Among the environmental signals affecting seed development, temperature is the most influential in the formation of seed dormancy in wheat. In this study, transcriptional profiling of the effects of temperature on seed dormancy formation identified MFT as a candidate gene for seed dormancy regulation.
Abstract
Seed dormancy is an adaptive mechanism and an important agronomic trait. Temperature during seed development strongly affects seed dormancy in wheat (Triticum aestivum) with lower temperatures producing higher levels of seed dormancy. To identify genes important for seed dormancy, we used a wheat microarray to analyze gene expression in embryos from mature seeds grown at lower and higher temperatures. We found that a wheat homolog of MOTHER OF FT AND TFL1 (MFT) was upregulated after physiological maturity in dormant seeds grown at the lower temperature. In situ hybridization analysis indicated that MFT was exclusively expressed in the scutellum and coleorhiza. Mapping analysis showed that MFT on chromosome 3A (MFT-3A) colocalized with the seed dormancy quantitative trait locus (QTL) QPhs.ocs-3A.1. MFT-3A expression levels in a dormant cultivar used for the detection of the QTL were higher after physiological maturity; this increased expression correlated with a single nucleotide polymorphism in the promoter region. In a complementation analysis, high levels of MFT expression were correlated with a low germination index in T1 seeds. Furthermore, precocious germination of isolated immature embryos was suppressed by transient introduction of MFT driven by the maize (Zea mays) ubiquitin promoter. Taken together, these results suggest that MFT plays an important role in the regulation of germination in wheat.
INTRODUCTION
Seed dormancy controls the timing of germination and provides an important adaptive strategy for survival of long periods of unsuitable environmental conditions. For agronomic applications, the regulation of seed dormancy is important to prevent preharvest sprouting and to obtain uniform germination (Bewley and Black, 1994). Particularly in wheat (Triticum aestivum), preharvest sprouting is a serious problem in the regions of the world where the rainy season tends to overlap with the harvest season (Gale, 1989; Gerjets et al., 2010). Thus, wheat breeding programs have had to meet contradictory demands for a high level of seed dormancy at harvest (to prevent preharvest sprouting) and no seed dormancy at sowing (to obtain uniform germination). To meet these demands, differences in the mechanisms controlling preharvest sprouting and subsequent germination of harvested seeds must be identified. Despite the identification of a number of genes involved in this process, our knowledge is still too limited to control seed dormancy (Kucera et al., 2005; Finch-Savage and Leubner-Metzger, 2006; Bentsink and Koornneef, 2008; Finkelstein et al., 2008; Holdsworth et al., 2008a, 2008b).
Seed dormancy is a complex trait influenced by environmental factors. In wheat, temperature is a key environmental factor that affects seed dormancy mainly at two developmental stages, during seed development and at germination (Black et al., 1987). Low temperatures during seed development generate a higher level of seed dormancy (i.e., a higher percentage of seeds that do not germinate) (Buraas and Skinnes, 1985; Reddy et al., 1985; Black et al., 1987). By contrast, low temperatures at germination stimulate seed germination (Nyachiro et al., 2002). To explain the effect of temperature on seed dormancy, Hilhorst (2007) proposed that membrane fluidity altered by temperature signals a change in dormancy. However, the molecular mechanism underlying the response to temperature, so far, remains to be elucidated. Because the same cultivar can show different levels of seed dormancy in response to different temperatures during seed development, this effect clearly does not result from genomic sequence differences. Instead, temperature-dependent transcriptional regulation may be involved in this response. Thus, by analyzing this phenomenon using a transcriptomic approach, we may find genes that regulate the level of seed dormancy and its response to temperature during seed development.
Several transcriptomic analyses on seed development and germination have been reported in wheat and barley (Hordeum vulgare; Wilson et al., 2005; Sreenivasulu et al., 2008; Wan et al., 2008). Numerous transcriptome analyses have examined expression profiles during seed development and germination in Arabidopsis thaliana. These analyses confirmed that abscisic acid (ABA) and gibberellin (GA) play important roles in the regulation of seed dormancy; however, the transcriptional effects of temperature during seed development on the formation of seed dormancy remain unclear. Identifying the genes that regulate seed dormancy will substantially advance our understanding of this process
Genetic approaches have identified several genes that affect seed dormancy. For example, positional cloning of seed dormancy quantitative trait loci (QTL) has successfully found genes that affect seed dormancy, including DELAY OF GERMINATION1 (DOG1) in Arabidopsis (Bentsink et al., 2006) and Seed dormancy4 (Sdr4) in rice (Oryza sativa; Sugimoto et al., 2010). Indeed, ectopic overexpression of wheat and barley DOG1 homologs in transgenic Arabidopsis increased seed dormancy (Ashikawa et al., 2010). DOG1 and Sdr4 encode proteins of unknown function, with no annotated domains and no similarity to proteins of known function. Both genes also show seed-specific expression, with higher expression levels in dormant seeds. Further analysis showed that DOG1 is also involved in sugar/ABA signaling (Teng et al., 2008), seed germination per se, and its regulation by ABA (Graeber et al., 2010).
Previous genetic studies on seed dormancy in wheat have detected a number of QTL (Flintham et al., 2002). Among them, two QTL that appear to have stable and large effects, QPhs.ocs-3A.1 and Phs1, have been found on chromosomes 3A and 4A (Mares et al., 2005; Mori et al., 2005; Torada et al., 2008). However, so far, the DNA sequences responsible for the wheat seed dormancy QTL and their natural variation have not been determined.
To identify genes involved in the regulation of seed dormancy, we screened for differentially expressed genes in embryos grown at different temperatures and identified a wheat MOTHER OF FT AND TFL1 (MFT) homolog that is transcriptionally upregulated after physiological maturity at lower temperature during seed development. MFT-like genes show seed-specific expression (Chardon and Damerval, 2005) and belong to the plant phosphatidyl ethanolamine binding protein (PEBP) family, which is divided into three subfamilies, FLOWERING LOCUS T (FT)-like, TERMINAL FLOWER1 (TFL1)-like, and MFT-like (Chardon and Damerval, 2005). In Arabidopsis, FT and TFL1 are thought to be molecular switches for reproductive development (Turck et al., 2008), and MFT is phylogenetically ancestral to them (Hedman et al., 2009). MFT has recently been shown to be a negative regulator of ABA sensitivity during seed germination in Arabidopsis (Xi et al., 2010). Moreover, in tomato (Solanum lycopersicum), modulation of the balance between the tomato FT homolog SINGLE FLOWER TRUSS and the TFL1 homolog SELF-PRUNING affects a variety of growth processes, including flowering response, reiterative growth, termination cycles, leaf maturation, and stem growth (Shalit et al., 2009). Thus, genes in the FT and TFL1 families are likely to act as general growth regulators, rather than just as the floral initiation regulator or florigen.
In this study, we provide evidence that MFT is one of the key factors regulating seed germination and temperature effects on the formation of seed dormancy during seed development in wheat. In addition, our data suggest that MFT is the causal gene for the wheat seed dormancy QTL QPhs.ocs-3A.1 and that the single nucleotide polymorphism (SNP) identified in the MFT promoter may be an important target for marker-assisted selection to improve preharvest sprouting resistance in wheat.
RESULTS
Temperature-Dependent Formation of Seed Dormancy
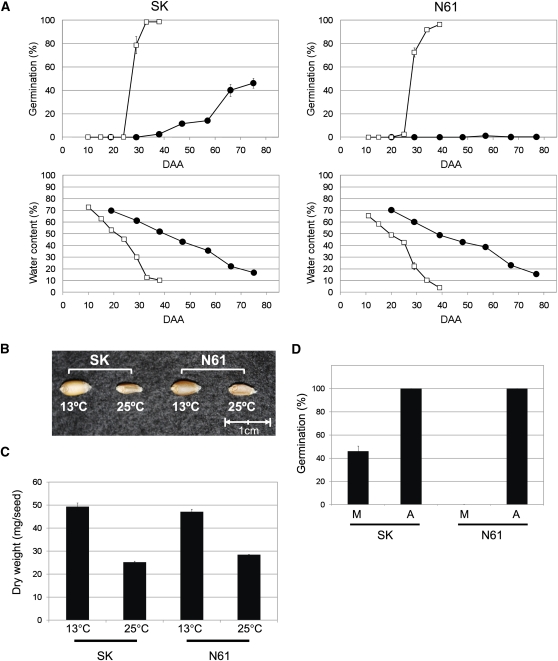
To assess temperature effects on seed dormancy in wheat, two cultivars, Shiroganekomugi (SK) and Norin61 (N61) were grown at 13 and 25°C after anthesis. Among Japanese wheat cultivars, SK is susceptible to preharvest sprouting and has a lower level of seed dormancy, and N61 is resistant to preharvest sprouting and has a high level of seed dormancy. There are no differences between the two cultivars in seed water content reduction during development (Figure 1A), indicating their seed maturation progresses similarly. Basically, the seed water content gradually decreased to around 40% (25 d after anthesis [DAA25] for 25°C and DAA55 for 13°C); this stage is considered the physiological maturity stage, when developing seeds reach their maximum dry weight. After that, the water content rapidly decreased to <20%, when maturation of the seeds was complete. Changes in the percentage of germination of seeds grown at 25°C were almost the same between the two cultivars, and the percentage of germination rapidly increased to around 100% after physiological maturity at DAA25 (Figure 1A). However, the changes in the percentage of germination of seeds grown at 13°C were different between the two cultivars. For SK grown at 13°C, germination percentages increased gradually after DAA30, and after physiological maturity, its germination percentages increased more rapidly to ~50%. However, the final germination percentage was half that of SK grown at 25°C. For N61 grown at 13°C, the seeds became extremely dormant, and the germination percentage was always around 0%. The greater response to temperature in N61 is consistent with the dormancy traits of the two cultivars.
