b–containing organisms have necessarily retained the chlorophyll cycle. In the cyanobacterial lineage, Prochlorococcus has chlorophyll b, but it does not contain genes for CBR, HCAR, or cyanobacterial DVR in its genome. Thus, the appearance of the chlorophyll cycle seems to be consistent with that of LHC in green lineages. This is reasonable because the chlorophyll cycle regulates the formation and degradation of the LHC.
With this study, all of the enzymes responsible for the chlorophyll cycle are now identified. This progress will enable the detailed molecular analysis of the regulation and function of the chlorophyll cycle in various physiological processes, such as greening, acclimation, and senescence.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana (Columbia ecotype) was grown for 4 to 6 weeks in a chamber equipped with white fluorescent lamps under continuous illumination at a light intensity of 100 μmol m–2 s–1 at 24°C. The plants were subsequently transferred to darkness for 3 to 9 d at 24°C for the chlorophyll degradation experiments. The leaves were harvested at the times indicated in Results. For the greening experiments, seedlings were grown on agar plates containing half-strength diluted Murashige and Skoog medium for 3 d in the dark. Etiolated seedlings were illuminated (100 μmol m–2 s–1) for 4 h and incubated in the dark for an additional 24 h. The T-DNA insertion mutants, lacking either AT1G04620 (SALK_018790C, hcar-1; CS908281, hcar-2), AT4G13250 (SALK_091664, nyc1), or AT5G04900 (AL759262, nol), were obtained from the ABRC (Ohio State University) and GABI-Kat, respectively. Every mutant was crossed, and the triple mutant was identified by PCR-based genotyping. Production of the pao mutant that overexpressed antisense RNA for the PaO gene was described in our previous report (Tanaka et al., 2003).
Pigment Analysis
Leaves were weighed and pulverized in acetone using a ShakeMaster grinding apparatus (BioMedical Science), and the extracts were centrifuged for 15 min at 22,000g. The pigments were separated on a Symmetry C8 column (150 × 4.6 mm; Waters), according to a method reported previously (Zapata et al., 2000). The elution profiles were monitored by measuring the absorbance at 653 nm (SPD-M10A; Shimadzu), and the pigments were identified by their retention times and spectral patterns. Pigment quantification was performed using the area of the peaks.
Enzymatic Assay
The coding region of HCAR lacking its transit peptide was amplified by PCR (PrimeSTAR Max DNA polymerase; Takara) using primers 5′-GGATCCTCCGTCGTTAACTCTTCTTC-3′ (the underlined region is an engineered BamHI site) and 5′-CTCGAGTTTCTTGGAGAGCATTTTAT-3′ (the underlined region is an engineered XhoI site) and cloned into the BamHI/XhoI sites of pET-30a(+) (Novagen). The recombinant protein obtained from this plasmid possesses His tags at the both the N and C termini. This recombinant protein did not exhibit enzymatic activity, and it was expected that the His tag at the N terminus disturbed the HCAR activity. Therefore, the His tag at the N terminus was removed by PCR using the primers 5′-ATGTCCGTCGTTAACTCTTCTTC-3′ and 5′-ATGTATATCTCCTTCTTAAA-3′ to amplify the plasmid DNA. The PCR products were phosphorylated by T4 polynucleotide kinase (Takara), and self-ligation was performed. The expression plasmid was introduced into Escherichia coli BL21 cells. The transformed cells were grown at 37°C with 200 mL Luria-Bertani medium containing kanamycin (50 μg/mL) and 100 μM of ammonium ferric citrate until the optical density at 600 nm reached at 1.3. The expression of the HCAR gene was induced with 0.2 mM isopropyl-ββ-d-thiogalactopyranoside for 3 h. After incubation, the culture was harvested by centrifugation, and the collected cells were resuspended in buffer (5 mM imidazole, 500 mM NaCl, and 20 mM Tris-HCl, pH 7.9) and disrupted by sonication. The recombinant HCAR contained in the soluble fraction was purified using a nickel column (His bind kit; Novagen); a desalting column (Mini Trap G-25; GE Healthcare) was used to remove the imidazole from the purified HCAR fraction.
HMChl was obtained by reducing chlorophyll b with 100 mM NaBH4 in methanol, whereas HMChlide was obtained from HMChl by hydrolysis with recombinant chlorophyllase (Tsuchiya et al., 1999).
Purified HCAR was diluted to a concentration of 160 μg/mL in buffer (50 mM Tris-HCl, pH 7.5, and 100 mM NaCl) containing 0.9 mg/mL Glc, 0.4 mg/mL Glc oxidase (Nacalai), and 0.04 mg/mL catalase (Sigma-Aldrich) to reduce the O2 level in the reaction mixture. Fifty microliters of the mixture was used for the reaction. HMChlide was dissolved in acetone to a concentration of 0.15 mM, and 1 μL of HMChlide solution, 1 μL of 50 mM NADPH, 1 μL of spinach (Spinacia oleracea) FNR (0.1 mg/mL; Sigma-Aldrich) and 1 μL of spinach ferredoxin (2.2 mg/mL; Sigma-Aldrich) were added to the reaction mixture.
When HMChl was used as the substrate, HCAR-expressing E. coli cells were lysed with 4 mL of BugBuster (Novagen), and 50 μL of the lysate and 1μL of HMChl (0.09 mM) dissolved in acetone were incubated with the an above-described reductant. The mixtures were incubated for 15 min at 37°C, after which, 200 μL of acetone was added. After centrifugation at 22,000g for 10 min, the supernatant containing the chlorophyll derivatives was analyzed by HPLC as described above.
Spectral Measurement of HCAR
The absorbance spectrum of the purified HCAR was measured at a concentration of 1 mg/mL (U-331 spectrophotometer; Hitachi). For the reduction of HCAR, dithionite was added to a concentration of 10 mM.
Determination of the Flavin Species in HCAR
Recombinant Arabidopsis HCAR protein that was overexpressed in E. coli and then purified as described above was incubated at 100°C for 7 min to release the flavin. After centrifugation at 22,000g for 10 min, the supernatant (containing the flavin) and FAD and FMN standards (Sigma-Aldrich) were analyzed by HPLC. An aliquot of the sample was subjected to a ODS column (6 × 150 mm, Shim-Pack ODS; Shimadzu) equilibrated with 5 mM ammonium acetate, pH 6.0 (buffer A). The column was eluted with a linear gradient of 0 to 30% buffer B (methanol containing 5 mM ammonium acetate, pH 6.0) for 9 min and linear gradient of 100% buffer B for 1 min at a flow rate of 0.75 mL/min. The effluent from the column was monitored at the wavelength of 450 nm (Stuehr et al., 1991).
Quantification of Iron in HCAR
The iron content was determined colorimetrically using bathophenanthroline as an iron-chelating agent. Three hundred microliters of the eluate from the nickel column that contains recombinant HCAR (see above) was incubation with 150 μL of 1 n HCl at 95°C for 2 min. To remove the insoluble material, 150 μL of trichloroacetic acid (100% [w/v]) was added. After centrifugation, 150 μL of ascorbic acid (0.16 mg/mL) was added to 300 μL of the supernatant. Bathophenanthroline solution (150 μL of 0.67 mg/mL) and 150 μL of sodium acetate solution (300 mg/mL) were added to the mixture. The samples were incubated at room temperature for 5 min, and the absorbance at 535 nm was measured. The concentration of ferrous ion was calculated using an iron standard solution (Wako).
Sequence Analysis
The database search was performed using the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/blast). Amino acid sequences were aligned using the ClustalW program (Thompson et al., 1994) in the BioEdit program (http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
Phylogenetic Analysis
For the phylogenetic analysis, the protein sequences were aligned using MEGA 4 software (Tamura et al., 2007), and the midpoint-rooted neighbor-joining tree was generated using the same software with the following parameters: Poisson model, uniform rates, complete deletion, and bootstrap (1000 replicates).