Figure 1.
The Developmental Status of Seeds for Microarray Analysis.
(A) Time course of germination percentages and water content during seed development. Wheat cultivars SK and N61 were grown at 13 or 25°C after anthesis. Results from triplicate independent biological samples (n = 3) are shown as lines with closed circles (grown at 13°C) and open squares (grown at 25°C). Error bars represent sd.
(B) Mature seeds grown at 13 or 25°C. N61 seeds are mature at DAA76 grown at 13°C or at DAA34 grown at 25°C, and SK seeds are mature at DAA80 grown at 13°C or at DAA 38 grown at 25°C.
(C) Average dry weight of mature seeds grown at 13 or 25°C. Results from triplicate independent biological samples (n = 3) are shown; error bars represent sd.
(D) Germination percentages of after-ripened mature seeds grown at 13°C. M, the mature seeds grown at 13°C during seed development. A, the after-ripened seeds: the mature seeds grown at 13°C stored for 6 months at 4°C. After-ripened 30 seeds were incubated for 5 d at 15°C. Results from triplicate independent biological samples (n = 3) are shown, and error bars represent sd. The germination percentages of the after-ripened seeds in SK and N61 were 100% in the triplicate repeats.
The maturation time of seeds grown at 25°C is ~40 d. This is half that of the seeds grown at 13°C (~80 d). This difference affected the size of the mature seeds (Figure 1B). The average dry weight of seeds grown at 25°C is approximately half that of seeds grown at 13°C (Figure 1C). This seems to be because they do not have enough time to synthesize storage materials (mainly starch) to fill in the endosperm.
Although mature N61 seeds grown at 13°C had a very low percentage of germination, the high level of seed dormancy was broken by after-ripening. After 6 months storage at 4°C, the N61 seeds grown at 13°C germinated at 100% (Figure 1D). The SK seeds also showed complete germination under these conditions (Figure 1D).
Identification of a Temperature-Responsive MFT Homolog
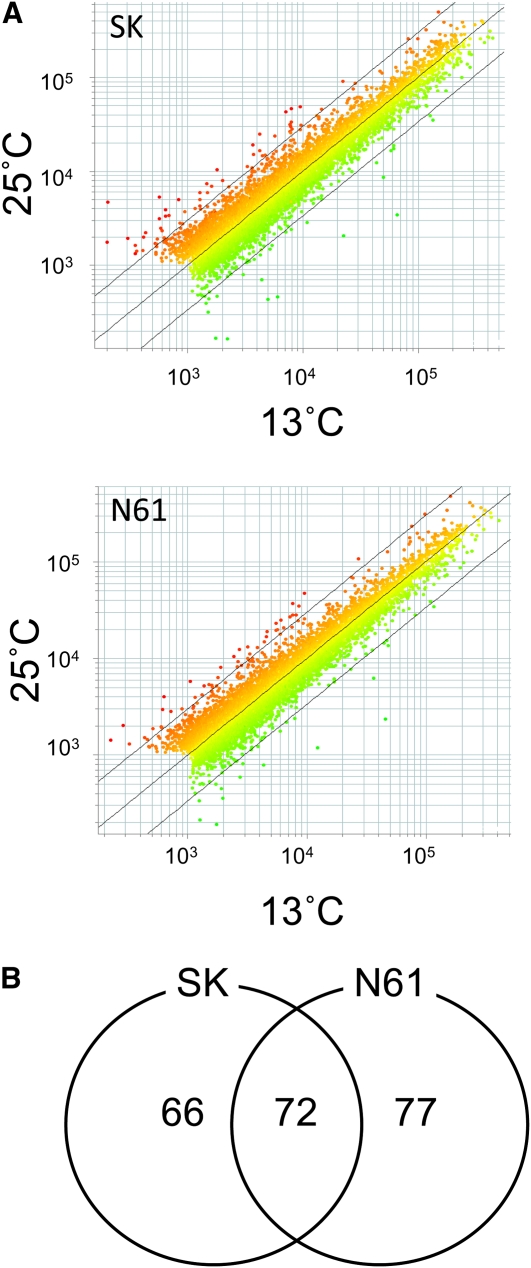
To identify candidate genes that encode regulators of seed dormancy and its temperature responsiveness, we next used transcription profiling, hypothesizing that such genes should show differential, seed-specific expression in different temperature conditions. Because the distinguishing difference in the germination percentages was between mature seeds grown at 13 or 25°C, we compared gene expression between embryos isolated from seeds grown under these conditions. We performed microarray analyses of ~38,000 wheat probes using a 60-mer oligo wheat DNA microarray from Agilent Technologies. Our microarray analysis showed that gene expression patterns in embryos grown at 13 or 25°C were highly similar in both SK and N61 (Figure 2A). To examine gene expression, we first selected the probes that showed detectable expression in at least one temperature condition and reproducible expression in the three biological replicates. There were 10,762 and 10,453 genes in SK and N61, respectively, that showed a signal intensity of more than the cutoff threshold of 1000 with either Cy3 (13°C) or Cy5 (25°C) and no flags (marks of microarray spots that have bad quality evaluated by Feature Extraction software) in all three biological replicates. Within this set, only 138 and 149 genes in SK and N61, respectively, showed greater than a twofold change in levels with a q-value of <0.05, which indicates statistical significance (Table 1; see Supplemental Data Sets 1 and 2 online). Thus, ~1.4% of genes with detectable expression showed more than a twofold change in expression between the two temperature conditions. Among them, 72 genes were detected in both SK and N61 (Figure 2B; see colored rows in Supplemental Data Sets 1 and 2 online). In SK, only six genes showed more than a 10-fold expression change, and only two genes did in N61 (Table 1, listed in Table 2). Among these, two genes were found in both SK and N61. By BLAST analysis (Altschul et al., 1990) using the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/), one shows high similarity (e-value: 2e-115) to the rice gene Os06g0708600, which is annotated as a zinc finger, C2H2-like domain–containing protein (RAP-DB, http://rapdb.dna.affrc.go.jp/), and the other shows high similarity (e-value: 2e-114) to Os01g0111600, which is annotated as similar to MFT. Because the previously identified seed dormancy regulators DOG1 and Sdr4 show seed-specific expression, we next evaluated tissue-specific expression of the rice homologs of each wheat gene using the RiceXPro database (http://ricexpro.dna.affrc.go.jp/) (Sato et al. 2011), assuming that expression would likely be similar in rice and wheat. Of the two candidate genes, only the rice gene corresponding to the wheat homolog of MFT showed seed-specific expression. This is supported by the fact that MFT-like genes were previously reported to show seed-specific expression (Chardon and Damerval, 2005; Danilevskaya et al., 2008; Xi et al., 2010). Thus, we selected MFT for further investigation in wheat.
Figure 2.
The Results of Microarray Analysis.
(A) Scatterplots illustrating the distributions of the microarray signal intensities. Wheat cultivars SK and N61 grown at 13 and 25°C after anthesis. Average signal intensities of three biological replicates are shown. There are 10,762 genes in SK and 10,453 genes in N61 that showed signal intensity of >1000 in either the 13 or 25°C samples and no flags (marks of microarray spots that have bad quality evaluated by Feature Extraction software). The diagonal black line in the middle of the points represents 1:1 signal ratio (no change), and the flanking lines on either side represent a threefold change.
(B) A Venn diagram of differentially expressed genes in SK and N61. The numbers of genes with the control or raw signal intensity >1000, fold change >2, and q-value <0.05 (average of three biological replicates) are shown.
Table 1.
Number of Differentially Expressed Genes
| No. of Genes |
||
| Fold Change | SK | N61 |
| >2 | 138 | 149 |
| >3 | 73 | 62 |
| >4 | 30 | 26 |
| >5 | 23 | 14 |
| >10 | 6 | 2 |
Number of differentially expressed genes identified by our microarray analysis with signal intensities at 13°C or 25°C of >1000 and q-values <0.05 (three independent biological replicates) is shown. N61 and SK are the wheat cultivars.
Table 2.
Summary of Microarray Data of the Genes Whose Expression Levels Were Strongly Affected by Temperature during Seed Development
| Signal Intensity |
||||||||
| Cultivar | Probe Name | (Cy3, 13°C) | (Cy5, 25°C) | Fold Change | FDR (q-Value) | Rice Homolog Locus ID | Annotation | Accession No. |
| SK | wheat0130Contig14075 | 201 | 4708 | 24.1 | 0.00310 | Os06g0726200 | Seed chitinase-c protein | AY973229 |
| wheat0130Contig12202 | 65781 | 3453 | 19.3 | 0.00310 | Os06g0708600 | Zinc finger protein | AK333024 | |
| MUGEST2003_23lib_Contig17196 | 6010 | 461 | 13.4 | 0.00310 | Os06g0254300 | EF-hand calcium binding protein | HQ020505 | |
| MUG002E09R990520 | 2186 | 166 | 13.0 | 0.00310 | Cold-regulated protein | AJ132439 | ||
| MUGEST2003_23lib_Contig13877 | 4964 | 433 | 11.4 | 0.00310 | Os02g0184700 | Cytochrome P450 family protein | AK356149 | |
| wheat0130Contig787 | 22469 | 2046 | 11.1 | 0.00310 | Os01g0111600 | Mother of FT and TFL1 protein | AK330655 | |
| N61 | wheat0130Contig12202 | 45608 | 2354 | 19.5 | 0.00424 | Os06g0708600 | Zinc finger protein | AK333024 |
| wheat0130Contig787 | 12236 | 1180 | 10.1 | 0.00424 | Os01g0111600 | Mother of FT and TFL1 protein | AK330655 | |
Results of differential expression between mature embryos grown at 13 and 25°C are shown with fold changes and q-values. N61 and SK are the wheat cultivars used for this microarray analysis. Samples from 13°C were labeled with Cy3, and samples from 25°C were labeled with Cy5. Probes with more than 10-fold changes in N61 and SK are listed. Accession numbers of wheat mRNA (except for AK356149, which is barley) identified by BLAST search using the probe nucleotide sequences with e-value 0 are listed. Data sets for all the differential expressed genes with control or raw signals >1000, fold changes >2, and q-values <0.05 can be found in Supplemental Data Sets 1 and 2 online. A complete set of microarray data was deposited to the GEO repository under accession number GSE22786. FDR is false discovery rate.
In Arabidopsis, MFT is directly regulated by ABA-INSENSITIVE3 (ABI3) and ABI5, and upregulated by DELLA proteins in the GA signaling pathway (Xi et al., 2010). Thus, we looked into the expression levels of the wheat genes corresponding to these loci in our microarray data (see Supplemental Table 1 online). The wheat ABI5 homolog ABA response element binding factor showed a small difference in its expression level between 13 and 25°C in SK and N61 with average fold changes of 1.6 in SK and 1.7 in N61 with P < 0.05. However, the wheat DELLA gene Reduced Height1 and the wheat ABI3 homolog Viviparous1 did not show significant differences in their expression levels between at 13 and 25°C.
Analysis of MFT Expression by Quantitative RT-PCR
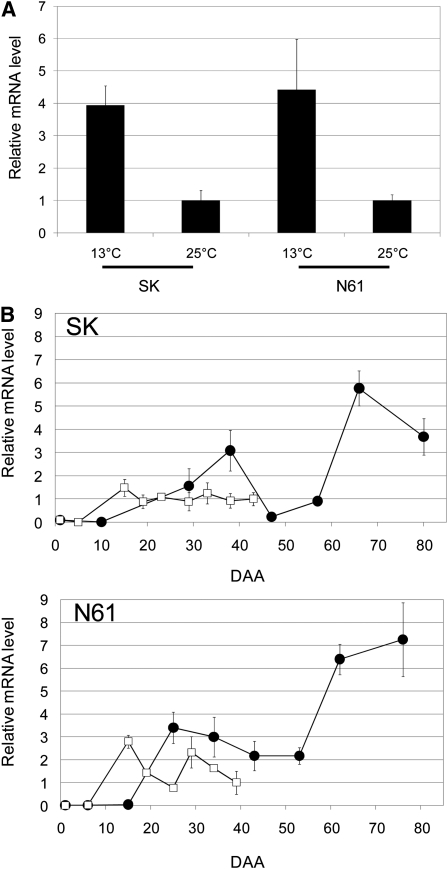
To confirm the differential expression of MFT seen in the microarray analysis, we analyzed MFT expression during seed development by quantitative RT-PCR (qRT-PCR). In mature seeds of both SK and N61, the expression level of MFT at 13°C was ~4 times higher than that at 25°C (Figure 3A). These results are consistent with the results of the microarray analysis.
Figure 3.
The Temperature Effect on the Expression of MFT.
(A) MFT expression levels in mature embryos. Wheat cultivars SK and N61 grown at 13 and 25°C after anthesis. qRT-PCR was performed. Mature seeds grown at 13°C were isolated at DAA80 in SK and at DAA76 in N61. Mature seeds grown at 25°C were isolated at DAA43 in SK and at DAA39 in N61. The relative mRNA levels at 25°C were set at a standard value of 1. Transcript levels were normalized using the actin gene as an internal control. Results from quintuple independent biological samples (n = 5) are shown, and error bars represent sd.
(B) Time course of MFT expression levels during seed development in SK and N61. qRT-PCR was performed with triplicate technical samples (n = 3), and equivalent results were obtained using duplicate independent biological samples. The error bars represent sd. Transcript levels were normalized using the actin gene as an internal control. In SK, the relative mRNA level at 25°C at DAA43 was set at a standard value of 1. In N61, the relative mRNA level at 25°C at DAA39 was set at a standard value of 1. Closed circles, grown at 13°C; open squares, grown at 25°C.
Next, we examined changes in MFT mRNA levels during seed development. In all conditions, the expression of MFT started at early stages of seed development and seemed to have two peaks, one during the immature embryo stage and another after the physiological maturity stage (Figure 3B). The marked difference in MFT expression levels between 13 and 25°C was observed after the physiological maturity stage. The wheat microarray analysis also showed the same changes in expression level of MFT in N61 during the seed maturation stage (see Supplemental Figure 1 online).
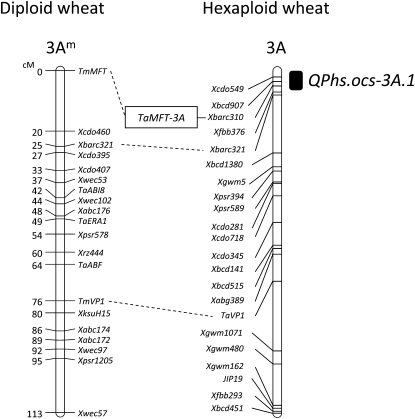
Mapping Diploid and Hexaploid Wheat MFT Homologs
We analyzed the location of the MFT homolog (Tm-MFT) of diploid wheat (Triticum monococcum) on a genetic map using recombinant inbred lines (RILs) derived from a cross between T. monococcum and Triticum boeoticum. Tm-MFT was mapped to the distal end of the short arm of chromosome 3Am, 25 centimorgans distal from Xbarc321 (Figure 4). In hexaploid wheat, a seed dormancy QTL, QPhs.ocs-3A.1, has been mapped to the terminal region of the short arm of chromosome 3A using RILs derived from a cross between the highly dormant wheat cultivar Zenkoujikomugi (Zen) and the less dormant cultivar Chinese Spring (CS). The simple sequence repeat (SSR) marker Xbarc310 is located at the logarithm of odds peak of QPhs.ocs-3A.1 (Mori et al., 2005). Thus, we analyzed the genotypes of Xbarc310 and Ta-MFT-3A using 2852 F2 plant lines derived from a cross between CS and CS(Zen3A), which is the Zen chromosome 3A substitution line. This analysis showed that Ta-MFT cosegregates with Xbarc310, indicating that Ta-MFT resides in the seed dormancy QTL QPhs.ocs-3A.1 (Figure 4).
Figure 4.
Genetic Map of Diploid and Hexaploid Wheat MFT Homologs.
In the diploid wheat genetic map, the marker order is shown on the right, with genetic distances (centimorgan [cM] scale) on the left. Tm-MFT: diploid wheat MFT homolog (see Supplemental Figure 4 online). In the hexaploid wheat genetic map, the marker order is shown on the left, with the location of seed dormancy QTL QPhs.ocs-3A.1 indicated on the right by a black rectangle. The construction of the hexaploid wheat genetic map using 125 RILs derived from a cross between Zen and CS was described previously by Mori et al. (2005). The locations of homologous genes and markers between diploid and hexaploid wheat are connected with dashed lines.
Expression of Ta-MFT in CS and CS(Zen3A) and Localization of Ta-MFT Expression during Seed Development
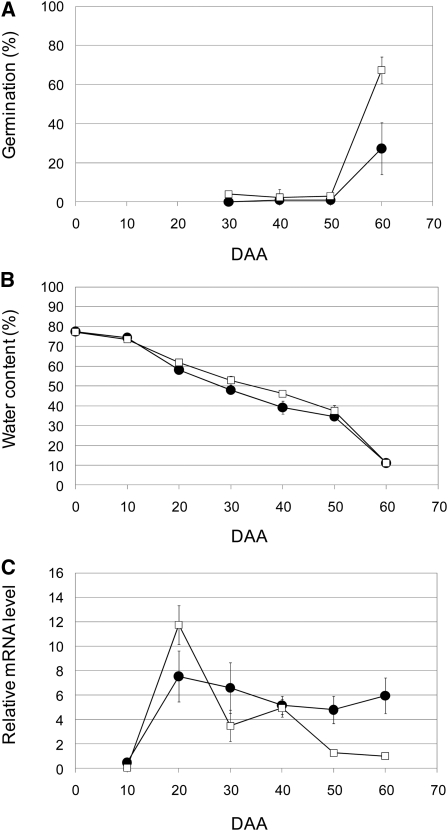
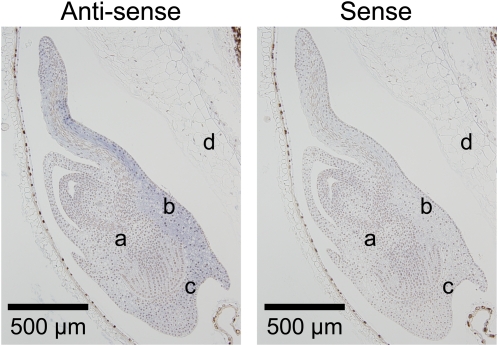
To further examine MFT function, we next isolated MFT cDNAs. In CS, we isolated and sequenced 12 cDNAs of MFT (see Supplemental Figure 2 online). They all have the same nucleotide sequence, and using the deduced amino acid sequence, MFT in CS was phylogenetically grouped into the MFT-like family (see Supplemental Figure 3 online). This cDNA sequence specifically matched the coding sequence of the partial genomic sequence assigned to chromosome 3A (see Supplemental Figures 4A and 4B online). This means it is likely that MFT-3A is the main MFT expressed in CS. Although QPhs.ocs-3A.1 has been found using RILs derived from a cross between Zen and CS, we used CS and CS(Zen3A) to analyze the association of MFT expression levels with seed dormancy levels. This approach minimizes the effects of differences in genetic background. CS and CS(Zen3A) showed ~0% germination at DAA30, 40, and 50 (Figure 5A). Under this growth condition, DAA50 was estimated to be the physiologically mature stage, based on changes in water content (Figure 5B). When seeds matured completely at DAA60, the germination percentage of CS was ~60%; by contrast, the germination percentage of CS(Zen3A) was ~30% (Figure 5A). Under these conditions, we analyzed the level of MFT expression during seed development by qRT-PCR. The level of MFT expression in both CS and CS(Zen3A) peaked at an early stage of seed development at DAA20. Subsequently, the level of MFT expression in CS decreased rapidly around the physiological maturity stage (i.e., DAA50; Figure 5C). By contrast, the level of MFT expression in CS(Zen3A) decreased gradually after its first peak at DAA20 but was significantly more highly expressed than in CS even after DAA50 (Figure 5C). Eventually, the level of MFT expression in CS(Zen3A) was ~6 times higher than CS at DAA60, the completely mature stage. Therefore, these results are consistent with an association of the expression level of MFT after physiological maturity with the level of seed dormancy in CS and CS(Zen3A). Our in situ hybridization analysis revealed that MFT was specifically expressed in the scutellum and coleorhizae (Figure 6). Our subcellular localization analysis showed TaMFT:DsRed was both nuclear and cytoplasmic in onion leaf epidermal cells (see Supplemental Figure 5 online) compared with wheat Flowering Locus T(TaFT):DsRed as a control.