Plasmid Construction and Plant Transformation
To express HCAR-GFP fusion proteins, we modified the pGreenII MH binary vector that was constructed in our previous study (Yamasato et al., 2005). The vector contained the cauliflower mosaic virus 35S promoter, the tobacco mosaic virus ω sequence, the GFP (S65T) sequence, and the nopaline terminator in the backbone of pGreenII-0029 (Hellens et al., 2000). The coding region of HCAR was amplified by PCR using the primers 5′- GTCGACATGATTACTGTCGTCACCTC- 3′ (the underlined section is an engineered SalI site) and 5′- CCATGGATTTCTTGGAGAGCATTTTAT-3′ (the underlined section is an engineered NcoI site). The GFP (S65T) gene in this plasmid was fused with the HCAR gene at the SalI and NcoI sites. The plasmids were subsequently transformed into an Agrobacterium tumefaciens (strain GV2260), and wild-type Arabidopsis was transformed.
Analysis of GFP Expression by Confocal Laser Scanning Microscopy
Fluorescence images were recorded on a C1si Spectral Imaging confocal laser scanning microscopy system with an ECLIPSE 80i microscope (Nikon). The microscope was equipped with a Nikon Plan Apo VC ×100 objective. An argon laser was used to generate an excitation source at 488 nm, and GFP fluorescence was recorded at 500 to 550 nm. A blue diode laser was used to generate an excitation source at 405 nm, and chlorophyll fluorescence was recorded at 650 to 680 nm. Images were processed with EZ-C1 Viewer 3.20 (Nikon).
Immunoblot Analysis
Total protein was extracted from leaves by grinding with extraction buffer (50 mM Tris, pH 6.8, 2 mM EDTA, 10% [w/v] glycerol, 2% [w/v] SDS, and 6% [v/v] 2-mercaptoethanol). Homogenates were centrifuged at 22,000g for 10 min. The supernatants were separated by 10% SDS-PAGE, and the resolved proteins were transferred onto a Hybond-P membrane (GE Healthcare). HCAR protein was detected with an anti-HCAR rabbit primary antiserum (diluted 1:10,000) that was raised against the recombinant Arabidopsis HCAR polypeptide used for the enzymatic assay. PaO protein was detected as previously reported (Hirashima et al., 2009). Anti-rabbit IgG linked to horseradish peroxidase (GE Healthcare) was used as a secondary antibody. The horseradish peroxidase activity was detected using the ECL Plus protein gel blotting detection system (GE Healthcare) following the manufacturer's protocol.
Accession Numbers
The HCAR homologous protein amino acid sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Anabaena variabilis ATCC 29413, YP_324708; Arabidopsis, NP_171956; Chlamydomonas reinhardtii, XP_001699546; Chlorobium phaeobacteroides DSM 266, YP_910679; Chloroflexus aurantiacus J-10-fl, YP_001636150; Gloeobacter violaceus PCC 7421, NP_923824; Methanosaeta thermophila PT, YP_842613; Micromonas sp RCC299, XP_002503439; Oryza sativa japonica Group, CAE01504; Ostreococcus lucimarinus CCE9901, XP_001416225; Ostreococcus tauri, XP_003080144; Physcomitrella patens subsp patens, XP_001770443; Populus trichocarpa, XP_002315601; Rhodopseudomonas palustris BisA53, YP_780232; R. palustris BisB5, YP_570899; Synechococcus elongatus PCC 6301, YP_170905; Synechococcus sp WH 5701, ZP_01086192; Synechococcus sp WH 7805, ZP_01124436; Synechocystis sp PCC 6803, NP_441896; Thalassiosira pseudonana CCMP1335, XP_002288079; Thermosynechococcus elongatus BP-1, NP_682635; and Vitis vinifera, XP_002285592.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. HCAR Accumulation in the Chloroplast.
Supplemental Figure 2. Immunoblot Analysis of HCAR and PaO in Wild-Type and hcar-1 Plants.
Supplemental Figure 3. Chlorophyll Retention during Dark Incubation.
Supplemental Figure 4. Electrolyte Leakage of the Leaves during Dark Incubation.
Supplemental Figure 5. SDS-PAGE Analysis of Purified HCAR.
Supplemental Figure 6. Hypothesized Reaction Pathway of Hydroxymethyl Reduction.
Supplemental Data Set 1. Alignment of HCAR Homolog for the Construction of Phylogenetic Tree in Figure 10.