Figure 5.
Time Course of MFT Expression during Seed Development in CS and CS(Zen3A).
Results from triplicate independent biological samples (n = 3) are shown as lines with closed circles for CS(Zen3A) and open squares for CS. Error bars represent sd.
(A) Time course of germination percentages.
(B) Time course of seed water content.
(C) Time course of the expression levels of MFT. Total RNA was isolated from whole seeds (DAA10) or embryos (DAA20 to 60) at the indicated DAA. Transcript levels were normalized using the actin gene as an internal control. The relative mRNA level of CS at DAA60 was set at a standard value of 1.
Figure 6.
Localization of MFT Transcripts in Immature Embryos by in Situ Hybridization.
Longitudinal sections of the embryos of CS at 21 DAA were probed with antisense or sense RNA. Positive hybridization signals are visible as a blue color in the scutellum and coleorhiza tissues. a, embryo; b, scutellum; c, coleorhiza; d, endosperm.
Comparison of the MFT-3A Genomic Sequences of CS and Zen
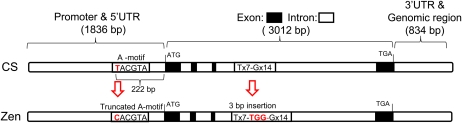
The expression level of MFT after physiological maturity was different between CS and CS(Zen3A), indicating that these lines might harbor nucleotide sequence polymorphisms in their MFT promoter regions. Thus, we examined the genomic sequences, including promoter regions of MFT-3A in CS and Zen. First, three CS wheat genomic BAC clones were isolated by PCR screening using MFT-3A–specific primers. The clone WCS1322O14, containing the longest insertion of ~184 kb, was sequenced and analyzed, and MFT-3A was identified in one of the sequenced contigs of this BAC clone. Using this information, the genomic MFT-3A sequences of CS and Zen were amplified by PCR using primers CS3A-5-R1 and CS3A-6-R2 (see Supplemental Table 2 online) and sequenced. A comparison of the genomic sequences of MFT-3A in CS and Zen (Figure 7) revealed polymorphisms at two sites. One SNP was located 222 bp upstream from the initiation codon of MFT-3A (Figure 7), and a SSR polymorphism was located in the 3rd intron (Figure 7); the SSR polymorphism was previously used for genotyping MFT-3A (see Supplemental Figure 6 online). The SNP in the promoter region had a T in CS and a C in Zen (Figure 7); in the dormant Zen cultivar, this SNP truncated the A-box motif, a bZIP transcription factor binding site (Izawa et al., 1993), TACGTA, by conversion of the 5′ nucleotide from T to C. Thus, a bZIP transcription factor may negatively regulate MFT expression in nondormant CS lines. A cleaved amplified polymorphic sequence (CAPS) marker was developed for this SNP (see Supplemental Figure 7A online). Using this CAPS marker, we genotyped the 136 RILs derived from a cross between CS and Zen, which includes the 125 RILs used for detection of QPhs.ocs-3A.1 (Mori et al., 2005). This analysis showed that the MFT CAPS marker always cosegregated with Xbarc310 (see Supplemental Figure 7B online), suggesting that both markers are tightly linked to the locus influencing seed dormancy. This result supports the mapping result (Figure 4; see Supplemental Figure 6 online) using the SSR polymorphism marker. Thus, we identified a candidate functional SNP in the promoter region of MFT-3A that may increase MFT expression and likely contributes to increased seed dormancy; we also developed a CAPS marker for this SNP.
Figure 7.
Comparison of the CS and Zen MFT-3A Genomic Sequences.
The red arrows indicate two loci in the genomic sequence of MFT-3A that differ between Zen and CS. The red bold nucleotides show polymorphism. ATG, initiation codon; TGA, stop codon; CS, less dormant wheat cultivar Chinese Spring; Zen, dormant wheat cultivar Zenkoujikomugi; Tx7, seven repeats of T; Gx14, 14 repeats of G.
Correlation between MFT Expression and Seed Dormancy in T1 Seeds
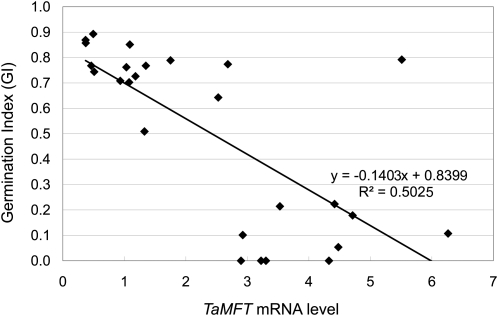
To determine whether the Zen MFT genomic sequence is enough to increase MFT expression and seed dormancy levels in CS, the entire 5685-bp genomic sequence of MFT-3A of Zen (Figure 7) was stably introduced into CS. By PCR screening for the bar selectable marker gene (which confers resistance to the herbicide bialaphos), we obtained 26 independent wheat transgenic T0 lines that likely have the MFT-3A genomic fragment of Zen and also had enough T1 seeds per spike for qRT-PCR expression and germination index (GI) analyses. Using these 26 lines, we analyzed MFT expression in mature embryos and GI in T1 seeds in the same spikes harvested ~1 month after seed maturation. We found a correlation between the expression level and GI in these lines (Figure 8). The value of the decision coefficient R2 between the analyzed lines was ~0.5. When the expression level is normalized to the expression level of MFT of CS at DAA 60 (Figure 5C), which was set as 1, the degree of the minimum and maximum relative mRNA levels in the T1 seeds was ~1 and 6 (Figure 8). This is consistent with the observed sixfold increase in MFT-3A mRNA levels in CS(Zen3A) at the mature stage DAA60 (Figure 5C). These results suggested that the introduced MFT-3A genomic fragment of Zen is sufficient to increase levels of MFT expression in mature embryos and thus increase seed dormancy. Environmental and position effects from transgene insertion may contribute to the fluctuations of MFT expression levels in T1 seeds. Even though these factors may alter MFT expression levels and affect the GI, the detection of the correlation between MFT levels and GI indicates the possibility that a strong association exists between them.
Figure 8.
Correlation between MFT Expression Level and Germination Index in T1 Seeds.
MFT mRNA levels and germination index were measured using T1 seeds from the same spikes of 26 independent T0 plants (n = 26) that were produced by transforming CS with the entire genomic sequence of Zen MFT3A. The MFT expression level was measured using qRT-PCR. Total RNA was extracted from embryos of 30 seeds. The expression level was normalized to the level of MFT of CS at DAA60 (Figure 5C), which was set as 1. The germination index was measured using 16 or 24 seeds incubated at 20°C for 7 d.
Transient Expression of MFT in Immature Embryos Induces Seed Dormancy
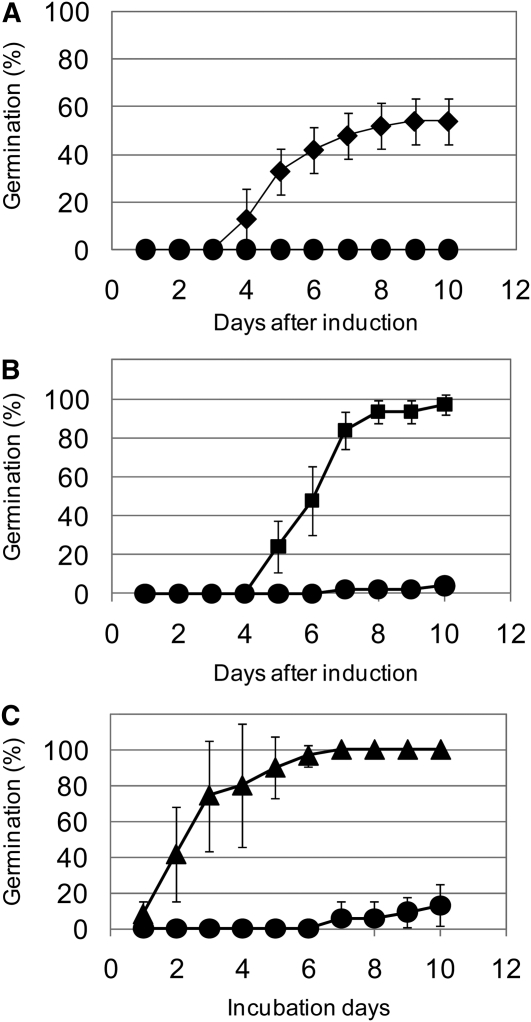
MFT is highly expressed in early seed development and during seed maturation (Figures 3 and 5C). In addition, MFT is expressed mainly in the scutellum and coleorhiza of immature embryos (Figure 6). To examine the function of MFT at early seed developmental stages, we transiently overexpressed MFT in immature embryos and examined their germination. MFT driven by the maize (Zea mays) ubiquitin promoter (Ubi:TaMFT) was directly bombarded into immature embryos isolated from CS seeds at approximately DAA10. In general, cultured immature wheat embryos germinate precociously (Williamson et al., 1985). However, we found that the immature embryos transformed with Ubi:TaMFT did not germinate even 10 d after transformation (Figure 9A; see Supplemental Figure 8 online). We also examined immature embryos transformed with the control construct β-glucuronidase (GUS) driven by the maize ubiquitin promoter (Ubi:GUS) and embryos transformed with Ubi:TaMFT(Stop), in which the 86th codon in MFT, which encodes a Trp, was changed to a stop codon; embryos transformed with these control constructs germinated well (Figures 9A and 9B; see Supplemental Figure 8 online). GA is known to promote germination and is used for breaking seed dormancy in wheat (Nakamura, 1962); thus, to test whether the embryos transformed with Ubi:TaMFT could germinate, we transferred the nongerminated embryos onto medium containing 1 μM GA and incubated them for 10 d. The immature embryos treated with exogenous GA eventually germinated (see Supplemental Figure 8 online). This indicates that the nongerminating immature embryos retained their ability to germinate but did not have enough endogenous active GA to overcome the suppression of germination caused by Ubi:TaMFT. Similar results were obtained using another relatively nondormant cultivar, Bobwhite (BW) (Figures 9B and 9C). The time courses of germination percentages after the introduction of Ubi:TaMFT, Ubi:GUS, or Ubi:TaMFT(Stop) into immature embryos of CS or BW are shown in Figures 9A to 9C.