Acknowledgments
We thank Hitoshi Tamiaki for valuable discussion and Sachiko Tanaka for technical assistance. This work was supported by a Grant-in-Aid for Scientific Research (No. 21370014 to A.T. and No. 68700307 to R.T.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
AUTHOR CONTRIBUTIONS
M.M., H.I., and A.Tanaka designed the research. M.M. and H.I. performed research. M.M., H.I., A.Takabayashi, R.T., and A.Tanaka analyzed data. H.I. and A.Tanaka wrote the article.
References
- Barber J., Morris E., Büchel C. (2000). Revealing the structure of the photosystem II chlorophyll binding proteins, CP43 and CP47. Biochim. Biophys. Acta 1459: 239–247 [DOI] [PubMed] [Google Scholar]
- Bollinger J.M., Jr., Jiang W., Green M.T., Krebs C. (2008). The manganese(IV)/iron(III) cofactor of Chlamydia trachomatis ribonucleotide reductase: structure, assembly, radical initiation, and evolution. Curr. Opin. Struct. Biol. 18: 650–657 [DOI] [PubMed] [Google Scholar]
- Chen M., Schliep M., Willows R.D., Cai Z.L., Neilan B.A., Scheer H. (2010). A red-shifted chlorophyll. Science 329: 1318–1319 [DOI] [PubMed] [Google Scholar]
- Deppenmeier U. (2004). The membrane-bound electron transport system of Methanosarcina species. J. Bioenerg. Biomembr. 36: 55–64 [DOI] [PubMed] [Google Scholar]
- Dym O., Eisenberg D. (2001). Sequence-structure analysis of FAD-containing proteins. Protein Sci. 10: 1712–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., von Heijne G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espineda C.E., Linford A.S., Devine D., Brusslan J.A. (1999). The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 10507–10511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folly P., Engel N. (1999). Chlorophyll b to chlorophyll a conversion precedes chlorophyll degradation in Hordeum vulgare L. J. Biol. Chem. 274: 21811–21816 [DOI] [PubMed] [Google Scholar]
- Fromme P., Melkozernov A., Jordan P., Krauss N. (2003). Structure and function of photosystem I: Interaction with its soluble electron carriers and external antenna systems. FEBS Lett. 555: 40–44 [DOI] [PubMed] [Google Scholar]
- Gray J., Janick-Buckner D., Buckner B., Close P.S., Johal G.S. (2002). Light-dependent death of maize lls1 cells is mediated by mature chloroplasts. Plant Physiol. 130: 1894–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B.R., Durnford D.G. (1996). The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 685–714 [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hirashima M., Tanaka R., Tanaka A. (2009). Light-independent cell death induced by accumulation of pheophorbide a in Arabidopsis thaliana. Plant Cell Physiol. 50: 719–729 [DOI] [PubMed] [Google Scholar]
- Horie Y., Ito H., Kusaba M., Tanaka R., Tanaka A. (2009). Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J. Biol. Chem. 284: 17449–17456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S. (2006). Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 57: 55–77 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S., Vicentini F., Matile P. (1995). Chlorophyll breakdown in senescent cotyledons of rape, Brassica napus L - Enzymatic cleavage of phaeophorbide a in vitro. New Phytol. 129: 237–246 [DOI] [PubMed] [Google Scholar]
- Islam M.R., Aikawa S., Midorikawa T., Kashino Y., Satoh K., Koike H. (2008). slr1923 of Synechocystis sp PCC6803 is essential for conversion of 3,8-divinyl(proto)chlorophyll(ide) to 3-monovinyl(proto)chlorophyll(ide). Plant Physiol. 148: 1068–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Ohtsuka T., Tanaka A. (1996). Conversion of chlorophyll b to chlorophyll a via 7-hydroxymethyl chlorophyll. J. Biol. Chem. 271: 1475–1479 [DOI] [PubMed] [Google Scholar]
- Ito H., Tanaka A. (1996). Determination of the activity of chlorophyll b to chlorophyll a conversion during greening of etiolated cucumber cotyledons by using pyrochlorophyllide b. Plant Physiol. Biochem. 34: 35–40 [Google Scholar]