Figure 9.
Time Course of the Germination Percentage in the Transient MFT Expression Assay.
(A) Time course of the germination percentage after transformation with Ubi:TaMFT (closed circles) or Ubi:GUS (closed diamonds). Wheat cultivar CS was used. Results from triplicate independent biological samples (n = 3) are shown, and error bars represent sd. Equivalent results were obtained in triplicate repeats of triplicate independent biological samples.
(B) Time course of germination percentage after transformation with Ubi:TaMFT (closed circles) or Ubi:TaMFT(Stop) (closed squares). Wheat cultivar Bobwhite (BW) was used. Results from triplicate independent biological samples (n = 3) are shown, and error bars represent sd. Equivalent results were obtained in triplicate repeats of triplicate independent biological samples.
(C) Time course of germination percentage after transferring the ungerminated embryos with Ubi:TaMFT onto medium with or without GA. The isolated immature embryos cultured for 10 d after induction of Ubi:TaMFT did not germinate. Subsequently, they were transferred and cultured for 10 d on medium with (closed triangles) or without (closed circles) 1 μM GA. Results from triplicate independent biological samples (n = 3) are shown, and error bars represent sd. Equivalent results were obtained in triplicate repeats of triplicate independent biological samples.
The growth and development of the CS immature embryos transformed with Ubi:GUS or Ubi:TaMFT is shown in Figure 10. The Ubi:TaMFT transformed immature embryos became larger and yellowish, but the emergence of the radicle or plumule from the transformed immature embryos was not observed even after 10 d of incubation. This indicated that the immature embryos continued the maturation process on the medium, but their germination was arrested. By contrast, in Ubi:GUS transformed embryos, the radicle emerged as the primary seminal root after ~7 d of incubation, primordia for a pair of first seminal roots developed on both sides of the embryos after 4 d, a pair of first seminal roots emerged at 8 d of incubation, and coleoptiles extended from the embryos from at least 7 d of incubation.
Figure 10.
Time Course of Isolated CS Immature Embryo Development after Transformation with Ubi:TaMFT or Ubi:GUS.
h, hours after transformation; d, days after transformation. Bar = 2 mm.
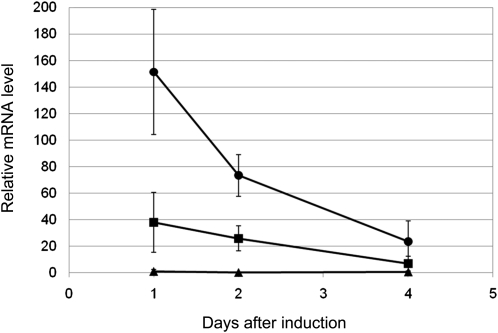
Using immature embryos from the BW cultivar, we checked the time course of MFT expression in the cultured immature embryos during the first 4 d (Figure 11). At 1 d after introduction, the introduced Ubi:TaMFT gave 150 times higher levels of MFT expression than that of MFT of CS at DAA60 (Figure 5C), which was used as the standard. The expression level gradually decreased but still maintained levels 20 times higher at 4 d after introduction. The introduced Ubi:TaMFT(Stop) induced 20 times higher levels of MFT expression than the standard at 1 d after introduction, and the expression level gradually decreased to ~7 times higher at 4 d after introduction. The levels of Ubi:TaMFT(Stop) were much lower than Ubi:TaMFT, possibly due to nonsense-mediated RNA decay or because MFT may act to increase its own expression.
Figure 11.
Time Course of MFT Expression after Transformation with Ubi:TaMFT or Ubi:TaMFT(Stop).
The expression level of MFT was analyzed by qRT-PCR using isolated immature embryos of Bobwhite (BW) cultured for 1, 2, and 4 d after transformation with Ubi:TaMFT or Ubi:TaMFT(Stop). Closed circles, Ubi:TaMFT; closed squares, Ubi:TaMFT(Stop); closed triangles, untransfomed, isolated immature embryos simply placed on the medium. Results from triplicate independent biological samples (n = 3) are shown, and error bars represent sd. The expression level was normalized to the level of MFT of CS at DAA60 (Figure 5C), which was set as 1.
DISCUSSION
Among the environmental signals affecting seed development, temperature is the most influential in the formation of seed dormancy in wheat (Reddy et al., 1985). In this study, transcriptional profiling of the effects of temperature on seed dormancy formation identified MFT as a candidate gene for seed dormancy regulation.
MFT was initially identified as a floral regulator in Arabidopsis (Yoo et al., 2004) because its amino acid sequence is highly similar to those of FT and TFL1. However, later studies using expression analysis and transformation analysis of MFT-like genes indicated that MFT-like genes function in seed development rather than in flowering (Chardon and Damerval, 2005; Danilevskaya et al., 2008; Kikuchi et al., 2009). Recent detailed analysis showed that MFT is a negative regulator of ABA sensitivity during seed germination in Arabidopsis (Xi et al., 2010). MFT expression attenuates ABA’s suppression of germination. By contrast, in wheat, our expression, transformation, and transient assay analyses indicate that Ta-MFT expression suppresses germination, opposite to the action of At-MFT.
In contrast with MFT-like genes, the functions of FT-like and TFL1-like genes have been studied extensively. Because PEBP family members are highly conserved, the known functions of FT-like and TFL1-like genes may provide us with some clue to the function of MFT. Recently, in addition to flowering time control, FT-like and TFL1-like genes have been found to be involved in more diverse functions involving meristem transitions, such as wild potato tuberization response to short-day conditions (Rodríguez-Falcón et al., 2006), stem elongation and transition to dormancy in poplar buds (Böhlenius et al., 2006; Ruonala et al., 2008), and attenuation of apical meristem growth before and independent of floral production (Lifschitz et al., 2006). Thus, FT-like and TFL1-like genes are now thought to be versatile regulators of responses to various environmental conditions (photoperiod, light quality, and temperature) for meristem transitions (Rohde and Bhalerao, 2007; Shalit et al., 2009). Therefore, it is tempting to speculate that MFT-like genes might also be involved in the regulation of embryogenic meristems in seeds in response to environmental conditions. The expression analysis of Ta-MFT supported part of this hypothesis, showing that MFT expression is regulated in response to temperature. Also, MFT seemed to be transmitting temperature signals to a downstream temperature signaling cascade to regulate the depth of seed dormancy. Our in situ hybridization experiment showed that MFT is specifically expressed in the scutellum and coleorhiza (Figure 6); however, we do not know whether MFT protein localizes only to the scutellum and coleorhiza or moves to other tissues. Two PEBP subfamilies are known to include mobile proteins. For example, At-FT and its rice ortholog Hd3a move from the leaf to the shoot apical meristem (Corbesier et al., 2007; Tamaki et al., 2007), and At-TFL1 moves from the inner to outer cells of the shoot meristem (Conti and Bradley, 2007). Thus, it is quite possible that Ta-MFT protein moves from the scutellum and coleorhiza to other parts of the seed. The effect of Ubi:TaMFT in the transient assay also supports this suggestion. The bombarded Ubi:TaMFT should function in the cells into which it was introduced. If no signals are transmitted from the transformed cells, it would be difficult to suppress other cells from undergoing precocious germination. Thus, it is likely that some signal is transmitted to other cells to achieve the coordinate suppression of precocious germination in immature embryos. The result that MFT expression in the Ubi:TaMFT(Stop)-bombarded embryos is much lower than that of the Ubi:TaMFT-bombarded embryos (Figure 11) also may indicate the possibility that MFT amplifies the signals by inducing MFT expression in the adjacent cells. We do not know whether this signal is MFT itself or if other signaling substances are produced as a consequence of the introduction of MFT. Another indication came from the result that a signal peptide prediction program, SOSUI (Gomi et al., 2004), predicted that Ta-MFT has a transmembrane signal peptide at the N terminus (see Supplemental Figure 2 online). This indicates that Ta-MFT may be a secreted protein because a hydrophobic signaling peptide at N terminus is often used as a secretion signal (Sakaguchi, 1997). If Ta-MFT is secreted from cells, it could transmit signals to other cells. The localization of Ta-MFT protein needs to be investigated empirically to determine whether Ta-MFT is a mobile protein. The subcellular localization analysis of Ta-MFT and Ta-FT showed a nuclear as well as cytoplasmic localization (see Supplemental Figure 5 online), similar to the results for FT:EGFP reported by Abe et al. (2005). These results showed that the subcellular localization of Ta-MFT appears to be similar to that of At-FT, which is known to be mobile, and to Ta-FT.
The germination of seeds mainly starts after the physiological maturity stage (Figures 1A, 5A, and 5B), indicating that a signal around or after physiological maturity releases the suppression of precocious germination. We speculate that the signal may be the decreasing water content of the seed itself. After physiological maturity, the water content of seeds decreased rapidly from ~40 to 10% (Figures 1A and 5B), and eventually the seeds became almost completely desiccated. This may promote the denaturation of MFT or other proteins that suppress precocious germination, thus releasing the inhibition effect of the protein and allowing germination. In the case of MFT, its expression level remained high in mature dormant embryos; therefore, the imbibed seeds might resynthesize MFT protein, which would suppress germination similar to its suppression of precocious germination demonstrated by our transient assay using Ubi:TaMFT.
The location of MFT on the genetic map also implicated it in regulation of seed dormancy. The mapped position of MFT-3A cosegregated with Xbarc310, a marker that is colocated at the logarithm of odds peak of the seed dormancy QTL QPhs.ocs-3A.1 (Mori et al., 2005). This suggests that MFT-3A may be the causal gene for the QTL. To determine genetically whether MFT-3A is the causal gene for the QTL, high-resolution mapping of MFT-3A is in progress. Comparison of the genomic sequences of MFT-3A showed that only two sites are different between CS and Zen. The transgenic wheat containing the 5685-bp Zen MFT-3A genomic fragment in the CS background showed the same degree of variation of MFT expression as the CS and CS(Zen3A) (Figure 8). This result strongly indicates that the higher expression of MFT in CS(Zen3A) is caused by the genomic fragment of MFT-3A in Zen and that the SNP in the promoter region (Figure 7) appears to be a promising functional SNP candidate for the cause of the higher level of expression of MFT in Zen. Since this SNP was located in an A-motif sequence, which is thought to be a binding site of bZIP transcription factors (Izawa et al., 1993), a bZIP transcription factor might be involved in the negative regulation of MFT. If the SNP is actually related to the regulation of MFT expression, the CAPS marker (see Supplemental Figure 7A online) may prove useful for marker-assisted selection in wheat breeding programs to improve the seed dormancy trait, though the effect of this SNP will need to be verified by further analysis.
To explore the functions of MFT in immature embryos, we transiently overexpressed MFT in isolated immature embryos using Ubi:TaMFT. Surprisingly, we found that the introduction of MFT into isolated immature embryos suppressed their precocious germination. This result supports the hypothesis that a higher level of MFT expression after physiological maturity inhibits germination in mature seeds. This effect of MFT resembles the inhibitory effect of ABA on the precocious germination of isolated immature embryos. As described by Triplett and Quatrano (1982), isolated immature embryos cultured with 1 to 100 μM ABA continue to undergo normal embryogenesis leading to a state of developmental arrest. The difference between the effects of MFT and ABA was the effective timing of the stimuli relative to the stage of the immature embryos. MFT should be introduced into earlier immature embryos at approximately DAA10 in our conditions. When MFT is introduced into more developed immature embryos, MFT is not able to arrest precocious germination. By contrast, ABA can effectively inhibit precocious germination in a broad range of stages in immature embryos. One possible explanation for the narrow window of action of MFT is that MFT needs time to establish its inhibitory effect and is not able to stop the germination process after a certain point.
Exogenously applied 1 μM GA released the suppression of precocious germination after transformation with Ubi:TaMFT. Exogenously applied GA is also known to effectively break wheat seed dormancy (Nakamura, 1962). This implies that endogenous GA may contribute to breaking seed dormancy and promoting subsequent germination in wheat. Whereas the main site of de novo GA biosynthesis in germinating wheat seeds was proposed to be the scutellum (Appleford and Lenton, 1997), a more precise analysis of the location of de novo GA biosynthesis in imbibed rice seeds has shown that the scutellar epithelium and shoot apex are the locations where bioactive GA is synthesized (Kaneko et al., 2003). Considering that MFT is expressed in the scutellum, MFT may be involved in suppression of GA synthesis in the scutellar epithelium. In addition, the inception of GA synthesis in the shoot apical meristem after imbibition appears to be important for inducing meristem growth leading to subsequent germination (Kaneko et al., 2003). If MFT is a mobile protein and involved in the regulation of meristem growth, it is intriguing to speculate that MFT may also be involved in the suppression of GA biosynthesis in the shoot apical meristem.
It is reported that, in barley, the coleorhiza plays a major role in conferring dormancy by acting as a barrier to radicle emergence; in Arabidopsis and Lepidium sativum, seed germination requires coordinated emergence of the radicle and weakening of the micropylar endosperm cap covering the radicle (Barrero et al., 2009; Linkies et al., 2009; Nambara et al., 2010). These results suggest that coordinated tissue interactions in seeds also regulate seed germination. In the immature embryos transiently expressing Ubi:TaMFT, the radicles did not emerge (Figure 10) and seemed to not rupture the coleorhizae. Ta-MFT might also act as a messenger in the coordinated action between tissues in seed germination.
In this study, analyzing temperature effects on wheat seed dormancy, we found that MFT is involved in regulation of germination. Our global analysis of temperature effects on gene expression therefore seems to be an effective way to identify genes that act on regulation of seed dormancy. We analyzed only one gene in detail in this study, but we identified ~70 genes that are differentially expressed between mature embryos grown at 13 and 25°C. Thus, further characterization of the remaining genes may reveal other genes that act in seed dormancy or germination.
METHODS
Plant Materials
Hexaploid wheat (Triticum aestivum) cultivars N61 and SK were grown in a greenhouse. After anthesis, they were grown under controlled temperature conditions at 13 or 25°C. Other hexaploid wheat cultivars or lines, CS, the single chromosome substitution line [CS(Zen3A)], Bobwhite (BW), and CS transformed with genomic Ta-MFT-3A were grown in a growth chamber with short-day conditions (day/night temperature; 17/10°C, with 8-h daylength) for ~3 months and subsequently were transferred to another growth chamber with long-day conditions (day/night temperature;17/12°C, with 14-h daylength). After ~1 month, they flowered and were harvested at 10 DAA intervals. CS(Zen3A), the single chromosome substitution line CS with a substituted chromosome 3A from Zen, was developed by conventional procedures as described by Law and Worland (1973) using a CS monotelosomic 3A line as the recurrent parent. In the comparison of wheat cultivars, generally Zen has an extremely high level of seed dormancy, N61 and CS(Zen3A) have high levels of seed dormancy, and CS and BW have low levels of seed dormancy.
Germination Tests and Dry Weight Measurement
All germination tests were performed on at least three independent biological replicates. In the germination test for SK and N61, five spikes were harvested from each sample and were imbibed in water for 24 h and then incubated in the dark for 10 d at 15°C under saturated humidity conditions. After that, the germinated and ungerminated seeds were counted and germination percentages were calculated. In the germination test for CS and CS(Zen3A), seeds from a harvested spike were sown on two sheets of No.2 filter paper in 9-cm Petri dishes. The dishes were incubated in the dark for 7 d at 20°C, and then the germinated and ungerminated seeds were counted and germination percentages were calculated. In the germination test for the transient assay, embryos on medium in 9-cm Petri dishes were incubated at 25°C, the number of germinated embryos was counted at daily intervals, and the GI was calculated as described by Nakamura et al. (2007). The dry weight of seeds was measured after incubating seeds from a spike at 80°C for 48 h. The water content of seeds was determined by (fresh weight − dry weight)/fresh weight. The water content was measured in three independent biological replicates.
Microarray Analysis
The embryos were isolated from completely mature seeds of a spike of N61 at DAA76 grown at 13°C or at DAA34 grown at 25°C, and of SK at DAA80 grown at 13°C or at DAA38 grown at 25°C. Total RNA was extracted from the embryos using Trizol reagents (Invitrogen), and its quality was checked using the Agilent 2100 Bioanalyzer (Agilent Technologies). The RNAs (400-ng aliquots) were labeled with a Quick Amp labeling kit (Agilent Technologies) according to the manufacturer’s instructions. Aliquots of Cy3-labeled (13°C) and Cy5-labeled (25°C) cRNAs (825 ng each) samples were used for hybridization to the Agilent Wheat Oligo Microarray (44K, custom-made; Agilent Technologies). Three biological replicate samples sets for both 13 and 25°C conditions in each SK and N61 were analyzed, for a total of six hybridizations. After hybridization, microarray slides were scanned (scanner model G2505B; Agilent Technologies), and data were analyzed using Feature Extraction software (version 9.5; Agilent Technologies) with the default settings. Subsequently, the data were analyzed using GeneSpring GX 7.3 software (Agilent Technologies). The false positives were controlled by measuring the false discovery rate (Benjamini and Hochberg, 1995). This Agilent Wheat Oligo microarray with 37,826 probes is registered as GPL9805 in the Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI). All expression tag contig sequences used for the design of probes have been published online (KOMUGI, http://www.shigen.nig.ac.jp/wheat/komugi/array/probe/download.jsp). A complete set of microarray data from this study was deposited to the GEO repository under the accession number GSE22786.
qRT-PCR
Total RNA was extracted using Trizol reagents (Invitrogen). The tissues and harvesting times of samples are summarized in Supplemental Table 3 online. All samples were treated with RNase-free DNase I (Invitrogen). First-strand cDNA synthesis was performed using a PrimeScript RT reagent kit (Takara). qRT-PCR was performed using a 7500 Real-Time PCR System (Applied Biosystems) and a SYBR Premix Ex Taq GC kit (Takara) in accordance with the manufacturers’ instructions. An actin gene was used as an internal control. This actin gene was selected from our microarray analysis based on the relatively small fluctuation of its expression levels during seed development (see Supplemental Figure 9D online). In addition, this actin gene did not show significantly differentially expression between embryos grown at 13 and 25°C (see Supplemental Table 4 online). In accordance with the manufacturer’s instructions, the amplification efficiency of the primers was determined. The obtained data using the ΔΔ cycle threshold analysis method were analyzed by RQ study software (Applied Biosystems). Primers for PCR used in this study are listed in Supplemental Table 2 online. The detailed qRT-PCR procedure is described in Supplemental Figure 9 online.