- Ito H., Tanaka Y., Tsuji H., Tanaka A. (1993). Conversion of chlorophyll b to chlorophyll a by isolated cucumber etioplasts. Arch. Biochem. Biophys. 306: 148–151 [DOI] [PubMed] [Google Scholar]
- Ito H., Yokono M., Tanaka R., Tanaka A. (2008). Identification of a novel vinyl reductase gene essential for the biosynthesis of monovinyl chlorophyll in Synechocystis sp. PCC6803. J. Biol. Chem. 283: 9002–9011 [DOI] [PubMed] [Google Scholar]
- Johnson E.F., Mukhopadhyay B. (2005). A new type of sulfite reductase, a novel coenzyme F420-dependent enzyme, from the methanarchaeon Methanocaldococcus jannaschii. J. Biol. Chem. 280: 38776–38786 [DOI] [PubMed] [Google Scholar]
- Kulkarni G., Kridelbaugh D.M., Guss A.M., Metcalf W.W. (2009). Hydrogen is a preferred intermediate in the energy-conserving electron transport chain of Methanosarcina barkeri. Proc. Natl. Acad. Sci. USA 106: 15915–15920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M., Ito H., Morita R., Iida S., Sato Y., Fujimoto M., Kawasaki S., Tanaka R., Hirochika H., Nishimura M., Tanaka A. (2007). Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19: 1362–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Yang Y.-T., Liu H.-H., Yang G.-D., Zhang N.-H., Zheng C.C. (2004). NTZIP antisense plants show reduced chlorophyll levels. Plant Physiol. Biochem. 42: 321–327 [DOI] [PubMed] [Google Scholar]
- Meskauskiene R., Nater M., Goslings D., Kessler F., op den Camp R., Apel K. (2001). FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98: 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray D.L., Kohorn B.D. (1991). Chloroplasts of Arabidopsis thaliana homozygous for the ch-1 locus lack chlorophyll b, lack stable LHCPII and have stacked thylakoids. Plant Mol. Biol. 16: 71–79 [DOI] [PubMed] [Google Scholar]
- Nagane T., Tanaka A., Tanaka R. (2010). Involvement of AtNAP1 in the regulation of chlorophyll degradation in Arabidopsis thaliana. Planta 231: 939–949 [DOI] [PubMed] [Google Scholar]
- Nagata N., Tanaka R., Satoh S., Tanaka A. (2005). Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell 17: 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- op den Camp R.G., Przybyla D., Ochsenbein C., Laloi C., Kim C., Danon A., Wagner D., Hideg E., Gobel C., Feussner I., Nater M., Apel K. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer M., Pierce B.S., Crawford J.A., Ray K., Helm R.F., Sobrado P. (2010). Recombinant expression, purification, and characterization of ThmD, the oxidoreductase component of tetrahydrofuran monooxygenase. Arch. Biochem. Biophys. 496: 123–131 [DOI] [PubMed] [Google Scholar]
- Oster U., Tanaka R., Tanaka A., Rüdiger W. (2000). Cloning and functional expression of the gene encoding the key enzyme for chlorophyll b biosynthesis (CAO) from Arabidopsis thaliana. Plant J. 21: 305–310 [DOI] [PubMed] [Google Scholar]
- Papenbrock J., Pfündel E., Mock H.P., Grimm B. (2000). Decreased and increased expression of the subunit CHL I diminishes Mg chelatase activity and reduces chlorophyll synthesis in transgenic tobacco plants. Plant J. 22: 155–164 [DOI] [PubMed] [Google Scholar]
- Pruzinská A., Tanner G., Anders I., Roca M., Hörtensteiner S. (2003). Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 100: 15259–15264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdiger W. (2002). Biosynthesis of chlorophyll b and the chlorophyll cycle. Photosynth. Res. 74: 187–193 [DOI] [PubMed] [Google Scholar]
- Schelbert S., Aubry S., Burla B., Agne B., Kessler F., Krupinska K., Hörtensteiner S. (2009). Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21: 767–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheumann V., Ito H., Tanaka A., Schoch S., Rüdiger W. (1996). Substrate specificity of chlorophyll(ide) b reductase in etioplasts of barley (Hordeum vulgare L.). Eur. J. Biochem. 242: 163–170 [DOI] [PubMed] [Google Scholar]