Mapping of Diploid and Hexaploid Wheat MFT Homologs
The general procedures for constructing the diploid wheat genetic map were described previously by Nakamura et al. (2007). The CAPS method used for genotyping the diploid wheat MFT homolog (Tm-MFT) is described in Supplemental Figure 7C online. Genetic maps were constructed using JoinMap, version 3.0 (Biometris; http://www.joinmap.nl) with the Kosambi function (Kosambi, 1944).
The hexaploid wheat mapping population consisted of 2852 F2 plants derived from a cross between CS and CS(Zen3A). The mapping of the SSR marker Xbarc310 was performed using primers Xbarc310-F and Xbarc310-R (see Supplemental Table 2 online) as described by Mori et al. (2005). The detailed mapping procedure for Ta-MFT-3A is described in Supplemental Figure 7 online.
A CAPS marker was developed for the SNP in the Ta-MFT-3A promoter region of Zen and CS. The detailed mapping procedure is described in Supplemental Figure 7A online.
Screening of a CS Wheat Genomic BAC Library
A 203-bp fragment was amplified from the Ta-MFT-3A genomic fragment in CS (CS6) (see Supplemental Figure 4 online) by PCR with primers CSZENSSR-F1 and CSZENSSR-R1 (see Supplemental Table 2 online). Using PCR screening with these primers, we isolated BAC clones that contain Ta-MFT-3A from CS genomic BAC library (Allouis et al., 2003).
Shotgun sequencing of the BAC was performed by the standard method (International Rice Genome Sequencing Project, 2005). The BAC sequence was analyzed using the Rice Genome Automated Annotation System (RiceGAAS) website (http://ricegaas.dna.affrc.go.jp/; Sakata et al., 2002).
In Situ Hybridization Analysis
A 278-bp PCR fragment corresponding to the 3′-untranslated region of Ta-MFT-3A was amplified using primers TaMFTishF and TaMFTishR (see Supplemental Table 2 online) and cloned into the pTAC-2 vector (BioDynamics Laboratory). The cloned fragment was sequenced and the confirmed clone was linearized using EcoRI and BamHI and then used as a template to generate antisense and sense probes using T7 and SP6 RNA polymerase. In situ hybridization with a digoxigenin-labeled RNA probe and immunological detection were conducted in accordance with the methods of Kouchi and Hata (1993), with thorough modifications to improve the reactions as described by Komatsuda et al. (2007).
Wheat Transformation and Transient Expression Assay
The entire of 5685-bp length of MFT-3A genomic sequence of Zen (Figure 7) was cloned into a pUC12 plasmid (Vieira and Messing, 1982) that contains the bar gene (which confers resistance to the herbicide bialaphos) as a selection marker under the control of the maize ubiquitin promoter (see Supplemental Figure 10 online). The plasmid was transformed by particle bombardment using immature embryos following the method of Pellegrineschi et al. (2002). The integration of the transgene was confirmed by PCR screening for the bar selection marker gene (see Supplemental Figure 10 online) using primers barF and barR (see Supplemental Table 2 online). The detailed procedure for the transient gene expression assay in wheat immature embryos is described in Supplemental Figure 8 online.
Phylogenetic Analysis
The amino acid alignment was conducted using ClustalW (Thompson et al., 1994) and then manually adjusted to optimize the alignment (see Supplemental Data Sets 3 and 4 online). The phylogenetic tree (see Supplemental Figure 3 online) was constructed by MEGA5 (Tamura et al., 2011) using the neighbor-joining method, Poisson model with rate uniformily among sites, and complete-deletion option for gaps/missing data treatment. Bootstrap analysis for 1000 replicates was preformed to provide confidence estimates for the tree topologies.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: genomic sequences for MFT-3A(CS) (AB571512) and MFT-3A(Zen) (AB571513) and the actin gene used as an internal control for qRT-PCR (GQ339780). Additional accession numbers are listed in Supplemental Figures 2 and 4 online. Wheat oligomicroarray with 37,826 probes is registered as GPL9805 in GEO at NCBI. A complete set of microarray data from this study was deposited to the GEO repository under the accession number GSE22786.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Time Course of Gene Expression Profiles during Seed Development Using the Wheat Microarray.
Supplemental Figure 2. Alignment of the Deduced Amino Acid Sequences of Ta-MFT Homologs.
Supplemental Figure 3. Phylogenetic Analysis of Ta-MFT with the PEBP Proteins from Wheat, Barley, Maize, Rice, and Arabidopsis.
Supplemental Figure 4. Phylogram of the Ta-MFT Partial Genomic Sequences.
Supplemental Figure 5. Subcellular Localization Analysis of Ta-MFT.
Supplemental Figure 6. The Ta-MFT-3A PCR Fragment Length Polymorphism.
Supplemental Figure 7. Agarose Gel Electrophoresis of Fragments of the CAPS Markers.
Supplemental Figure 8. The Effect of Introduction of Ubi:TaMFT into CS Immature Embryos.
Supplemental Figure 9. The Amplification Efficiency of qRT-PCR Primers.
Supplemental Figure 10. Constructs Generated in This Study.
Supplemental Table 1. Summary of Microarray Data for the Rht1, VP1, and ABF Genes.
Supplemental Table 2. Primers Used in This Study.
Supplemental Table 3. Days after Anthesis of Embryos for Total RNA Extraction for Microarray Analysis.
Supplemental Table 4. Summary of Microarray Data for the Internal Control Actin Gene.
Supplemental Data Set 1. Differentially Expressed Genes in SK in Microarray Experiments.
Supplemental Data Set 2. Differentially Expressed Genes in N61 in Microarray Experiments.
Supplemental Data Set 3. Text File of the Alignment Used to Generate the Phylogenetic Tree Shown in Supplemental Figure 3.
Supplemental Data Set 4. Text File of the Alignment Used to Generate the Phylogenetic Tree Shown in Supplemental Figure 4A.
Acknowledgments
We thank H. Morishige (National Institute of Crop Science [NICS]), Y. Watanuki (NICS), and Y. Naito (NICS) for technical assistance, K. Mochida (RIKEN Plant Science Center) for microarray data analysis, Y. Nagamura (National Institute of Agrobiological Sciences) and R. Motoyama (National Institute of Agrobiological Sciences) for technical help on microarray analysis, and R.R. Finkelstein (University of California, Santa Barbara) and C.M. Steber (Washington State University) for critical reading of the manuscript. This research was supported by a “Development of innovative crops through the molecular analysis of useful genes” project grant from the National Agriculture and Food Research Organization and by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation, TRC, TRG-1001, and GMZ-1001).
AUTHOR CONTRIBUTIONS
S.N. designed the research, performed research, analyzed the data, and wrote the article. F.A., H.K., K.N., T.O., H.H., and H.M. designed and performed the research and analyzed the data. S.U. and T.M. performed the research and analyzed data. K.K. and Y.O. analyzed data. A.T., H.I., and M.M. performed the research.
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Allouis S., Moore G., Bellec A., Sharp R., Faivre-Rampant P., Mortimer K., Pateyron S., Foote T.N., Griffiths S., Caboche M., Chaloub B. (2003). Construction and characterization of a hexaploid wheat BAC library from the reference germplasm ‘Chinese Spring’. Cereal Res. Commun. 31: 331–338 [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Appleford N.E.J., Lenton J.R. (1997). Hormonal regulation of -amylase gene expression in germinating wheat (Triticum aestivum) grains. Physiol. Plant. 100: 534–542 [Google Scholar]
- Ashikawa I., Abe F., Nakamura S. (2010). Ectopic expression of wheat and barley DOG1-like genes promotes seed dormancy in Arabidopsis. Plant Sci. 179: 536–542 [DOI] [PubMed] [Google Scholar]
- Barrero J.M., Talbot M.J., White R.G., Jacobsen J.V., Gubler F. (2009). Anatomical and transcriptomic studies of the coleorhiza reveal the importance of this tissue in regulating dormancy in barley. Plant Physiol. 150: 1006–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. (Ser. B [Methodol.]) 57: 289–300 [Google Scholar]
- Bentsink L., Jowett J., Hanhart C.J., Koornneef M. (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L., Koornneef M. (2008). Seed dormancy and germination. In The Arabidopsis Book 6: e0119, doi/10.1199/tab.0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley J.D., Black M. (1994). Seeds: Physiology of Development and Germination. (New York: Plenum; ). [Google Scholar]
- Böhlenius H., Huang T., Charbonnel-Campaa L., Brunner A.M., Jansson S., Strauss S.H., Nilsson O. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312: 1040–1043 [DOI] [PubMed] [Google Scholar]
- Black M., Butler J., Hughes M. (1987). Control and development of dormancy in cereals. In 4th International Symposium on Pre-Harvest Sprouting in Cereals, Mares D.J., (Boulder, CO: Westview Press; ), pp. 379–392 [Google Scholar]
- Buraas T., Skinnes H. (1985). Development of seed dormancy in barley, wheat and triticale under controlled conditions. Acta Agric. Scand. 35: 233–244 [Google Scholar]
- Chardon F., Damerval C. (2005). Phylogenomic analysis of the PEBP gene family in cereals. J. Mol. Evol. 61: 579–590 [DOI] [PubMed] [Google Scholar]
- Conti L., Bradley D. (2007). TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell 19: 767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Danilevskaya O.N., Meng X., Hou Z., Ananiev E.V., Simmons C.R. (2008). A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 146: 250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage W.E., Leubner-Metzger G. (2006). Seed dormancy and the control of germination. New Phytol. 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R., Reeves W., Ariizumi T., Steber C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Flintham J., Adlam R., Bassoi M., Holdsworth M., Gale M. (2002). Mapping genes for resistance to sprouting damage in wheat. Euphytica 126: 39–45 [Google Scholar]
- Gale M.D. (1989). The genetics of pr-harvest sprouting in cereals, particularly in wheat. In Pre-Harvest Field Sprouting in Cereals, Derera N.F., (Boca Raton, FL: CRC Press; ), pp. 85–110 [Google Scholar]
- Gerjets T., Scholefield D., Foulkes M.J., Lenton J.R., Holdsworth M.J. (2010). An analysis of dormancy, ABA responsiveness, after-ripening and pre-harvest sprouting in hexaploid wheat (Triticum aestivum L.) caryopses. J. Exp. Bot. 61: 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi M., Sonoyama M., Mitaku S. (2004). High performance system for signal peptide prediction: SOSUIsignal. Chem-Bio. Inform. J. 4: 142–147 [Google Scholar]
- Graeber K., Linkies A., Müller K., Wunchova A., Rott A., Leubner-Metzger G. (2010). Cross-species approaches to seed dormancy and germination: conservation and biodiversity of ABA-regulated mechanisms and the Brassicaceae DOG1 genes. Plant Mol. Biol. 73: 67–87 [DOI] [PubMed] [Google Scholar]
- Hedman H., Källman T., Lagercrantz U. (2009). Early evolution of the MFT-like gene family in plants. Plant Mol. Biol. 70: 359–369 [DOI] [PubMed] [Google Scholar]
- Hilhorst H.W.M. (2007). Definitions and hypotheses of seed dormancy. In Seed Development, Dormancy and Germination, Bradford K.J., Nonogaki H., (Sheffield, UK: Blackwell; ), pp. 50–71 [Google Scholar]
- Holdsworth M.J., Bentsink L., Soppe W.J.J. (2008a). Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Holdsworth M.J., Finch-Savage W.E., Grappin P., Job D. (2008b). Post-genomics dissection of seed dormancy and germination. Trends Plant Sci. 13: 7–13 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Izawa T., Foster R., Chua N.H. (1993). Plant bZIP protein DNA binding specificity. J. Mol. Biol. 230: 1131–1144 [DOI] [PubMed] [Google Scholar]
- Kaneko M., Itoh H., Inukai Y., Sakamoto T., Ueguchi-Tanaka M., Ashikari M., Matsuoka M. (2003). Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J. 35: 104–115 [DOI] [PubMed] [Google Scholar]
- Kikuchi R., Kawahigashi H., Ando T., Tonooka T., Handa H. (2009). Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 149: 1341–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsuda T., et al. (2007). Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. USA 104: 1424–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosambi D.D. (1944). The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175 [Google Scholar]
- Kouchi H., Hata S. (1993). Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 238: 106–119 [DOI] [PubMed] [Google Scholar]
- Kucera B., Cohn M.A., Leubner-Metzger G. (2005). Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 15: 281–307 [Google Scholar]
- Law C.N., Worland A.J. (1973). Aneuploidy in wheat and its uses in genetic analysis. Plant Breeding Institute, Annual Report 1972: 26–65 [Google Scholar]
- Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z., Alvarez J.P., Eshed Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103: 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A., Müller K., Morris K., Turecková V., Wenk M., Cadman C.S.C., Corbineau F., Strnad M., Lynn J.R., Finch-Savage W.E., Leubner-Metzger G. (2009). Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: A comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 21: 3803–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mares D., Mrva K., Cheong J., Williams K., Watson B., Storlie E., Sutherland M., Zou Y. (2005). A QTL located on chromosome 4A associated with dormancy in white- and red-grained wheats of diverse origin. Theor. Appl. Genet. 111: 1357–1364 [DOI] [PubMed] [Google Scholar]
- Mori M., Uchino N., Chono M., Kato K., Miura H. (2005). Mapping QTLs for grain dormancy on wheat chromosome 3A and the group 4 chromosomes, and their combined effect. Theor. Appl. Genet. 110: 1315–1323 [DOI] [PubMed] [Google Scholar]
- Nakamura S. (1962). Germination of grass seeds. Proc. Int. Seed Test Ass. 27: 710–729 [Google Scholar]
- Nakamura S., Komatsuda T., Miura H. (2007). Mapping diploid wheat homologues of Arabidopsis seed ABA signaling genes and QTLs for seed dormancy. Theor. Appl. Genet. 114: 1129–1139 [DOI] [PubMed] [Google Scholar]
- Nambara E., Okamoto M., Tatematsu K., Yano R., Seo M., Kamiya Y. (2010). Abscisic acid and the control of seed dormancy and germination. Seed Sci. Res. 20: 55–67 [Google Scholar]
- Nyachiro J.M., Clarke F.R., DePauw R.M., Knox R.E., Armstrong K.C. (2002). Temperature effects on seed germination and expression of seed dormancy in wheat. Euphytica 126: 123–127 [Google Scholar]
- Pellegrineschi A., Noguera L.M., Skovmand B., Brito R.M., Velazquez L., Salgado M.M., Hernandez R., Warburton M., Hoisington D. (2002). Identification of highly transformable wheat genotypes for mass production of fertile transgenic plants. Genome 45: 421–430 [DOI] [PubMed] [Google Scholar]
- Reddy L.V., Metzger R.J., Ching T.M. (1985). Effect of temperature on seed dormancy of wheat. Crop Sci. 25: 455–458 [Google Scholar]
- Rodríguez-Falcón M., Bou J., Prat S. (2006). Seasonal control of tuberization in potato: Conserved elements with the flowering response. Annu. Rev. Plant Biol. 57: 151–180 [DOI] [PubMed] [Google Scholar]
- Rohde A., Bhalerao R.P. (2007). Plant dormancy in the perennial context. Trends Plant Sci. 12: 217–223 [DOI] [PubMed] [Google Scholar]
- Ruonala R., Rinne P.L.H., Kangasjärvi J., van der Schoot C. (2008). CENL1 expression in the rib meristem affects stem elongation and the transition to dormancy in Populus. Plant Cell 20: 59–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M. (1997). Eukaryotic protein secretion. Curr. Opin. Biotechnol. 8: 595–601 [DOI] [PubMed] [Google Scholar]
- Sakata K., Nagamura Y., Numa H., Antonio B.A., Nagasaki H., Idonuma A., Watanabe W., Shimizu Y., Horiuchi I., Matsumoto T., Sasaki T., Higo K. (2002). RiceGAAS: An automated annotation system and database for rice genome sequence. Nucleic Acids Res. 30: 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Antonio B.A., Namiki N., Takehisa H., Minami H., Kamatsuki K., Sugimoto K., Shimizu Y., Hirochika H., Nagamura Y. (2011). RiceXPro: A platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 39 (Database issue): D1141–D1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalit A., Rozman A., Goldshmidt A., Alvarez J.P., Bowman J.L., Eshed Y., Lifschitz E. (2009). The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA 106: 8392–8397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N., Usadel B., Winter A., Radchuk V., Scholz U., Stein N., Weschke W., Strickert M., Close T.J., Stitt M., Graner A., Wobus U. (2008). Barley grain maturation and germination: Metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol. 146: 1738–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Takeuchi Y., Ebana K., Miyao A., Hirochika H., Hara N., Ishiyama K., Kobayashi M., Ban Y., Hattori T., Yano M. (2010). Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. USA 107: 5792–5797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (May 4, 2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. http://dx.doi.org/10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S., Rognoni S., Bentsink L., Smeekens S. (2008). The Arabidopsis GSQ5/DOG1 Cvi allele is induced by the ABA-mediated sugar signalling pathway, and enhances sugar sensitivity by stimulating ABI4 expression. Plant J. 55: 372–381 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torada A., Koike M., Ikeguchi S., Tsutsui I. (2008). Mapping of a major locus controlling seed dormancy using backcrossed progenies in wheat (Triticum aestivum L.). Genome 51: 426–432 [DOI] [PubMed] [Google Scholar]
- Triplett B.A., Quatrano R.S. (1982). Timing, localization, and control of wheat germ agglutinin synthesis in developing wheat embryos. Dev. Biol. 91: 491–496 [DOI] [PubMed] [Google Scholar]
- Turck F., Fornara F., Coupland G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. (1982). The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19: 259–268 [DOI] [PubMed] [Google Scholar]
- Wan Y., Poole R.L., Huttly A.K., Toscano-Underwood C., Feeney K., Welham S., Gooding M.J., Mills C., Edwards K.J., Shewry P.R., Mitchell R.A.C. (2008). Transcriptome analysis of grain development in hexaploid wheat. BMC Genomics 9: 121–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson J.D., Quatrano R.S., Cuming A.C. (1985). Em polypeptide and its messenger RNA levels are modulated by abscisic acid during embryogenesis in wheat. Eur. J. Biochem. 152: 501–507 [DOI] [PubMed] [Google Scholar]
- Wilson I.D., Barker G.L., Lu C., Coghill J.A., Beswick R.W., Lenton J.R., Edwards K.J. (2005). Alteration of the embryo transcriptome of hexaploid winter wheat (Triticum aestivum cv. Mercia) during maturation and germination. Funct. Integr. Genomics 5: 144–154 [DOI] [PubMed] [Google Scholar]
- Xi W., Liu C., Hou X., Yu H. (2010). MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22: 1733–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.Y., Kardailsky I., Lee J.S., Weigel D., Ahn J.H. (2004). Acceleration of flowering by overexpression of MFT (MOTHER OF FT AND TFL1). Mol. Cells 17: 95–101 [PubMed] [Google Scholar]