- Scheumann V., Schoch S., Rüdiger W. (1998). Chlorophyll a formation in the chlorophyll b reductase reaction requires reduced ferredoxin. J. Biol. Chem. 273: 35102–35108 [DOI] [PubMed] [Google Scholar]
- Scheumann V.V., Schoch S., Rudiger W. (1999). Chlorophyll b reduction during senescence of barley seedlings. Planta 209: 364–370 [DOI] [PubMed] [Google Scholar]
- Shimada Y., Tanaka A., Tanaka Y., Takabe T., Takabe T., Tsuji H. (1990). Formation of chlorophyll-protein complexes during greening 1. Distribution of newly synthesized chlorophyll among apoproteins. Plant Cell Physiol. 31: 639–647 [Google Scholar]
- Stuehr D.J., Cho H.J., Kwon N.S., Weise M.F., Nathan C.F. (1991). Purification and characterization of the cytokine-induced macrophage nitric oxide synthase: An FAD- and FMN-containing flavoprotein. Proc. Natl. Acad. Sci. USA 88: 7773–7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugishima M., Kitamori Y., Noguchi M., Kohchi T., Fukuyama K. (2009). Crystal structure of red chlorophyll catabolite reductase: enlargement of the ferredoxin-dependent bilin reductase family. J. Mol. Biol. 389: 376–387 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tanaka A., Ito H., Tanaka R., Tanaka N.K., Yoshida K., Okada K. (1998). Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc. Natl. Acad. Sci. USA 95: 12719–12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., Tanaka R. (2006). Chlorophyll metabolism. Curr. Opin. Plant Biol. 9: 248–255 [DOI] [PubMed] [Google Scholar]
- Tanaka A., Yamamoto Y., Tsuji H. (1991). Formation of chlorophyll-protein complexes during greening 2. Redistribution of chlorophyll among apoproteins. Plant Cell Physiol. 32: 195–204 [Google Scholar]
- Tanaka R., Hirashima M., Satoh S., Tanaka A. (2003). The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: inhibition of the pheophorbide a oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis. Plant Cell Physiol. 44: 1266–1274 [DOI] [PubMed] [Google Scholar]
- Tanaka R., Koshino Y., Sawa S., Ishiguro S., Okada K., Tanaka A. (2001). Overexpression of chlorophyllide a oxygenase (CAO) enlarges the antenna size of photosystem II in Arabidopsis thaliana. Plant J. 26: 365–373 [DOI] [PubMed] [Google Scholar]
- Tanaka R., Oster U., Kruse E., Rüdiger W., Grimm B. (1999). Reduced activity of geranylgeranyl reductase leads to loss of chlorophyll and tocopherol and to partially geranylgeranylated chlorophyll in transgenic tobacco plants expressing antisense RNA for geranylgeranyl reductase. Plant Physiol. 120: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R., Tanaka A. (2005). Effects of chlorophyllide a oxygenase overexpression on light acclimation in Arabidopsis thaliana. Photosynth. Res. 85: 327–340 [DOI] [PubMed] [Google Scholar]
- Tanaka R., Tanaka A. (2007). Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58: 321–346 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T., Ohta H., Okawa K., Iwamatsu A., Shimada H., Masuda T., Takamiya K.i. (1999). Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: Finding of a lipase motif and the induction by methyl jasmonate. Proc. Natl. Acad. Sci. USA 96: 15362–15367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoni M.A., Dossena L., van den Heuvel R.H., Curti B. (2005). Structure-function studies on the complex iron-sulfur flavoprotein glutamate synthase: The key enzyme of ammonia assimilation. Photosynth. Res. 83: 219–238 [DOI] [PubMed] [Google Scholar]
- Yamasato A., Nagata N., Tanaka R., Tanaka A. (2005). The N-terminal domain of chlorophyllide a oxygenase confers protein instability in response to chlorophyll b accumulation in Arabidopsis. Plant Cell 17: 1585–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata M., Rodríguez F., Garrido J.L. (2000). Separation of chlorophylls and carotenoids from marine phytoplankton: a new HPLC method using a reversed phase C8 column and pyridine-containing mobile phases. Mar. Ecol. Prog. Ser. 195: 29–45 [Google Scholar]